Abstract
Acidic mammalian chitinase (AMCase) is a mammalian chitinase that has been implicated in allergic asthma. One of only two active mammalian chinases, AMCase, is distinguished from other chitinases by several unique features. Here, we present the novel structure of the AMCase catalytic domain, both in the apo form and in complex with the inhibitor methylallosamidin, determined to high resolution by X-ray crystallography. These results provide a structural basis for understanding some of the unique characteristics of this enzyme, including the low pH optimum and the preference for the β-anomer of the substrate. A triad of polar residues in the second-shell is found to modulate the highly conserved chitinase active site. As a novel target for asthma therapy, structural details of AMCase activity will help guide the future design of specific and potent AMCase inhibitors.
Keywords: chitinase, structure, crystallography, asthma, inhibitor, chitin degradation, 3D-structure
Introduction
Allergic asthma is a chronic inflammatory disorder of the airways. The incidence of allergic asthma has increased dramatically over the last 2 decades in westernized countries. The immune responses that cause elements of asthma mirror beneficial immune responses to helminth infection. Acidic mammalian chitinase (AMCase), a member of family 18 glycosyl hydrolases, is expressed in lung and stomach tissue1 and is up-regulated in human asthma,2 as well as murine asthma models.2–4 Inhibition of AMCase by either a small molecule antagonist or an anti-AMCase antibody ameliorates airway inflammation and hyper-responsiveness in murine asthma models.2 Certain polymorphisms and haplotypes of AMCase are associated with bronchial asthma in humans.5 Additional polymorphisms of the AMCase gene are associated with atopic asthma and total serum IgE levels in the Indian population.6 These observations suggest that AMCase may be an important target for the treatment of human asthma.
Here, we describe the experimentally determined high resolution crystal structures of AMCase, in the apo form as well as in complex with the broad spectrum chitinase inhibitor methyllallosamidin. We compare this structure with other chitinase structures and explore the structural basis of characteristic features of AMCase. Chitinase enzymatic activity is well understood from extensive studies in other systems, as well as mutational analysis and homology modeling in AMCase.7 These structures expand our understanding of the structural basis of characteristics unique to AMCase, the most striking of which is the low pH optimum for catalysis. A triad of polar residues in the second-shell is found to modulate the highly conserved chitinase active site. An intricate network of hydrogen bonds linking surrounding charged residues to the key active site acidic residues is observed. The derived contacts are consistent with amino acid residues His-208, His-269, and Arg-145 most likely accounting for the low pH optimum of the enzyme. The structural basis of AMCase's preference for the β-anomer of short chitin-like sugars is also elucidated. As the only chitinase implicated in asthma, the information derived from its crystal structure will provide valuable insights in the potential discovery of drugs that will specifically target AMCase and the abnormalities causing allergic asthma.
Results
Structure determination
The structure of the AMCase datalytic domain was determined to a resolution of 2.0 Å in the apo form and to 1.7 Å in complex with methylallosamidin by molecular replacement using human chitotriosidase a search model (Table I). The construct that was successfully crystallized was a C-terminal truncation including the catalytic domain, from the natural N-terminus at amino acid 22 through amino acid 408. We chose to mutate the Ser originally at position 354 to Phe, a polymorphism that is at least as common as the Ser, to avoid N-linked glycosylation. The apo protein crystallized in space group P21, and contains four molecules of AMCase in the asymmetric unit, implying a solvent content of 39.5%. The resulting map showed good electron density for the residues 22 to 400 (monomer A, B, C, and D, with the exception of amino acids 231–236 in molecule B). Residues C-terminal to 400, including the His tag, are not seen in the electron density maps and are presumably disordered. AMCase in complex with methylallosamidin crystallized in space group P212121, and contains two molecules of AMCase in the asymmetric unit, implying a solvent content of 39.7%. The resulting map shows good electron density for residues 22 through 398 in both molecules A and B, as well as for the bound methylallosamidin (Fig. 1). Residues C-terminal to 398, including the His tag, are not seen in the electron density maps and are presumably disordered.
Table I.
Data Collection and Refinement Statisticsa
| Apo AMCase | AMCase + methyallosamidin | |
|---|---|---|
| Wavelength (Å) | 1.000 | 1.000 |
| Resolution range of data (Å) | 50.0–2.0 | 20.0–1.7 |
| Rmergeb (%) | 12.7 (45.3) | 10.1 (59.8) |
| Completeness (%) | 97.9 (91.4) | 90.0 (91.1) |
| Redundancy | 3.8 | 6.3 |
| Total reflections | 343,748 | 406,206 |
| Unique reflections | 91,479 | 64,057 |
| I/σ(I) | 9.77 (2.04) | 15.21 (2.1) |
| Resolution range of refinement (Å) | 30.0–2.0 | 20.0–1.7 |
| No. of molecules in asymmetric unit | 4 | 2 |
| No. of atoms in asymmetric unit | 24,391 | 12,570 |
| No. of water molecules in asymmetric unit | 497 | 582 |
| B average | ||
| Protein atoms (Å2) | 5.2 | 6.4 |
| Water molecules (Å2) | 12.4 | 21.4 |
| MALLO (Å2) | – | 10.0 |
| Rworkc (%) | 19.71 | 17.48 |
| Rfreed (%) | 22.84 | 19.39 |
| RMS deviations from ideal geometry for: | ||
| Bonds (Å) | 0.008 | 0.008 |
| Angles (°) | 1.060 | 1.058 |
Values in parenthesis are for the highest resolution shell.
Rmerge= |Ih−<Ih>|/Ih, where <Ih> is the average intensity over symmetry equivalents. Numbers in parentheses reflect statistics for the last shell.
Rwork = ||Fobs|−|Fcalc||/|Fobs|.
Rfree is equivalent to Rwork, but calculated for a randomly chosen 5% of reflections omitted from the refinement process.
Figure 1.
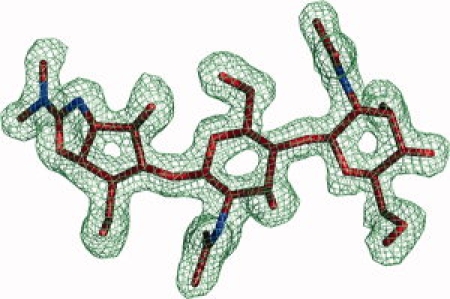
Methylallosamidin and surrounding |2Fo−Fc| electron density map contoured at 1.0 σ.
Overall structure and interaction with methylallosamidin
The structures of AMCase alone and in complex with the inhibitor methylallosamidin reveal an eight stranded beta-barrel structure (TIM-barrel) with an α/β lobe inserted between β-sheet 7 and α-helix I, that is composed of a 5-stranded anti-parallel beta sheet and an alpha-helix [Fig. 2(A)]. The overall tertiary structure is very similar to the structure of the related human chitotriosidase8,9 (PDB accession codes 1HKK, 1GUV, 1LG1, 1LG2). A long cleft, which is lined with solvent-exposed aromatic residues, including Trp-71, Tyr-34, Trp-31, Trp-360, Trp-99, Trp-218, is the site of chitin binding8–10 [Fig. 2(B)]. These aromatic residues are highly conserved across species.11 In the liganded structure presented here, methylallosamidin is bound in part of this groove [Figs. 2(A and B)]. The active site is dominated by the acidic residues featuring the common chitinase motif DXXDXDXE [Figs. 3(A and B)]. Disulfide bonds are observed between Cys-26 and Cys-51, Cys-307 and Cys-372, and Cys-49 and Cys-394. The disulfide bond between Cys-49 and Cys-394, which does not have an equivalent in chitotriosidase, affixes the C-terminal to the base of the beta-barrel [Fig. 2(A)] and likely contributes to the overall stability of the catalytic domain.
Figure 2.
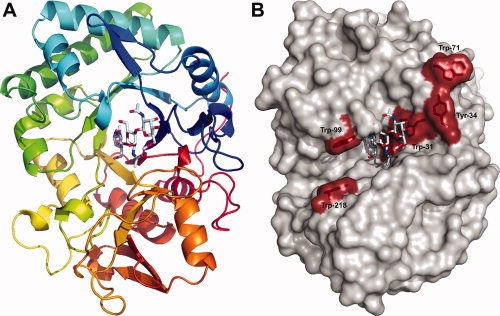
(A) A ribbons diagram of the AMCase catalytic domain is color-ramped blue to red, looking down the central β-barrel. Methylallosamidin is show in pale cyan, bound at the top of the barrel. A disulfide bond between Cys-49 and Cys-394 can be seen near the back, connecting the C-terminal end (in red) to an N-terminal loop (in blue). (B) A surface rendition in the same orientation as 2A shows the aromatic residues implicated in chitin binding in red. Methylallosamidin is shown in pale cyan, the subunits of which correspond to subsite positions −1, −2, and −3, from left to right. Positive subsite positions extend towards the left in this view, with subsite +1 just under Trp-99 and subsite +2 just above Trp-218.
Figure 3.
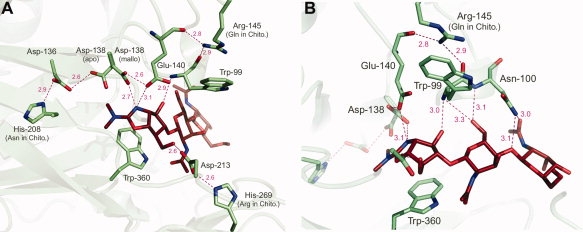
(A) A ribbons diagram of the AMCase active site depicts side chains of significant residues. Asp-136, Asp-138, Glu-140, Asp-213, and Trp-360 are highly conserved active site residues. Arg-145, His-208, and His-269 are residues specific to AMCase. Dotted pink lines indicate hydrogen bonds, with the distance in angstroms indicated. (B) Arg-145 is shown in the context of the active site. Arg-145 affects substrate binding via a network of hydrogen bonds mediated by Trp-99, Asn-100, and Glu-140.
Allosamidin and its variant methylallosamidin are broad-spectrum natural product inhibitors. They are pseudo-trisaccharides with two NAG sugars coupled to allosamizoline. In the complex structure with methylallosamidin, the inhibitor is well defined, with the allosamizoline binding in the active site at the −1 subsite, and the attached NAG sugars reaching into the −2 and −3 subsites [Figs. 2(A,B), 3(A,B), and 4]. The methyl group, differentiating this inhibitor from allosamidin, is clearly seen in the electron density maps (Fig. 1). As it is solvent-exposed and not making significant contacts with the protein, methylallosamidin is not expected to bind differently than allosamidin. An ordered water molecule (HOH-346 and HOH-84 for AMCase chains A and B) is observed 3.5 Å from allosamizoline C17 and 2.8 Å from Glu-140. This interaction mimics that of the water which effects the hydrolysis of the oxazolinium ion intermediate during the final step of chitin cleavage.12
Figure 4.
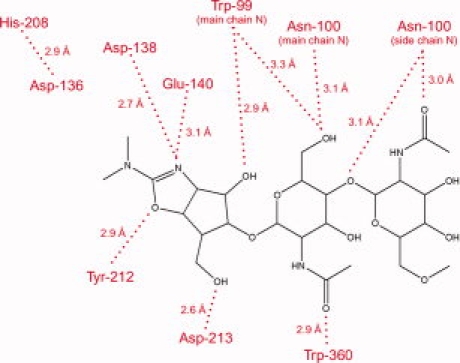
A summary of interactions between methylallosamidin and AMCase active site residues.
In the apo AMCase structure, we see an invasion of the active site of one molecule by the C-terminal tail of a neighboring molecule (Fig. 5). There are four molecules in the crystallographic asymmetric unit, and the exact position taken on by the invading tail is slightly different between two molecules from the other two (molecules A and C are nearly identical, as are B and D). In either case, Gln-399 roughly occupies the −2 subsite and reaches towards the −1 subsites, and Pro-400 roughly occupies the −3 subsite. It is likely that this interaction is not physiologically relevant, but is a result of the packing in this particular crystal form.
Figure 5.
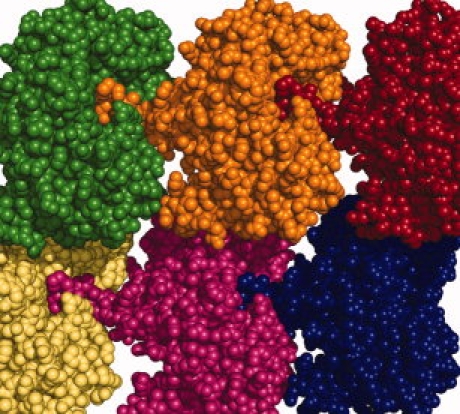
A space-filled representation of AMCase molecules shows the crystal packing of Apo AMCase. The C-terminal end of one molecule binds in the active site cleft of the neighboring molecule.
Comparison of apo and methylallosamidin bound structures
Overall differences between APO and MALLO-bound structures are minimal in the αC trace, including a small movement around loop 98–102 (between β-strand 3 and α-helix), which accommodates the movement of Asn-100 to form hydrogen-bonds with methylallosamidin in multiple places through both main chain and side chain atoms. The most striking difference between these structures is the rotamer of Asp-138. In the unliganded enzyme, Asp-138 is in a rotamer that allows a hydrogen bond with Asp-136. In the methylallosamidin-bound enzyme, the Asp-138 side chain swings nearly 180° around, breaking the bond with Asp-136 and taking up a position within 2.6 Å of Glu-140 and forming H-bonds with Glu-140 and methylallosamidin at the same time [Fig. 3(A)]. These alternative rotamers of equivalent Asp residues have been well documented in other chitinase structures. Furthermore, there is a rotation of the Glu-140 side chain from the apo-structure where it is pointing towards the main chain N of Trp-99 (3.3 Å) and the side chain hydroxyl of Tyr-141. Upon binding of methylallosamidin, Glu-140 rotates about 60° to form hydrogen-bonds with Asp-138 and with the allosamizoline N. It is the protonation of Glu-140 that is the final step of chitin hydrolysis.
Preference for β-anomer of substrate
Models of α-and β-chitopentose bound to AMCase, respectively, are in agreement with and provide an explanation for the previously reported preference of AMCase for the β-anomer of the substrate.13 In studies with hexa-, penta- and tetra-saccharide substrates, AMCase has been shown to cleave disaccharide and trisaccharide units from the nonreducing end.13 This indicates that a penta-saccharide substrate can productively occupy subsites −3 to +2, as well as subsites −2 to +3.
Alpha- and β-chitopentose were modeled into AMCase occupying subsites −3 through +2, and elucidate the structural basis for a preference for the β-anomer at the +2 subsite. In the models, the fourth and fifth N-acetylglucosamine (NAG) units of chitopentose pack between Trp-99 and Trp-218 (Fig. 6). The substrate NAG in the +2 position is packing against Trp-218, and the +1 position NAG packs against Trp-99. This stacking presumably limits the mobility of this part of the substrate. In this position, the terminal hydroxyl of the alpha anomer makes an unfavorable electrostatic interaction with the —NH of the indole ring of Trp-218, whereas the equatorial position of the terminal hydroxyl of the beta anomer does not. On the basis of the models, the interaction energy of β-chitopentose with AMCase is 2.95 kcal/mol more favorable than that of the α-chitopentose. This offer a structural basis for AMCase's preference for the β-anomer of chitopentose at the +2 subsite. The structural basis of a preference for the β-anomer at the +3 subsite is not clear from these structures, however, given the presence of both α-and β-anomers in both the 2-mer and 3-mer hydrolysis products,13 it is possible that there is no additional preference at the +3 position.
Figure 6.
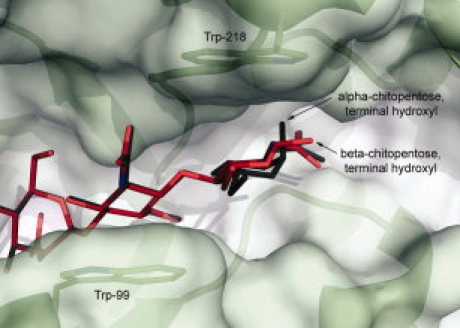
The modeled binding of the α-anomer versus the β-anomer of a chitin fragment (chitopentose) shows the structural basis of the preference for the β-anomer of the substrate by AMCase. The terminal hydroxyl of the α-anomer, shown in dark gray, makes an unfavorable electrostatic interaction with the —NH of the indole ring of Trp-218 because of partial charges on the atoms, and thus binding of the β-anomer (shown in red) is energetically more favorable.
Chitotetrose is cleaved exclusively into 2-mers, positioning the terminal reducing-end NAG at the +2 subsite, thus the above arguments are equally applicable in the structural basis of the preference for the β-anomer in chitotetrose.
Although there is no apparent reason for a preference in anomer for a chitohexose bound at the −2 to +4 position, a hydrolysis and processivity event places the reducing end at the +2 subsite. As the β-anomer is clearly preferred in that position, it is possible that this drives the hydrolysis of β-anomers of chitohexose and retards the hydrolysis of α-anomers of chitohexose, translating into an apparent preference for the β-anomer of chitohexose.
Labeling analysis
In an attempt to determine the role of the conserved histidines found within the active site of AMCase, labeling studies done with diethyl pyrocarbonate (DEPC) were initiated. DEPC is shown to react in a selective manner with histidines, but reactions with other amino acids, such as tyrosine, are possible.14 Difference spectrum of 1 μM AMCase and AMCase treated with DEPC show an increase in absorbance at 245 nm, in accordance with labeling of histidines. A lack of a decrease in absorbance at 278 nm demonstrates that very few if any tyrosine are labeled. From this data, it has been determined that ∼3 histidines are labeled. The number of modified histidines is based on the maximum absorbance of the difference spectrum at 245 nm using the extinction coefficient of 3200 M−1 cm−1 for a modified His.
Treatment of AMCase with varying amounts of DEPC inactivated the enzyme, but complete loss of activity was not achieved. This lowering of AMCase activity without complete loss suggest that histidine is not essential for activity, but may be required for optimum activity at low pH. However, assays were done a pH range of 5 to 6. AMCase activity has been demonstrated at a pH of 3. Assays done at lower pH, such as 3, may give more insight into the function of the histidine at lower pH. The pH dependence on DEPC inactivation shows a pKa of 6.9, corresponding to an imidazole group of histidine. Kinetics analysis of AMCase that was labeled with DEPC show a decrease in Vmax, consistent with activity data, but no change in the Km for the substrate, indicating that the observed lower activity is not due to interference in substrate binding (Table II). Together these enzymatic data support the hypothesis that His-208 and His-269 are playing a role in the activity of this enzyme.
Table II.
Effects of DEPC Labelinga
| AMCase (un-labeled) | DEPC labeled AMCase | |
|---|---|---|
| Km | 11.1 ± 3.1 μM | 12.2 ± 2.2 μM |
| kcat | 8.8 min−1 | 3.5 min−1 |
Concentration of AMCase used in the assay was 2.25 nM.
Discussion
Comparison of AMCase and chitotriosidase: pH optimum
Interestingly, although both AMCase and the other human chitinase, chitotriosidase, are secreted enzymes, only AMCase is implicated in asthma. Among the most notable differences between AMCase and human chitotriosidase are three residue differences near the active site. Two of these three residues, His-208 and His-269, are conserved in AMCase genes across species11 but are different in human chitotriosidase, suggesting that they together have an important structural role in AMCases. An α-carbon alignment of other chitinase/allosamidin complex structures available in the PDB including Chitinase A from Serratia marcescens (ChiA-PDBID 1FFQ), Chitinase B from S. marcescens (ChiB-PDBID 1E6R), Chitinase 1 from Coccidioides immitis (Chi1-PDBID 1LL4) and Hevamine from Hevea brasiliensis (Hev-PDBID 1LLO) reveals that the interactions made by these three residues influencing the active site are not duplicated in any of the other structures. One of these residues, His-208, has previously been proposed to have a role in determining the pH optimum of this enzyme.7 We postulate that rather than this His alone, it is the three together that are in position to address the structural basis of AMCase's low pH optimum as compared with chitotriosidase. It is well established that the pKa values of active site residues can be modified by adjacent residues. Two additional resides are observed to influence the active site's key acidic residue system (the Asp-138/Glu-140 system). His-208, His-269, and Arg-145 all contact key conserved residues within the active site and are in a position to change the pH optimum of the enzyme by influencing the pKa of the Asp-138/Glu-140 system.
In both apo and liganded structures, AMCase His-208 is a hydrogen-bonding partner of the conserved Asp-136 [Figs. 3(A) and 4], part of the DXXDXDXE motif common to all chitinases. By contrast, human chitotriosidase has an Asn at this position. ChiB and Chi1 also have an Asn at the equivalent position whereas ChiA has a Phe and Hev has a Trp. Mutation of this His-208 to Asn in AMCase has been shown to play an important role in the acidic pH optimum of AMCase.7 These studies suggest that a protonated His in this position contributes to the destabilization of the Asp-136/Asp-138 H-bond, which must take place to allow the binding of substrate. The destabilization of this bond allows Asp-138 to shuttle back and forth (rotating about the Cα—Cβ bond) between Asp-136 in the unliganded enzyme and the catalytic Glu-140 when methylallosamidin, and presumably substrate, is bound. On the basis of the studies in ChiB,15 the pKa of Asp-136 is extremely low, ensuring that Asp-138 carries the H as it swings from Asp-136 in the apo protein to Asp-138 in the liganded structure. The protonated Glu-140 then participates in the cleavage at the β(1,4) linkage between NAG sugars. In AMCase His-208 is 2.8/2.9 Å from Asp-136 and is well within H-bonding range. Although the position of chitotriosidase's Asn-208 is very similar, the respective H donor and acceptor atoms slightly further apart 3.2/3.4 Å apart. The other residues contacting Asp-136 are essentially identical between AMCase and chitotriosidase; H-bond donor groups from Tyr-27 and Thr-181 (Ser-181 in chito).
His-269 has not previously been remarked upon, yet represents another key difference between AMCase and chitotriosidase. His-269 is observed to form a hydrogen-bond with Asp-213, a conserved and essential residue in the active site [Figs. 3(A) and 4]. Asp-213 lies on the bottom of the active site and coordinates the boat conformation of the substrate sugar in the −1 subsite by accepting a hydrogen bond from a hydroxyl of the substrate sugar in the −2 subsite, in preparation for hydrolysis. In studies in ChiB it has been shown to play a role in increasing the pKa values in the Asp-138/Glu-140 system.15 It the ChiB system, Asp-215 (the equivalent to 213 in AMCase) has a very low pKa in part due to its salt-bridge with the Arg present in both the ChiB and chitotriosidase at the position equivalent to AMCase's 269. ChiA and Chi1 also have an Arg at this position, whereas in Hev the comparison can't be make as this section of the chain is very different. In AMCase we see replacement of this Arg with His-269, with the side chain N of His being positioned to H-bond with Asp-213 as the Arg side chain guanidyl group does. As a result of coordinating with a weaker base, the pKa of Asp-213 is likely not as low. This, consequently, would render Asp-213 less able to increase the pKa of the Asp-138/Glu-140 system. The net result of a lower pKa in the Asp-138/Glu-140 system would be manifest as a lower pH optimum for activity. This amino acid change is likely to play a significant role in this difference between AMCase and other chitinases with pH optimums closer to neutral pH. In an alignment of vertebrate chitotriosidase and AMCase genes, His at this position tracks perfectly with low-pH optimum enzymes.11
Another novel feature revealed by these structures is the packing of Arg-145 side chain against Trp-99, one of the key aromatic residues coordinating substrate binding. The charged Arg-145 side chain forms H-bonds with the main chain carbonyls of Trp-99 and Glu-140. Glu-140, Trp-99, and the neighboring Asn-100 in turn H-bond with methylallosamidin (and by extension, the substrate) in five separate places, transmitting the effects of Arg-145 [Fig. 3(B)]. Having a strongly basic residue influencing these active site residues would have the effect of lowering the pKa of the Asp-138/Glu-140 system, again lowering the optimal pH of the enzyme. By contrast, in human chitotriosidase the Gln at this position is not spatially within range to make these contacts; thus a water molecule makes the equivalent interactions with the exposed main chain atoms of these active site residues in that enzyme. The chitotriosidase gene of some other species, however, also have an Arg at this position.11 Thus, this residue most likely serves to modulate the pKa of the Asp-138/Glu-140 system rather than being the primary determinant responsible for the pH optimum. ChiA, ChiB, Chi1, and Hev are all also absent residues that makes H-bonds with the main chain carbonyls of Trp-99 and Glu-140 in this way.
In conclusion, the structure determination of unliganded and methylallosamidin-bound AMCase has allowed further insights into the activity of this enzyme. We have presented a detailed analysis of the active site and comparison with the related enzyme chitotriosidase, and have concluded that three second-shell amino acid residues (His-208, His-269, and Arg-145) modulate the highly conserved chitinase active site, and establish the low pH optimum of this enzyme. We have presented enzymatic data supporting the hypothesis that these two histidines play a role in the activity of this enzyme. Finally, we have elucidated the structural basis for the observed preference of the enzyme for the beta-anomer of the substrate,13 finding that the indole ring of Try-218 has an unfavorable interaction with the terminal hydroxyl of alpha-pentose, but not with the terminal hydroxyl of beta-pentose. Together these findings elucidate the complete mechanism of achieving specificity for low pH while guarding an active site that is highly conserved among all chitinases, and broaden our understanding of this enzyme. These structures will help guide the future design of AMCase inhibitors for the potential treatment of asthma.
Materials and Methods
AMCase human cloning
A nucleotide sequence encoding truncated human AMCase, starting at amino acid #1 with the natural leader sequence and ending at amino acid #408, including a mutation at position 354 to replace a serine by a phenylalanine (S354F) was cloned into pTMED vector. A histidine tag is attached at the C-terminal end to facilitate protein purification. The amino acid sequence of the mature truncated human AMCase protein used for the crystallization corresponds to AMCase amino acids 22-408, and is as follows:

The S354 mutated human AMCase was generated because the cloned human cDNA had a serine residue at position 354, which resulted in an N-linked glycosylation site. In examining other cDNA encoding human AMCase, it was discovered that position 354 is a polymorphism, and that a phenylalanine at that position is at least as prevalent as the S. Therefore, when the truncated form was made for crystallization, the serine was changed to a phenylalanine at position 354 to avoid an N-linked glycosylated protein.
CHO cell development
As described previously.13
Purification of AMCase
As previously described.13
Methylallosamidin production and isolation
A strain of streptomyces from the proprietary Wyeth culture collection was grown in 10-L fermentation vessels on a medium containing nutrasoy (12.5 g/L), dextrose (12.5 g/L), N-Z amine (12.5 g/L), NH4Cl (1.5 g/L), andCaCl2 (1.0 g/L) with the pH adjusted to 6.8, to produce mainly methylallosamidin and other allosamidins16 at titers of ∼5 mg/L after 5 days. The titers of the allosamidins and the progress of methylallosamidin isolation and purification was followed by LCMS using positive ion ms-peaks in the 609–637 range (details of this method will be published elsewhere).
Because of the occurrence of methylallosamidin in both the medium and pellet, the fermentation batch was stored at 4°C for 3 to 5 days following fermentation. The mash was then centrifuged to remove the pellet cell paste. The pH of the supernatant was subsequently adjusted to pH 9.2 using NH4OH and passed over an open HP-20 column (4 × 36 cm) and the column was washed with 2 L of 0.01M NH4OH. A methylallosamidin-rich fraction was then eluted from the column using 1 L of 0.01M CH3COOH solution containing 20% methanol. This fraction was concentrated under vacuum to remove methanol. The pH of the concentrated fraction (∼250 mL) was adjusted to pH 8 with NaOH and this was added to a prepared bulk C18-silica-gel column (3 × 13 cm) and washed with 100 mL water. Absorbed methylallosamidin was then eluted with 0.1% CH3COOH. Cuts of 40 mL fractions were taken with the bulk of the methylallosamidin eluting in fractions 5 and 6. These were pooled and concentrated to ca. 5 mL and added to a prepared LH-20 column (3 × 30 cm) packed in a methanol/water 50:50. The column was developed with the same solvent system yielding methylallosamidin in fractions 7 and 8. Methanol was evaporated and the aqueous solution was freeze-dried to yield 27.1 mg of a white, fluffy powder, containing mostly methylallosamidin (87% by LCMS).
Crystallization
Apo AMCase crystals were grown by hanging drop vapor diffusion at 18°C in drops containing 1.0 μL protein stock solution (21 mg/mL protein, 25 mM TRIS pH 7.5, 50 mM NaCl) mixed with 1.0 μL well solution (15% PEG 3350, 120 mM ammonium sulfate, 60 mM sodium acetate pH 4.6) and equilibrated against 0.5 mL well solution. Rectangular crystals grew in 1 week, measuring ∼80 μm across.
AMCase/MALLO crystals were grown by hanging drop vapor diffusion at 18°C in drops containing 0.2 μL protein stock solution (21 mg/mL protein, 25 mM TRIS pH 7.5, 50 mM NaCl, 1 mM methylallosamidin from a 10 mg/mL solution in PBS) mixed with 0.2 μL well solution (20% PEG 3350, 200 mM ammonium formate) and equilibrated against 0.2 mL well solution. Slate-like crystals grew in 1 week, measuring ∼80 μm across.
Data collection and processing
Apo AMCase crystals belong to the space group P21 with unit cell parameters a = 69.94 Å, b = 92.36 Å, c = 111.67 Å, α = γ = 90°, β = 95.92°, and contain four molecules of AMCase in the asymmetric unit, implying a solvent content of 39.5%. Crystals were drawn through a solution of 25% glycerol and 75% well solution, and cooled rapidly in liquid nitrogen. Diffraction data were recorded at ALS beamline 5.0.1 on a q-210 ccd camera. Intensities were integrated and scaled using the programs Denzo and Scalepack.17
AMCase/MALLO co-crystals belong to the space group P212121 with unit cell parameters a = 63.69 Å, b = 89.29 Å, c = 126.69 Å, and α = β = γ = 90°, and contain two molecules of AMCase in the asymmetric unit, implying a solvent content of 39.5%. Crystals were drawn through a solution of 25% glycerol and 75% well solution, and cooled rapidly in liquid nitrogen. Diffraction data were recorded at ALS beamline 5.0.1 on a q-210 ccd camera. Intensities were integrated and scaled using the programs Denzo and Scalepack.17
Phasing, model building, and refinement
The apo structure was determined by molecular replacement using the protein model of the human chitotriosidase8 (PDBID 1GUV, 55% sequence identity) as the search model. After several iterative cycles of refinement using CNX18 and Refmac5,19 and model improvement in Coot,20 final Rwork and Rfree values of 19.71% and 22.84% were obtained (Table I). The AMCase/MALLO co-structure was determined by the same procedure using the apo protein as a starting molecular replacement model, and obtaining final Rwork and Rfree values of 17.48% and 19.39% for AMCase/MALLO.
Molecular modeling
Models of α-and β-chitopentose bound to AMCase, respectively, were generated using the docking program Glide and the MacroModel eMBrAcE minimization procedure in Maestro (Schrodinger, New York, NY, 2007). In a first step, one NAG unit was docked into the active site of the AMCase + MALLO protein structure using GlideSP version 4.5. Next, the structure of Chitinase B with a pentamer bound12 (PDBID = 1E6N) was superimposed onto the AMCase + MALLO protein structure by aligning the sequences in Maestro. The +2 and +1 NAG units were built into the AMCase structure based on their positions in the Chitinase B structure and the −1 and −2 NAG units were added based on their position in the allosamidin bound to AMCase. Beta-chitopentose with central NAG unit in a boat-like conformation mimicking the transition state12 was then minimized using eMBrAcE with the OPLS force field21 and implicit water solvent.22 More specifically, the substrate and all residues with an atom within 3 Å of the ligand were subject to up to 1000 steps of full minimization, the next shell of residues within 5 Å of the ligand was subject to harmonic constraints during the minimization, and a second shell of residues within 7 Å were fixed.
Labeling with diethyl pyrocarbonate difference spectrum
A large excess of DEPC, 200 μM, was added to 1 μM AMCase and allowed to incubate for 10 to 15 min in a citric acid/phosphate buffer pH 6.9. The spectrum of unlabeled AMCase was compared with spectrum of labeled AMCase. A spectral change at 278 nm represents modifications of tyrosines whereas a spectral change at 242 nm represents modified histidines. Modified His has extinction coefficient of 3.2 × 103M−1 cm−1. The change in absorbance at 242 nm is 0.059 AU, corresponding to 18 μM His labeled.
Enzyme assays
Assays were done with 3 nM enzyme in Assay Buffer (35 mM Citrate, 128 mM sodium Phosphate pH 6.0, 0.005% Brij-35) in a volume of 40 μL. Results were read on Tecan Safire on kinetics mode with Ex wavelength of 319 nm and Em wavelength of 441 nm. Rates were determined from the first several minutes of the linear part of the curve.
Inactivation activity analysis
The activity of AMCase was determined after incubation with varying amounts of DEPC (0 to 1000 μM). Inactivation was done at pH 6.9 with 0.75 μM AMCase in which the reaction was quenched by dilution (1:200) in buffer pH 6.0. Activity was measured with 30 μM substrate and change in activity (Ao/A) was plotted against ratio of DEPC to AMCase ([DEPC]/[AMCase]). Reaction time was 5 and 15 min. This shows that there is a 40% loss in AMCase activity after 15 min even at 1000 μM DEPC.
pH of inactivation
This was done to confirm that the loss in activity seen was due to labeling of His. The pKa of the imidazole of His is 6.9, therefore the pH dependence of inactivation should mirror the pKa of His. The inactivation assay was done with 0.75 μM AMCase and 200 μM DEPC at varying pH from 5.7 to 7.8 and quenched at different times from 0 to 15 min in assay buffer pH 6.0. The rate of inactivation at each pH was determined and fit to the pH dependence equation [v = Vmax/(1 + 10⁁(pKa−pH))].
The pKa determined was 6.9 ± 0.1, which corresponds to a His.
Effect of DEPC labeling on the kinetics of AMCase
The kinetic constants of the di-sugar substrate were determined for labeled and non-labeled AMCase. This will show what effect the labeling of AMCase has on the enzyme. In this labeling reaction 10 μM DEPC was used with 0.75 μM AMCase. The reaction was quenched by dilution and the kinetics was determined. Resulting data was plotted in a double-reciprocal plot and shows that there is an effect seen on Vmax but not on Km. The values are for control (Vmax = 380 ± 38 RFU/min with a Km = 11.1 ± 3.1 μM) while the labeled AMCase (Vmax = 153 ± 11 RFU/min with a Km = 12.2 ± 2.2 μM). This represents a loss in activity of ∼59%.
pH rate profile: labeled versus unlabeled AMCase
The pH dependence of the kinetic parameters was determined for the labeled and non-labeled AMCase. The samples were treated the same with the exception of the addition of 250 μM DEPC in the inactivation assay. The inactivation assay was performed at pH 6.9 and then quenched by dilution into buffer at varying pH (pH 2.8 to 7.3). The steady state kinetic parameters were then determined at each pH and the log Vmax and log V/K were plotted against pH. There was no significant difference observer in the pK values for the labeled and un-labeled AMCace. The log (Vmax) versus pH did show the loss in activity in AMCase when the enzyme is labeled. Detailed kinetic analysis will have to be done to determine the mechanism behind the loss in activity when AMCase is labeled.
Data Deposition
The atomic coordinates have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, New Jersey (3FXY and 3FY1).
Acknowledgments
The authors acknowledge X-ray resources provided by the Advanced Light Source (ALS) at the Lawrence Berkeley National Laboratory, Beamline 5.0.1., Berkeley, CA 94720.
Glossary
Abbreviations
- AMCase
acidic mammalian chitinase
- DEPC
diethyl pyrocarbonate
- LCMS
liquid chromatography mass spectrometry
- MALLO
methylallosamidin
- NAG
N-acetylglucosamine
- PBS
phosphate-buffered saline
- PEG
polyethylene glycol
- TRIS
tris(hydroxymthyl)aminomethane.
References
- 1.Boot RG, Blommaart EF, Swart E, Ghauharali-van der Vlugt K, Bijl N, Moe C, Place A, Aerts JM. Identification of a novel acidic mammalian chitinase distinct from chitotriosidase. J Biol Chem. 2001;276:6770–6778. doi: 10.1074/jbc.M009886200. [DOI] [PubMed] [Google Scholar]
- 2.Zhu Z, Zheng T, Homer RJ, Kim YK, Chen NY, Cohn L, Hamid Q, Elias JA. Acidic mammalian chitinase in asthmatic Th2 inflammation and IL-13 pathway activation. Science. 2004;304:1678–1682. doi: 10.1126/science.1095336. [DOI] [PubMed] [Google Scholar]
- 3.Zimmermann N, Mishra A, King NE, Fulkerson PC, Doepker MP, Nikolaidis NM, Kindinger LE, Moulton EA, Aronow BJ. Rothenberg ME transcript signatures in experimental asthma: identification of STAT6-dependent and -independent pathways. J Immunol. 2004;172:1815–1824. doi: 10.4049/jimmunol.172.3.1815. [DOI] [PubMed] [Google Scholar]
- 4.Follettie MT, Ellis DK, Donaldson DD, Hill AA, Diesl V, DeClercq C, Sypek JP, Dorner AJ, Wills-Karp M. Gene expression analysis in a murine model of allergic asthma reveals overlapping disease and therapy dependent pathways in the lung. Pharmacogenomics J. 2006;6:141–152. doi: 10.1038/sj.tpj.6500357. [DOI] [PubMed] [Google Scholar]
- 5.Bierbaum S, Nickel R, Koch A, Lau S, Deichmann KA, Wahn U, Superti-Furga A, Heinzmann A. Polymorphisms haplotypes of acid mammalian chitinase are associated with bronchial asthma. Am J Respir Crit Care Med. 2005;172:1505–1509. doi: 10.1164/rccm.200506-890OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chatterjee R, Batra J, Das S, Sharma SK, Ghosh B. Genetic association of acidic mammalian chitinase with atopic asthma and serum total IgE levels. J Allergy Clin Immunol. 2008;122:202–208. e1–e7. doi: 10.1016/j.jaci.2008.04.030. [DOI] [PubMed] [Google Scholar]
- 7.Bussink AP, Vreede J, Aerts JM, Boot RG. A single histidine residue modulates enzymatic activity in acidic mammalian chitinase. FEBS Lett. 2008;582:931–935. doi: 10.1016/j.febslet.2008.02.032. [DOI] [PubMed] [Google Scholar]
- 8.Fusetti F, von Moeller H, Houston D, Rozeboom HJ, Dijkstra BW, Boot RG, Aerts JM, van Aalten DM. Structure of human chitotriosidase. Implications for specific inhibitor design and function of mammalian chitinase-like lectins. J Biol Chem. 2002;277:25537–25544. doi: 10.1074/jbc.M201636200. [DOI] [PubMed] [Google Scholar]
- 9.Rao FV, Houston DR, Boot RG, Aerts JM, Sakuda S, van Aalten DM. Crystal structures of allosamidin derivatives in complex with human macrophage chitinase. J Biol Chem. 2003;278:20110–20116. doi: 10.1074/jbc.M300362200. [DOI] [PubMed] [Google Scholar]
- 10.Houston DR, Shiomi K, Arai N, Omura S, Peter MG, Turberg A, Synstad B, Eijsink VG, van Aalten DM. High-resolution structures of a chitinase complexed with natural product cyclopentapeptide inhibitors: mimicry of carbohydrate substrate. Proc Natl Acad Sci USA. 2002;99:9127–9132. doi: 10.1073/pnas.132060599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bussink AP, Speijer D, Aerts JM, Boot RG. Evolution of mammalian chitinase(-like) members of family 18 glycosyl hydrolases. Genetics. 2007;177:959–970. doi: 10.1534/genetics.107.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Aalten DM, Komander D, Synstad B, Gaseidnes S, Peter MG, Eijsink VG. Structural insights into the catalytic mechanism of a family 18 exo-chitinase. Proc Natl Acad Sci USA. 2001;98:8979–8984. doi: 10.1073/pnas.151103798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chou YT, Yao S, Czerwinski R, Fleming M, Krykbaev R, Xuan D, Zhou H, Brooks J, Fitz L, Strand J, Presman E, Lin L, Aulabaugh A, Huang X. Kinetic characterization of recombinant human acidic mammalian chitinase. Biochemistry. 2006;45:4444–4454. doi: 10.1021/bi0525977. [DOI] [PubMed] [Google Scholar]
- 14.Miles EW. Modification of histidyl residues in proteins by diethylpyrocarbonate. Methods Enzymol. 1977;47:431–442. doi: 10.1016/0076-6879(77)47043-5. [DOI] [PubMed] [Google Scholar]
- 15.Synstad B, Gaseidnes S, Van Aalten DM, Vriend G, Nielsen JE, Eijsink VG. Mutational and computational analysis of the role of conserved residues in the active site of a family 18 chitinase. Eur J Biochem. 2004;271:253–262. doi: 10.1046/j.1432-1033.2003.03923.x. [DOI] [PubMed] [Google Scholar]
- 16.Sakuda S, Isogai A, Matsumoto S, Suzuki A. Search for microbial insect growth regulators. Part II. Allosamidin, a novel insect chitinase inhibitor. J Antibiot (Tokyo) 1987;40:296–300. doi: 10.7164/antibiotics.40.296. [DOI] [PubMed] [Google Scholar]
- 17.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276(Macromol Crystallogr Part A) 1997:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 18.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54(Part 5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 19.CCP4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50(Part 5):760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 20.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60(Part 12 and Part 1):2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 22.Ghosh A, Rapp CS, Friesner RA. Generalized born model based on a surface integral formulation. J Phys Chem B. 1998;102:10983–10990. [Google Scholar]


