Abstract
The link between ERG rearrangement and PTEN deletion is unclear in prostate cancer progression. Using fluorescence in situ hybridization, we systematically validated the frequency and distribution of ERG and PTEN aberrations in a cohort of 73 benign prostate tissues, 59 high grade prostatic intraepithelial neoplasia (HGPIN) foci, 281 localized prostate cancer and 47 androgen-independent metastatic prostate cancer patients. Overall, ERG rearrangement was present in 15% (5/33) of HGPIN, 45% (121/267) of localized cancers and 35% (15/43) of metastases. By contrast, PTEN deletion was identified in 9% (3/33) of HGPIN, 17% (42/251) of localized cancers and 54% (22/41) of metastases, of which 0%, 40% (17/42) and 45% (10/22) were homozygous, respectively. Concomitance of ERG rearrangement and PTEN deletion was observed in a subset of HGPIN. Significantly, association between PTEN deletion and ERG rearrangement was present both in localized cancers (p=0.0008) and metastases (p=0.02). Further, immunohistochemistry revealed significant correlation of decreased PTEN protein expression with PTEN genomic deletion both in localized and metastatic cancer. Of note, ERG aberration, but not PTEN deletion, was consistently identical both in localized cancer and adjacent HGPIN. Similarly, whereas all metastases (41/41, 100%) shared the same ERG status across multiple sites from the same patient, 5% (2/41) of cases showed discordance for PTEN deletion status across multiple sites. Collectively, our data support PTEN deletion as a late genetic event in human prostate cancer, presumably a “second hit” after ERG rearrangement. PTEN deletion and ERG rearrangement may cooperate, but contribute at different stages, in prostate cancer progression.
Keywords: PTEN, Deletion, ERG, Rearrangement, Prostate cancer, Fluorescence in situ hybridization
Introduction
Prostate cancer is the second leading cause of cancer-related death among North American men.1 It proceeds through a putative precursor lesion, termed high grade prostatic intraepithelial neoplasia (HGPIN), to hormone naïve clinically localized cancer and finally to androgen-independent metastatic cancer.1, 2 Despite its high prevalence, the molecular basis of prostate cancer progression remains unclear.2
Recently, recurrent gene fusions involving the ETS family of transcription factors,ERG, ETV1, ETV4, and ETV5, fused to TMPRSS2 or other upstream partners, have been identified in the majority of prostate cancers.3-10 Among these aberrations, TMPRSS2-ERG fusion is the most prevalent, occurring in approximately 50% of localized prostate cancers and 30% of androgen-independent metastatic cancers.10-12 As TMPRSS2 and ERG are located ∼3 Mb apart on chromosome 21, the rearrangement between them occurs either through translocation or by an interstitial deletion.13 Emerging data suggests that TMPRSS2-ERG fusion plays an important role in carcinogenesis in vitro and in vivo.14, 15 Clinically, ERG rearrangement has been observed in 10∼20% of HGPIN.16-18 Mosquera et al. showed that of 143 HGPIN cases, 16% (23 of 143) were ERG rearrangement positive, and in all cases the paired prostate cancer was ERG rearrangement positive through the same mechanism.18 These observations suggest that ERG rearrangement may be an early event in prostate cancer. Additionally, studies in watchful waiting cohorts suggest that ETS rearrangement cancers is associated with a more aggressive phenotype, however conflicting reports have been described in prostatectomy series.12, 19-22
PTEN (phosphatase and tensin homolog deleted on chromosome 10) is a key tumor suppressor gene in prostate cancer.23 Loss of PTEN function results in increased PIP3 (Phosphatidylinositol (3,4,5)-triphosphate) levels and subsequent AKT phosphorylation and modulation of its downstream molecular oncogenic processes.24 A series of in vivo studies have demonstrated the role of PTEN in prostate carcinogenesis with prostate-specific deletion.25, 26 Clinically, deletion or mutation of at least one PTEN allele was reported to occur in 20-40% of localized cancers27-29 and up to 60% of metastases.30 Fluorescent in situ hybridization (FISH) and immunohistochemical studies demonstrated that PTEN genomic deletion and absence of PTEN expression are associated with unfavorable clinical outcome measures.29-31 Recent studies also showed that PTEN inactivation plays an important role in prostate cancer during progression to androgen-independence.32, 33
Whereas ERG rearrangement and PTEN deletion are strongly implicated in prostate cancer development, little is known about the link between these two genomic events. Most recently, Yoshimoto et al. reported that TMPRSS2-ERG fusion could be accompanied by PTEN deletion in localized prostate cancer.34 However, there has been no systematic FISH validation on these genomic aberrations in the context of prostate cancer progression. Hence, we comprehensively evaluated a wide spectrum of benign tissues, premalignant and malignant lesions to characterize ERG rearrangement and PTEN deletion during prostate cancer progression.
Materials and Methods
Study population, clinical data, and tissue microarray (TMA)
A total of six TMAs were interrogated in this study that represents: 1) 281 clinically localized prostate cancer patients who underwent radical prostatectomy as a monotherapy between 1995-1996 and 2004-2006 at the University of Michigan Hospital. 2) 47 androgen-independent metastatic prostate cancer patients with multiple metastatic sites and tumors in the prostate (when present) from a rapid autopsy program described previously.12 3) 20 benign prostate hyperplasia, 18 atrophy and 35 benign prostate tissues derived from the peripheral zone of prostate containing cancer. Morphology was confirmed by three pathologists (B.H., R.B.S and R.M.), and three cores (0.6 mm in diameter) were taken from each representative area of interest. Patient demographics of localized prostate cancer are shown in Supplementary Table 1. The detailed clinical, pathological, and TMA data were maintained on a secure relational database as previously described.11 This study was approved by the Institutional Review Board at the University of Michigan Medical School and all patients provided written informed consent. Both radical prostatectomy series and the rapid autopsy program were part of the University of Michigan Prostate Cancer Specialized Program of Research Excellence Tissue Core.
Case selection for high grade prostatic intraepithelial neoplasia (HGPIN)
A total of 59 HGPIN present in prostatectomy specimens from 56 localized prostate cancers represented in this cohort were also included in the study. Initially, we randomly chose 34 HGPIN lesions from equal number of localized cancer patients to assess the frequency of PTEN deletion and/or ERG rearrangement. We further reviewed localized cancer patients that harbored known ERG and/or PTEN genomic aberrations and selected 20 cases containing HGPIN to analyze association of PTEN and ERG aberrations. All HGPIN lesions were selected by three pathologists (B.H., R.M. and R.B.S.) by consensus and divided into two categories: those adjacent to cancer (distance less than 3 mm from the edge of the cancer, HGPINadj) or those away from cancer (distance more than 3 mm from the closest caner in any single section and 4 mm from the closest cancer on the adjacent section above or below, HGPINaway).35 Although HGPINadj cases selected for the study may potentially represent an intraductal spread of prostate cancer, none of the HGPINadj lesions included in the study morphologically contained high-grade pleomorphic nuclei which are 6× size of normal nuclei and/or intraluminal comedonecrosis, features usually considered characteristic of an intraductal spread of cancer.36
Fluorescence in situ hybridization (FISH)
Interphase FISH was performed as described.8, 11 Bacterial artificial chromosomes (BACs) were obtained from the BACPAC Resource Center (Oakland, CA), and probes were prepared as described.3, 11 For detection of ERG rearrangement, RP11-95I21 (5′ to ERG) and RP11-476D17 (3′ to ERG) were used with a break-apart probe strategy.11 To detect PTEN deletion, a combination of PTEN gene locus-specific probe (RP11-165M8) and 10q11.1-specific probe (RP11-351D16) for chromosome identification were utilized. Schematic BACs for ERG and PTEN are shown in Figure 1A. The integrity and correct localization of all probes were verified by hybridization to metaphase spreads of normal peripheral lymphocytes. Slides were examined using an ImagingZ1 microscope (Carl Zeiss, Oberkochen, Germany). FISH signals were scored manually (100× oil immersion) in morphologically intact and non-overlapping nuclei by two pathologists (B.H. and R.M.), and a minimum of 50 cancer cells from each site were recorded. Cancer sites with very weak or no signals were recorded as insufficiently hybridized. Cases lacking tumor tissue in all three cores were excluded.
Figure 1. FISH probe design and representative ERG aberrations and PTEN deletions detected in prostate cancer.

A, Schematic of BACs located 5′ and 3′ to ERG and locus/control for PTEN used as probes for interphase FISH. Chromosomal coordinates are from the March 2006 build of the human genome using the UCSC Genome Browser. BACs are indicated as numbered rectangles, with the number identifying the BAC as described below and the color indicating the probe color in the accompanying images. Genes are shown with the direction of transcription indicated by the arrowhead and exons indicated by bars. B, B1-B2, FISH was performed using BACs as indicated with the corresponding fluorescent label on formalin-fixed paraffin-embedded tissue sections for break-apart FISH of the ERG gene. Green and red arrows showed individual signals, whereas yellow signals were indicated as colocalized probes. B1, ERG rearrangement positive (with deletion) case showed loss of one red labeled probe 5′ to ERG. B2, ERG rearrangement positive (translocation) case showed one pair of split 5′ and 3′ signals. B3-B4, Representative images of hemizygous and homozygous PTEN deletion in prostate cancer. B3, Representative case with PTEN hemizygous deletion showed one red signals (10q23/PTEN locus) and pairs of green signals (10q11.1) in tumor cells. B4, Representative case with PTEN homozygous deletion showed absence of red signals (10q23/PTEN locus) but retained pairs of green signals (PTEN control). For all assays, at least 50 cancer cell nuclei were evaluated. C, Frequency of PTEN deletion and ERG rearrangement of benign tissues, HGPIN, localized and metastatic prostate cancers is indicated. Color legend signifies respective aberrations.
For validation of PTEN deletion, we utilized a previously documented method with minor modification.37 Briefly, based on hybridization in five control cores (data not shown), hemizygous deletion of PTEN gene was defined as >50% nuclei (mean±3 standard deviations in nonneoplastic controls) containing either one signal of locus probe and ≥ 2 signals of reference probe (absolute deletion), or two signals of locus probe and ≥ 4 signals of reference probe (relative deletion). Homozygous deletion of PTEN was exhibited by the simultaneous lack of the both PTEN locus signals and the presence of control signals in > 30% of cells.29, 34, 37 Representative FISH images of PTEN deletion and ERG rearrangement are demonstrated in Figure 1B.
Immunohistochemistry (IHC)
IHC was performed using a rabbit polyclonal antibody against PTEN (Ab9552, Cell Signaling, MA) on TMA using 1:100 dilution, and incubated overnight at 4°C following standard LSAB immunohistochemical staining protocol.38 The slides were evaluated blind by three independent observers (B.H., K. S. and R.M.). Triplicate cores from each specimen were scored separately, and the presence of tumor tissue in at least two interpretable cores was required to include a case for analysis. According to the cytoplasmic staining intensity, the tumors were divided into three categories as previously described:39, 40 grade 2 showed increased or equal staining intensity compared to the corresponding normal tissue; grade 1 had decreased staining intensity; and grade 0 demonstrated complete absence of staining.
Statistical analysis
Fisher's exact test was used to test the statistical significance of associations between PTEN deletion status and ERG rearrangement status, as well as association between PTEN genomic deletion status and PTEN protein expression level, with p-values < 0.05 being considered statistically significant. Statistical analyses were performed using the R software package, version 2.7.2 (www.r-project.org).
Results
Frequency of PTEN deletion and ERG rearrangement
To determine the frequency of PTEN and ERG genomic aberrations in different prostate tissue types, we screened a wide spectrum of prostate lesions and benign prostate tissues represented on multiple TMAs. As shown in Table 1, PTEN deletion was found in 17% (42/251) of localized prostate cancer patients, out of which 60% (25/42) exhibited hemizygous deletion. In androgen-independent metastatic prostate cancer, PTEN deletion was present in 54% (22/41) of cases, among which 55% (12/22) showed hemizygous deletion. By contrast, only 9% (3/33) of HGPIN showed PTEN deletion, all of which were hemizygous.
Table 1. Summary of ERG rearrangement and PTEN deletion status*.
| ERG | PTEN | |||||
|---|---|---|---|---|---|---|
| Tissue types | No rearrangement | Translocation | 5′ deletion | Not deleted | Hemizygous | Homozygous |
| Normal prostate tissue | 35 (100%) | 0 (0%) | 0 (0%) | 32 (100%) | 0 (0%) | 0 (0%) |
| BPH | 20 (100%) | 0 (0%) | 0 (0%) | 19 (100%) | 0 (0%) | 0 (0%) |
| Atrophy | 18 (100%) | 0 (0%) | 0 (0%) | 18 (100%) | 0 (0%) | 0 (0%) |
| HGPIN | 28 (85%) | 2 (6%) | 3 (9%) | 30 (91%) | 0 (0%) | 3 (9%) |
| Localized prostate cancer | 146 (55%) | 54 (20%) | 67 (25%) | 209 (83%) | 25 (10%) | 17 (7%) |
| Androgen-independent metastasis | 28 (65%) | 1 (2%) | 14 (33%) | 19 (46%) | 12 (29%) | 10 (25%) |
BPH:Benign prostate hyperplasia.
HGPIN: High grade prostatic intraepithelial neoplasia.
Not all cases included are informative for both ERG rearrangement and PTEN deletion status
Overall, ERG was rearranged in 45% (121/267) of localized prostate cancer cases, of which 55% (67/121) demonstrated deletion of the 5′end of ERG (Table 1). A similar frequency of ERG rearrangement (35%, 15/43) was observed in androgen-independent metastatic cancers as previously reported,12 the majority demonstrating deletion of the 5′ end of ERG. By contrast, ERG aberrations were identified in 15% (5/33) of HGPIN, and as expected, were not detected at all in non-neoplastic prostate tissues. Of note, part of our FISH data of ERG aberrations represented on three TMAs have been published before as part of the University of Michigan cohort.9, 11, 12
Association of PTEN deletion and ERG rearrangement in localized and androgen-independent metastatic prostate cancer
Because ERG rearrangement and PTEN deletion are among the most common genomic aberrations in prostate cancer,10, 23 we next explored the association of these two genomic events in this cohort. As shown in Table 2, the ERG rearrangement was present in approximately 71% (29/41) of localized cancers with PTEN deletion (hemizygous or homozygous). Likewise, PTEN deletion occurred more frequently in cases that harbored ERG rearrangement (26%, 29/110) as compared to those ERG rearrangement negative cases (9%, 12/127). Of the androgen-independent metastases, co-existence of the PTEN deletion and ERG rearrangement was present in 28% (11/39) of cases. Overall, a significant association between PTEN deletion and ERG rearrangement was observed both in localized prostate cancer (p = 0.0008) and androgen-independent metastatic prostate cancer (p = 0.02) (Table 2). Interestingly, for localized cancers with PTEN deletion, ERG fusion positive cases were more likely to show homozygous deletion of PTEN, although this association did not reach statistical significance (p = 0.08). Of note, a total of 12 localized cancer cases harboring rearrangement of ETV1 or ETV4 in the current study. Overall, eight of them are informative for PTEN genetic status, in which 50% (4/8) are with PTEN deletions.
Table 2. Distribution of ERG rearrangement and PTEN deletion status in localized and metastatic prostate cancer patients.
| ERG | ||||||
|---|---|---|---|---|---|---|
| Localized cancer* | Metastatic cancer* * | |||||
| PTEN | No rearrangement | Translocation | 5′ deletion | No rearrangement | Translocation | 5′ deletion |
| No deletetion | 115 | 38 | 43 | 15 | 1 | 2 |
| Hemizygous | 10 | 5 | 9 | 7 | 0 | 5 |
| Homozygous | 2 | 5 | 10 | 3 | 0 | 6 |
p = 0.0008
p = 0.02
PTEN deletion and ERG rearrangement in HGPIN
HGPIN is a putative precancerous lesion, and previous studies have shown that PTEN genomic deletion as well as ERG rearrangement could occur in subset of HGPIN. 16-18, 41 Additionally, Perner et al. demonstrated that ERG rearrangement displayed a different picture in HGPINadj vs. HGPINaway.16 However, it is unclear whether the PTEN deletion and ERG rearrangement could co-exist in HGPIN and what their association is to cancer. To address this, we analyzed 25 HGPIN on prostatectomy specimens from 20 selected localized cancers which harbored genomic aberrations of ERG and/or PTEN. As shown in Figure 2A, 100% (11/11) HGPINadj shared the same ERG aberrations with the paired cancer foci (T1-T9, T13, T20), which is comparable with previous studies.16, 18 By contrast, 60% (6/10) HGPINadj shared the same PTEN genomic aberrations with the paired cancer foci (T7-T11, T13), whereas the remaining four cases were not (T6, T12, T19-20). Of note, concomitance of PTEN deletion and ERG rearrangement were observed in HGPINadj intermingling with cancer foci from four localized cancer patients (T7-T9, T13). Interestingly, no PTEN deletion or ERG rearrangement was observed in HGPINaway. Figure 2B represents a reconstructed map of the prostatectomy sections in index cases T6 and T9. In total, these findings suggest co-existence of ERG rearrangement and PTEN deletion in a subset of HGPINadj.
Figure 2. Genomic aberrations of ERG and PTEN in HGPIN.

A, Matrix representation of ERG and PTEN genomic aberrations in selected localized prostate cancer patients with paired HGPIN (Adj: HGPIN adjacent to cancer; Away: HGPIN away from cancer) assessed by FISH. Patient case numbers are shown on the left of the matrix map. Each column represents one case and each row represents FISH evaluation for aberrations of ERG or PTEN. Color legend signifies respective aberrations or availability. B, Representative reconstructed maps of the prostectomy sections in case T9 (B1) and T6 (B2). Tumor is represented as T; HGPINadj and HGPINaway are represented as PIN1 and PIN2, respectively. A summary of genomic aberrations of ERG and PTEN for each of these foci is presented in the boxes.
Homogeneity of ERG rearrangement, but not PTEN deletion in multiple metastatic sites of androgen-independent prostate cancer
Previously, we validated TMPRSS2-ETS aberrations in 30 androgen-independent metastatic prostate cancer patients. In patients exhibiting ERG aberrations, we observed that multiple metastatic sites from an individual case harbored the same TMPRSS2-ERG rearrangement, all of which occurred through intrachromosomal deletion.12 In this study, we extrapolated our initial findings to 47 metastatic prostate cancer patients representing the University of Michigan warm autopsy cohort and evaluated ERG status in 156 tumor foci from different organs as well as the prostate (when present). Similar to our previous study,12 ERG was rearranged in 35% (15/43) of cases, and cases were rearranged through deletion of 5′ end of ERG, except M39, in which ERG split (translocation) was observed (Figure 3A). Notably, homogeneous ERG aberrations were present in all metastatic sites and primary tumors (when present) within an individual patient. These results support the concept that ERG rearrangement occurs at the clinically localized stage before progression to an androgen-independent metastatic stage in prostate cancer.
Figure 3. Genomic aberrations of ERG and PTEN in androgen-independent metastatic prostate cancer.
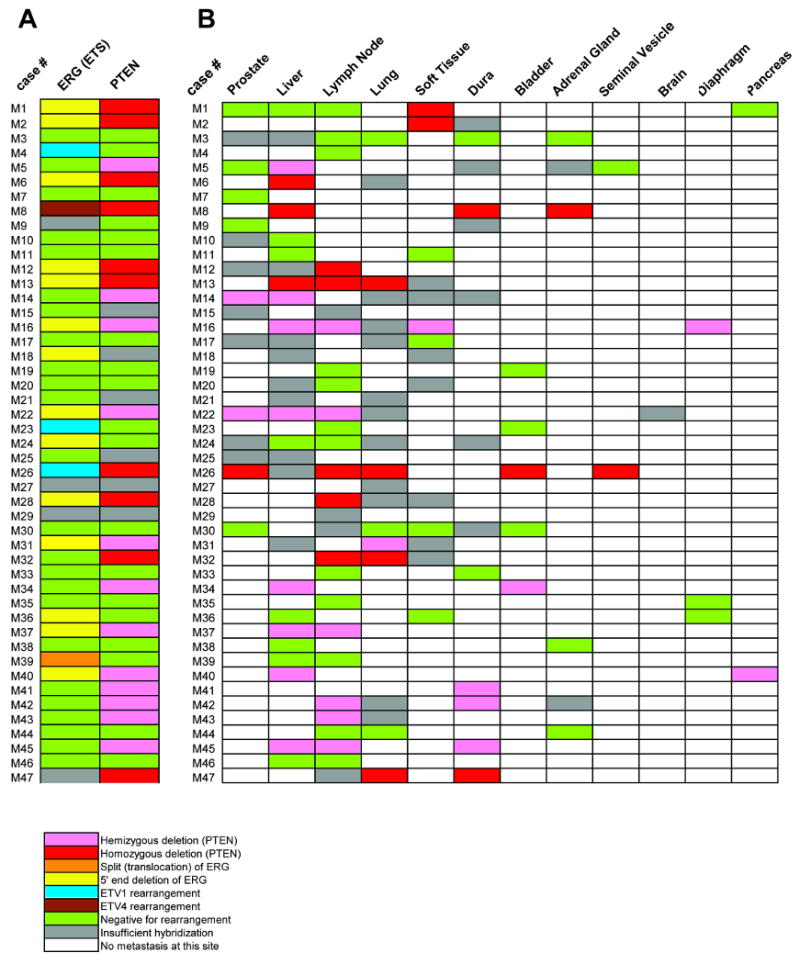
A, Matrix representation of the genomic aberrations of ERG and PTEN in metastatic prostate cancers. Patient case numbers are shown on the left of the map. Each column represents one case and each row represents FISH evaluation for ERG or PTEN aberration. Rearrangements of ETV1 and ETV4 in this cohort that were previously published are indicated. B, Matrix representation of PTEN deletion status of the metastatic sites and residual tumor in the prostate (when present) as evaluated in this cohort. Patient case numbers are shown on the left of the map. Each column represents one case and each row represents FISH evaluation for PTEN deletion at each organ site. Color legend signifies respective aberrations or availability.
We further analyzed PTEN deletion status across all metastatic sites in these 47 cases. As shown in Figure 3B, of 41 interpretable warm autopsy cases, 39 showed concordant PTEN deletion status across all metastatic sites and primary tumors in prostate (when present). By contrast, in two cases (M1 and M5), hemizygous PTEN deletion was identified in metastatic foci of soft tissue (M1) or liver (M5), but no deletion was present in the prostate and other metastatic sites. We further examined all available paraffin tissue specimens for all the metastatic sites and primary tumors but did not find any additional cancer foci harboring PTEN deletion in these two cases (data not shown). Thus these results suggested that PTEN deletion could occur after dissemination of tumor in a small subset of androgen-independent metastatic cancer patients.
Comparison of PTEN genomic deletion and PTEN protein expression by IHC
We previously reported that more than 95% of prostate cancer cases with ERG over-expression harbor TMPRSS2-ERG gene fusions.3 By contrast, multiple mechanisms account for loss of PTEN protein expression, including genomic deletion, mutation, and promoter methylation.23 To explore the association of PTEN gene status with PTEN protein expression, we analyzed 207 prostate cancer patients in this cohort. Predominantly cytoplasmic, with occasional nuclear, staining of PTEN was observed by IHC (Figure 4A). As shown in Table 3, statistically significant associations were observed between PTEN deletion and decreased PTEN protein expression in both localized (p < 0.0001) and metastatic (p = 0.045) cancer. Of note, 26 (19%) localized cancer cases revealed decreased PTEN protein expression, but were negative for PTEN genomic deletion, suggesting that reduced PTEN expression might be due to other mechanisms. Additionally, we detected PTEN protein expression in selected HGPINadj with known PTEN genomic aberrations in localized cancer T7-T11 and T13 (Figure 2A). As expected, all but one (HGPINadj in T9) exhibited decreased or absent PTEN expression.
Figure 4. PTEN expression in prostate cancer by immunohistochemistry.

A, Representative case of prostate cancer exhibits positive staining for PTEN graded as 2 (greater than or equivalent to normal adjacent tissues in the same section) (A1, H&E stained section. A2, immunohistochemical staining). Original magnification, 200 X. B, Representative case of prostate cancer exhibits weak staining for PTEN graded as 1 (B1, H&E staining. B2, immunohistochemical staining). C, Representative case of prostate cancer exhibits absence staining for PTEN graded as 0. (C1, H&E staining. C2, immunohistochemical staining). Original magnification, 200 X.
Table 3. The correlation between PTEN deletion status and PTEN protein expression in prostate cancer.
| PTEN protein (IHC) | ||||
|---|---|---|---|---|
| PTEN gene | 0 | 1+ | 2+ | P-value |
| Localized prostate cancer | ||||
| No deletion | 6 | 20 | 93 | p < 0.0001 |
| Hemizygous | 7 | 3 | 1 | |
| Homozygous | 4 | 6 | 0 | |
| Metastatic prostate cancer | ||||
| No deletion | 3 | 5 | 9 | p = 0.045 |
| Hemizygous | 3 | 7 | 2 | |
| Homozygous | 4 | 5 | 1 | |
Concordance between PTEN deletion status and PTEN protein expression was also observed in 28 out of 39 (72%) androgen-independent metastases (Table 3). Out of 11 discordant cases, decreased (grade 1) or absence (grade 0) of PTEN protein expression was observed in 8 cases that were negative for PTEN deletion. In the remaining 3 cases, normal PTEN protein expression was identified, although PTEN hemizygous deletion was observed.
Discussion
ERG rearrangement and PTEN deletion are two of the most common genomic events in human prostate cancer. In this initial study, we used rearrangements in ERG as a marker for ETS rearrangements in prostate cancer, as >90% of all ETS rearrangements involve ERG.10 Additionally, as only TMPRSS2 and SLC45A3 have been identified as 5′fusion partners of ERG, with SLC45A3-ERG being extremely rare in our PSA-screened radical prostatectomy series,9 we can conclude that most of the ERG rearranged prostate cancers are TMPRSS2-ERG fusions (>97%). In the current study, for the first time, we have observed significant association of ERG rearrangement and PTEN deletion in clinically localized (n=281) and androgen-independent metastatic prostate cancers (n=47). Biologically, we and others have reported that overexpression of ERG resulted in increased cellular invasion in vitro.14, 15 By contrast, PTEN deletion was associated with a gain of transformation potential and marked increase in cellular proliferation in prostate cancer through its negative regulation of the PI3K pathway.24 Of note, many studies have demonstrated that PTEN can synergize with other oncogenic factors or related genes in mouse models, including NKx3.1, p27 and p53 to promote cancer development and androgen independence.25, 42, 43 Although it is unknown whether crosstalk exists between ERG and the PTEN-PI3K pathway, we hypothesize that the interactions between these two genomic aberrations may be synergic or additive. Indeed, while this study was in preparation, Yoshimoto et al. reported that concurrent PTEN deletion and TMPRSS2-ERG fusions was present in a subset of prostate cancer cases and associated with an unfavorable outcome.34 Using Oncomine, they further attempted to explore the potential signaling pathways involved for synergy between these genomic events. How ERG overexpression interacts with PTEN deletion is still unclear and compound transgenic mice recapitulating these lesions will likely be ideal models to further explore this relationship. Using IHC, we also identified significant correlation between decreased PTEN expression and PTEN genomic deletion status, supporting deletion as a major mechanism leading to decreased PTEN protein expression.
Further, we attempted to investigate the roles of ERG and PTEN genomic aberrations in prostate cancer progression. Of note, in line with previous studies, no benign prostate glands, atrophy, or BPH harbored ERG rearrangements.13, 16 ERG rearrangement was present in ∼15% of HGPIN lesions in this series. In addition, in 11 selected ERG rearranged cancer cases, 100% (11/11) HGPIN shared the same ERG rearrangement status as the paired cancer foci, and all of these HGPIN foci were adjacent to the cancer. Although we can not exclude the possibility that HGPINadj lesions may represent an intraepithelial spread from the adjacent invasive cancer or evolved together temporally and developed shared genetic abnormities at the same time, our data, in line with previous studies,16 strongly supported ERG aberrations as an early molecular event in prostate cancer development. Previously, we and other groups have reported that TMPRSS2-ERG gene fusion could induce HGPIN in transgenic mice.14, 15 It is still not clear whether aberrant ERG overexpression in human induces HGPIN or may drive cancer progression during the transition from HGPIN to localized cancer in human. Further functional characterization, especially using animal models, would be helpful in addressing this issue. By contrast, PTEN deletion was not consistently identical both in localized cancer and adjacent HGPIN. These findings suggested that PTEN deletion may not play a significant role in a subset of HGPIN lesions, which develop to localized cancer eventually. In comparison with localized prostate cancers, the prevalence of ERG aberration was similar in androgen-independent metastatic cancer (35% vs 47%) in this study. If ERG aberrations were a later genomic event in prostate cancer progression, one would expect a higher percentage of metastatic prostate cancers to be rearrangement positive. Consistent with previous findings, all metastatic foci from an individual case were uniformly ERG rearrangement negative or positive, indicating that ERG rearrangement occurred before progression to metastatic disease.12
Although a series of in vivo studies have showed that PTEN haploinsufficiency results in the HGPIN in transgenic mouse,23, 24, 42, 43 in the current study, only 9% (3/33) of human HGPIN lesions harbored PTEN hemizygous deletion. By contrast, we observed a strikingly increase of prevalence for PTEN deletion in androgen-independent metastatic cancer (54%), which is comparable with those reported in recent studies.31, 33 These findings suggest that PTEN deletion is more likely a late event in prostate cancer progression, although this genetic aberration occurs earlier in tumor evolution in a subset of cases. Further, discordance of PTEN status in different organ sites was observed in a subset of androgen-independent metastatic cancers. That is, PTEN genomic deletion was detected in only one of multiple metastases in cases M1 and M5, but absent in the primary tumor and other organs. This suggests that PTEN deletion occurred during or after the formation of metastatic foci, possibly due to more generalized genomic instability seen in disseminated cancer. Similarly, Suzuki et al. has also observed heterogeneity of PTEN genetic aberrations between different metastatic sites within the same patient.30 Considering the important role of PTEN and high frequency of PTEN aberration in late-stage prostate cancer, one could assume that PTEN deletion may be a critical “second hit” after ERG rearrangement in a subset of ERG fusion prostate cancer cases. Supporting this concept, by array CGH, Lapointe et al. have reported that TMRPSS2-ERG fusions appears to occur prior to PTEN genomic deletion.44
It has been reported that PTEN dose is a key determinant in prostate cancer progression in mouse models.26 However, our study did not observe a significant difference of PTEN deletion patterns (hemizygous vs homozygous) between localized and metastatic cancers. Therefore, these data suggest that human localized and metastatic prostate cancers do not seem to select for homozygous deletion. Alternatively, hemizygous deletion of PTEN may cooperate with other oncogenic factors including ERG rearrangements during prostate cancer progression, manifesting a phenotype that is equivalent to PTEN homozygous deletion.
Progression to androgen independence is a complex process including clonal selection and adaptation. Recent evidence suggested that PTEN deletion was associated with androgen independence and could functionally control “two” hits: cell transformation and androgen-independent growth.32, 33 In the current study, we showed that almost all androgen-independent metastatic cancers harboring ERG rearrangement were associated with the 5′ deletion of ERG. Thus, we could expect that concomitance of PTEN deletion and ERG rearrangement (through 5′ end deletion) may select for more aggressive cancers that are able to progress to androgen-independent metastatic cancer. Notably, the co-existence of the PTEN deletion and the ERG rearrangement was present in nearly one third (11/39) of metastatic cases, underscoring the combinatorial roles of aberrant ERG and PTEN in metastatic cancer.
Collectively, our data support PTEN deletion as a late genetic event in human prostate cancer, possibly as a “second hit” after ERG rearrangement. ERG rearrangement and PTEN deletion may cooperate, but contribute at different stages in prostate cancer progression. Although future work is needed to test this notion, understanding the molecular crosstalk between these two genetic events may provide insight into understanding prostate cancer development. Also, it suggests that simultaneous therapeutic targeting of PTEN and ETS gene fusions may be important for treating a subset of advanced prostate cancers.
Supplementary Material
Acknowledgments
We thank Jill Granger for editorial review of this manuscript. Supported in part by Department of Defense (PC040517 to R.M., W81XWH-06-1-0224), the National Institutes of Health (Prostate SPORE P50CA69568, R01 CA102872), the Early Detection Research Network (UO1 CA111275-01), the Prostate Cancer Research Foundation and a sponsored research agreement from Gen-Probe, Inc. A.M.C. is supported by a Clinical Translational Research Award from the Burroughs Welcome Foundation. S.A.T. is a Fellow of the Medical Scientist Training Program and is supported by a Young Investigator Award from the Prostate Cancer Foundation.
Footnotes
Disclosure: The University of Michigan has filed a patent on ETS gene rearrangements in prostate cancer, on which R.M., S.A.T., and A.M.C. are co-inventors, and the diagnostic field of use has been licensed to Gen-Probe Incorporated. Gen-Probe has not played a role in the design and conduct of the study, nor in the collection, analysis, or interpretation of the data, and no involvement in the preparation, review, or approval of the manuscript. A.M.C. serves as a consultant to Gen-Probe Inc.
Contributor Information
Bo Han, Email: boh@umich.edu.
Rohit Mehra, Email: mrohit@umich.edu.
Robert J. Lonigro, Email: lonigro@umich.edu.
Lei Wang, Email: wanglei@umich.edu.
Khalid Suleman, Email: khalidsu@umich.edu.
Anjana Menon, Email: amenon@umich.edu.
Nallasivam Palanisamy, Email: nallasiv@umich.edu.
Scott A. Tomlins, Email: tomlinss@umich.edu.
Arul M. Chinnaiyan, Email: arul@umich.edu.
Rajal B. Shah, Email: rajshah@umich.edu.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 2.Abate-Shen C, Shen MM. Molecular genetics of prostate cancer. Genes Dev. 2000;14:2410–34. doi: 10.1101/gad.819500. [DOI] [PubMed] [Google Scholar]
- 3.Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–8. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- 4.Tomlins SA, Mehra R, Rhodes DR, et al. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- 5.Helgeson BE, Tomlins SA, Shah N, et al. Characterization of TMPRSS2:ETV5 and SLC45A3:ETV5 gene fusions in prostate cancer. Cancer Res. 2008;68:73–80. doi: 10.1158/0008-5472.CAN-07-5352. [DOI] [PubMed] [Google Scholar]
- 6.Hermans KG, Bressers AA, van der Korput HA, Dits NF, Jenster G, Trapman J. Two unique novel prostate-specific and androgen-regulated fusion partners of ETV4 in prostate cancer. Cancer Res. 2008;68:3094–8. doi: 10.1158/0008-5472.CAN-08-0198. [DOI] [PubMed] [Google Scholar]
- 7.Attard G, Clark J, Ambroisine L, et al. Heterogeneity and clinical significance of ETV1 translocations in human prostate cancer. Br J Cancer. 2008;99:314–20. doi: 10.1038/sj.bjc.6604472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tomlins SA, Laxman B, Dhanasekaran SM, et al. Distinct classes of chromosomal rearrangements create oncogenic ETS gene fusions in prostate cancer. Nature. 2007;448:595–9. doi: 10.1038/nature06024. [DOI] [PubMed] [Google Scholar]
- 9.Han B, Mehra R, Dhanasekaran SM, et al. A fluorescence in situ hybridization screen for E26 transformation-specific aberrations: identification of DDX5-ETV4 fusion protein in prostate cancer. Cancer Res. 2008;68:7629–37. doi: 10.1158/0008-5472.CAN-08-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kumar-Sinha C, Tomlins SA, Chinnaiyan AM. Recurrent gene fusions in prostate cancer. Nat Rev Cancer. 2008;8:497–511. doi: 10.1038/nrc2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mehra R, Tomlins SA, Shen R, et al. Comprehensive assessment of TMPRSS2 and ETS family gene aberrations in clinically localized prostate cancer. Mod Pathol. 2007;20:538–44. doi: 10.1038/modpathol.3800769. [DOI] [PubMed] [Google Scholar]
- 12.Mehra R, Tomlins SA, Yu J, et al. Characterization of TMPRSS2-ETS gene aberrations in androgen-independent metastatic prostate cancer. Cancer Res. 2008;68:3584–90. doi: 10.1158/0008-5472.CAN-07-6154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res. 2006;66:8337–41. doi: 10.1158/0008-5472.CAN-06-1482. [DOI] [PubMed] [Google Scholar]
- 14.Tomlins SA, Laxman B, Varambally S, et al. Role of the TMPRSS2-ERG gene fusion in prostate cancer. Neoplasia. 2008;10:177–88. doi: 10.1593/neo.07822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klezovitch O, Risk M, Coleman I, et al. Proc Natl Acad Sci U S A. 2008;105:2105–10. doi: 10.1073/pnas.0711711105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perner S, Mosquera JM, Demichelis F, et al. TMPRSS2-ERG fusion prostate cancer: an early molecular event associated with invasion. Am J Surg Pathol. 2007;31:882–8. doi: 10.1097/01.pas.0000213424.38503.aa. [DOI] [PubMed] [Google Scholar]
- 17.Cerveira N, Ribeiro FR, Peixoto A, et al. TMPRSS2-ERG gene fusion causing ERG overexpression precedes chromosome copy number changes in prostate carcinomas and paired HGPIN lesions. Neoplasia. 2006;8:826–32. doi: 10.1593/neo.06427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosquera JM, Perner S, Genega EM, et al. Characterization of TMPRSS2-ERG fusion high-grade prostatic intraepithelial neoplasia and potential clinical implications. Clin Cancer Res. 2008;14:3380–5. doi: 10.1158/1078-0432.CCR-07-5194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rajput AB, Miller MA, De Luca A, et al. Frequency of the TMPRSS2:ERG gene fusion is increased in moderate to poorly differentiated prostate cancers. J Clin Pathol. 2007;60:1238–43. doi: 10.1136/jcp.2006.043810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demichelis F, Fall K, Perner S, et al. TMPRSS2:ERG gene fusion associated with lethal prostate cancer in a watchful waiting cohort. Oncogene. 2007;26:4596–9. doi: 10.1038/sj.onc.1210237. [DOI] [PubMed] [Google Scholar]
- 21.Gopalan A, Leversha MA, Satagopan JM, et al. TMPRSS2-ERG gene fusion is not associated with outcome in patients treated by prostatectomy. Cancer Res. 2009;69:1400–6. doi: 10.1158/0008-5472.CAN-08-2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saramaki OR, Harjula AE, Martikainen PM, Vessella RL, Tammela TL, Visakorpi T. TMPRSS2:ERG fusion identifies a subgroup of prostate cancers with a favorable prognosis. Clin Cancer Res. 2008;14:3395–400. doi: 10.1158/1078-0432.CCR-07-2051. [DOI] [PubMed] [Google Scholar]
- 23.Di Cristofano A, Pandolfi PP. The multiple roles of PTEN in tumor suppression. Cell. 2000;100:387–90. doi: 10.1016/s0092-8674(00)80674-1. [DOI] [PubMed] [Google Scholar]
- 24.Sansal I, Sellers WR. The biology and clinical relevance of the PTEN tumor suppressor pathway. J Clin Oncol. 2004;22:2954–63. doi: 10.1200/JCO.2004.02.141. [DOI] [PubMed] [Google Scholar]
- 25.Chen Z, Trotman LC, Shaffer D, et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–30. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trotman LC, Niki M, Dotan ZA, et al. Pten dose dictates cancer progression in the prostate. PLoS Biol. 2003;1:E59. doi: 10.1371/journal.pbio.0000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCall P, Witton CJ, Grimsley S, Nielsen KV, Edwards J. Is PTEN loss associated with clinical outcome measures in human prostate cancer? Br J Cancer. 2008;99:1296–301. doi: 10.1038/sj.bjc.6604680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li J, Yen C, Liaw D, et al. PTEN, a putative protein tyrosine phosphatase gene mutated in human brain, breast, and prostate cancer. Science. 1997;275:1943–7. doi: 10.1126/science.275.5308.1943. [DOI] [PubMed] [Google Scholar]
- 29.Yoshimoto M, Cunha IW, Coudry RA, et al. FISH analysis of 107 prostate cancers shows that PTEN genomic deletion is associated with poor clinical outcome. Br J Cancer. 2007;97:678–85. doi: 10.1038/sj.bjc.6603924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Suzuki H, Freije D, Nusskern DR, et al. Interfocal heterogeneity of PTEN/MMAC1 gene alterations in multiple metastatic prostate cancer tissues. Cancer Res. 1998;58:204–9. [PubMed] [Google Scholar]
- 31.McMenamin ME, Soung P, Perera S, Kaplan I, Loda M, Sellers WR. Loss of PTEN expression in paraffin-embedded primary prostate cancer correlates with high Gleason score and advanced stage. Cancer Res. 1999;59:4291–6. [PubMed] [Google Scholar]
- 32.Jiao J, Wang S, Qiao R, et al. Murine cell lines derived from Pten null prostate cancer show the critical role of PTEN in hormone refractory prostate cancer development. Cancer Res. 2007;67:6083–91. doi: 10.1158/0008-5472.CAN-06-4202. [DOI] [PubMed] [Google Scholar]
- 33.Bertram J, Peacock JW, Fazli L, et al. Loss of PTEN is associated with progression to androgen independence. Prostate. 2006;66:895–902. doi: 10.1002/pros.20411. [DOI] [PubMed] [Google Scholar]
- 34.Yoshimoto M, Joshua AM, Cunha IW, et al. Absence of TMPRSS2:ERG fusions and PTEN losses in prostate cancer is associated with a favorable outcome. Mod Pathol. 2008 doi: 10.1038/modpathol.2008.96. [DOI] [PubMed] [Google Scholar]
- 35.Mehra R, Han B, Tomlins SA, et al. Heterogeneity of TMPRSS2 gene rearrangements in multifocal prostate adenocarcinoma: molecular evidence for an independent group of diseases. Cancer Res. 2007;67:7991–5. doi: 10.1158/0008-5472.CAN-07-2043. [DOI] [PubMed] [Google Scholar]
- 36.Guo CC, Epstein JI. Intraductal carcinoma of the prostate on needle biopsy: Histologic features and clinical significance. Mod Pathol. 2006;19:1528–35. doi: 10.1038/modpathol.3800702. [DOI] [PubMed] [Google Scholar]
- 37.Korshunov A, Sycheva R, Gorelyshev S, Golanov A. Clinical utility of fluorescence in situ hybridization (FISH) in nonbrainstem glioblastomas of childhood. Mod Pathol. 2005;18:1258–63. doi: 10.1038/modpathol.3800415. [DOI] [PubMed] [Google Scholar]
- 38.Han B, Mori I, Wang X, Nakamura M, Nakamura Y, Kakudo K. Combined small-cell carcinoma of the stomach: p53 and K-ras gene mutational analysis supports a monoclonal origin of three histological components. Int J Exp Pathol. 2005;86:213–8. doi: 10.1111/j.0959-9673.2005.00424.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Perren A, Weng LP, Boag AH, et al. Immunohistochemical evidence of loss of PTEN expression in primary ductal adenocarcinomas of the breast. Am J Pathol. 1999;155:1253–60. doi: 10.1016/S0002-9440(10)65227-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schmitz M, Grignard G, Margue C, et al. Complete loss of PTEN expression as a possible early prognostic marker for prostate cancer metastasis. Int J Cancer. 2007;120:1284–92. doi: 10.1002/ijc.22359. [DOI] [PubMed] [Google Scholar]
- 41.Yoshimoto M, Cutz JC, Nuin PA, et al. Interphase FISH analysis of PTEN in histologic sections shows genomic deletions in 68% of primary prostate cancer and 23% of high-grade prostatic intra-epithelial neoplasias. Cancer Genet Cytogenet. 2006;169:128–37. doi: 10.1016/j.cancergencyto.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 42.Kim MJ, Cardiff RD, Desai N, et al. Cooperativity of Nkx3.1 and Pten loss of function in a mouse model of prostate carcinogenesis. Proc Natl Acad Sci U S A. 2002;99:2884–9. doi: 10.1073/pnas.042688999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Di Cristofano A, De Acetis M, Koff A, Cordon-Cardo C, Pandolfi PP. Pten and p27KIP1 cooperate in prostate cancer tumor suppression in the mouse. Nat Genet. 2001;27:222–4. doi: 10.1038/84879. [DOI] [PubMed] [Google Scholar]
- 44.Lapointe J, Li C, Giacomini CP, et al. Genomic profiling reveals alternative genetic pathways of prostate tumorigenesis. Cancer Res. 2007;67:8504–10. doi: 10.1158/0008-5472.CAN-07-0673. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


