Abstract
The molecular diversity of phospholipids is essential for their structural and signaling functions in cell membranes. In the current work, we present, the results of mass spectrometric characterization of individual molecular species in major classes of phospholipids -phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn), phosphatidylserine (PtdSer), phosphatidylinositol (PtdIns), sphingomyelin (CerPCho), and cardiolipin (Ptd2Gro) - and their oxidation products during apoptosis induced in neurons by staurosporine (STS). The diversity of molecular species of phospholipids in rat cortical neurons followed the order Ptd2Gro > PtdEtn ≫ PtdCho ≫ PtdSer > PtdIns > CerPCho. The number of polyunsaturated oxidizable species decreased in the order Ptd2Gro ≫ PtdEtn > PtdCho > PtdSer > PtdIns > CerPCho. Thus a relatively minor class of phospholipids, Ptd2Gro, was represented in cortical neurons by the greatest variety of both total and peroxidizable molecular species. Quantitative fluorescence HPLC analysis employed to assess the oxidation of different classes of phospholipids in neuronal cells during intrinsic apoptosis induced by staurosporine (STS) revealed that three anionic phospholipids — Ptd2Gro ≫ PtdSer > PtdIns — underwent robust oxidation. No significant oxidation in the most dominant phospholipid classes – PtdCho and PtdEtn – was detected. MS-studies revealed the presence of hydroxy-, hydroperoxy- as well as hydroxy-/hydroperoxy-species of Ptd2Gro, PtdSer, and PtdIns. Experiments in model systems where total cortex Ptd2Gro and PtdSer fractions were incubated in the presence of cytochrome c (cyt c) and H2O2, confirmed that molecular identities of the products formed were similar to the ones generated during STS-induced neuronal apoptosis. The temporal sequence of biomarkers of STS induced apoptosis and phospholipid peroxidation combined with recently demonstrated redox catalytic properties of cyt c realized through its interactions with Ptd2Gro and PtdSer suggest that cyt c acts as a catalyst of selective peroxidation of anionic phospholipids yielding Ptd2Gro and PtdSer peroxidation products. These oxidation products participate in mitochondrial membrane permeability transition and in PtdSer externalization leading to recognition and uptake of apoptotic cells by professional phagocytes.
INTRODUCTION
Phospholipids are the major building blocks of the membrane bilayer of plasma and intracellular membranes. Latest characterizations by soft ionization mass spectrometry revealed that their diversified molecular speciation and asymmetry define many essential characteristics of membrane identity and physiology (Watson 2006, Schwab et al. 2007).
In mammals, the fatty acid residues in sn-2 position of phospholipids are polyunsaturated hence they are highly prone to oxygenation (Kagan 1988, Spiteller 2006). The significance of this is that phospholipids are precursors of important signaling molecules whereby their hydrolytic or oxidative metabolism, in particular eicosanoid and docosanoid pathways, are entirely dependent on availability of phospholipid substrates for phospholipase A2 (PLA2) hydrolysis, release of fatty acids and their subsequent oxygenation (Nigam & Schewe 2000, Lambert et al. 2006, McGinley & van der Donk 2003, Schneider et al. 2007). A variety of cyclooxygenases, lipooxygenases and myeloperoxidases are involved in these metabolic pathways (Phillis et al. 2006, Schneider et al. 2007, Heinecke 2007, Serhan et al. 2008). Lately, neuroprotectins and resolvins have been identified as signaling molecules formed by multistage oxygenation of docosahexaenoic acid (DHA) and eicosapentaenoic acid (EPA) (Bazan 2005, Serhan et al. 2008). An alternative pathway may include initiating peroxidative metabolism of phopsholipids followed by hydrolytic reactions to release oxygenated fatty acids (Kagan 1988). This pathway is much less explored yet may represent a substantial source of regulatory oxygenated fatty acid molecules. Particularly, enzymatic mechanisms involved in the initial peroxidation stage of polyunsaturated phospholipids have not been identified. Studies initiated in the early 1970s and continued in the 1980s demonstrated that peroxidized phospholipids are preferred substrates of PLA2 reactions leading to release of oxidatively modified fatty acids (Kagan et al. 1978, Sevanian et al. 1988, Rashba-Step et al. 1997). Further, the peroxyredoxin family of enzymes catalyzes reduction and subsequent hydrolysis of peroxidized phospholipids yielding oxygenated fatty acids (Manevich et al. 2007).
Finally, non-hydrolyzed oxidized phospholipids have been also demonstrated to act as signals in monocyte activation, programmed cell death and phagocytotic clearance of apoptotic cells (Maskrey et al. 2007, Walton et al. 2003, Greenberg et al. 2006, Tyurina et al. 2004b). Molecular characterization of the latter pathway is just beginning to emerge mostly due to employment of mass spectrometric protocols.
Lately two anionic phospholipids – cardiolipin (Ptd2Gro) in mitochondria and phosphatidylserine (PtdSer) in extra-mitochondrial compartments - have been identified as oxidation substrates of cytochrome c (cyt c) catalyzed reactions (Kagan et al. 2004, Kagan et al. 2005). Accumulation of their oxidation products has been associated with the release of pro-apoptotic factors from mitochondria into the cytosol and externalization of PtdSer on the cell surface, respectively. Thus two major physiological processes, programmed cell death and clearance of apoptotic cells, are dependent on the coordinated oxidation of membrane phospholipids. However details of molecular characterization of individual oxidized species of Ptd2Gro and PtdSer remain to be elucidated. Notably accumulation of oxidized species of Ptd2Gro and PtdSer and their molecular identity have been documented in cerebral cortex of rats exposed to traumatic brain injury (Bayir et al. 2007). Association of the anionic phospholipid oxidation products with apoptosis as well as their confinement to neurons, while suggested, have not been directly established. Recently recognized great diversity of polyunsaturated phospholipids molecular species in neurons (Cheng et al. 2008) suggests that their oxidation products may play key signaling roles.
In the current work, we present the results of mass spectrometric characterization of molecular diversity of major classes of phospholipids and their oxidation products during apoptosis induced in neurons by staurosporine (STS).
MATERIALS and METHODS
Reagents
1,2-Diheptadecanoyl-sn-glycero-3-[phospho-L-serine] (sodium salt), 1,1′,2,2′-tetramyristoyl-cardiolipin (sodium salt), and 1,1′,2,2′-tetralinoleoyl-cardiolipin (sodium salt) were from Avanti Polar Lipids Inc. (Alabaster, AL). Chloroform, methanol, acetone, ammonium hydroxide, acetic acid glacial, CaCl2, ethylene diamine tetraacetic acid (EDTA), ethylene glycol tetraacetic acid (EGTA), diethylene triamine pentaacetic acid (DTPA), sodium dodecyl sulfate (SDS), cyt c, microperoxidase-11, STS, PLA2, butylated hydroxytoluene (BHT) and triphenylphosphine (TPP) were from Sigma-Aldrich (St. Louis, MO). HPTLC silica G plates were purchased from Whatman, (Schleicher & Schuell, England). N-acetyl-3, 7-dihydroxyphenoxazine (Amplex Red) resorufin were from Molecular Probes (Eugene, OR).
Cell culture
Neurons were harvested from fetuses of Sprague-Dawley rats (at embryologic day 16–18) as described previously (Du et al. 2004). Dissociated cell suspensions were filtered through a 70μm Falcon nylon cell strainer then seeded in 96-well plates (at a density of 5×104 cells/cm2), or plastic dishes coated with 100 μg/mL poly-D-lysine (at a density of 1.3×107 cells/cm2). Each contained Neurobasal™ medium supplemented with B27™ (Gibco) and GlutaMaxl™ (Sigma) for neuron-enriched cultures (Brewer et al. 1993). Rat cortical neurons were grown in Neurobasal medium containing 2 mM L-glutamine, 100 U/mL penicillin/streptomycin supplemented with 2% B27 at 37°C in a humidified atmosphere (5% CO2 plus 95% air). On the 2nd and 6th day in vitro the culture media was replaced with fresh media. Experiments were done at 8–12 day in vitro, when cultures consist primarily of neurons (>95% MAP2 immunopositive cells, <5% GFAP immunopositive cells). Neuronal cultures were incubated in the presence of STS (1 μM) in culture media for 2, 4 and 8 hours.
Biomarkers of apoptosis
Cytochrome c ELISA
Cells at DIV 8–10 were untreated or treated with STS for different time intervals as described. After treatment, cells were collected and lysed in preparation buffer (250 mM sucrose, 20 mM HEPES-KOH (pH 7.4), 10 mM NaCl, 1.5 mM MgCl2, 1mM EDTA, 1 mM EGTA, 1 mM DTT, 2 μg/mL aprotinin and 1 mM PMSF) for 10 min on ice followed by centrifugation at 4°C for 20 min at 10,000g to separate the cytosolic and mitochondrial fraction. Cyt c concentrations were determined by ELISA in 96-well plates as per manufacturer’s instruction (R&D Systems, Minneapolis, MN). Protein concentration was measured by BioRad kit. The ELISA was repeated in triplicate to calculate the average cyt c (ng) in cytosolic or mitochondrial fractions per mg of protein.
PtdSer externalization was determined by annexin V-FITC apoptosis detection kit (BioVision, Mountain View, CA) using a FACScan flow cytometer (Becton Dickinson, San Jose, CA).
Caspase 3 Activity Assays
Caspase-3/7 activity was measured using a luminescence Caspase Glo® 3/7 assay kit (Promega, Madison, WI). Luminescence was measured using a plate reading chemiluminometer ML1000 (Dynatech Laboratories). Activity of caspase-3/7 was expressed as luminescence arbitrary units (LAU) per mg protein. In addition, Caspase-3 activity was determined using a fluorometric method using EnzChek caspase-3 Assay kit (Invitrogen, Carlsbad, CA) according to the manufacturer’s instructions. The fluorescence of free aminomethylcoumarin (AMC), generated by cleavage of z-DEVD-AMC was measured by a Fusion Plate Reader (PerkinElmer, Boston, MA) at excitation wavelength of 365/50 nm and emission wavelength of 460/35 nm. Fluorescent units were converted to pmol of free AMC using a standard curve generated with reagent AMC. Specificity for caspase-3-like activity was confirmed by inhibition with Ac-DEVD-CHO.
Western blot analysis of active caspase 3
Following treatment, cells were scraped to harvest and lyzed in RIPA buffer with protease inhibitors. Proteins (50 μg) were resolved by 12% SDS-PAGE and detected by Western blot using an antibody to the larger (p17) subunit of active caspase 3 (1:250, Abcam, Cambridge, MA). Horseradish peroxidase conjugated goat anti-mouse immunoglobulin G (KPL, Gaithersburg, MA) was used as a secondary antibody at a dilution of 1 in 5000. To confirm equal protein concentrations in samples, membranes were stripped and re-blotted with a primary antibody to β-actin (Novus, 1:5000) and processed as described above.
Lipid extraction and 2D-HPTLC analysis
Total lipids were extracted from cells by Folch procedure (Folch et al. 1957). Lipid extracts were separated and analyzed by 2D-HPTLC (Rouser et al. 1970). Special measures were taken to prevent oxidative modification of phospholipids during their processing and separation. To bind adventitious transition metals from silica, plates were treated with methanol containing 1 mM EDTA, 100 μM DTPA prior to application and separation of phospholipids by 2D-HPTLC. Then the plates were first developed with a solvent system consisting of chloroform: methanol: 28% ammonium hydroxide (65:25:5 v/v). After the plates were dried with a forced N2 blower to remove the solvent, they were developed in the second dimension with a solvent system consisting of chloroform:acetone:methanol:glacial acetic acid:water (50:20:10:10:5 v/v). The phospholipids were visualized by exposure to iodine vapors and identified by comparison with authentic phospholipid standards.
Plasmalogens are liable to hydrolysis under acidic conditions; some investigators use acetic acid (90%, 40–100°C, 4–15 h, in the presence of 0.1 N HCl) for hydrolysis of vinyl bonds. Although several reports alert about possible interferences by using acidic conditions during TLC of phospholipids (Murphy et al. 1993), separation of phospholipids (including plasmalogens) generally accepted protocols for TLC, HPLC, ESI-MS include low amounts of acetic acid (1–10%) (Kerwin et al. 1994, Koivusalo et al. 2001, Albert et al. 2001, Pulfer & Murphy 2003). In our experiments, the second solvent system employed for TLC of phospholipids (5×5 cm TLC plates, 5 min run) contained acetic acid (10%). We did not observe any evidence of hydrolysis of vinyl ether lipids after iodine exposure. Isolated PtdEtn and PtdCho gave single spots on TLC.
For electrospray ionization mass spectrometry (ESI-MS) and analysis of phospholipid hydroperoxides by fluorescence HPLC using Amplex Red, the phospholipid spots on the silica plates were visualized by spraying the plates with deionized water. After this, the spots were scraped from the silica plates and phospholipids were extracted by choloroform:methanol:water (20:10:2 v/v). Extracted phospholipids were divided into aliquots for phosphorus, Amplex Red and ESI-MS analysis. Lipid phosphorus was determined by a micro-method (Böttcher et al. 1961). Phospholipid hydroperoxides were determined by fluorescence HPLC of products formed in MP-11 catalyzed reaction with a fluorogenic substrate, Amplex Red (Kagan et al. 2005). To reduce Ptd2Gro –OOH and PtdSer -OOH into Ptd2Gro –OH and PtdSer -OH, respectively, oxidized Ptd2Gro and PtdSer (50 μg/mL) were incubated in ethanol with a known reductant of organic hydroperoxides, TPP (1 mg/mL, for 20 min at 4°C) (Tanaka et al. 2006). Ptd2Gro –OH and PtdSer -OH formed were analyzed by ESI–MS.
Electrospray ionization tandem mass spectrometry
ESI-MS was utilized for qualitative analysis of complex phospholipids (Pulfer & Murphy 2003, Forrester et al. 2004). ESI-MS analysis was performed by direct infusion into linear ion-trap mass spectrometer LXQ™ with the Xcalibur operating system (Thermo Fisher Scientific, San Jose, CA). Samples collected after 2D-HPTLC separation were evaporated under N2, re-suspended in chloroform:methanol 1:1 v/v (20 pmol/μL) and directly utilized for acquisition of negative-ion or positive-ion ESI mass spectra at a flow rate of 5 μL/min. The ESI probe was operated at a voltage differential of 3.5–5.0 kV in the negative or positive ion mode. Capillary temperature was maintained at 70 or 150°C. Using full range zoom (200–2000 m/z) in positive and negative ion mode, the profile spectra were acquired. Tandem mass spectrometry (MS/MS analysis) of individual phospholipid species was employed to determine the fatty acid composition. The MS/MS spectra were acquired using isolation width 1.0 m/z. For analysis of Ptd2Gro structure, the scan time setting of ion trap for full MS (range 1400–1600 m/z) was set at 50 microscans with maximum injection time 1000 msec. MSn analysis ms. Two ion activation techniques were used for MS analysis: collision-induced dissociation (CID, Q=0.25, low mass cut off at 28% of the precursor m/z) and pulsed-Q dissociation technique (PQD), with Q=0.7, and no low mass cut off for analysis of low molecular weight fragment ions (Schwartz et al. 2005). MS/MS fragmentation of ether-linked alkenyl species resulted in the formation of two typical product ions formed after loss of fatty acid in sn-2 position: mono-lyso-alkenyl species and mono-lyso-acyl-species. To further confirm the identity of glycerophospholipids, they were exposed to HCl fumes known to hydrolyze alkenyl-acyl-glycerophospholipids to yield their lyso-acyl-derivatives. The reaction products were subjected to HPTLC and spots corresponding to glycerophospholipids were analyzed by ESI-MS. This treatment resulted in disappearance of molecular ions corresponding to molecular species of alkenyl-acyl- glycerophospholipids while those of diacyl- phospholipids remained unchanged. Product ions representing mono-lyso-alkyl species have mass differences of 14 compared with the product ions of corresponding mono-lyso-acyl species.
Analysis of PL oxidized molecular species (hydroperoxy- and hydroxy-) was performed as described by (MacMillan & Murphy 1995). Additionally, we treated the reaction products with a reductant, triphenyl-phosphine, to confirm the conversion of Ptd2Gro–OOH and PtdSer-OOH into Ptd2Gro–OH and PtdSer-OH, respectively. This resulted in the reduction of hydroperoxy-Ptd2Gro and hydroperoxy-PtdSers to their respective hydroxy-derivatives detectable in the spectra. To account for isotopic interferences, we performed isotopic corrections by entering the chemical composition of each species into the Qual browser of Xcalibur (operating system) and using the simulation of the isotopic distribution to make adjustments for the major peaks. To minimize isotopic interferences between isolated masses M+2, the MS/MS spectra were acquired using isolation width of 1.0 m/z.
Based on MS fragmentation data, chemical structures of lipid molecular species were obtained using ChemDraw and confirmed by comparing with the fragmentation patterns presented in Lipid Map Data Base (www.lipidmaps.org and www.byrdwell.com).
Quantitation of lipid hydroperoxides
Lipid hydroperoxides were determined by fluorescence HPLC of resorufin formed in peroxidase-catalyzed reduction of specific phospholipid hydroperoxides (PL-OOH) with Amplex Red. Phospholipids were hydrolyzed by porcine pancreatic PLA2 (1 U/μL) in 25 mM phosphate buffer containing 1.0 mM Ca, 0.5 mM EGTA and 0.5 mM SDS (pH 8.0 at RT for 30 min), prior to exposure to reagents for the peroxidase reaction (MP-11/Amplex Red). This resulted in the production of lyso-phospholipids and release of hydroperoxide group-containing fatty acids (FA-OOH). While MP-11 can catalyze reduction of some PL-OOH without pretreatment with PLA2, non-deesterified Ptd2Gro-OOH did not effectively react with MP-11/Amplex Red. For the peroxidase reaction, 50 μM Amplex Red and MP-11 (1.0 μg/μL) were added to hydrolyzed lipids, and the samples were incubated at 4°C for 40 min. The reaction was started by addition of 1 μL of solution MP-11 (1.0 μg/μL) and terminated by a stop reagent (100 μL of solution of 10 mM HCl, 4 mM BHT in ethanol). The samples were centrifuged at 10,000 g for 5 min and the supernatant was used for HPLC analysis. Aliquots (5 μL) were injected into a C-18 reverse phase column (Eclipse XDB-C18, 5 μm, 150 × 4.6 mm) and eluted using a mobile phase composed of 25 mM KH2PO4 (pH 7.0)/methanol (60:40 v/v) at a flow rate of 1 mL/min. The resorufin fluorescence was measured at 590 nm after excitation at 560 nm. Shimadzu LC-100AT vp HPLC system equipped with fluorescence detector (model RF-10Axl) and autosampler (model SIL-10AD vp) was used (Kagan et al. 2005).
Statistics
The results are presented as means ± SD values from at least three independent experiments, and statistical analyses were performed by one-way ANOVA. The statistical significance of differences was set at P< 0.05.
RESULTS
Biomarkers of apoptosis in STS-treated neurons
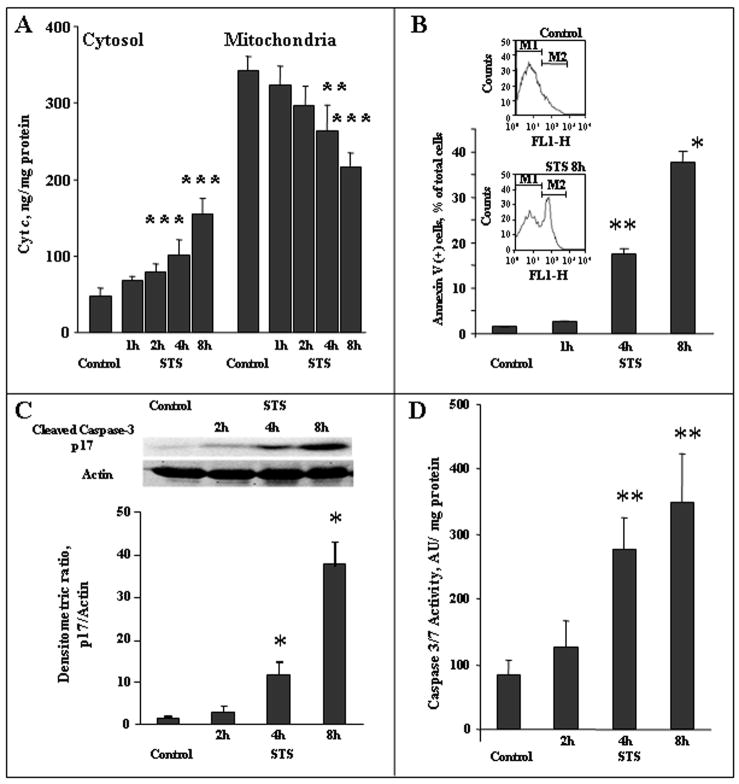
Treatment of neurons with STS triggered the intrinsic apoptotic cell death program. This was evidenced by: 1) release of cyt c from mitochondria into the cytosol (Fig. 1A, a, b), 2) activation of caspase-3 (Fig. 1B), and 3) externalization of PtdSer on the cell surface detected by Annexin V (Fig. 1, C). We assessed caspase-3 activation by western blot analysis using an antibody against the larger subunit (p17) of activated caspase-3 (Fig. 1). The level of p17 subunit was slightly increased upon 2-h incubation with STS. The appearance of p17 subunit became significant (~8 fold over control, p<0.01) after 4-h treatment. Accordingly, chemiluminescence assay showed a marked increase in enzymatic activity for caspase-3/7 activity at 4-h and further elevation at 8-h after STS exposure. Additionally, we analyzed the caspase-3 like activity using a fluorometric method. Compared with non-treated neurons, a 7.5-fold increase in the fluorescence was detected in the lysate of neurons after 8-h treatment with STS.
Figure 1.
Tine course of biomarkers of apoptosis in primary cortical neurons treated with STS. A - Cytosolic and mitochondrial cyt c levels measured by ELISA. ***p<0.001, **p<0.05 vs control, respectively, n=6. B - Externalization of PtdSer assessed by annexin V binding. Insert -Flow cytometric data from representative experiments: cell distribution by annexin V-FITC fluorescence in log scale. **p<0.05, *p<0.01 vs control, respectively, n=4. C - Western blot analysis of larger p17 subunit of activated caspase-3 (n=3). Equal amount of protein (50 μg) was subjected to 12% of SDS-PAGE detected by Western blot using an antibody to the larger (p17) subunit of active caspase 3 (1:250, abcam, Cambridge, MA). To confirm equal protein concentrations in samples, membranes were stripped and re-blotted with a primary antibody to β-actin (1:5000, Novus, Littleton, CO). Semi-quantitation of the bands was carried out by densitometry using Labworks Image Acquisition and Analysis Software (UVP, Upland, CA). The level of p17 was expressed as the mean densitometry ratio of p17 over actin, n=3, independent measurements, *p<0.01 vs control. D. Caspase 3/7 activity, n=4, **p<0.05 vs control.
These results are in line with earlier reports on effectiveness of STS in inducing intrinsic pathways of neuronal apoptosis (D’Sa-Eipper & Roth 2000, Gil et al. 2003, Pong et al. 2001). Based on assessments of PtdSer externalization, STS induced apoptosis in 18% and 36% of neuronal cells after 4h and 8 h exposures, respectively. Next, we determined whether execution of apoptotic program was associated with changes of phospholipid molecular species, particularly those containing polyunsaturated acyls whose oxygenation is relevant to apoptotic signaling.
Analysis of phospholipids in intact and apoptotic rat cortical neurons
We performed lipidomics analysis of major classes of phospholipids – phosphatidylcholine (PtdCho), phosphatidylethanolamine (PtdEtn), phosphatidylinositol (PtdIns), PtdSer, sphingomyelin (CerPCho), and Ptd2Gro - in cultured rat cortical control and apoptotic neurons. The molecular diversity of phospholipids was assessed by ESI-MS after preliminary separation of phospholipids by 2D-HPTLC.
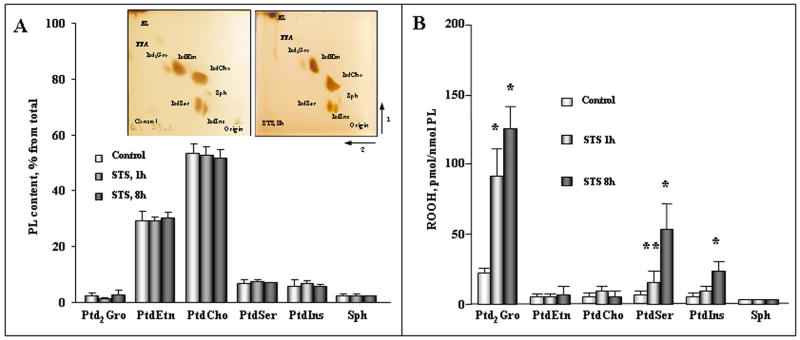
A typical 2D-HPTLC profile and the abundance of major phospholipid classes from the primary neuronal cells before and after incubation with STS are shown in Figure 2A. Six different phospholipid spots were detected by 2D-HPTLC of lipids extracted from either control or STS-exposed cortical neuronal cells (Fig. 2, A). PtdCho and PtdEtn represented the two dominant phospholipid classes of the total phospholipids. Additionally, other phospholipids in the order of their abundance – PtdSer > PtdIns > CerPCho ≫ Ptd2Gro – were detected on the plates. These data are in agreement with previously reported results on the relative abundance of phospholipids in cortical neurons (Gasull et al. 2003). Based on assessments of phosphorus in the spots, no significant changes in the phospholipid distribution were found in neurons following exposure to STS (Fig. 2A).
Figure 2.
Effect of STS on the phospholipid content (A) and accumulation of phospholipid hydroperoxides (B) in primary cortical neurons before and after STS treatment. Insert shows typical 2D-HPTLC of total lipids extracted from cortical neurons before and after treatment of cells by STS. NL -neutral lipids, Ptd2Gro – cardiolipin, PtdEtn - phosphatidylethanolamine, PtdCho- phosphatidylcholine, CerPCho – sphingomyelin, PtdIns – phosphatidylinositol, PtdSer – phosphatidylserine, FFA - free fatty acids. PL content, % from total refers to the percentage of phospholipid phosphorus recovered from the respective thin layer chromatograms. In the case of Ptd2Gro, calculation takes into account 2 phosphates. Lipid hydroperoxides were determined by Amplex Red protocol and values were normalized to the individual PL content. *p<0.01 versus control, **p<0.05 versus control, n=8 independent experiments. For incubation conditions see the legend of Fig. 1.
ESI–MS analysis of molecular species of phospholipids
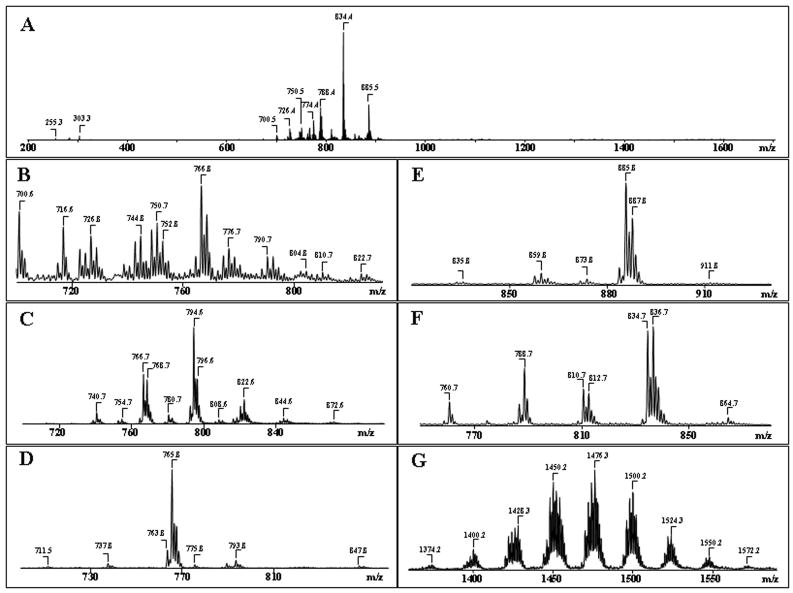
We used ESI–MS analysis to characterize individual molecular species of neuronal phospholipids with particular emphasis on those containing polyunsaturated fatty acid residues, which undergo oxidative modification. Direct infusion of total lipid extracts from neuronal cells yielded only poorly resolved spectra in negative ionization mode regimen with only 4–5 clusters of signals corresponding to major classes of phospholipids (Fig. 3, A), likely due to significant mutual suppression of ionization by non-separated mixtures of phospholipids (Mallet et al. 2004, Marchese et al. 1998). In contrast, after pre-separation of lipids by 2D-HPTLC, the MS spectra of major phospholipid classes displayed rich diversity of molecular species. Full-scan ESI–MS analysis in the negative ionization mode was employed for all phospholipids classes (Fig. 3). Additionally, MS-spectra were recorded in positive mode for PtdCho and CerPCho. In general, detection of phospholipids in the positive ionization mode provides information about the phospholipid head groups, whereas tandem MS/MS analysis in the negative mode is a source of structural information relevant to the identity and positional distribution at sn-1 and sn-2 of individual acyl chains. Typical ESI mass spectra of different molecular species of several major classes of phospholipids from neuronal cells are shown in Fig. 3. To identify the phospholipid molecular species, MS2 experiments were performed using the CID or PQD techniques (Tables 1, 2, 3).
Figure 3.
Typical negative ion ESI-MS of molecular species of major phospholipids obtained from cortical neuronal cells before (A) and after (B–G) their separation by 2D-HPTLC. A- Total lipid extract; B- PtdEtn; C- PtdCho; D - CerPCho; E- PtdIns; F- PtdSer; G- Ptd2Gro.
Table 1.
Major phospholipid molecular species from cortical neuronal cells.
| Molecular species | [M-H]− m/z | Identified Acyl Chains | Molecular species | [M-H]− m/z | Identified Acyl Chains |
|---|---|---|---|---|---|
| Phosphatidylethanolamine | Phosphatidylcholine | ||||
| Diacyl species | Diacyl species | ||||
| 34:2 | 714.5 | (C16:1)/(C18:1) | 28:0 | 712.6** | (C14:0)/(C14:0) |
| 34:1 | 716.5 | (C16:0)/(C18:1) | 30:0 | 740.6** | (C16:0)/(C14:0) |
| 34:0 | 718.5 | (C16:0)/(C18:0) | 32:1 | 766.6** | (C16:1)/(C16:0); (C18:1)/(C14:0)* |
| 36:4 | 738.5 | (C16:0)/(C20:4) | 32:0 | 768.6** | (C16:0)/(C16:0); (C14:0)/(C18:0)* |
| 36:3 | 740.5 | (C16:0)/(C20:3); (C18:1)/(C18:2) | 33:0 | 782.6** | (C16:0)/(C17:0); (C15:0)/(C18:0) |
| 36:2 | 742.5 | (C18:1)/(C18:1); (C16:1)/(C20:1) | 34:4 | 788.6** | (C14:0)/(C20:4) |
| 36:1 | 744.5 | (C18:1)/(C18:0); (C16:0)/(C20:1) | 34:2 | 792.6** 756.6 |
(C16:0)/(C18:2); (C18:1)/(C16:1); (C16:0)/(C18:2); (C18:1)/(C16:1) |
| 36:0 | 746.5 | (C18:0)/(C18:0) | 34:1 | 794.6** | (C18:1)/(C16:0) |
| 38:5 | 764.5 | (C18:1)/(C20:4); (C16:0)/(C22:5)* | 34:0 | 796.6** | (C16:0)/(C18:0) |
| 38:4 | 766.5 | (C18:0)/(C20:4); (C18:1)/(C20:3)*; (C16:0)/(C22:4)* | 35:0 | 810.6** | (C17:0)/(C18:0) |
| 38:3 | 768.5 | (C18:0)/(C20:3); (C16:0)/(C22:3)* | 36:4 | 816.6** | (C16:0)/(C20:4); (C18:1)/(C18:3); (C18:2)/(C18:2) |
| 38:2 | 770.5 | (C18:0)/(C20:2); (C18:1)/(C20:1)* | 36:3 | 818.6** 782.6 |
(C16:0)/(C20:3) (C18:0)/(C18:3) |
| 38:1 | 772.5 | (C18:0)/(C20:1) | 36:2 | 820.6** 784.6 |
(C18:1)/(C18:1); (C18:0)/(C18:2) (C18:1)/(C18:1); (C18:0)/(C18:2) |
| 39:4 | 780.5 | (C20:4)/(C19:0) | 36:1 | 822.6** 786.6 |
(C18:0)/(C18:1): (C16:0)/(C20:1) (C16:1)/(C20:0)*; (C14:1)/(C22:0)* |
| 40:7 | 788.5 | (C18:1)/(C22:6) | 36:0 | 788.6 | (C18:0)/(C18:0); (C16:0)/(C20:0) |
| 40:6 | 790.5 | (C18:0)/(C22:6); (C18:1)/(C22:5)* | 38:5 | 842.6** 806.6 |
(C16:0)/(C22:5); (C18:0)/(C20:5); (C18:1)/(C20:4); (C20:3)/(C18:2) |
| 40:5 | 792.5 | (C18:0)/(C22:5); (C18:1)/(C22:4)*; (C20:4)/(C20:1)* | 38:4 | 844.6** 808.6 |
(C18:0)/(C20:4); (C16:0)/(C22:4); (C20:2)/(C18:2) |
| 40:4 | 794.5 | (C18:0)/(C22:4); (C18:1)/(C22:3)*; (C20:4)/(C20:0)*; (C20:3)/(C20:1)* | 40:1 | 842.6 | (C18:1)/(C22:0) |
| Alkenyl-acyl species | Alkenyl-acyl species | ||||
| 34:2 | 700.5 | (C16:1p)/(C18:1) | 32:1 | 752.6** | (C16:0p#)/(C16:1) |
| 36:4 | 722.5 | (C16:0p)/(C20:4) | 32:0 | 754.6** | (C18:0p)/(C14:0) |
| 36:3 | 724.5 | (C16:0p)/(C20:3) | 34:1 | 744.6 | (C16:0p)/(C18:1); (C18:1p)/(C16:0) |
| 36:2 | 726.5 | (C18:1p)/(C18:1);(C16:0p)/(C20:2)* | 36:4 | 766.6 | (C16:0p)/(C20:4) |
| 36:1 | 728.5 | (C16:0p)/(C20:1);(C18:0p)/(C18:1) | 36:2 | 770.6 | (C18:1p)/(C18:1) |
| 38:6 | 746.5 | (C16:0p)/(C22:6) | 36:1 | 808.6** | (C18:0p)/(C18:1); (C18:1p)/(C18:0); (C16:1p)/(C20:0) |
| 38:5 | 748.5 | (C16:0p)/(C22:5);(C18:1p)/(C20:4) | 36:0 | 810.6** | (C16:0p)/(C20:0); (C18:0p)/(C18:0) |
| 38:4 | 750.5 | (C16:0p)/(C22:4); (C18:0p)/(C20:4) | 38:7 | 788.6 | (C16:1p)/(C22:6) |
| 38:3 | 752.5 | (C18:0p)/(C20:3); (C16:0p)/(C22:3) | 38:6 | 826.6** | (C16:1p)/(C22:5)* |
| 38:2 | 754.5 | (C16:0p)/(C22:2); (C18:0p)/(C20:2);(C18:1p)/(C20:1) | 38:5 | 792.6 | (C18:1p)/(C20:4); (C16:0p)/(C22:5) |
| 38:1 | 756.5 | (C18:0p)/(C20:1) | 38:4 | 794.6 | (C16:0p)/(C22:4); (C18:0p)/(C20:4) |
| 40:7 | 772.5 | (C18:1p)/(C22:6) | 38:3 | 796.6 | (C18:0p)/(C20:3); (C18:1p)/(C20:2) |
| 40:6 | 774.5 | (C18:0p)/(C22:6); (C18:1p)/(C22:5) | 38:2 | 798.6 | (C16:0p)/(C22:2); (C18:0p)/(C20:2) |
| 40:5 | 776.5 | (C18:1p)/(C22:4); (C18:0p)/(C22:5) | Sphingomyelin | ||
| 40:4 | 778.5 | (C18:0p)/(C22:4); (C18:1p)/(C22:3) | Sphingoid base/Acyl | ||
| 40:3 | 780.5 | (C18:0p)/(C22:3) | 34:1 | (C18:1)/(C16:0) | |
| Phosphatidylserine | 36:2 | (C18:1)/(C18:1) | |||
| Diacyl species | 36:1 | (C18:1)/(C18:0) | |||
| 34:2 | 758.6 | (C18:1)/(C16:1) | 38:1 | (C18:1)/(C20:0) | |
| 34:1 | 761.6 | (C18:0)/(C16:1); (C16:0)/(C18:1) | 42:2 | (C18:1)/(C24:1) | |
| 34:0 | 760.6 | (C18:0)/(C16:0) | Phosphatidylinositol | ||
| 35:1 | 774.6 | (C18:0)/(C17:1) | Diacyl species | ||
| 36:2 | 786.6 | (C18:1)/(C18:1); (C18:0)/(C18:2)*; (C16:1)/(C20:1)** | 34:3 | 831.5 | (C16:1)/(C18:2); (C16:0)/(C18:3)* |
| 36:1 | 788.6 | (C18:0)/(C18:1) | 34:2 | 833.5 | (C16:0)/(C18:2); (C16:1)/(C18:1) |
| 38:5 | 808.6 | (C18:1)/(C20:4) | 34:1 | 835.5 | (C16:0)/(C18:1) |
| 38:4 | 810.6 | (C18:0)/(C20:4) | 35:0 | 851.5 | (C17:0)/(C18:0) |
| 40:6 | 834.6 | (C18:0)/(C22:6) | 36:4 | 857.5 | (C16:0)/(C20:4) |
| 40:5 | 836.6 | (C18:0)/(C22:5) | 36:3 | 859.5 | (C16:0)/(C20:3) |
| 40:4 | 838.6 | (C18:0)/(C22:4) | 36:2 | 861.5 | (C16:0)/(C20:2) |
| 40:3 | 840.6 | (C18:0)/(C22:3) | 37:3 | 873.5 | (C17:0)/(C20:3) |
| 40:2 | 842.6 | (C18:0)/(C22:3) | 38:5 | 883.5 | (C18:1)/(C20:4) |
| 41:5 | 850.6 | (C19:0)/(C22:5) | 38:4 | 885.5 | (C18:0)/(C20:4) |
| 40:6 | 856.6& | (C18:0)/(C22:6) | 38:3 | 887.5 | (C18:0)/(C20:3) |
| 42:7 | 860.6 | (C20:1)/(C22:6); (C20:3)/(C22:4); (C20:4)/(C22:3) | 38:2 | 889.5 | (C18:0)/(C20:2) |
| 42:6 | 862.8 | (C18:0)/(C24:6); (C20:3)/(C22:3); (C20:1)/(C22:5) | 40:5 | 891.5 | (C18:0)/(C22:5) |
| 42:5 | 864.6 | (C20:0)/(C22:5); (C20:4)/(C22:1); (C18:0)/(C24:5); (C20:3)/(C22:2) | 40:4 | 893.5 | (C18:0)/(C22:4) |
| 42:4 | 866.6 | (C20:0)/(C22:4); (C18:0)/(C24:4) | 40:3 | 895.5 | (C18:0)/(C22:3) |
Minor
[M+Na]−
[M+Cl]−
p - an sn-1 vinyl ether (alkenyl- or plasmalogen) linkage;
Phospholipids are designated as follows: diacyl 38:4 PE, where 38 indicates the summed number of carbon atoms at both the sn-1, sn-2, positions and 4 designates the summed number of double bonds at both positions. These m/z values indicate ratios of mass to charge for singly charged [M-H]− ions (and doubly charged [M-2H]−2 ions for CL, see Table 2), respectively.
Table 2.
Major cardiolipin molecular species from cortical neuronal cells.
| Molecular species |
[M-2H]2− m/z |
[M-H]− m/z |
Identified Acyl Chains |
|---|---|---|---|
| Cardiolipin | |||
| Diacyl species | |||
| 62:1 | 660.5 | 1322.0 | (C14:0)2/(C16:1)1/(C18:0)1 |
| 64:4 | 671.5 | 1344.0 | (C16:1)4 |
| 66:4 | 685.5 | 1372.0 | (C16:1)3/(C18:1)1 |
| 68:7 | 696.5 | 1394.0 | (C16:1)3/(C20:4)1 |
| 68:5 | 698.5 | 1398.0 | (C16:1)2/(C18:1)1/(C18:2)1 |
| 68:4 | 699.5 | 1400.0 | (C16:1)2/(C18:1)2 |
| 68:2 | 700.5 | 1402.0 | (C16:1)1/(C16:0)1/(C18:1)1/(C18:0)1 |
| 70:7 | 710.5 | 1422.0 | (C16:1)1/(C18:2)3 |
| 70:6 | 711.5 | 1424.0 | (C16:1)1/(C18:1)1/(C18:2)2 |
| 70:5 | 712.5 | 1426.0 | (C16:1)1/(C18:2)1/(C18:1)2 |
| 70:4 | 713.5 | 1427.9 | (C16:1)1/(C18:1)3 |
| 70:2 | 715.5 | 1432.0 | (C16:0)1/(C18:0)1//(C18:1)2 |
| 72:9 | 722.6 | 1446.2 | (C16:1)1/(C16:0)1/(C18:3)1/(C22:5)1; (C16:1)1/(C18:1)1/(C18:3)1/(C20:4)1 |
| 72:8 | 723.5 | 1448.0 | (C18:2)4; (C16:1)1/(C18:1)1/(C18:2)1/(C20:4)1; (C16:0)1/(C18:1)1/(C18:3)1/(C20:4)1* |
| 72:7 | 724.5 | 1450.0 | (C16:1)1/(C18:1)1/(C18:2)1/(C20:3)1; (C18:1)1/(C18:2)3 |
| 72:6 | 725.5 | 1452.0 | (C18:1)2/(C18:2)2; (C16:1)1/(C18:1)2/(C20:3)1 |
| 72:5 | 726.5 | 1454.0 | (C16:0)1/(C18:1)2/(C20:3)1; (C18:1)3/(C18:2)1 |
| 72:4 | 727.5 | 1456.0 | (C18:1)4; (C16:0)1/(C18:1)2/(C20:2)1 |
| 74:9 | 736.5 | 1474.0 | (C18:1)1/(C18:2)2/(C20:4)1; (C16:0)1/(C18:1)1/(C18:2)1/(C22:6)1 |
| 74:8 | 737.5 | 1476.0 | (C16:1)1/(C18:1)2/(C22:5)1; (C16:1)1/(C18:2)1/(C20:3)1/(C20:2)1; (C16:0)1/(C18:1)2/(C22:6)1; (C16:0)1/(C18:1)1/(C20:4)1/(C20:3)1 |
| 74:7 | 738.5 | 1478.0 | (C18:1)3/(C20:4)1; (C16:1)1/(C18:2)1/(C20:3)1/(C20:1)1* |
| 74:6 | 739.5 | 1480.0 | (C18:1)2/(C18:0)1/(C20:4)1; (C16:0)1/(C18:1)1/(C20:4)1/(C22:4)1; (C16:0)1/(C18:0)1/(C20:4)1/(C22:5)1 |
| 76:13 | 746.7 | 1494.4 | (C16:1)1/(C18:2)1/(C20:4)1/(C22:6)1 |
| 76:12 | 747.7 | 1496.4 | (C16:1)1/(C18:2)1/(C20:3)1/(C22:6)1 |
| 76:11 | 748.9 | 1498.8 | (C18:1)1/(C18:2)1/(C20:4)2 |
| 76:10 | 749.5 | 1500.0 | (C18:1)2/(C20:4)2; (C18:0)1/(C18:2)2/(C22:6)1 |
| 76:9 | 750.5 | 1501.9 | (C18:1)2/(C20:4)1/(C20:3)1 |
| 76:8 | 751.5 | 1503.9 | (C18:1)1/(C18:2)1/(C20:3)1/(C20:2)1; (C16:0)1/(C18:2)1/(C20:2)1/(C22:4)1 |
| 76:7 | 752.5 | 1506.0 | (C16:0)1/(C18:1)1/(C20:3)1/(C22:3)1; (C16:0)1/(C18:0)1/(C20:1)1/(C22:6)1 |
| 78:15 | 758.9 | 1518.9 | (C16:1)1/(C18:2)1/(C22:6)2 |
| 78:13 | 759.5 | 1520.0 | (C18:2)1/(C20:4)3 |
| 78:13 | 760.5 | 1521.9 | (C18:1)1/(C20:4)3; (C18:1)1/(C18:2)1/(C20:4)1/(C22:6)1 |
| 78:12 | 761.5 | 1524.0 | (C18:1)2/(C20:4)1/(C22:6)1 |
| 78:11 | 762.5 | 1526.0 | (C18:1)2/(C20:3)1/(C22:6)1 |
| 78:8 | 765.4 | 1531.8 | (C16:1)1/(C18:1)1/(C20:3)1/(C22:3)1;(C18:1)2/(C20:0)1/(C22:6)1 |
| 80:15 | 772.0 | 1545.9 | (C18:1)1/(C20:4)2/(C22:6)1 |
| 80:14 | 773.5 | 1548.0 | (C18:0)1/(C20:4)2/(C22:6)1 |
| 80:13 | 774.5 | 1550.0 | (C18:1)1/(C18:0)1/(C22:6)2; (C18:1)2/(C22:5)1/(C22:6)1; (C18:0)1/(C20:4)2/(C22:5)1 |
| 80:12 | 776.0 | 1553.0 | (C18:0)2/(C22:6)2 |
| 80:11 | 776.5 | 1554.1 | (C18:1)2/(C22:5)1/(C22:4)1;(C18:1)1/(C18:0)1/(C22:4)1/(C22:6)1 |
| 82:17 | 784.5 | 1569.9 | (C18:1)1/(C20:4)1/(C22:6)2 |
| 82:16 | 785.5 | 1572.0 | (C18:1)1/(C20:3)1/(C22:6)2 |
| 82:11 | 790.5 | 1582.0 | (C18:0)1/(C20:1)1/(C22:5)2 |
| 82:10 | 792.5 | 1586.0 | (C20:4)1/(C20:0)1/(C20:2)1/(C22:4)1 |
Minor
Phospholipids are designated as follows: tetra-acyl 74:8 cardiolipin (Ptd2Gro), where 74 indicates the summed number of carbon atoms at the sn-1, sn-2, and sn-1′, sn-2′ positions and 8 designates the summed number of double bonds at both the sn-1, sn-2, and sn-1, sn-2 positions. These individual Ptd2Gro molecular species were detected by ESI as deprotonated species of Ptd2Gro in the negative ionization mode at m/z ratios of 737.5 and 1,476.0. These m/z values indicate ratios of mass to charge for singly charged [M-H]− ions and doubly charged [M-2H]−2 ions, respectively.
Table 3.
Major oxidized phospholipid molecular species by STS from cortical neuronal cells.
| Molecular species |
[M-H]− m/z |
Identified Acyl Chains | Molecular species |
[M-H]− m/z |
Identified Acyl Chains |
|---|---|---|---|---|---|
| Phosphatidylethanolamine | |||||
| Diacyl species | Oxidized Diacyl species by STS | ||||
| 38:6 | 762.5 | (C16:0)/(C22:6) | 38:6 | 794.5 | (C16:0)/(C22:6+OO) |
| 40:6 | 790.5 | (C18:0)/(C22:6); (C18:1)/(C22:5)* | 40:6 | 822.6 | (C18:0)/(C22:6+OO) |
| 40:5 | 792.5 | (C18:0)/(C22:5) (C18:1)/(C22:4)*; (C20:4)/(C20:1)* | 40:5 | 824.6 | (C18:0)/(C22:5+OO) |
| Alkenyl-acyl species | Oxidized Alkenyl-acyl species by STS | ||||
| 36:4 | 722.5 | (C16:0p)/(C20:4) | 36:4 | 738.5 | (C16:0p)/(C20:4+O) |
| 38:6 | 746.5 | (C16:0p)/(C22:6) | 38:6 | 778.5 | (C16:0p)/(C22:6+OO) |
| 38:5 | 748.5 | (C16:0p)/(C22:5); (C18:1p)/(C20:4) | 38:5 38:5 |
764.5 780.5 |
(C18:1p)/(C20:4+O) (C16:0p)/(C22:6+OO) |
| 40:7 | 772.5 | (C18:1p)/(C22:6) | 40:7 | 804.5 | (C18:1p)/(C22:6+OO) |
| 40:6 | 774.5 | (C18:0p)/(C22:6); (C18:1p)/(C22:5) | 40:6 40:6 |
806.6 822.6 |
(C18:0p)/(C22:6+OO) (C18:0p)/(C22:6+OOO) |
| 40:5 | 776.5 | (C18:1p)/(C22:4); (C18:0p)/(C22:5) | 40:5 | 808.6 | (C18:0p)/(C22:5+OO) |
| Phosphatidylinositol | |||||
| Diacyl species | Oxidized Diacyl species by STS | ||||
| 38:4 | 885.5 | (C18:0)/(C20:4) | 38:4 | 901.5 917.5 |
(C18:0)/(C20:4+O) (C18:0)/(C20:4+OO) |
| Phosphatidylserine | |||||
| Diacyl species | Oxidized Diacyl species by STS | ||||
| 38:4 | 810.6 | (C18:0)/(C20:4) | 38:4 | 826.6 842.6 |
(C18:0)/(C20:4+O) (C18:0)/(C20:4+OO) |
| 40:6 | 834.6 | (C18:0)/(C22:6) | 40:6 | 850.6 866.6 |
(C18:0)/(C22:6+O); (C18:0)/(C22:6+OO) |
| 40:5 | 836.6 | (C18:0)/(C22:5) | 40:5 | 868.6 | (C18:0)/(C22:5+OO) |
| 40:4 | 838.6 | (C18:0)/(C22:4) | 40:4 | 870.6 | (C18:0)/(C22:4+OO) |
| Cardiolipin | |||||
| Diacyl species | Oxidized Diacyl species by STS | ||||
| 72:8 | 1448.0 | (C18:2)4 | 72:8 | 1559.9 | (C18:2+O)1/(C18:2+OO)3 |
| 74:9 | 1474.0 | (C18:1)1/(C18:2)2/(C20:4)1 (C16:0)1/(C18:1)1/(C18:2)1/(C22:6)1 |
74:9 | 1506.0 | (C18:1)1/(C18:2)1/(C18:2+O)1/(C20:4+O)1; (C16:0)1/(C18:1)1/(C18:2)1/(C22:6+OO)1 |
| 74:8 | 1476.0 | (C16:0)1/(C18:1)1/(C20:4)1/(C20:3)1; (C16:1)1/(C18:2)1/(C20:3)1/(C20:2)1; (C16:0)1/(C18:1)2/(C22:6)1 | 74:8 | 1492.0 | (C16:0)1/(C18:1)1/(C20:4+O)1/(C20:3)1; (C16:0)1/(C18:1)2/(C22:6+O)1; (C16:1)1/(C18:2+O)1/(C20:3)1/(C20:2)1 |
| 74:7 | 1478.0 | (C18:1)3/(C20:4)1 | 74:7 | 1494.0 | (C18:1)3/(C20:4+O)1 |
| 76:10 | 1500.0 | (C18:1)2/(C20:4)2; (C18:0)1/(C18:2)2/(C22:6)1 | 76:10 | 1516.0 | (C18:1)2/(C20:4)1/(C20:4+O)1; (C18:0)1/(C18:2)2/(C22:6+O)1 |
| 78:12 | 1524.0 | (C18:1)2/(C20:4)1/(C22:6)1 | 78:12 | 1556.0 1588.0 |
(C18:1)2/(C20:4)1/(C22:6+2O)1;(C18:1)2/(C20:4+OO)1/(C22:6)1; (C18:1)2/(C20:4)1/(C22:6+4O)1 |
| 80:13 | 1550.0 | (C18:1)2/(C22:5)1/(C22:6)1 | 80:13 | 1582.0 | (C18:1)2/(C22:5)1/(C22:6+OO)1 |
| 80:11 | 1554.1 | (C18:1)1/(C18:0)1/(C22:4)1/(C22:6)1 (C18:1)2/(C22:5)1/(C22:4)1 |
80:11 | 1586.0 | (C18:1)1/(C18:0)1/(C22:4)1/(C22:6+OO)1 (C18:1)2/(C22:4)1/(C22:5+OO)1 |
The distribution of fatty acid molecular species within each major ion cluster for all major phospholipid classes of neuronal cells are shown in Table 1. The molecular species of PtdEtn were represented by deprotonated ions [M-H]− in negative mode that included two groups of molecular species: diacyl-PtdEtn (m/z 716, 744, 766, 790) and alkenyl-PtdEtn (m/z 700, 726, 750, 776) (Fig. 3, B). Molecular species of both diacyl-PtdEtn and alkenyl-PtdEtn contained C20:4 and C22:6 fatty acid residues as confirmed by MS2 analysis (Table 1). For example, after fragmentation of the PtdEtn dominant molecular ion with m/z 766, two prominent ions with m/z 283 and 303, that correspond to stearic (C18:0) and arachidonic (C20:4) fatty acids respectively, were obtained (Table 1). Typical product ions of PtdEtn with m/z 140 and 196 derived from [HPO4CH2CH2NH2]+ and [CH2C(OH)CH2PO4CH2CH2NH2]+ were observed after fragmentation of molecular species of PtdEtn in a positive mode (data not shown).
Molecular species of PtdCho in negative ion mode (Fig. 3C) demonstrated the presence of the major molecular species predominantly as chlorinated adducts [M+Cl]− with m/z 740, 766, 794 and 822 corresponding to multiple individual species containing C16:0/C14:0, C16:0/C16:1, C18:1/C16:0 and C18:1/C18:0 fatty acids, respectively. Some part of PtdCho molecular species in negative mode was presented by deprotonated ions [M-H]− with m/z 766, 792, 794 that contained alkenyl- PtdCho rich in arachidonic (C20:4) acid residues (Table 1).
Alkenyl-acyl- and diacyl-species of glycerophospholipids were detectable in ESI-MS spectra. Clusters of these signals which have the same fatty acid compositions are characterized with a mass difference of 14 due to their attachment to the glycerol backbone of glycerophospholipid by vinyl ether or ester bond (Taguchi et al. 2000).
Most CerPCho molecular species in negative ion mode demonstrated the presence of ions as chlorinated adducts [M+Cl]− with dominant peak at m/z 765 corresponding to the sphingosine long-chain base C18:1 and stearic acid C18:0 which are not susceptible to oxidation (Fig. 3D, Table 1). In positive ion mode, this molecular species was dominant as well and corresponded to ion at m/z 731.
ESI–MS analysis of PtdIns in negative mode revealed a predominant molecular ion represented by the deprotonated ion [M-H] − with m/z 885 (Fig. 3E) corresponding to molecular species, containing C18:0 and C20:4 fatty acids (Table 1). A molecular ion at m/z 856 that corresponds to molecular species, containing palmitic (C16:0) and arachidonic (C20:4) fatty acids was present in significantly lower amounts (Fig. 3E, Table 1).
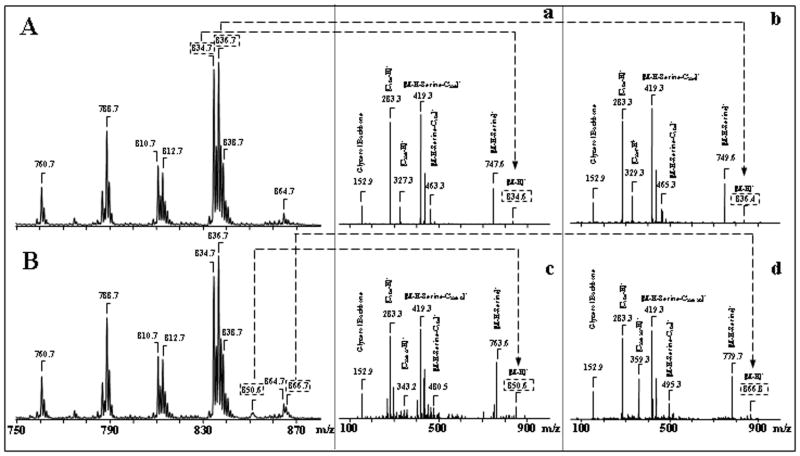
Analysis of PtdSer revealed four molecular clusters with m/z 760, 788, 810, 834 and 836 (Fig. 3F). Ion clusters at m/z 834 and 836, containing C22:6 and C22:5 fatty acids, respectively, dominated among other PtdSer molecular species (Fig. 3F, Table 1). The main peaks with m/z 747 and 749 formed after fragmentation of PtdSer parent ions with m/z 834 and 836, respectively, were due to the loss of the serine group. These fragmentation product ions, together with the deprotonated ions (m/z 834 and 836), effectively determined the molecular mass of PtdSer (Fig. 4A, a, b). There were also ions characteristic of the head group or its fragments (Fig. 4A, a, b), as well as common fragments for phospholipids: m/z 79 for [PO3]−, m/z 153 for [glycerophosphate - H2O-H]−. Molecular fragments with m/z 283, 327 and 329 correspond to carboxylate anions of stearic (C18:0), docosahexaenoic (C22:6) and docosapentaenoic (C22:5) fatty acids, respectively. Similar to PtdCho, PtdEtn and PtdIns smaller amounts of molecular species containing C20:4 fatty acid were also observed in PtdSer (Table 1).
Figure 4.
Identification of PtdSer oxidation products in primary cortical neurons incubated with STS. Typical negative ion ESI-MS of molecular species detected after 2D-HPTLC isolation of PtdSer from primary cortical neurons (A- control and B- after STS exposure). Tandem MS-MS experiments confirmed the structures of non-oxidized and oxidized PtdSer species (panels A, a, b; B, c, d). Identification of individual oxidized molecular species of PS containing C22:6+O (c) and C22:6+2O (d).
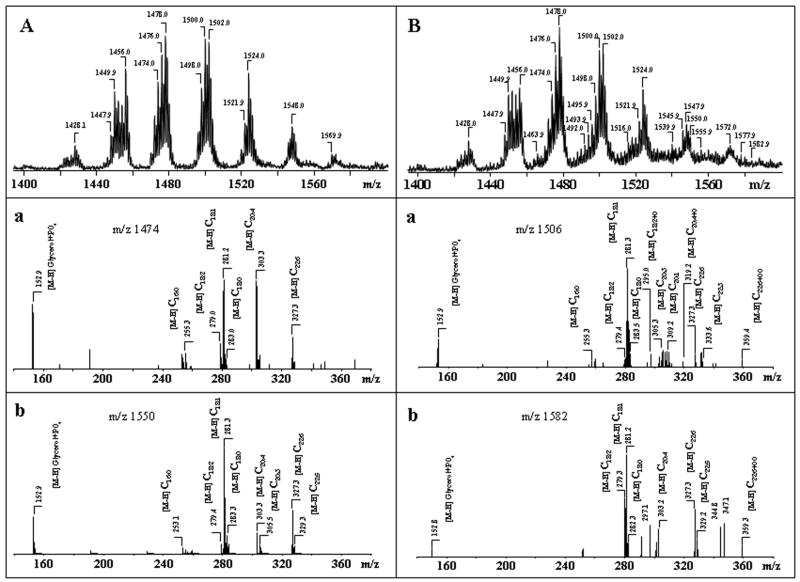
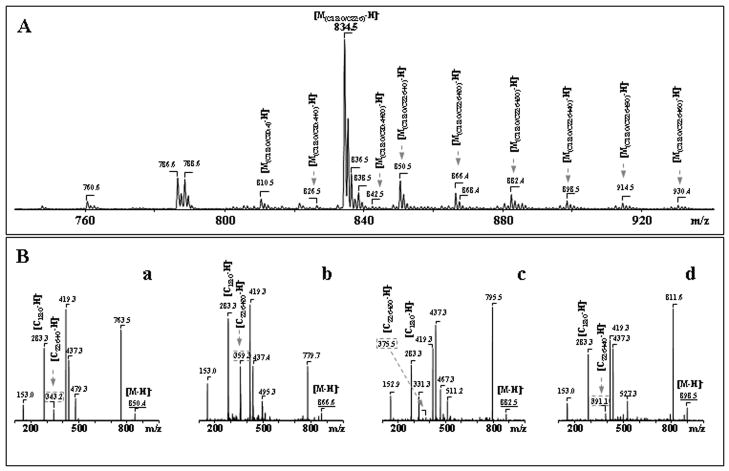
Ptd2Gro possesses two negative charges resulting in either singly charged [M-H]− or doubly charged [M-2H]−2 ions. As shown in Fig. 3, G singly charged ions of Ptd2Gro in negative mode were represented by at least 9 different molecular clusters with m/z 1374, 1400, 1428, 1450, 1476, 1500, 1524, 1550 and 1572 with a variety of fatty acid residues (from C14:0 to C22:6), including polyunsaturated fatty acids highly susceptible to oxidation - C20:4, C22:5 and C22:6 (Table 2). One of the major Ptd2Gro molecular clusters with m/z 1474, includes at least three different Ptd2Gro molecular species, containing C18:2, C20:4, C22:5 and C22:6 fatty acids, as follows: (C18:1)1/(C18:2)2/(C20:4)1; (C16:1)1/(C18:1)2/(C22:6)1; and (C16:1)1/(C18:1)1/(C18:2)1/(C22:5)1 (Table 2). Typical ions formed during fragmentation process of Ptd2Gro (a, b, a+56, or b+136) were identified in MS2 spectra as described by Hsu and co-authors (Hsu et al. 2004). Then MS3 was performed on each of a or b ions to assign fatty acids and their positions. Using pulsed-Q-dissociation technique, we also obtained MS spectrum of Ptd2Gro ion fragments including a, b, a+56; a+136; b+56; b+136 and fatty acid carboxylate anions. For example, MS2 fragmentation of a parent ion with m/z 1500 resulted in a major ion with m/z 721 with characteristic ion fragments (a+56, a+136), with m/z 777 and 857. MS3 fragmentation of ion with m/z 721 yielded carboxylate ions with m/z 281 and 303 leading to identification of the symmetric structure (C18:1/C20:4/C18:1/C20:4)-Ptd2Gro. However, Ptd2Gro from neuronal cells are represented by multi-molecular clusters with more than 60 different molecular species which overlap with oxidized molecular species in apoptotic samples. Therefore, the data presented in Tables 2 and 3 did not include stereo-specific position of Ptd2Gro fatty acids -(C18:1)2/(C20:4)2-Ptd2Gro.
Overall, the diversity of molecular species of phospholipids in rat cortical neurons followed the order Ptd2Gro > PtdEtn ≫ PtdCho ≫ PtdSer > PtdIns > CerPCho. With the focus of this study on phospholipid peroxidation, it is also important that the number of polyunsaturated oxidizable species decreased in the order Ptd2Gro ≫ PtdEtn >PtdCho > PtdSer > PtdIns > CerPCho. Thus a relatively minor class of phospholipids, Ptd2Gro, was represented in cortical neurons by the greatest variety of both total and peroxidizable molecular species.
Selective STS induced oxidation of phospholipids in neurons
Neuronal apoptosis induced by STS is accompanied by the production of ROS (Pong et al. 2001) suggesting a possibility of peroxidative modification of one or more classes of phospholipids. In fact, indirect indications of phospholipid peroxidation during neuronal apoptosis, particularly of Ptd2Gro, have been reported (Kirkland et al. 2002). However, neither definitive quantitative assessments nor identification of individual molecular species of phospholipids involved, to the best of our knowledge, have been documented. Therefore, we used oxidative lipidomics analysis to assess the oxidation of different classes of phospholipids in neuronal cells during intrinsic apoptosis induced by STS. To characterize the diversity of phospholipid oxidation products generated during apoptosis we employed quantitative assessments of lipid hydroperoxides using fluorescence HPLC-based protocol and identification of oxidation products of polyunsaturated phospholipid molecular species by ESI-MS analysis (Pulfer & Murphy 2003, Forrester et al. 2004, Spickett & Dever 2005).
Analysis of total amounts of hydroperoxides in major classes of phospholipids revealed that three anionic phospholipids — Ptd2Gro ≫ PtdSer > PtdIns — underwent robust oxidation in neuronal cells after exposure to STS (Fig. 2B). No significant oxidation in the most dominant phospholipid classes – PtdCho and PtdEtn - was detected after STS incubation as evidenced by only slight elevations of the PtdEtn OOH and PtdCho OOH (Fig. 2B).
Identification of oxidized phospholipid molecular species can be performed by their mass-charge ratio (MS1) and fragmentation properties (MS2). Exposure of primary neurons to STS did not cause marked changes in diversity of individual molecular species in the major classes of phospholipids (with the notable exceptions of PtdSer - see below). It should be noted, however, that accurate quantitative assessments of the amounts of each molecular species are ambiguous as each of the m/z signals usually contains more than one molecular species. For example, MS2 analysis of molecular species of PtdSer signal with m/z 866 after exposure of neurons to STS revealed the presence of several individual species: (C20:0)1/(C22:4)1, (C18:0)1/(C24:4)1, as well as oxygenated PtdSer species (C18:0)1/(C22:6+OO)1. Similarly, fragmentation analysis of Ptd2Gro at m/z 1506 identified the following contributing species: (C16:0)1/(C18:1)1/(C20:3)1/(C22:3)1 and (C16:0)1/(C18:0)1/(C20:1)1/(C22:6)1, along with oxygenated species (C18:1)1/(C18:2)1/(C18:2+O)1/(C20:4+O)1 and (C16:0)1/(C18:1)1/(C18:2)1/(C22:6+OO)1.
We found the appearance of new Ptd2Gro molecular clusters corresponding to species containing oxidized C22:6 acid, (Table 3) in MS-spectra of STS exposed neuronal cells which were not present in Ptd2Gro of non-exposed cells. In full MS spectra, these signals overlapped with the signals from non-oxidized Ptd2Gro molecular species (Table 2). To distinguish between oxygenated and non-oxygenated species, MS2 analysis was performed. For example, fragmentation of two ion clusters with m/z 1506 and 1582 in non-STS treated neurons revealed dominant ion peaks that corresponded to (C16:1)1/(C18:0)1/(C20:3)1/(C22:3)1 and (C18:0)1/(C20:1)1/(C22:5)2, respectively (Table 2). However, after the exposure of the cells to STS, MS2 analysis of these ion clusters revealed oxygenated C22:6 molecular species (Table 3). Detailed analysis of the peak with m/z 1506 demonstrated that this cluster corresponds to multiple Ptd2Gro species with a dominant isomer of (C16:1)1/(C18:1)2/(C22:6+OO)1 originating from the ion at m/z 1474 containing (C16:1)1/(C18:1)2/(C22:6)1. Similarly, MS2 analysis of the peak at m/z 1582 showed that the [M-H]− ion included molecular Ptd2Gro species with a component of (C18:1)1/(C22:6)1/(C18:0)1/(C22:6+OO)1 originating from the ion at m/z 1550 (Table 3).
In the case of PtdSer, the effect of STS showed distinct differences between treated and control cells (Fig. 4). In STS exposed cells, elevated levels of PtdSer molecular species with oxidized C22:6, PtdSer-OH and PtdSer-OOH with m/z 850 and 866, respectively, were detected compared with control cells (Fig. 4A, a, b; 4B, c, d). Detailed analysis of these peaks by MS/MS demonstrated that the [M-H]− ions at m/z 850 and 866 corresponded to the species (C18:0/C22:6+O) and (C18:0/C22:6+OO); these types of PtdSer molecules were produced via oxygenation of the PtdSer species with the corresponding ion at m/z 834 (C18:0/C22:6) (Table 1, 3). Additionally, molecular species of PtdSer with arachidonic (C20:4), docosatetraenoic (C22:4) and docosapentaenoic (C22:5) acyls also underwent oxidation and formed oxidized clusters with m/z 826, 842, 868 and 870 (Table 3).
Further, exposure of cells to STS resulted in oxidation of mostly C20:4 containing molecular species of PtdIns at m/z 901 and 917 which corresponded to PtdIns-OH and PtdIns-OOH with the dominant products of (C18:0/C20:4+O) and (C18:0/C20:4+OO); the oxygenated PtdIns molecular species originated from the ion at m/z 885 (C18:0/C20:4) (Table 1, 3).
Several oxygenation products such as hydroxy- and hydroperoxy-derivatives formed from different molecular species of the two most abundant phospholipids - PtdCho and PtdEtn - were also observed after STS treatment (Table 3). However, quantitatively their amounts were markedly less as compared with oxidized molecular species of Ptd2Gro, PtdSer and PtdIns. As shown in Table 3, STS induced formation of PtdEtn-OH and PtdEtn-OOH occurred predominantly in PtdEtn-alkenyl molecular species.
Oxidized Molecular species formed from neuronal Ptd2Gro and PtdSer in the presence of Cyt c/H2O2
Selective predominant peroxidation of anionic phospholipids in cortical neurons treated with STS suggests that there may be a specific mechanism involved in the oxidative metabolism. Recently, we reported that Ptd2Gro and PtdSer avidly bind to cyt c and form a complex in which the partially unfolded protein functions as a peroxidase, catalyzing oxidation of these anionic phospholipids (Kagan et al. 2004, Kagan et al. 2005, Belikova et al. 2006, Kapralov et al. 2007). This new peroxidase activity of cyt c towards Ptd2Gro and PtdSer was found to take place both in model biochemical systems as well as in cells undergoing apoptosis. However, molecular identification of the products formed was not performed. We reasoned that selective peroxidation of anionic phospholipids in STS-treated neurons could be due, at least in part, to cyt c-dependent reactions. To better understand possible involvement of cyt c in STS oxidation of anionic phospholipids in neurons, we incubated Ptd2Gro and PtdSer isolated from rat brain cortex with cyt c in the presence and in the absence (control) of H2O2 for 30 min. Assessments of the total amount of phospholipids hydroperoxides demonstrated the accumulation of peroxidized Ptd2Gro (58.4 ±10.2 pmol Ptd2Gro -OOH/nmol Ptd2Gro vs control 3.6 ± 0.6 pmol Ptd2Gro -OOH/nmol Ptd2Gro, respectively) and peroxidized PtdSer (35.8 ±2.7 pmol PtdSer -OOH/nmol PtdSer vs control 1.6 ± 0.6 pmol PtdSer -OOH/nmol PtdSer, respectively).
ESI-MS analysis revealed a variety of oxygenated species (Fig. 5, 6). Typical ESI-MS spectra of singly-charged molecular ions of non-oxidized Ptd2Gro and those obtained after Ptd2Gro incubation with cyt c/H2O2 are presented in Fig. 5, A, B. Oxidation of Ptd2Gro resulted in the appearance of several hydroxy- and hydroperoxy-Ptd2Gro (Fig. 5, B). Ions at m/z 1463, 1490, 1492, 1494, 1512, 1514, 1516 and 1518 corresponded to hydroxy-Ptd2Gro molecular species which were formed from C18:2, C20:4, C22:5 and C22:6 acids (and originated from ions at m/z 1447, 1474, 1476, 1478, 1496, 1498, 1500, 1502, respectively). MS2-fragmentation of two ion clusters with m/z 1506 and 1582 detectable in non-oxidized Ptd2Gro showed dominant ion peaks that corresponded to (C16:1)1/(C18:0)1/(C20:3)1/(C22:3)1 and (C18:0)1/(C20:1)1/(C22:5)2, respectively (Table 2). MS2- analysis of peaks at m/z 1506 and 1582 after CL oxidation by cyt c/H2O2 showed that the [M-H]− ions included both non-oxidized (see above) and oxidized molecular Ptd2Gro species (C16:0)1/(C18:1)1/(C18:2)1/(C22:6+OO)1 and (C18:1)1/(C22:6)1/(C18:0)1/(C22:6+OO)1 originating from the ions at m/z 1474 and 1550 (Fig. 5A, a, b, Fig. 5B, a, b, Table 3). These oxidized Ptd2Gro were also observed in STS treated neurons.
Figure 5.
Characterization of oxidation products formed during incubation of total brain cortical Ptd2Gro in the presence of cyt c/H2O2. Typical negative ion ESI-MS of cortical Ptd2Gro molecular species (A- control and B- after cyt c/H2O2 treatment). MS-MS experiments confirmed the structures of non-oxidized and oxidized Ptd2Gro species (panels A, a, c, B, b, e).
Figure 6.
Characterization of oxidation products formed during incubation of total brain cortical PtdSer in the presence of cyt c/H2O2. Typical negative ion ESI-MS of cortical PtdSer molecular species are shown. MS-MS experiments confirmed the structures of non-oxidized and oxidized PtdSer species (panels A, B, a, b, c, d).
Oxidized PtdSer contained several molecular species that were represented by deprotonated ions [M-H]− with m/z 850; 866; 882 and 898 corresponding to hydroxy-, hydroperoxy-, hydroxy-hydroperoxy- and dihydroperoxy- PtdSer (Fig. 6A, B). MS2 analysis of oxidized PtdSer molecular species showed that they originated from the molecular cluster of PtdSer with m/z 834 (C18:0/C22:6) and contained hydroxy-DHA (C22:6+O), hydroperoxy-DHA (C22:6+OO), hydroxy-hydroperoxy-DHA (C22:6+O+OO) and di-hydroperoxy-DHA (C22:6+2OO) at m/z 343; 359; 375 and 391, respectively (Fig. 6B). To confirm and structurally characterize the conversion of Ptd2Gro–OOH and PtdSer-OOH into Ptd2Gro–OH and PtdSer-OH, respectively, we treated the reaction products with a reductant, TPP. This resulted in the reduction of hydroperoxy-Ptd2Gro and hydroperoxy-PtdSers to their respective hydroxy-derivatives (data not shown).
DISCUSSION
Compartmentalization and integration of metabolic processes as well as coordination of biochemical reactions in membranes are two essential functions of phospholipids (van Meer et al. 2008). While inherently complex, these functions may be fulfilled with a relatively limited number of molecular species of several classes of phospholipids (Dikarev et al. 1982). Recent lipidomics studies, however, indicate that the molecular diversity of phospholipids in tissues, particularly for some of them, may be very high. An interesting example is Ptd2Gro, which may be represented predominantly by one or only few molecular species in some tissues (eg, heart, liver or intestines) (Schlame et al. 2005, Han et al. 2006, Tyurina et al. 2008) but may be enriched with over hundreds of individual species in others (eg, brain) (Cheng et al. 2008, Kiebish et al. 2008). The underlying reasons for the diversity or uniformity of molecular speciation of Ptd2Gro and other phospholipids are still largely unknown.
Another role that phospholipids play is signaling that commonly utilizes phospholipid oxygenation products or hydrolysis intermediates (Kim 2007, Bazan 2005, Bazan 2007). Interestingly, the signaling and structural roles are separated such that oxygenated or hydrolyzed phospholipids are not normally common components of membrane bilayers (Kagan 1988). Conversely, effective reconstitution of functions of membrane proteins – while potentially phospholipid class-specific – is not usually achieved by peroxidized phospholipids (Casey 1995, Epand 2007, Lemmon 2008). For example, depletion of mitochondrial complexes (cyt c oxidase) of Ptd2Gro results in the loss of activity that can be re-gained by the addition of non-oxidized Ptd2Gro but not of oxidized CL (Musatov 2006).
This paper reports the molecular diversity of major classes of phospholipids in rat cortical neurons. Notably, Ptd2Gro – the least dominant phospholipid – was most diversified, followed by PtdEtn and PtdCho while PtdSer, PtdIns and CerPCho were represented by a relatively smaller numbers of individual molecular species. For comparison, at least 60 species of Ptd2Gro were revealed after analysis of our ESI-MS data as compared with 50 for PtdEtn, 45 for PtdCho, and 30, 15, and 5, for PtdSer, PtdIns, and CerPCho, respectively. Pathways leading to diversified molecular species of Ptd2Gro, particularly in the brain, include Ptd2Gro synthesized through the condensation of PtdGro and CDP-diacylglycerol (so-called immature Ptd2Gro) as well as Ptd2Gro deacylation to monolyso-Ptd2Gro and reacylation (to yield mature Ptd2Gro) (Hauff & Hatch 2006, Kiebish et al. 2008, Cheng et al. 2008). Formation of the mature molecular species of Ptd2Gro is reportedly realized via the trans-acylation from the sn-2 position of donor choline (ChoGpl) and ethanolamine (EtnGpl) glycerophospholipids (Xu et al. 2003, Schlame & Ren 2006). In a recent work, Cheng et al (Cheng et al. 2008) found that molecular profile of Ptd2Gro of cultured neuronal cells from the neocortices of mice of 15–17 days gestation was essentially identical to immature molecular speciation of Ptd2Gro in the brain of prenatal mice. Our current study demonstrates that the pattern of molecular species of Ptd2Gro in cultured cortical neurons obtained from 17 days old rat embryos was similar to the diversified profile of cortex Ptd2Gro characteristic of postnatal (17 days old) animals (Bayir et al. 2007). These differences in the diversification of Ptd2Gro molecular profiles between the primary neuronal cultures obtained from mice (Cheng et al. 2008) or rats (the current study) may be attributed to species differences as well as different culture conditions (Dulbecco’s minimum Eagle’s medium supplemented with 20 mM glucose, 26 mM sodium bicarbonate, 2 mM L-glutamine vs Neurobasal(tm) medium supplemented with B27(tm) (Gibco) and GlutaMaxl(tm) (Sigma) (Brewer et al. 1993). Our experiments were conducted at 8–12 day in vitro and previous studies have revealed that neuronal maturation can be achieved by day 8–12 in primary rat cortical neuronal cultures (Choi et al. 1987, Katayama et al. 1997). As a result of these different conditions during subculturing, the primary rat cortical neurons employed in our study elicited a more advanced differentiation associated with a higher content of Ptd2Gro molecular species containing long-chain polyunsaturated acyl chains (“signaling fatty acids” such as arachidonic acid and docosahexaenoic acid (Cheng et al. 2008).
Thus, the diversity of different phospholipid classes is not determined by their abundance. This suggests that structural roles of phospholipids, likely associated with most abundant phospholipids, do not likely require multiple molecular species of phospholipids with the same polar head. It is tempting to speculate that involvement of specific classes of phospholipids in signaling dictates their diversity. This becomes even more clear when one compares the diversity of phospholipids with highly oxidizable polyunsaturated fatty acid residues. Again, three phospholipids Ptd2Gro, PtdEtn, and PtdCho are mostly enriched with polyunsaturated molecular species. It is possible that oxidation of these phospholipids is associated with their participation in signaling either directly or after hydrolysis of oxygenated fatty acids. Because our 2D-HPTLC analysis did not reveal accumulation of any significant amounts of lyso-forms of any of phospholipids in control or STS-treated samples, we suggest that oxidized phospholipids could be either directly involved in signaling processes or underwent rapid and effective remodeling from their lyso-derivatives.
We did not detect marked changes in phospholipid composition or molecular speciation of major phospholipids classes after exposure of cells to a pro-apoptotic agent, STS. Given that under the conditions used 20 and 40% of cells were apoptotic after 4 and 8 hrs, this indicates that quantitatively only a small fraction of phospholipids – representing only a minimal percentage in each of the molecular species, could be involved in signaling processes. This prompted us to focus on detailed analysis of oxidatively modified phospholipids.
This work demonstrates, that apoptosis in cortical neurons was associated with selective peroxidation of anionic phospholipids Ptd2Gro, PtdSer, and PtdIns, and identified molecular species of peroxidized lipids in each class. Multiple products may be formed in the course of lipid peroxidation, particularly in metal-catalyzed reactions. Low abundance of these numerous products makes their analysis very difficult. The diversity of the oxidized phospholipids derivatives, however, stems from the same primary molecular product, phospholipid hydroperoxides (Yoshida et al. 2005, Yoshida et al. 2006, Kitano et al. 2007). This led us to choose phospholipid hydroperoxides as the biomarker of peroxidation. Our approach was based on quantitative assessments of phospholipid hydroperoxides in each of major phospholipids classes followed by subsequent identification of individual peroxidized substrates and products in them. We found that accumulation of hydroperoxides was most prominent in Ptd2Gro followed by PtdSer and PtsIns. No significant accumulation of hydroperoxides was detected in two most abundant phospholipids – PtdCho and PtdEtn.
Recent comprehensive examination of the brain mitochondrial lipidome established heterogeneity of Ptd2Gro and PtdSer distribution in highly purified synaptic and non-synaptic mitochondria obtained from C57BL/6J mouse cerebral cortex (Kiebish et al. 2008). These studies established that higher levels of Ptd2Gro were characteristic of non-synaptic mitochondria vs synaptic mitochondria; on the contrary levels of PtdSer were higher in synaptic than in non-synaptic mitochondria. The contents of PtdIns were not different in synaptic and non-synaptic mitochondria. All three of these anionic phospholipids form complexes with cyt c and confer peroxidase activity on the protein whereby the resulting peroxidase complex readily oxidizes phospholipids (Kapralov et al. 2007). Thus all three anionic phospholipids – Ptd2Gro, PtdSer, and PtdIns – can undergo peroxidation catalyzed by cyt c in mitochondria. However, the binding constant of Ptd2Gro with cyt c is about two orders of magnitude higher than those for PtdSer and PtdIns (Kapralov et al. 2007). Considering also that the content of Ptd2Gro in brain mitochondria is more than 5-fold higher than that of PtdSer (Kiebish et al. 2008) it seems logical to conclude that Ptd2Gro represents the major substrate for cyt c catalyzed peroxidation in mitochondria (see below).
ESI-MS analysis established that hydroperoxides were not the only products of anionic phospholipids oxygenation during STS-induced neuronal apoptosis: significant amounts of hydroxy- as well as hydroxy-/hydroperoxy-derivatives were identified in peroxidized phospholipids. This implies that not only di-oxygenation but also subsequent reduction of phospholipid hydroperoxides took place in STS-treated cells. While accurate quantitative assessments of the amounts of hydroxy- and peroxy- derivatives of different phospholipids molecular species are limited by the lack of the appropriate standards for peroxidized phospholipids, estimates (using standards for non-peroxidized phospholipids) show that approximately one third of oxygenated phospholipids were represented by their hydroxy-derivatives while two thirds were found as hydroperoxides. These ESI-MS data are in good agreement with our assessments of phospholipid hydroperoxides based on fluorescence HPLC-based protocol with Amplex Red. To better characterize the structure of oxidation products formed from Ptd2Gro in vivo, we performed experiments in which we oxidized Ptd2Gro isolated from neurons by cyt c/H2O2 system in vitro. We found that Ptd2Gro molecular species containing C22:6 fatty acids were predominantly oxidized both in cells and in the model system; more abundant molecular species containing C20:4 were oxidized to a lesser extent. This indicates that the level of unsaturation rather than abundance determines Ptd2Gro oxidation in STS treated neurons.
Because different hydroxy-fatty acids formed from hexaenoic and pentaenoic fatty acids – neuroprotectins, resolvins (Serhan et al. 2008, Schwab et al. 2007, Bazan 2008, Bazan 2007) – may act as potent regulators of neuronal processes it will be important to explore the possibility that peroxidized phospholipids may be used as sources of these and other physiologically active oxygenated fatty acid molecules. Notably, PtdSer has been identified as an important starting molecule in biosynthesis of neuroprotectins (Bazan 2005, Kim 2007).
Several earlier reports indicated that Ptd2Gro oxidation occurred during neuronal apoptosis (Kirkland et al. 2002, Fernandez-Gomez et al. 2005). Most of them, however, employed fluorogenic indicator, NAO; the applicability and validity of this approach for quantitative assessments of Ptd2Gro in cells have been rigorously debated (Garcia Fernandez et al. 2004, Kaewsuya et al. 2007). Our results not only quantitatively assess Ptd2Gro peroxidation during STS induced apoptosis but also present molecular identification of different oxygenated Ptd2Gro molecular species in apoptotic cortical neurons.
Finally, very small amounts of oxygenated derivatives were detected in PtdEtn and PtdCho. These were mostly represented by hydroxy-homologues and were not reliably detectable in our fluorescence HPLC-based assay of phospholipid hydroperoxides.
Our previous studies demonstrated that Ptd2Gro and its oxidation products are important participants and signaling molecules in the execution of the mitochondrial stage of intrinsic apoptosis (Kagan et al. 2005). Current study identifies individual molecular species of Ptd2Gro and PtdSer containing hydroperoxy- and hydroxy-groups and accumulating during apoptosis induced in neurons by staurosporine (STS). In a preliminary study, we found that stretched neurons undergoing apoptosis - similar to STS-induced apoptosis – respond by selective oxidation of Ptd2Gro and PtdSer. Further, we found that γ-irradiation caused both apoptotic cell death and significant oxidation of Ptd2Gro in vitro (Belikova et al. 2007) and in vivo (Tyurina et al. 2008). In these cases too, the major phospholipid molecular species involved in γ-irradiation triggered apoptosis and peroxidation of phospholipid included hydroperoxy-, hydroxy/hydroperoxy- and hydroxy- molecular species of Ptd2Gro and PtdSer. The “uniformity” of the oxidation pattern may due to the common catalytic mechanism – peroxidase function of cyt c (Kagan et al. 2004, Kagan et al. 2005, Tyurina et al. 2004a, Tyurina et al. 2008, Jiang et al. 2003, Belikova et al. 2007, Kapralov et al. 2007). Thus, selective accumulation of these oxygenated species of Ptd2Gro and PtdSer may be a characteristic oxidative fingerprint of apoptosis. It is possible that necrotic cell death associated with the disruption of plasma membrane may involve oxidation of other classes of phospholipids, particularly PtdCho in the outer leaflet of the membrane. This scenario is likely to occur when oxidation is driven by exogenous sources of ROS such as myeloperoxidase or NADPH oxidase of inflammatory cells.
The question about possible catalytic redox mechanisms of apoptosis-associated peroxidation of anionic phospholipids needs further investigations. Previous studies demonstrated STS induced disruption of mitochondrial electron transport (Cai & Jones 1998) followed by the generation of superoxide (Gil et al. 2003). Our previous work has identified cyt c as a potentially important catalyst of peroxidation of anionic phospholipids during apoptosis (Kagan et al. 2004, Kagan et al. 2005).
The folded native structure of cyt c, optimal for maintenance of its hexacoordinate arrangement, is essential for its function as a mobile electron carrier. In the native conformation, the iron atom of cyt c is protected against interaction with H2O2, by coordination with porphyrin and two axial ligands, Met80 and His18, resulting in its very weak peroxidase activity (Florence 1985, Radi et al. 1991, Kagan et al. 2005, Tyurina et al. 2006). Interaction of cyt c with Ptd2Gro induces partial unfolding of cyt c (as evidenced by an increased fluorescence of Trp59), weakening and disruption of the Fe-S(Met80) bond (detectable by disappearance of characteristic absorbance at 695 nm), and facilitation of H2O2 penetration to the catalytic site (Vlasova et al. 2006, Basova et al. 2007, Kapralov et al. 2007). As a result, the peroxidase activity of cyt c towards typical phenolic substrates (2′,7′-dichlorofluorescin or Amplex Red) is increased many-fold (up to 100 times) upon binding with Ptd2Gro (Kagan et al. 2005). In the absence of alternative substrates, peroxidase activity of cyt c complexes with polyunsaturated Ptd2Gro species is realized in oxidation of both its components. Oxidation of the protein results in its oligomerization via recombination of protein-immobilized (possibly tyrosyl) radicals, while oxidation of polyunsaturated Ptd2Gro yields its hydroperoxy- and hydroxy-derivatives. As with other peroxidases, two major catalytic mechanisms may be potentially involved in the cleavage of H2O2 – the homolytic and the heterolytic pathways (Barr et al. 1996, Banci 1997, Gebicka 2001, Diederix et al. 2002, Furtmüller et al. 2006) - leading to Ptd2Gro oxidation. Detailed analysis of these catalytic mechanisms, to the best of our knowledge, has not been reported for cyt c/polyunsaturated Ptd2Gro complexes. It should be noted, however, that a phenolic lipid radical scavenger – etoposide – demonstrated low efficiency in inhibiting tetralinoleoyl-Ptd2Gro oxidation induced by cyt c/H2O2 compared to its markedly higher antioxidant activity when peroxidation of tetralinoleoyl-Ptd2Gro was induced by a lipid-soluble azo-initiator, 2,2′-azobis(2,4-dimethyl-valeronitrile (Tyurina et al. 2006). This suggests that random non-enzymatic propagation of the peroxidation process is not a major contributor to cyt c-catalyzed oxidation of polyunsaturated Ptd2Gro and that radical intermediates are not readily accessible to phenolic radical scavengers. This interpretation is in line with previous findings demonstrating selective peroxidation of Ptd2Gro (rather than other more abundant polyunsaturated phospholipids such as PtdCho and PtdEtn) during early apoptosis in different cell lines (Kagan et al. 2005). However, molecular speciation of the Ptd2Gro substrates involved, has not been characterized, particularly as it relates to neuronal cells. In the current work, we performed analysis of Ptd2Gro individual molecular species involved in the peroxidation process that takes place during apoptosis induced in cortical neurons by STS.
Using a number of cell lines, we demonstrated that Ptd2Gro and PtdSer are the major targets of cyt c catalyzed utilization of ROS (superoxide and its dismutations product, H2O2) to generate oxygenated derivatives of these phospholipids first in mitochondria and subsequently (after the release of cyt c into the cytsol) in extra-mitochondrial compartments (Kagan et al. 2004, Kagan et al. 2005, Belikova et al. 2007, Jiang et al. 2008). The significance of these redox reactions is that Ptd2Gro peroxidation products are essential for the release of pro-apopotic factors from mitochondria whereas PtdSer oxidation products stimulate PtdSer externalization (Tyurina et al. 2004c) and recognition of apoptotic cells by professional phagocytes (Gao et al. 2006, Greenberg et al. 2006, Borisenko et al. 2004). This new role of cyt c as a redox catalyst of Ptd2Gro and PtdSer peroxidation agrees well with the known facts on the involvement of peroxidized Ptd2Gro in detachment of cyt c from the mitochondrial membrane (Ott et al. 2007), and participation in pro-apoptotic proteins Bax/Bak-dependent mitochondrial permeability transition (Jiang et al. 2008, Lucken-Ardjomande et al. 2008). Results of our experiments in model systems with total Ptd2Gro and PtdSer fractions from neurons confirmed a very similar molecular identity of the products formed when they were incubated in the presence of cyt c and H2O2. It is tempting to speculate that selective peroxidation of Ptd2Gro and PtdSer was due, at least in part, to their catalytic redox interactions with cyt c during STS induced apoptosis in neurons. Importantly, our previous work established that Ptd2Gro and PtdSer, predominantly their molecular species containing C22:6, are specific and prominent targets for peroxidative acute brain injury in vivo (Bayir et al. 2007). Consequently, potent mitochondria-targeted strategies can be applied to regulate Ptd2Gro peroxidation to either inhibit (in neurodegenerative diseases) or stimulate (in neuro-tumors) as a promising new paradigm in drug design.
Acknowledgments
Supported by grants from NIH HL70755, HD057587-01A2, DAMD 17-01-2-637, Pennsylvania Department of Health SAP 4100027294, AHA-0535365N, Human Frontier Science Program.
References
- Albert CJ, Crowley JR, Hsu FF, Thukkani AK, Ford DA. Reactive chlorinating species produced by myeloperoxidase target the vinyl ether bond of plasmalogens. Identification of 2-chlorohexadecanal. J Biol Chem. 2001;276:23733–23741. doi: 10.1074/jbc.M101447200. [DOI] [PubMed] [Google Scholar]
- Banci L. Structural properties of peroxidases. J Biotechnol. 1997;53:253–263. doi: 10.1016/s0168-1656(97)01677-5. [DOI] [PubMed] [Google Scholar]
- Barr DP, Gunther MR, Deterding LJ, Tomer KB, Mason RP. ESR spin-trapping of a protein-derived tyrosyl radical from the reaction of cytochrome c with hydrogen peroxide. J Biol Chem. 1996;271:15498–15503. doi: 10.1074/jbc.271.26.15498. [DOI] [PubMed] [Google Scholar]
- Basova LV, Kurnikov IV, Wang L, et al. Cardiolipin switch in mitochondria: shutting off the reduction of cytochrome c and turning on the peroxidase activity. Biochemistry. 2007;46:3423–3434. doi: 10.1021/bi061854k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayir H, Tyurin VA, Tyurina YY, et al. Selective early cardiolipin peroxidation after traumatic brain injury: an oxidative lipidomics analysis. Ann Neurol. 2007;62:154–169. doi: 10.1002/ana.21168. [DOI] [PubMed] [Google Scholar]
- Bazan NG. Neuroprotectin D1 (NPD1): a DHA-derived mediator that protects brain and retina against cell injury-induced oxidative stress. Brain Pathol. 2005;15:159–166. doi: 10.1111/j.1750-3639.2005.tb00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazan NG. Omega-3 fatty acids, pro-inflammatory signaling and neuroprotection. Curr Opin Clin Nutr Metab Care. 2007;10:136–141. doi: 10.1097/MCO.0b013e32802b7030. [DOI] [PubMed] [Google Scholar]
- Bazan NG. Neurotrophins induce neuroprotective signaling in the retinal pigment epithelial cell by activating the synthesis of the anti-inflammatory and anti-apoptotic neuroprotectin D1. Adv Exp Med Biol. 2008;613:39–44. doi: 10.1007/978-0-387-74904-4_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belikova NA, Jiang J, Tyurina YY, Zhao Q, Epperly MW, Greenberger J, Kagan VE. Cardiolipin-specific peroxidase reactions of cytochrome C in mitochondria during irradiation-induced apoptosis. Int J Radiat Oncol Biol Phys. 2007;69:176–186. doi: 10.1016/j.ijrobp.2007.03.043. [DOI] [PubMed] [Google Scholar]
- Belikova NA, Vladimirov YA, Osipov AN, et al. Peroxidase activity and structural transitions of cytochrome c bound to cardiolipin-containing membranes. Biochemistry. 2006;45:4998–5009. doi: 10.1021/bi0525573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisenko GG, Iverson SL, Ahlberg S, Kagan VE, Fadeel B. Milk fat globule epidermal growth factor 8 (MFG-E8) binds to oxidized phosphatidylserine: implications for macrophage clearance of apoptotic cells. Cell Death Differ. 2004;11:943–945. doi: 10.1038/sj.cdd.4401421. [DOI] [PubMed] [Google Scholar]
- Böttcher CSF, Van Gent CM, Fries C. A rapid and sensitive sub-micro phosphorus determination. Anal Chim Acta. 1961;24:203–204. [Google Scholar]
- Brewer GJ, Torricelli JR, Evege EK, Price PJ. Optimized survival of hippocampal neurons in B27-supplemented Neurobasal, a new serum-free medium combination. J Neurosci Res. 1993;35:567–576. doi: 10.1002/jnr.490350513. [DOI] [PubMed] [Google Scholar]
- Cai J, Jones DP. Superoxide in apoptosis. Mitochondrial generation triggered by cytochrome c loss. J Biol Chem. 1998;273:11401–11404. doi: 10.1074/jbc.273.19.11401. [DOI] [PubMed] [Google Scholar]
- Casey PJ. Protein lipidation in cell signaling. Science. 1995;268:221–225. doi: 10.1126/science.7716512. [DOI] [PubMed] [Google Scholar]
- Cheng H, Mancuso DJ, Jiang X, Guan S, Yang J, Yang K, Sun G, Gross RW, Han X. Shotgun Lipidomics Reveals the Temporally Dependent, Highly Diversified Cardiolipin Profile in the Mammalian Brain: Temporally Coordinated Postnatal Diversification of Cardiolipin Molecular Species with Neuronal Remodeling. Biochemistry. 2008 doi: 10.1021/bi7023282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi DW, Maulucci-Gedde M, Kriegstein AR. Glutamate neurotoxicity in cortical cell culture. J Neurosci. 1987;7:357–368. doi: 10.1523/JNEUROSCI.07-02-00357.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Sa-Eipper C, Roth KA. Caspase regulation of neuronal progenitor cell apoptosis. Dev Neurosci. 2000;22:116–124. doi: 10.1159/000017433. [DOI] [PubMed] [Google Scholar]
- Diederix RE, Ubbink M, Canters GW. Peroxidase activity as a tool for studying the folding of c-type cytochromes. Biochemistry. 2002;41:13067–13077. doi: 10.1021/bi0260841. [DOI] [PubMed] [Google Scholar]
- Dikarev VP, VI S, VE V. Noctiluca miliaris - one more protozoan with unusual lipid composition. Comp Biochem Physiol. 1982;72B:137–140. [Google Scholar]
- Du L, Bayir H, Lai Y, Zhang X, Kochanek PM, Watkins SC, Graham SH, Clark RS. Innate gender-based proclivity in response to cytotoxicity and programmed cell death pathway. J Biol Chem. 2004;279:38563–38570. doi: 10.1074/jbc.M405461200. [DOI] [PubMed] [Google Scholar]
- Epand RM. Membrane lipid polymorphism: relationship to bilayer properties and protein function. Methods Mol Biol. 2007;400:15–26. doi: 10.1007/978-1-59745-519-0_2. [DOI] [PubMed] [Google Scholar]
- Fernandez-Gomez FJ, Gomez-Lazaro M, Pastor D, Calvo S, Aguirre N, Galindo MF, Jordan J. Minocycline fails to protect cerebellar granular cell cultures against malonate-induced cell death. Neurobiol Dis. 2005;20:384–391. doi: 10.1016/j.nbd.2005.03.019. [DOI] [PubMed] [Google Scholar]
- Florence TM. The degradation of cytochrome c by hydrogen peroxide. J Inorg Biochem. 1985;23:131–141. doi: 10.1016/0162-0134(85)83017-8. [DOI] [PubMed] [Google Scholar]
- Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- Forrester JS, Milne SB, Ivanova PT, Brown HA. Computational lipidomics: a multiplexed analysis of dynamic changes in membrane lipid composition during signal transduction. Mol Pharmacol. 2004;65:813–821. doi: 10.1124/mol.65.4.813. [DOI] [PubMed] [Google Scholar]
- Furtmüller PG, Zederbauer M, Jantschko W, Helm J, Bogner M, Jakopitsch CCO. Active site structure and catalytic mechanisms of human peroxidases. Arch Biochem Biophys. 2006;445:199–213. doi: 10.1016/j.abb.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Gao S, Zhang R, Greenberg ME, Sun M, Chen X, Levison BS, Salomon RG, Hazen SL. Phospholipid hydroxyalkenals, a subset of recently discovered endogenous CD36 ligands, spontaneously generate novel furan-containing phospholipids lacking CD36 binding activity in vivo. J Biol Chem. 2006;281:31298–31308. doi: 10.1074/jbc.M604039200. [DOI] [PubMed] [Google Scholar]
- Garcia Fernandez MI, Ceccarelli D, Muscatello U. Use of the fluorescent dye 10-N-nonyl acridine orange in quantitative and location assays of cardiolipin: a study on different experimental models. Anal Biochem. 2004;328:174–180. doi: 10.1016/j.ab.2004.01.020. [DOI] [PubMed] [Google Scholar]
- Gasull T, Sarri E, DeGregorio-Rocasolano N, Trullas R. NMDA receptor overactivation inhibits phospholipid synthesis by decreasing choline-ethanolamine phosphotransferase activity. J Neurosci. 2003;23:4100–4107. doi: 10.1523/JNEUROSCI.23-10-04100.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebicka L. Peroxidase-like activity of cytochrome c in the presence of anionic surfactants. Res Chem Intermed. 2001;27:717–723. [Google Scholar]
- Gil J, Almeida S, Oliveira CR, Rego AC. Cytosolic and mitochondrial ROS in staurosporine-induced retinal cell apoptosis. Free Radic Biol Med. 2003;35:1500–1514. doi: 10.1016/j.freeradbiomed.2003.08.022. [DOI] [PubMed] [Google Scholar]
- Greenberg ME, Sun M, Zhang R, Febbraio M, Silverstein R, Hazen SL. Oxidized phosphatidylserine-CD36 interactions play an essential role in macrophage-dependent phagocytosis of apoptotic cells. J Exp Med. 2006;203:2613–2625. doi: 10.1084/jem.20060370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, Yang J, Cheng H, Gross RW. Shotgun lipidomics of cardiolipin molecular species in lipid extracts of biological samples. J Lipid Res. 2006;47:864–879. doi: 10.1194/jlr.D500044-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauff KD, Hatch GM. Cardiolipin metabolism and Barth Syndrome. Prog Lipid Res. 2006;45:91–101. doi: 10.1016/j.plipres.2005.12.001. [DOI] [PubMed] [Google Scholar]
- Heinecke JW. The role of myeloperoxidase in HDL oxidation and atherogenesis. Curr Atheroscler Rep. 2007;9:249–251. doi: 10.1007/s11883-007-0027-2. [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J, Rhoades ER, Russell DG, Shi YX, Groisman EA. Structural characterization of cardiolipin by tandem quadrupole and multiple-stage quadrupole ion-trap mass spectrometry with electrospray ionization. J Am Soc Mass Spectr. 2004;16:491–504. doi: 10.1016/j.jasms.2004.12.015. [DOI] [PubMed] [Google Scholar]
- Jiang J, Huang Z, Zhao Q, Feng W, Belikova NA, Kagan VE. Interplay between bax, reactive oxygen species production, and cardiolipin oxidation during apoptosis. Biochem Biophys Res Commun. 2008;368:145–150. doi: 10.1016/j.bbrc.2008.01.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang J, Serinkan BF, Tyurina YY, Borisenko GG, Mi Z, Robbins PD, Schroit AJ, Kagan VE. Peroxidation and externalization of phosphatidylserine associated with release of cytochrome c from mitochondria. Free Radic Biol Med. 2003;35:814–825. doi: 10.1016/s0891-5849(03)00429-5. [DOI] [PubMed] [Google Scholar]
- Kaewsuya P, Danielson ND, Ekhterae D. Fluorescent determination of cardiolipin using 10-N-nonyl acridine orange. Anal Bioanal Chem. 2007;387:2775–2782. doi: 10.1007/s00216-007-1135-0. [DOI] [PubMed] [Google Scholar]
- Kagan VE. Lipid peroxidation in biomembranes. CRC Press; 1988. [Google Scholar]
- Kagan VE, Borisenko GG, Tyurina YY, Tyurin VA, Jiang J, Potapovich AI, Kini V, Amoscato AA, Fujii Y. Oxidative lipidomics of apoptosis: redox catalytic interactions of cytochrome c with cardiolipin and phosphatidylserine. Free Radic Biol Med. 2004;37:1963–1985. doi: 10.1016/j.freeradbiomed.2004.08.016. [DOI] [PubMed] [Google Scholar]
- Kagan VE, Shvedova AA, Novikov KN. [Participation of phospholipases in the “repair” of photoreceptor membranes subjected to peroxidation] Biofizika. 1978;23:279–284. [PubMed] [Google Scholar]
- Kagan VE, Tyurin VA, Jiang J, et al. Cytochrome c acts as a cardiolipin oxygenase required for release of proapoptotic factors. Nat Chem Biol. 2005;1:223–232. doi: 10.1038/nchembio727. [DOI] [PubMed] [Google Scholar]
- Kapralov AA, Kurnikov IV, Vlasova II, et al. The hierarchy of structural transitions induced in cytochrome c by anionic phospholipids determines its peroxidase activation and selective peroxidation during apoptosis in cells. Biochemistry. 2007;46:14232–14244. doi: 10.1021/bi701237b. [DOI] [PubMed] [Google Scholar]
- Katayama M, Mizuta I, Sakoyama Y, Kohyama-Koganeya A, Akagawa K, Uyemura K, Ishii K. Differential expression of neuroD in primary cultures of cerebral cortical neurons. Exp Cell Res. 1997;236:412–417. doi: 10.1006/excr.1997.3757. [DOI] [PubMed] [Google Scholar]
- Kerwin JL, Tuininga AR, Ericssont LH. Identification of molecular species of glycerophosp holipids and sphingomyelin using electrospray mass spectrometry. Lipid Res. 1994;35:1102–1114. [PubMed] [Google Scholar]
- Kiebish MA, Han X, Cheng H, Lunceford A, Clarke CF, Moon H, Chuang JH, Seyfried TN. Lipidomic analysis and electron transport chain activities in C57BL/6J mouse brain mitochondria. J Neurochem. 2008 doi: 10.1111/j.1471-4159.2008.05383.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HY. Novel metabolism of docosahexaenoic acid in neural cells. J Biol Chem. 2007;282:18661–18665. doi: 10.1074/jbc.R700015200. [DOI] [PubMed] [Google Scholar]
- Kirkland RA, Adibhatla RM, Hatcher JF, Franklin JL. Loss of cardiolipin and mitochondria during programmed neuronal death: evidence of a role for lipid peroxidation and autophagy. Neuroscience. 2002;115:587–602. doi: 10.1016/s0306-4522(02)00512-2. [DOI] [PubMed] [Google Scholar]
- Kitano S, Yoshida Y, Kawano K, Hibi N, Niki E. Oxidative status of human low density lipoprotein isolated by anion-exchange high-performance liquid chromatography--assessment by total hydroxyoctadecadienoic acid, 7-hydroxycholesterol, and 8-iso-prostaglandin F(2alpha) Anal Chim Acta. 2007;585:86–93. doi: 10.1016/j.aca.2006.12.032. [DOI] [PubMed] [Google Scholar]
- Koivusalo M, Haimi P, Heikinheimo L, Kostiainen R, Somerharju P. Quantitative determination of phospholipid compositions by ESI-MS: effects of acyl chain length, unsaturation, and lipid concentration on instrument response. Lipid Res. 2001;42:663–672. [PubMed] [Google Scholar]
- Lambert IH, Pedersen SF, Poulsen KA. Activation of PLA2 isoforms by cell swelling and ischaemia/hypoxia. Acta Physiol (Oxf) 2006;187:75–85. doi: 10.1111/j.1748-1716.2006.01557.x. [DOI] [PubMed] [Google Scholar]
- Lemmon MA. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol. 2008;9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- Lucken-Ardjomande S, Montessuit S, Martinou JC. Contributions to Bax insertion and oligomerization of lipids of the mitochondrial outer membrane. Cell Death Differ. 2008 doi: 10.1038/cdd.2008.9. [DOI] [PubMed] [Google Scholar]
- MacMillan DK, Murphy RC. Analysis of lipid hydroperoxides and long-chain conjugated keto acids by negative ion electrospray mass spectrometry. J Am Soc Mass Spectrom. 1995;6:1190–1201. doi: 10.1016/1044-0305(95)00505-6. [DOI] [PubMed] [Google Scholar]
- Mallet CR, Lu Z, Mazzeo JR. A study of ion suppression effects in electrospray ionization from mobile phase additives and solid-phase extracts. Rapid Commun Mass Spectrom. 2004;18:49–58. doi: 10.1002/rcm.1276. [DOI] [PubMed] [Google Scholar]
- Manevich Y, Reddy KS, Shuvaeva T, Feinstein SI, Fisher AB. Structure and phospholipase function of peroxiredoxin 6: identification of the catalytic triad and its role in phospholipid substrate binding. J Lipid Res. 2007;48:2306–2318. doi: 10.1194/jlr.M700299-JLR200. [DOI] [PubMed] [Google Scholar]
- Marchese A, McHugh C, Kehler J, Bi H. Determination of Pranlukast and its metabolites in human plasma by LC/MS/MS with PROSPEKT on-line solid-phase extraction. J Mass Spectrom. 1998;33:1071–1079. doi: 10.1002/(SICI)1096-9888(1998110)33:11<1071::AID-JMS719>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Maskrey BH, Bermudez-Fajardo A, Morgan AH, et al. Activated platelets and monocytes generate four hydroxyphosphatidylethanolamines via lipoxygenase. J Biol Chem. 2007;282:20151–20163. doi: 10.1074/jbc.M611776200. [DOI] [PubMed] [Google Scholar]
- McGinley CM, van der Donk WA. Enzymatic hydrogen atom abstraction from polyunsaturated fatty acids. Chem Commun (Camb) 2003:2843–2846. doi: 10.1039/b311008g. [DOI] [PubMed] [Google Scholar]
- Murphy EJ, Stephens R, Jurkwitz-Alexander M, Horrocks LA. Acidic hydrolysis of plasmalogens followed by high-performance liquid chromatography. Lipids. 1993;28:565–568. doi: 10.1007/BF02536090. [DOI] [PubMed] [Google Scholar]
- Musatov A. Contribution of peroxidized cardiolipin to inactivation of bovine heart cytochrome c oxidase. Free Radic Biol Med. 2006;41:238–246. doi: 10.1016/j.freeradbiomed.2006.03.018. [DOI] [PubMed] [Google Scholar]
- Nigam S, Schewe T. Phospholipase A(2)s and lipid peroxidation. Biochim Biophys Acta. 2000;1488:167–181. doi: 10.1016/s1388-1981(00)00119-0. [DOI] [PubMed] [Google Scholar]
- Ott M, Zhivotovsky B, Orrenius S. Role of cardiolipin in cytochrome c release from mitochondria. Cell Death Differ. 2007;14:1243–1247. doi: 10.1038/sj.cdd.4402135. [DOI] [PubMed] [Google Scholar]
- Phillis JW, Horrocks LA, Farooqui AA. Cyclooxygenases, lipoxygenases, and epoxygenases in CNS: their role and involvement in neurological disorders. Brain Res Rev. 2006;52:201–243. doi: 10.1016/j.brainresrev.2006.02.002. [DOI] [PubMed] [Google Scholar]
- Pong K, Doctrow SR, Huffman K, Adinolfi CA, Baudry M. Attenuation of staurosporine-induced apoptosis, oxidative stress, and mitochondrial dysfunction by synthetic superoxide dismutase and catalase mimetics, in cultured cortical neurons. Exp Neurol. 2001;171:84–97. doi: 10.1006/exnr.2001.7747. [DOI] [PubMed] [Google Scholar]
- Pulfer M, Murphy RC. Electrospray mass spectrometry of phospholipids. Mass Spectrom Rev. 2003;22:332–364. doi: 10.1002/mas.10061. [DOI] [PubMed] [Google Scholar]
- Radi R, Thomson L, Rubbo H, Prodanov E. Cytochrome c-catalyzed oxidation of organic molecules by hydrogen peroxide. Arch Biochem Biophys. 1991;288:112–117. doi: 10.1016/0003-9861(91)90171-e. [DOI] [PubMed] [Google Scholar]
- Rashba-Step J, Tatoyan A, Duncan R, Ann D, Pushpa-Rehka TR, Sevanian A. Phospholipid peroxidation induces cytosolic phospholipase A2 activity: membrane effects versus enzyme phosphorylation. Arch Biochem Biophys. 1997;343:44–54. doi: 10.1006/abbi.1997.0134. [DOI] [PubMed] [Google Scholar]
- Rouser G, Fkeischer S, Yamamoto A. Two dimensional then layer chromatographic separation of polar lipids and determination of phospholipids by phosphorus analysis of spots. Lipids. 1970;5:494–496. doi: 10.1007/BF02531316. [DOI] [PubMed] [Google Scholar]
- Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Schlame M, Ren M, Xu Y, Greenberg ML, Haller I. Molecular symmetry in mitochondrial cardiolipins. Chem Phys Lipids. 2005;138:38–49. doi: 10.1016/j.chemphyslip.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Schneider C, Pratt DA, Porter NA, Brash AR. Control of oxygenation in lipoxygenase and cyclooxygenase catalysis. Chem Biol. 2007;14:473–488. doi: 10.1016/j.chembiol.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab JM, Chiang N, Arita M, Serhan CN. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447:869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz JC, Syka JEP, Quarmby ST. Improving the fundamentals of MSn on 2D linear uon traps: new ion activation and isolation techniques. Proccedings Amer. Soc. MS; San Antonio, Texas. 2005. [Google Scholar]
- Serhan CN, Chiang N, Van Dyke TE. Resolving inflammation: dual anti-inflammatory and pro-resolution lipid mediators. Nat Rev Immunol. 2008;8:349–361. doi: 10.1038/nri2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sevanian A, Wratten ML, McLeod LL, Kim E. Lipid peroxidation and phospholipase A2 activity in liposomes composed of unsaturated phospholipids: a structural basis for enzyme activation. Biochim Biophys Acta. 1988;961:316–327. doi: 10.1016/0005-2760(88)90079-3. [DOI] [PubMed] [Google Scholar]
- Spickett CM, Dever G. Studies of phospholipid oxidation by electrospray mass spectrometry: from analysis in cells to biological effects. Biofactors. 2005;24:17–31. doi: 10.1002/biof.5520240103. [DOI] [PubMed] [Google Scholar]
- Spiteller G. Peroxyl radicals: inductors of neurodegenerative and other inflammatory diseases. Their origin and how they transform cholesterol, phospholipids, plasmalogens, polyunsaturated fatty acids, sugars, and proteins into deleterious products. Free Radic Biol Med. 2006;41:362–387. doi: 10.1016/j.freeradbiomed.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Taguchi R, Hayakawa J, Takeuchi Y, Ishida M. Two-dimensional analysis of phospholipids by capillary liquid chromatography/electrospray ionization mass spectrometry. J Mass Spectrom. 2000;35:953–966. doi: 10.1002/1096-9888(200008)35:8<953::AID-JMS23>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Tanaka R, Hatate H, Ito M, Nakamura T. Elevation of lipid peroxide level and production of hydroxy lipids in cultures Hepa-T1 cells by oxidative stress. Fish Sci. 2006;72:665–672. [Google Scholar]
- Tyurina YY, Kawai K, Tyurin VA, Liu SX, Kagan VE, Fabisiak JP. The plasma membrane is the site of selective phosphatidylserine oxidation during apoptosis: role of cytochrome c. Antioxid Redox Signal. 2004a;6:209–225. doi: 10.1089/152308604322899288. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Kini V, Tyurin VA, et al. Mechanisms of cardiolipin oxidation by cytochrome c: relevance to pro- and antiapoptotic functions of etoposide. Mol Pharmacol. 2006;70:706–717. doi: 10.1124/mol.106.022731. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Serinkan FB, Tyurin VA, Kini V, Yalowich JC, Schroit AJ, Fadeel B, Kagan VE. Lipid antioxidant, etoposide, inhibits phosphatidylserine externalization and macrophage clearance of apoptotic cells by preventing phosphatidylserine oxidation. J Biol Chem. 2004b;279:6056–6064. doi: 10.1074/jbc.M309929200. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Tyurin VA, Epperly MW, Greenberger JS, Kagan VE. Oxidative lipidomics of gamma-irradiation-induced intestinal injury. Free Radic Biol Med. 2008;44:299–314. doi: 10.1016/j.freeradbiomed.2007.08.021. [DOI] [PubMed] [Google Scholar]
- Tyurina YY, Tyurin VA, Zhao Q, Djukic M, Quinn PJ, Pitt BR, Kagan VE. Oxidation of phosphatidylserine: a mechanism for plasma membrane phospholipid scrambling during apoptosis? Biochem Biophys Res Commun. 2004c;324:1059–1064. doi: 10.1016/j.bbrc.2004.09.102. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlasova II, Tyurin VA, Kapralov AA, Kurnikov IV, Osipov AN, Potapovich MV, Stoyanovsky DA, Kagan VE. Nitric oxide inhibits peroxidase activity of cytochrome c. cardiolipin complex and blocks cardiolipin oxidation. J Biol Chem. 2006;281:14554–14562. doi: 10.1074/jbc.M509507200. [DOI] [PubMed] [Google Scholar]
- Walton KA, Cole AL, Yeh M, et al. Specific phospholipid oxidation products inhibit ligand activation of toll-like receptors 4 and 2. Arterioscler Thromb Vasc Biol. 2003;23:1197–1203. doi: 10.1161/01.ATV.0000079340.80744.B8. [DOI] [PubMed] [Google Scholar]
- Watson AD. Thematic review series: systems biology approaches to metabolic and cardiovascular disorders. Lipidomics: a global approach to lipid analysis in biological systems. J Lipid Res. 2006;47:2101–2111. doi: 10.1194/jlr.R600022-JLR200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. J Biol Chem. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Hayakawa M, Niki E. Total hydroxyoctadecadienoic acid as a marker for lipid peroxidation in vivo. Biofactors. 2005;24:7–15. doi: 10.1002/biof.5520240102. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Itoh N, Hayakawa M, Habuchi Y, Inoue R, Chen ZH, Cao J, Cynshi O, Niki E. Lipid peroxidation in mice fed a choline-deficient diet as evaluated by total hydroxyoctadecadienoic acid. Nutrition. 2006;22:303–311. doi: 10.1016/j.nut.2005.07.020. [DOI] [PubMed] [Google Scholar]