Abstract
The rodent barrel cortex is a useful system to study the role of genes and neuronal activity in the patterning of the nervous system. Several genes encoding either intracellular signaling molecules or neurotransmitter receptors are required for barrel formation. Neurofibromin is a tumor suppressor protein that has Ras GTPase activity, thus attenuating the MAPK (mitogen-activated protein kinase) and and PI-3 kinase (phosphatidylinositol 3-kinase) pathways, and is mutated in humans with the condition neurofibromatosis type 1 (NF1). Neurofibromin is widely expressed in the developing and adult nervous system, and a common feature of NF1 is deficits in intellectual development. In addition, NF1 is an uncommonly high disorder among individuals with autism. Thus, NF1 may have important roles in normal CNS development and function. To explore roles for neurofibromin in the development of the CNS, we took advantage of a mouse conditional allele. We show that mice that lack neurofibromin in the majority of cortical neurons and astrocytes fail to form cortical barrels in the somatosensory cortex, whereas segregation of thalamic axons within the somatosensory cortex appears unaffected.
Keywords: NF1, Ras-GAP, hGFAP-Cre, thalamocortical axons, development, conditional mutant
Introduction
In the rodent CNS, the cortex is innervated by dorsal thalamic axons that segregate the cortex into different sensory regions such as visual, auditory, and somatosensory. Axons from the ventral posterior medial nucleus (VPM) of the thalamus innervate the somatosensory cortex where they segregate into discrete regions (Miller et al., 2001). Neurons within cortical layer IV form rings termed cortical barrels by arranging their cell bodies around and dendrites toward the incoming thalamic axons (Woolsey and Van der Loos, 1970; Woolsey et al., 1975).
Many studies have contributed to elucidation of factors required for barrel formation. Transplantation of visual cortex tissue in place of somatosensory cortex permitted barrel-like formation, suggesting that VPM thalamic axons specify barrel formation independent of the target (Schlaggar and O'Leary, 1991). Mouse knock-out studies have shown that barrel formation requires both ionotropic (NMDA) and metabotropic glutamate receptors, as well as various intracellular signaling molecules such as adenylyl cyclase type I, phospholipase C-β1 (PLC-β1), protein kinase A (PKA), and synaptic Ras GTPase activating protein (SynGAP) (Erzurumlu and Kind, 2001; Barnett et al., 2006; Inan et al., 2006; Watson et al., 2006).
The neurofibromatosis type 1 (NF1) gene encodes a large protein that contains a Ras GTPase-activating (GAP) domain that negatively regulates Ras activity. Inactivating mutations in NF1 cause neurofibromatosis type 1 (Cawthon et al., 1990; Viskochil et al., 1990; Wallace et al., 1990), a common genetic disease that affects 1 in 3000 individuals (Cichowski and Jacks, 2001; Zhu and Parada, 2001). Aside from the more common tumor phenotypes, a significant number of individuals with NF1 display intellectual deficits (Costa and Silva, 2003). In addition, it has been estimated that as many as 1% of children with autism spectrum disorders are subsequently diagnosed with NF1 (Mbarek et al., 1999; Marui et al., 2004). Mouse models of neurofibromatosis have been generated, and complete loss of NF1 is lethal in mice because of defects in cardiac development (Brannan et al., 1994; Jacks et al., 1994). In the peripheral nervous system, loss of NF1 allows sensory neurons to survive in the absence of neurotrophin signaling (Vogel et al., 1995) because of increased Ras and phosphatidylinositol 3-kinase (PI-3 kinase) activity (Klesse and Parada, 1998).
Although a few NF1 patients show abnormalities in brain structure (Korf et al., 1999; Balestri et al., 2003), little is known about the basis for NF1-associated intellectual deficits and possible roles in development of the CNS. To circumvent the early embryonic lethality of NF1 mutations, we have used cre/lox technology to generate a conditional allele of NF1 (Zhu et al., 2001). Mice lacking NF1 in the majority of neurons displayed a thin cortex, while maintaining a normal number of neurons and normal dendritic arborization (Zhu et al., 2001). In the current study, we characterize a mouse line that has a deletion of NF1 in the majority of cortical neurons and astrocytes. We find that these mice lack cortical barrels from early in development. Thalamic axons retain NF1 and segregate within cortical layer IV, although their appearance is not completely normal.
Materials and Methods
Animals.
The generation and genotyping of NF1flox/flox animals have been described previously (Zhu et al., 2001). Mice expressing Cre under the control of the human GFAP promoter hGFAP-Cre were a gift from Dr. A. Messing (University of Wisconsin, Madison, WI) (Zhuo et al., 2001) and were used to generate NF1flox/flox;hGFAP-Cre mice (Zhu et al., 2005b). No difference was seen in NF1flox/flox, NF1flox/+, and NF1flox/+;hGFAP-Cre, so they are grouped together as controls. Rosa26R (R26) and Z/EG mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All mice are on a mixed 129 and C57 background. All animal procedures conformed to National Institutes of Health and University of Texas Southwestern Medical Center Institutional guidelines for the care and use of animals.
5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside staining and Nissl staining.
Adult mice were anesthetized and perfused transcardially with PBS followed by 4% paraformaldehyde (PFA). The dissected brains and skulls were postfixed in PFA at 4°C for 2–24 h. For the analysis of trigeminal ganglia neurons, skulls from postnatal day 0 (P0) mice were dissected and postfixed in PFA, followed by decalcification in CalRite (Richard-Allan Scientific, Kalamazoo, MI). We performed 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-gal) staining on floating brain sections from adult R26;hGFAP-Cre and Z/EG;hGFAP-Cre mice as described previously (Kwon et al., 2006). Whole-mount X-gal staining was performed on the skulls, containing trigeminal ganglion, of adult R26;hGFAP-Cre mice. For Nissl staining, paraffin sections from P0 or adult mice were prepared as described previously (Kwon et al., 2006) and stained with 0.1% (w/v) toluidine blue (Poly Scientific, Bay Shore, NY) or cresyl violet for 5 min.
Cytochrome oxidase staining.
Cytochrome oxidase staining was performed as described previously (Wong-Riley and Welt, 1980). The brains were removed and left intact or the cortex was flattened between glass slides for tangential sections and postfixed for 4 h in PFA at 4°C. Brains were sectioned at 40 μm with a vibratome or, alternatively, cryoprotected for 2 d at 4°C in 30% sucrose/PBS, imbedded in O.C.T. (optimal cutting temperature) compound (Tissue-Tek; Sakura Finetek, Torrance, CA), and sectioned at 40 μm with a cryostat (Leica, Wetzlar, Germany). Sections were washed in PBS, placed in staining solution (0.55 mg/ml DAB, 0.3 mg/ml cytochrome C, and 45 mg/ml sucrose; Sigma, St. Louis, MO), and incubated in the dark at 37°C until the staining gave strong signal (2–4 h). Sections were washed in PBS, mounted onto slides, dehydrated through ethanol and xylene, and coverslipped with p-xylene-bis-pyridinium bromide (DPX) (Sigma).
4,6-Diamidino-2-phenylindole staining.
Intact or flattened cortices were sectioned at 40 μm and mounted onto slides to dry. Slides were washed in PBS-T (PBS/0.3% Triton X-100) and stained for 30 min in PBS-T plus 4′,6-diamidino-2-phenylindole (DAPI; 1 μg/ml; Sigma) to visualize cell nuclei. Slides were washed briefly in PBS and coverslipped with Immuno-mount (Thermo Shandon, Pittsburgh, PA) mounting medium. Images were taken with a Nikon (Tokyo, Japan) CCD camera using the MetaView program (Universal Imaging, West Chester, PA).
Immunohistochemistry.
P4 or P8 pups were anesthetized and perfused with PBS followed by cold 4% PFA, and the brains were removed and postfixed overnight in PFA. Brains were sectioned at 50 μm with a Vibratome (Leica). Serotonin immunohistochemistry was performed as follows. Sections were washed in PBS-T, blocked for 1 h in PBS-T/6% goat serum (Sigma), and incubated overnight at room temperature in a blocking buffer with the addition of rabbit anti-serotonin antibody (1:10,000; Immunostar, Hudson, WI). Sections were washed in PBS and incubated with a biotinalyated secondary antibody (goat anti-rabbit, 1:400; Vector Laboratories, Burlingame, CA) for 45 min. Antibody staining was visualized using the ABC kit (Vector Laboratories) with DAB. Sections were mounted onto slides, dried, dehydrated through ethanol and xylene, and coverslipped with DPX. Fluorescent immunostaining for serotonin was performed as above, except the primary antibody was used at a dilution of 1:5000. Antibody staining was visualized with Cy3-conjugated goat anti-rabbit antibody (1:400; Jackson ImmunoResearch, West Grove, PA). To visualize cell nuclei, sections were counterstained with DAPI (1 μg/ml). Sections were mounted onto slides and coverslipped with Immuno-mount mounting medium.
Measuring barrel formation.
To quantify the degree of barrel formation, a scoring system was used such that a score of 0 equaled no barrel segregation and a score of 3 equaled normal barrel segregation. Four consecutive DAPI-stained tangential sections were scored from each of four controls and three mutant animals from P30 to P35. An individual blind to the genotype determined scores. Statistical analysis was performed with a Student's t test.
Area measurements.
All analyses were performed blind to genotype. Somatosensory (S1) and posterior medial barrel subfield (PMBSF) areas were measured on tangential sections stained with cytochrome oxidase or anti-serotonin antibody as described above. Digital images taken with a Nikon CCD camera were analyzed using the MetaView program (Universal Imaging). The size of individual patches and the width of septa were determined from B1–B3, C1–C3, and D1–D3 barrels.
Cell count.
To quantify the number of neurons in the trigeminal ganglia, 5 μm continuous sagittal sections were collected from P0 mice and stained with cresyl violet. The images of every 10th section were used to count the number of neurons manually. The neuron number of the whole trigeminal ganglion was calculated by multiplying the sum of counts from individual sections by 10. To determine the density and wall/hollow distribution of nuclei within the barrel cortex, DAPI-stained tangential sections were viewed with a Leica confocal microscope. All sections were costained with anti-5-HT antibody to confirm the location within PMBSF. Three adjacent sections containing the barrel field were used for each animal. Seven micrometer optical images of B2, B3, C2, C3, D2, and D3 barrels were obtained, and each centered in a rectangle encompassing only the individual barrel. Cell counts were normalized to the area of the rectangle and presented as mean ± SEM. To determine the overall cell number in the barrel cortex, DAPI-stained tangential sections from P30–P35 animals were used. Five 50 μm sections were counted throughout the barrel cortex from six control and three mutant brains. Images were imported into NIH ImageJ software, and a particle count macro was used to determine the number of DAPI-positive puncta per section. Statistical analysis was performed with a Student's t test.
Immunoblotting.
Control and mutant littermates were collected at P0 and P4 (n = 3 for each genotype and time point). The entire somatosensory cortex was dissected out and immediately frozen in liquid nitrogen. Tissues were homogenized in PLC buffer [50 mm HEPES, pH 7.5, 150 mm NaCl, 10% glycerol, 0.1% Triton X-100, 1.5 mm MgCL2, and Complete proteinase inhibitor (Roche, Welwyn Garden City, UK)]. The extract was centrifuged, and protein concentration of the supernatant was quantified using the BCA method following the manufacturer's protocol (Pierce, Rockford, IL). Extracts (10 μg) were separated on SDS-PAGE gels and transferred to nitrocellulose. The following antibodies and concentrations were used: phospho-Erk (1:1000; Cell Signaling Technology, Danvers, MA), Erk (1:10,000; Santa Cruz Biotechnology, Santa Cruz, CA), phospho-Akt (1:1000; Cell Signaling Technology), Akt (1:1000; Cell Signaling Technology), phospho-PKA-RIIβ (1:1000; BD Transduction Laboratories, Lexington, KY), PKA-RIIβ (1:10,000; BD Transduction Laboratories), NMDA-R1 (1:500; Upstate Biotechnology, Lake Placid, NY), NF1GRD (1:500; Santa Cruz Biotechnology), SynGAP (1:2000; Affinity BioReagents, Golden, CO), PLC-β1 (1:4000; Santa Cruz Biotechnology), and actin (1:1000; Chemicon, Temecula, CA). Membranes were incubated in primary antibodies overnight at 4°C in blocking buffer (5% nonfat milk and 0.1% Tween 20 in TBS (TBS-T), or 5% BSA for phospho-antibodies). After washing in TBS-T, blots were incubated with appropriate secondary antibodies (1:10,000; Santa Cruz Biotechnology) for 1 h in TBS-T. Membranes were washed, developed using ChemiGlow West reagent (Alpha Innotech, San Leandro, CA), and exposed to film or imaged using Kodak Image Station 2000r (Eastman Kodak, Rochester, NY). As a loading control, blots were stripped and reprobed either with antibodies recognizing nonphosphorylated forms of the protein for the phosphor-specific antibodies and for total protein levels using actin antibody or Sypro Ruby (Invitrogen, Eugene, OR). Quantification of bands was performed essentially as described previously (Luikart et al., 2005). Briefly, the net band intensity for both the Western blots and the Sypro Ruby staining was determined using Kodak 1D Image Analysis software. Values for the phosphorylated blots were normalized to the levels of unphosphorylated controls. Values for the other P4 data were normalized to the Sypro Ruby staining, and values for P0 data were normalized to actin. Data from control animals was set to 100%. Student's t test was used to determine statistical differences.
Results
To explore the role of NF1 in cortical development, we used a mouse line containing a conditional allele of NF1 (Zhu et al., 2001) crossed to a transgenic line expressing Cre under control of the human GFAP promoter (Zhu et al., 2001; Luikart et al., 2005). In this line, Cre is expressed in cortical progenitors as early as embryonic day 12.5 (Zhuo et al., 2001; Malatesta et al., 2003; Zhu et al., 2005a) with continued expression in glia and neural progenitor cells in older animals. This early expression in cortical progenitors results in NF1 deletion in both neurons and glia (Zhu et al., 2005a). hGFAP-Cre mice were crossed to the Rosa26-LacZ (R26) reporter line (Soriano, 1999), and X-gal staining of adult brains revealed expression in the cortex but little staining in the thalamus (Fig. 1 a–c). This observation was further confirmed when hGFAP-Cre mice were crossed to the Z/EG reporter line (Novak et al., 2000), demonstrating that the majority of cells in the cortex had undergone recombination, whereas Cre expression in the thalamus was minimal (Fig. 1 d–f). Cortical neurons that remained unrecombined are thought to be interneurons (Malatesta et al., 2003). Additionally, Cre is also expressed in the spinal cord, cerebellum, and trigeminal ganglion (supplemental Fig. 1, available at www.jneurosci.org as supplemental material, and data not shown). NF1flox/flox;hGFAP-Cre mice are born in normal numbers but become noticeably smaller than their littermates as they age (Fig. 1 j), and most die by 4 months of age (Zhu et al., 2005b). Young adult NF1 mutant brains appear grossly normal (Fig. 1 g,h).
Figure 1.
Cre-mediated recombination is restricted to the cortex in hGFAP-Cre mice. a–f, X-gal staining of adult R26;hGFAP-Cre and Z/EG;hGFAP-Cre mice reveals recombination in the barrel cortex (a, b, d, e) with little expression in the thalamus (th) (c, f). g, h, Nissl staining of adult control (g) and NF1 conditional mutant (h) brains reveals no obvious differences in brain structure. i, Total brain weight is normal from P4 to P35 in neurofibromin mutant animals. j, Total body weight is normal until P35 in neurofibromin mutant animals. Scale bars, 100 μm. Error bars indicate SD.
To visualize the somatosensory cortex, we first performed cytochrome oxidase staining of brain sections in young adult mice (P35). Segregation of cytochrome oxidase staining in cortical layer IV throughout the barrel cortex is seen in coronal and tangential sections in both control (n = 13) and mutant (n = 11) mice (Fig. 2 a–d). No differences were seen in the overall size of S1 (controls, 4.725 ± 0.201 mm2; mutants, 4.673 ± 0.298 mm2) or PMBSF (controls, 1.053 ± 0.042 mm2; mutants, 1.011 ± 0.087 mm2) areas. However, the morphology of the individual patches was slightly altered, reflected in a reduction in size (controls, 22341.51 ± 732.22 μm2; mutants, 19803.49 ± 892.71 μm2; p < 0.05) and an increase in the width of septa between patches (controls, 24.537 ± 0.791 μm; mutants, 32.732 ± 1.417 μm; p < 0.01).
Figure 2.

Adult neurofibromin conditional mutant mice lack cortical barrels. a–d, Cytochrome oxidase staining of adult cortex in coronal (a, b) and tangential (c, d) orientation. Both control (a, c) and mutant (b, d) cortex show segregation of staining. e–j, DAPI staining of adult cortex in coronal (e, f) and tangential (g–j) orientation. Cortical barrels are seen in the control (e, arrows; g) but absent in conditional (f, h) knock-outs. Low-magnification images show no cortical segregation in any cortical region in mutants (j) compared with controls (i).
To directly examine local organization of cortical barrels, we labeled cell nuclei using DAPI staining. In either coronal or tangential sections, segregation of cortical cells into barrels is clearly seen in the control brain (Fig. 2 e, arrows, g,i) (n = 16). In the NF1 mutant mice, however, patterning of cortical cells into barrels is dramatically reduced or completely absent (n = 12) (Fig. 2 f,h,j; and supplemental Fig. 3a, available at www.jneurosci.org as supplemental material) despite normal lamination of the six cortical layers (Fig. 2 f). To determine the extent of cortical patterning deficit, we examined tangential sections throughout the entire cortex. We observed that loss of cortical NF1 resulted in reduced cellular aggregation throughout the entire somatosensory area (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). Scoring for degree of barrel formation revealed a significant decrease in mutants compared with controls (supplemental Fig. 3a, available at www.jneurosci.org as supplemental material). These data indicate that loss of NF1 in cortical cells has little effect on wild-type thalamic axons but impairs the local cellular organization of cells in the cortex. No defects were observed in NF1flox/+;hGFAP-Cre animals, so these were grouped together with controls (data not shown). The hGFAP-Cre transgene is expressed in trigeminal ganglion (supplemental Fig. 1a, available at www.jneurosci.org as supplemental material). To ascertain that unanticipated neuronal loss in trigeminal ganglion could contribute to the cortical barrel phenotype, we examined overall morphology and neuron number and found no difference in this ganglion in mutant animals (supplemental Fig. 1b–d, available at www.jneurosci.org as supplemental material). The defects in barrel formation are unlikely to be the result of general ill health of mutant mice because no difference in brain weight is seen in NF1mutant animals from P4 to P35, although total body weight lags behind as the mutant ages (Fig. 1 i). Consistent with this view, quantification of DAPI-positive puncta in tangential sections throughout the barrel cortex revealed no significant difference in numbers between controls and mutants (supplemental Fig. 3b, available at www.jneurosci.org as supplemental material). In addition, as described below, the deficit in mutant barrel cortex is evident at P8 when no differences in body weight are yet detectable.
To determine the timing of the defects in barrel cortex, we examined control and mutant littermates at P4, a time before cortical segregation has completed, and at P8 when cortical segregation is almost complete (Miller et al., 2001). Immunohistochemical staining for serotonin has been shown to label thalamic axons entering the barrel cortex in early postnatal mice (Cases et al., 1996). Thalamic axons, as visualized by serotonin immunohistochemistry, appear normal at P4 (Fig. 3 a,b) (n = 2), and clear segregation is seen by P8 in the mutant cortex (Fig. 3 c,d) (n = 8). In the P8 control brain, DAPI staining and immunohistochemistry for serotonin on tangential sections shows segregation of cortical barrels and thalamic axons, respectively (Fig. 3 e,g,i) (n = 8). In comparison, segregation of thalamic axons is also present in the mutant cortex, but cortical barrels are not observed (Fig. 3 f,h,j) (n = 8). Low-power micrographs of the serotonin immunostaining show segregation of multiple sensory regions (supplemental Fig. 4, available at www.jneurosci.org as supplemental material). Similar to observations from the adult mutant mice, the size of the S1 and PMBSF areas was unchanged, whereas the size of individual patches was reduced (significant differences between controls and mutants were found in the sizes of B1, B2, C1, and C3 patches). To quantify the change in segregation, DAPI-stained tangential sections across the PMBSF area were imaged as 7 μm optical slices. The density of DAPI-labeled nuclei in NF1 mutant mice was normal (controls, 55.68 ± 1.61; mutants, 55.94 ± 2.82), whereas the ring-like nuclei distribution was completely absent (wall-to-hollow ratios: controls, 1.81 ± 0.18; mutants, 1.03 ± 0.14; p < 0.01). Similar results were also seen at P10 with no barrel pattern of cortical cells seen in mutants (data not shown). Thus, cellular organization of cortical neurons requires NF1 function to respond appropriately to thalamocortical projection cues.
Figure 3.
Thalamic axon development in neurofibromin conditional mutant mice. a–d, Serotonin immunohistochemistry reveals both thalamic axon innervation and segregation into the cortex of mutant animals at P4 (b) and P8 (d) when compared with controls (a, c). e–j, Double-fluorescent staining for serotonin (red) and nuclei with DAPI (blue) of P8 tangential sections. There is clear thalamic axon segregation in the knock-out (h) similar to controls (g). DAPI staining shows cortical segregation into barrels in control animals (i) that is absent in NF1 mutant mice (j). k–n, High-magnification view of individual patches shows normal nuclei density in the NF1 mutant mice (l, n) compared with controls (k, m). Scale bars: k, l, 100 μm; m, n, 10 μm.
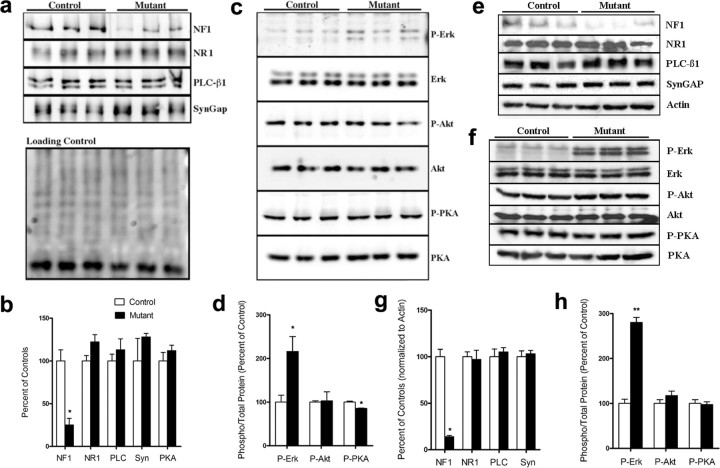
Several signaling molecules and neurotransmitters are required for barrel formation (Erzurumlu and Kind, 2001; Barnett et al., 2006). To explore the possible mechanism for the absence of cortical barrels in the NF1 mutants, we tested whether the expression of some of these proteins known to be required for barrel formation were affected by loss of NF1. Western blot analysis was performed on somatosensory cortex tissue extracts at P0 and P4, when cortical barrels are forming. No significant difference was found in the levels of NMDA receptor 1 (NR1), PLC-β1, SynGAP, or PKA-RIIβ at either age (Fig. 4 a–c,e,f) (n = 3 for each genotype and time point). Western blot analysis for NF1 shows an 80% reduction in the NF1 mutant animals (Fig. 4 a,b,e). The remaining NF1 expression likely comes from both interneurons and afferent axonal projections from non-Cre-expressing regions.
Figure 4.
Western blot analysis of somatosensory cortex shows normal levels of protein expression for other proteins required for cortical barrel formation. In the somatosensory cortex of P4 (a–d) and P0 (e–h) NF1 mutant mice, immunoblots show ∼80% reduction in NF1 (a, b, e, g). No significant difference was found in NR1, PLC-β1 (PLC), SynGAP (Syn), or the PKA-RIIβ (PKA) subunit at P4 (a, b) or P0 (e–g). Phospho-Erk levels are increased in mutant cortices at both ages (c, d, f, h). *p < 0.05; **p < 0.01. Error bars indicate SE. P, Phosphorylated.
Both the Ras–mitogen-activated protein kinase (MAPK) pathway and the PI-3 kinase–Akt pathway are known to be elevated in the absence of NF1 (Klesse and Parada, 1998). We examined the levels of phosphorylated Erk and Akt in NF1 mutant cortex at P0 and P4. Phospho-Erk showed a significant increase in levels, whereas phospho-Akt levels were unchanged (Fig. 4 c,d,f,h). We also examined the phosphorylation levels PKA-RIIβ subunit and found it to be unchanged (P0) (Fig. 4 f,h) or slightly decreased (P4) (Fig. 4 c,d) in the mutants, whereas the levels of total PKA-RIIβ were unaffected (Fig. 4 b,f).
Discussion
NF1 regulates cortical barrel formation
Since its initial discovery by Woolsey and Van der Loos (1970), the rodent barrel cortex has been intensely studied as a well defined system that accommodates the interplay of developmental, physiological, and behavioral neuroscience. The precise one-to-one correspondence of the large whiskers with layer IV cortical neuron segregations (“barrels”) provides an exquisite model for the studying of somatotopic mapping of sensory surfaces, as well as general mechanisms underlying the formation and functioning of the nervous system.
Development of the barrel cortex is regulated by an intricate network of genetic factors, many of which are only beginning to be unveiled (Erzurumlu and Kind, 2001; Petersen, 2007). In the present study, we demonstrate that the tumor suppressor neurofibromin 1 is also required for the proper formation of cortical barrels. Mice with a conditional deletion of NF1 in the cortex fail to form barrels postnatally (P8) and as adults. Thalamic neurons are devoid of Cre-mediated recombination, and their axonal segregation occurs with proper overall patterns. Mild morphological changes exist that are likely secondary effects of cortical NF1 ablation. This defect in cortical barrel segregation does not appear to be a result of gross brain development abnormality but rather a local consequence of NF1 deletion. Thus, the lack of cortical barrel formation can be attributed directly to NF1 loss in the cortex, supporting a cell-autonomous requirement of NF1 in this cellular compartment.
The bundling of thalamic axons into barrel-like patterns is thought to be instructive for cortical neurons to reposition their cell bodies and dendrites. This is best exemplified by transplantation studies demonstrating formation of barrels in visual cortex explants when they are placed in the somatosensory cortex and innervated by VPM thalamic axons (Schlaggar and O'Leary, 1991). Several lines of mouse genetic mutants have been generated that exhibit defects in the development of cortical barrels despite partial (Barnett et al., 2006; Hannan et al., 2001) or normal (Iwasato et al., 2000; Inan et al., 2006; Lu et al., 2006; Watson et al., 2006) thalamic axonal segregation. The phenotypes of the NF1 conditional knock-outs are similar to these mutants, further confirming the notion that signaling activities in the postsynaptic cortical neurons are critical in their rearrangement in response to thalamic cues.
Postsynaptic signaling and cortical mapping
Neurofibromin, the protein encoded by NF1, contains a Ras-GAP domain that serves as a checkpoint for Ras-mediated signaling events. Loss of NF1 leads to disinhibition of Ras and, consequently, increased activation of the MAPK and PI-3 kinase pathways when the specific cellular and environmental signaling capacity permits (Klesse and Parada, 1998). In the NF1 mutant cortex, we observed an upregulation of phospho-Erk, although the level of phospho-Akt remained unchanged. This finding, in conjunction with a recent report that germline mutant mice of SynGAP, another Ras-GAP-containing protein, also lack cortical barrels (Barnett et al., 2006), strongly implicates an essential role of Ras signaling in barrel formation. The physiological substrates of NF1 and SynGAP in the barrel cortex, as well as the mechanism by which Ras signaling may mediates cortical neuron repositioning remains to be elucidated.
Our findings that Erk is constitutively activated in the barrel cortex of NF1 mutant mice suggest the precise on/off regulation of the Ras–MAPK pathway by NF1 is required for cortical neurons to react to signal inputs from the thalamic axon terminals. Although the exact nature of these upstream signaling molecules remains to be identified, previous studies have associated NF1 with a number of proteins and pathways that are involved in the thalamocortical synaptic communication. Both NF1 and SynGAP have been shown to interact with the NMDA receptor complex (Chen et al., 1998; Kim et al., 1998; Husi et al., 2000). This coincides with evidence showing lack of cortical barrels in cortex-specific conditional NR1 mutant mice (Iwasato et al., 2000), suggesting possible NMDA-related deficits in the NF1 mutant mice that contribute to the absence of barrel formation. Alternatively, there has been substantial evidence linking NF1 to adenylyl cyclase and PKA, both of which have been reported to be required for proper barrel formation (Welker et al., 1996; Abdel-Majid et al., 1998; Watson et al., 2006). In Drosophila, loss of NF1 leads to defects in body size as well as learning and memory, which are attributable to a decrease in adenylyl cyclase and PKA activities, not Ras and MAPK signaling (Guo et al., 1997, 2000; The et al., 1997). In mice, it has been demonstrated that loss of NF1 can also lead to decreases in adenylyl cyclase activation in the embryonic brain (Tong et al., 2002), but as yet no in vivo defects have been attributed to the impairments of this pathway. Together, further delineation of the NF1 signaling mechanism in the postsynaptic compartment will provide vital insights into the development of barrel cortex and patterning in the CNS in general.
Clinical implication in NF1
Aside from the various forms of benign and malignant tumors, NF1 patients exhibit high incidence of learning disabilities and mental retardation (North, 2000). In mouse mutants, it has been shown that the loss of Ras-GAP-dependent regulation constitutes a molecular basis for spatial learning deficits (Costa et al., 2002). This coincides with human genetic studies showing a missense mutation that specifically abolishes the Ras-GAP function of NF1 leads to cognitive disabilities, among groups of other symptoms (Klose et al., 1998). However, little is known about the cellular and circuitry abnormalities responsible for the wide range of mental impairments observed in NF1 patients. In the present study, we identified a significant neuronal patterning deficit in mutant mice lacking both alleles of NF1 in the cortex. Although the behavioral ramifications of such deficit in mutant animals do not immediately reflect clinical symptoms in NF1 patients, it suggests loss of NF1 in human likely disturbs pattern formation in neocortical columns and the CNS in general. It is also likely that differences in the timing and cell types that undergo loss of the remaining wild-type allele contribute greatly to the variability in the behavioral and neuroanatomical phenotypes seen in individuals with NF1. Future studies using more specific spatial and temporal knock-outs of NF1 should continue to uncover new roles for NF1 in neuronal development. Thus, our findings provide novel insights into the etiology of mental impairments associated with neurofibromatosis.
Footnotes
This work was supported by National Institute of Neurological Disorders and Stroke Grants R37NS33199 and NS34296 and the American Cancer Society to L.F.P. We thank Yanjiao Li, Linda McClellan, Alemji Taku, and Alvin Chandra for technical assistance and all members of the Parada laboratory for helpful ideas and discussions.
References
- Abdel-Majid RM, Leong WL, Schalkwyk LC, Smallman DS, Wong ST, Storm DR, Fine A, Dobson MJ, Guernsey DL, Neumann PE. Loss of adenylyl cyclase I activity disrupts patterning of mouse somatosensory cortex. Nat Genet. 1998;19:289–291. doi: 10.1038/980. [DOI] [PubMed] [Google Scholar]
- Balestri P, Vivarelli R, Grosso S, Santori L, Farnetani MA, Galluzzi P, Vatti GP, Calabrese F, Morgese G. Malformations of cortical development in neurofibromatosis type 1. Neurology. 2003;61:1799–1801. doi: 10.1212/01.wnl.0000099080.90726.ba. [DOI] [PubMed] [Google Scholar]
- Barnett MW, Watson RF, Vitalis T, Porter K, Komiyama NH, Stoney PN, Gillingwater TH, Grant SG, Kind PC. Synaptic Ras GTPase activating protein regulates pattern formation in the trigeminal system of mice. J Neurosci. 2006;26:1355–1365. doi: 10.1523/JNEUROSCI.3164-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Cases O, Vitalis T, Seif I, De Maeyer E, Sotelo C, Gaspar P. Lack of barrels in the somatosensory cortex of monoamine oxidase A-deficient mice: role of a serotonin excess during the critical period. Neuron. 1996;16:297–307. doi: 10.1016/s0896-6273(00)80048-3. [DOI] [PubMed] [Google Scholar]
- Cawthon RM, Weiss R, Xu GF, Viskochil D, Culver M, Stevens J, Robertson M, Dunn D, Gesteland R, O'Connell P, White R. A major segment of the neurofibromatosis type 1 gene: cDNA sequence, genomic structure, and point mutations. Cell. 1990;62:193–201. doi: 10.1016/0092-8674(90)90253-b. [DOI] [PubMed] [Google Scholar]
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20:895–904. doi: 10.1016/s0896-6273(00)80471-7. [DOI] [PubMed] [Google Scholar]
- Cichowski K, Jacks T. NF1 tumor suppressor gene function: narrowing the GAP. Cell. 2001;104:593–604. doi: 10.1016/s0092-8674(01)00245-8. [DOI] [PubMed] [Google Scholar]
- Costa RM, Silva AJ. Mouse models of neurofibromatosis type I: bridging the GAP. Trends Mol Med. 2003;9:19–23. doi: 10.1016/s1471-4914(02)00008-4. [DOI] [PubMed] [Google Scholar]
- Costa RM, Federov NB, Kogan JH, Murphy GG, Stern J, Ohno M, Kucherlapati R, Jacks T, Silva AJ. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature. 2002;415:526–530. doi: 10.1038/nature711. [DOI] [PubMed] [Google Scholar]
- Erzurumlu RS, Kind PC. Neural activity: sculptor of “barrels” in the neocortex. Trends Neurosci. 2001;24:589–595. doi: 10.1016/s0166-2236(00)01958-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo HF, The I, Hannan F, Bernards A, Zhong Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science. 1997;276:795–798. doi: 10.1126/science.276.5313.795. [DOI] [PubMed] [Google Scholar]
- Guo HF, Tong J, Hannan F, Luo L, Zhong Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila . Nature. 2000;403:895–898. doi: 10.1038/35002593. [DOI] [PubMed] [Google Scholar]
- Hannan AJ, Blakemore C, Katsnelson A, Vitalis T, Huber KM, Bear M, Roder J, Kim D, Shin HS, Kind PC. PLC-beta1, activated via mGluRs, mediates activity-dependent differentiation in cerebral cortex. Nat Neurosci. 2001;4:282–288. doi: 10.1038/85132. [DOI] [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Inan M, Lu HC, Albright MJ, She WC, Crair MC. Barrel map development relies on protein kinase A regulatory subunit II beta-mediated cAMP signaling. J Neurosci. 2006;26:4338–4349. doi: 10.1523/JNEUROSCI.3745-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasato T, Datwani A, Wolf AM, Nishiyama H, Taguchi Y, Tonegawa S, Knopfel T, Erzurumlu RS, Itohara S. Cortex-restricted disruption of NMDAR1 impairs neuronal patterns in the barrel cortex. Nature. 2000;406:726–731. doi: 10.1038/35021059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T, Shih TS, Schmitt EM, Bronson RT, Bernards A, Weinberg RA. Tumour predisposition in mice heterozygous for a targeted mutation in Nf1. Nat Genet. 1994;7:353–361. doi: 10.1038/ng0794-353. [DOI] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20:683–691. doi: 10.1016/s0896-6273(00)81008-9. [DOI] [PubMed] [Google Scholar]
- Klesse LJ, Parada LF. p21 ras and phosphatidylinositol-3 kinase are required for survival of wild-type and NF1 mutant sensory neurons. J Neurosci. 1998;18:10420–10428. doi: 10.1523/JNEUROSCI.18-24-10420.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klose A, Ahmadian MR, Schuelke M, Scheffzek K, Hoffmeyer S, Gewies A, Schmitz F, Kaufmann D, Peters H, Wittinghofer A, Nurnberg P. Selective disactivation of neurofibromin GAP activity in neurofibromatosis type 1. Hum Mol Genet. 1998;7:1261–1268. doi: 10.1093/hmg/7.8.1261. [DOI] [PubMed] [Google Scholar]
- Korf BR, Schneider G, Poussaint TY. Structural anomalies revealed by neuroimaging studies in the brains of patients with neurofibromatosis type 1 and large deletions. Genet Med. 1999;1:136–140. doi: 10.1097/00125817-199905000-00004. [DOI] [PubMed] [Google Scholar]
- Kwon CH, Zhou J, Li Y, Kim KW, Hensley LL, Baker SJ, Parada LF. Neuron-specific enolase-cre mouse line with cre activity in specific neuronal populations. Genesis. 2006;44:130–135. doi: 10.1002/gene.20197. [DOI] [PubMed] [Google Scholar]
- Lu HC, Butts DA, Kaeser PS, She WC, Janz R, Crair MC. Role of efficient neurotransmitter release in barrel map development. J Neurosci. 2006;26:2692–2703. doi: 10.1523/JNEUROSCI.3956-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luikart BW, Nef S, Virmani T, Lush ME, Liu Y, Kavalali ET, Parada LF. TrkB has a cell-autonomous role in the establishment of hippocampal Schaffer collateral synapses. J Neurosci. 2005;25:3774–3786. doi: 10.1523/JNEUROSCI.0041-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malatesta P, Hack MA, Hartfuss E, Kettenmann H, Klinkert W, Kirchhoff F, Gotz M. Neuronal or glial progeny: regional differences in radial glia fate. Neuron. 2003;37:751–764. doi: 10.1016/s0896-6273(03)00116-8. [DOI] [PubMed] [Google Scholar]
- Marui T, Hashimoto O, Nanba E, Kato C, Tochigi M, Umekage T, Ishijima M, Kohda K, Kato N, Sasaki T. Association between the neurofibromatosis-1 (NF1) locus and autism in the Japanese population. Am J Med Genet B Neuropsychiatr Genet. 2004;131:43–47. doi: 10.1002/ajmg.b.20119. [DOI] [PubMed] [Google Scholar]
- Mbarek O, Marouillat S, Martineau J, Barthelemy C, Muh JP, Andres C. Association study of the NF1 gene and autistic disorder. Am J Med Genet. 1999;88:729–732. [PubMed] [Google Scholar]
- Miller B, Blake NMJ, Woolsey TA. Barrel cortex: structural organization, development, and early plasticity. In: Kossut M, editor. Plasticity of adult barrel cortex. Johnson City, TN: Graham; 2001. pp. 3–45. [Google Scholar]
- North K. Neurofibromatosis type 1. Am J Med Genet. 2000;97:119–127. doi: 10.1002/1096-8628(200022)97:2<119::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Novak A, Guo C, Yang W, Nagy A, Lobe CG. Z/EG, a double reporter mouse line that expresses enhanced green fluorescent protein upon Cre-mediated excision. Genesis. 2000;28:147–155. [PubMed] [Google Scholar]
- Petersen CC. The functional organization of the barrel cortex. Neuron. 2007;56:339–355. doi: 10.1016/j.neuron.2007.09.017. [DOI] [PubMed] [Google Scholar]
- Schlaggar BL, O'Leary DD. Potential of visual cortex to develop an array of functional units unique to somatosensory cortex. Science. 1991;252:1556–1560. doi: 10.1126/science.2047863. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- The I, Hannigan GE, Cowley GS, Reginald S, Zhong Y, Gusella JF, Hariharan IK, Bernards A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science. 1997;276:791–794. doi: 10.1126/science.276.5313.791. [DOI] [PubMed] [Google Scholar]
- Tong J, Hannan F, Zhu Y, Bernards A, Zhong Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat Neurosci. 2002;5:95–96. doi: 10.1038/nn792. [DOI] [PubMed] [Google Scholar]
- Viskochil D, Buchberg AM, Xu G, Cawthon RM, Stevens J, Wolff RK, Culver M, Carey JC, Copeland NG, Jenkins NA, White R, O'Connell P. Deletions and a translocation interrupt a cloned gene at the neurofibromatosis type 1 locus. Cell. 1990;62:187–192. doi: 10.1016/0092-8674(90)90252-a. [DOI] [PubMed] [Google Scholar]
- Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results in neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–742. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- Wallace MR, Marchuk DA, Andersen LB, Letcher R, Odeh HM, Saulino AM, Fountain JW, Brereton A, Nicholson J, Mitchell AL, Brownstein BH, Collins FS. Type 1 neurofibromatosis gene: identification of a large transcript disrupted in three NF1 patients. Science. 1990;249:181–186. doi: 10.1126/science.2134734. [DOI] [PubMed] [Google Scholar]
- Watson RF, Abdel-Majid RM, Barnett MW, Willis BS, Katsnelson A, Gillingwater TH, McKnight GS, Kind PC, Neumann PE. Involvement of protein kinase A in patterning of the mouse somatosensory cortex. J Neurosci. 2006;26:5393–5401. doi: 10.1523/JNEUROSCI.0750-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker E, Armstrong-James M, Bronchti G, Ourednik W, Gheorghita-Baechler F, Dubois R, Guernsey DL, Van der Loos H, Neumann PE. Altered sensory processing in the somatosensory cortex of the mouse mutant barrelless. Science. 1996;271:1864–1867. doi: 10.1126/science.271.5257.1864. [DOI] [PubMed] [Google Scholar]
- Wong-Riley MT, Welt C. Histochemical changes in cytochrome oxidase of cortical barrels after vibrissal removal in neonatal and adult mice. Proc Natl Acad Sci USA. 1980;77:2333–2337. doi: 10.1073/pnas.77.4.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolsey TA, Van der Loos H. The structural organization of layer IV in the somatosensory region (SI) of mouse cerebral cortex. The description of a cortical field composed of discrete cytoarchitectonic units. Brain Res. 1970;17:205–242. doi: 10.1016/0006-8993(70)90079-x. [DOI] [PubMed] [Google Scholar]
- Woolsey TA, Dierker ML, Wann DF. Mouse SmI cortex: qualitative and quantitative classification of golgi-impregnated barrel neurons. Proc Natl Acad Sci USA. 1975;72:2165–2169. doi: 10.1073/pnas.72.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Parada LF. Neurofibromin, a tumor suppressor in the nervous system. Exp Cell Res. 2001;264:19–28. doi: 10.1006/excr.2000.5138. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Romero MI, Ghosh P, Ye Z, Charnay P, Rushing EJ, Marth JD, Parada LF. Ablation of NF1 function in neurons induces abnormal development of cerebral cortex and reactive gliosis in the brain. Genes Dev. 2001;15:859–876. doi: 10.1101/gad.862101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Guignard F, Zhao D, Liu L, Burns DK, Mason RP, Messing A, Parada LF. Early inactivation of p53 tumor suppressor gene cooperating with NF1 loss induces malignant astrocytoma. Cancer Cell. 2005a;8:119–130. doi: 10.1016/j.ccr.2005.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Harada T, Liu L, Lush ME, Guignard F, Harada C, Burns DK, Bajenaru ML, Gutmann DH, Parada LF. Inactivation of NF1 in CNS causes increased glial progenitor proliferation and optic glioma formation. Development. 2005b;132:5577–5588. doi: 10.1242/dev.02162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo L, Theis M, Alvarez-Maya I, Brenner M, Willecke K, Messing A. hGFAP-cre transgenic mice for manipulation of glial and neuronal function in vivo. Genesis. 2001;31:85–94. doi: 10.1002/gene.10008. [DOI] [PubMed] [Google Scholar]