Abstract
Nitric Oxide Reductase (NOR) is an integral membrane protein performing the reduction of NO to N2O. NOR is composed of two subunits: the large one (NorB) is a bundle of 12 transmembrane helices (TMH). It contains a b type heme and a binuclear iron site, which is believed to be the catalytic site, comprising a heme b and a non-hemic iron. The small subunit (NorC) harbors a cytochrome c and is attached to the membrane through a unique TMH. With the aim to perform structural and functional studies of NOR, we have immunized dromedaries with NOR and produced several antibody fragments of the heavy chain (VHHs, also known as nanobodies™). These fragments have been used to develop a faster NOR purification procedure, to proceed to crystallization assays and to analyze the electron transfer of electron donors. BIAcore experiments have revealed that up to three VHHs can bind concomitantly to NOR with affinities in the nanomolar range. This is the first example of the use of VHHs with an integral membrane protein. Our results indicate that VHHs are able to recognize with high affinity distinct epitopes on this class of proteins, and can be used as versatile and valuable tool for purification, functional study and crystallization of integral membrane proteins.
Keywords: nitric oxide reductase, camelid antibodies, VHH domain, SPR, phage display
Introduction
In the past 12 years, camelid VHH domains (or nanobodies)1,2 have been widely used for basic research studies, as potential reagents3–7 or drugs.5,8–11 A wealth of crystal structures against protein antigens4,12–22 or haptens23,24 has revealed the amazing versatility of their binding modes. They can recognize enzyme clefts (lysozyme, amylase), use a unique CDR for providing interactions, interact with haptens laterally, or use part of their framework for proteins binding (amylase, etc). To date, however, all the complexes reported were from camelid Igs obtained through llama or dromedary immunization with soluble proteins.
During our study of the enzymes from the denitrification cascade, catalyzing the reduction of 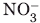 to N2,25 we have undertaken experiments aiming at a better understanding of Nitric Oxide Reductase (NOR), the last of the four enzymes whose structure still remains unknown.26–31 NOR is composed by two subunits: NorB, a 12 transmembrane helix b-type cytochrome harboring two heme groups and a non-heme Fe center, and NorC, a membrane bound mono-hemic c-type cytochrome25 (see Fig. 1). It catalyses the reduction of NO to N2O: 2NO + 2e− + 2H+ → N2O + H2O. NOR resembles strikingly bacterial cytochrome c oxidase (COX), which possess a Cu ion instead of the Fe center32 in the catalytic site (see Fig. 1). NOR and COX sequences are similar: Ps. nautica NOR (the enzyme studied here) large subunit shares 56% sequence identity (72% similarity) with the large subunit of Paracoccus denitrificans COX, of known structure.32 On the basis of this similarity, it has been proposed that they might be evolutionary related.33
to N2,25 we have undertaken experiments aiming at a better understanding of Nitric Oxide Reductase (NOR), the last of the four enzymes whose structure still remains unknown.26–31 NOR is composed by two subunits: NorB, a 12 transmembrane helix b-type cytochrome harboring two heme groups and a non-heme Fe center, and NorC, a membrane bound mono-hemic c-type cytochrome25 (see Fig. 1). It catalyses the reduction of NO to N2O: 2NO + 2e− + 2H+ → N2O + H2O. NOR resembles strikingly bacterial cytochrome c oxidase (COX), which possess a Cu ion instead of the Fe center32 in the catalytic site (see Fig. 1). NOR and COX sequences are similar: Ps. nautica NOR (the enzyme studied here) large subunit shares 56% sequence identity (72% similarity) with the large subunit of Paracoccus denitrificans COX, of known structure.32 On the basis of this similarity, it has been proposed that they might be evolutionary related.33
Figure 1.
Schematic representation of nitric oxide reductase (NOR, upper left) and cytochrome c oxidase (COX, lower left). The heme groups and iron ions are colored red and the copper ion blue. Right side: compact model of Paracoccus denitrificans COX (1AR1).32 The large subunit is green, the cytochrome c small subunit is orange, and the VH domain from the bound Fv is yellow. [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
With in mind to study Ps. nautica NOR structure and function, we have immunized a dromedary with the purified protein. We have obtained six VHHs against this enzyme with four of them displaying Kd values in the nM range. Using Surface Plasmon Resonance (SPR) and electron transfer assays, we could determine that several epitopes have been targeted. These nanobodies are presently used as tools towards Ps. nautica NOR crystallization. Finally, we report here, for the first time, that the complete and efficient procedure for immunization, VHHs selection and purification documented on soluble proteins can be adapted readily to integral membrane proteins.
Results
NOR purification
The NOR two-subunits complex has been successfully extracted and purified from DDM solubilized membranes of Ps. nautica cells (Timóteo CG, et al. in preparation). Calculated from the absorption measurement at 410 nm, this procedure yielded ∼10 mg of purified protein per 200 L of Ps. nautica fermentor culture. Protein content, purity and homogeneity were estimated by SDS-PAGE and MALDI-TOF mass spectrometry. NorB, which is an integral membrane protein with twelve predicted transmembrane helices, appears on gel as a band with an apparent mass of 38 kDa, smaller than the expected MW of 54.4 kDa, and it is not detectable by mass spectrometry, as often noticed with integral membrane proteins. NorC appears on gel as a band of the expected MW (17.6 kDa) and in mass spectrometry as a sharp symmetrical peak at the expected MW, indicating the absence of proteolysis (see Fig. 2). Purified protein in 100 mM K phosphate buffer at pH 7.0 with 0.02% DDM and 0.01% phenyl ethanol could be concentrated up to 30 mg/mL with no evidence of aggregation. The electron transfer activity of purified NOR has been tested both with cytochrome c552 and ascorbate as electron donors (see later).
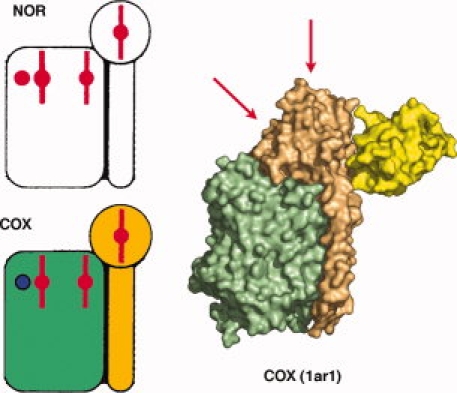
Figure 2.
Characterization of NOR and its complex with VHHn03 a) SDS-PAGE and anti-His Western blot; Lane 1: MW markers, lane 2: purified NOR, lane 3: purified NOR-VHHn03 complex, lane 4: purified VHHn03. b) MALDI-TOF mass spectrometry analysis of NOR-VHHn03 complex.
VHHs selection and production
Dromedary immunization was performed with purified NOR. Lymphocytes were isolated from blood samples, and a phage display library of the VHHs was generated using standard procedures (see Methods). Recombinant antibody fragments were selected from this phage display library by panning against NOR. Six VHHs binders (01, 02, 03, 15, 27, and 41) were obtained and sequenced (see Fig. 3). Binder sequences were aligned with the human POT vH35 and the dromedary VHHLys3, an inhibitor of hen egg-white lysozyme.12 Several features confirm the heavy-chain antibody origin of the antigen-specific binders2: (1) they show substitutions Leu11Ser, Val37Phe/Tyr, Gly44Glu, Leu45Arg, and Trp47Gly, characteristic of VHHs; (2) the three hypervariable regions (CDRs), with the exception of VHHn03, have an average length of the CDR3 loop of 16 amino acids; (3) as frequently observed in VHHs, all NOR binders excepted for VHHn03 and VHHn41 contain two extra-cysteines in CDR1 and CDR3 allowing the formation of an interloop disulfide bond.
Figure 3.

Alignment of VHH amino-acid sequences (lysozyme and NOR binders) and of one vH human variable domain of heavy chain (POT vH). The hypervariable regions (CDR) are shown in red, green and blue. Residues substitutions are in pink, Cys residues involved in an intradomain (C22 and C92) are in orange, extra Cys residues possibly involved in an interloop disulfide bond are in purple. Kabat numbering is used.34 [Color figure can be viewed in the online issue, which is available at www.interscience.wiley.com.]
The six VHHs binders (01, 02, 03, 15, 27, and 41) were expressed in the periplasm of E. coli WK6 as fusion proteins, with a C-terminal His6-tag. After two successive chromatographic steps, Ni affinity column and gel filtration, the VHHs fragments were at least 95% pure, as judged by SDS-PAGE (see Fig. 4). The purification yield varied between 0.5 and 9 mg/L of culture, depending on the clone. All purified proteins were monomeric, soluble and showed the expected molecular mass on SDS-PAGE (see Fig. 4) and by mass spectrometry.
Figure 4.
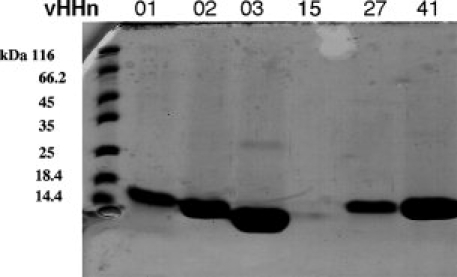
SDS-PAGE of purified VHHn01, 02, 03, 15, 27 and 41 of respectively 15.3, 14.3, 13.5, 14.5, 15.0, and 14.7 kDa (Lane1: MM marker).
NOR-VHH complexes purification and characterization
NOR-VHH complexes were prepared in two different ways. In the first protocol, purified NOR was mixed with the appropriate VHH and separated from the excess of VHH by gel filtration. In the second protocol, the His-tagged recombinant VHHs were used as powerful tool for the preparation and purification of NOR-VHH complexes. A VHH was mixed directly with Ps. nautica solubilized membrane fraction to form the complex, which was further purified in two-steps, Ni affinity-chromatography followed by gel filtration. This last procedure for obtaining the complex is much faster, since NOR has not to be prepurified. Moreover, the yield and purity of the resulting NOR-VHH complexes, analyzed by gel filtration chromatography, SDS-PAGE, Western blot, MALDI-TOF mass spectrometry (see Fig. 2), and HPLC-Wyatt analysis, were very satisfying. The complexes remained monodisperse in DDM buffer for several weeks as judged by analytical gel filtration chromatography on HPLC-Wyatt. This is illustrated in Figure 5 for the mass determination of the complex NOR-VHHn03 in DDM: the total mass is ∼182 kDa. The molecular mass measured for the complex is 87.6 kDa, which is close to the theoretical value of 85.7 kDa (71.8 kDa for NOR and 13.9 kDa for VHHn03), and indicates that NOR is present in solution as a monomer. The mass determined for the detergent is ∼100 kDa (see Fig. 5), indicating that ∼196 molecules of DDM (510.6 Da) maintain NOR in solution. NOR-VHH complexes purified by either method showed identical kinetic behavior (see enzymatic activity).
Figure 5.
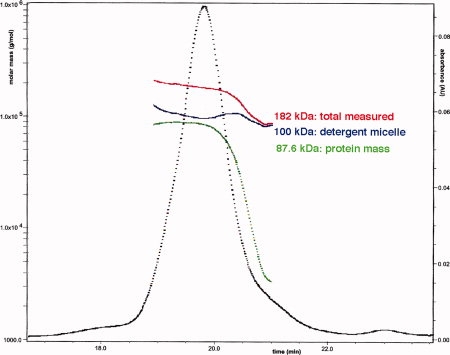
HPLC-Wyatt analysis of NOR-VHHn03 complex. The abscissa indicates the time scale of the HPLC injection, the left ordinate indicates the molar mass in g/mol (Da) and the right ordinate the UV absorbance at 280 nm. Absorption peak is in black and the curves indicating the molar mass for the complex in detergent, the protein alone and the detergent are in red, green, and blue, respectively. [Color figure can be viewed in the online issue, which is available at www.interscience. wiley.com.]
Affinity and epitope mapping of VHHs
We have determined the dissociation constants of the VHH binders for NOR by Surface Plasmon Resonance (SPR, BIAcore) experiments. Different VHHs were covalently linked to a BIAcore CM5 chip by amine coupling. We chose to attach the VHHs to the chip as they are more stable than NOR, making, thus, easy the regeneration procedure with ethylenglycol and/or SDS. Moreover, the larger molecular mass of NOR makes it a more favorable candidate to be used as analyte. In a first step, the equilibrium dissociation constants of five individual VHH binders (01, 02, 03, 27, and 41) for NOR were determined and found to be in the nanomolar range (1–50 nM) (Table I). In a second step, epitope mapping of the five VHHs versus VHHn03 was performed, using as analyte the preformed binary complex of NOR with VHHn03. The binary complex was able to bind only to VHHn41 and VHHn27, with Kd values close to those determined for NOR alone (7 and 5.8 nM for VHHn41 and 11 and 45 nM for VHHn27). These results clearly show that VHHn03, VHHn41, and VHHn27 interact with NOR at independent sites. As a positive control, the preformed ternary complex NOR-VHHn03-VHHn41 was passed as analyte over VHHn27 covalently bound. The quaternary complex is formed; the affinity of VHHn27 for the ternary complex is two times higher than that for the binary complex (5.9 and 11 nM) and one order of magnitude higher than that for NOR alone (5.9 and 45 nM). On the basis of the results of these binding experiments, we decided to use the binary, ternary, and quaternary complexes in further crystallization trials.
Table I.
Kinetic Rate Constants and Equilibrium Dissociation Constant of NOR Antibody Fragments as Determined by BIAcore. Purified NOR or NOR in Complex with VHHn Were Injected over the Sensor Chip Coated with the Corresponding VHHn Fragments
| kon105 M−1s−1 | koff 10−4s−1 | Kdiss 10−9M | |
|---|---|---|---|
| NOR+ | |||
| VHHn01 | 0.7 ± 0.2 | 6.2 ± 1.2 | 9.2 ± 1.1 |
| VHHn02 | 1.4 ± 0.3 | 6.7 ± 1.6 | 5.2 ± 1.7 |
| VHHn03 | 1.5 ± 0.3 | 1.8 ± 0.2 | 1.3 ± 0.25 |
| VHHn41 | 1.1 ± 0.35 | 5.6 ± 2.0 | 5.8 ± 1.6 |
| VHHn27 | 0.3 ± 0.1 | 15 ± 6 | 45 ± 5 |
| NOR-VHHn03+ | |||
| VHHn01 | nc | nc | nc |
| VHHn02 | nc | nc | nc |
| VHHn03 | nc | nc | nc |
| VHHn41 | 1.2 ± 0.4 | 8.3 ± 2.0 | 7.0 ± 1.5 |
| VHHn27 | 2.2 ± 0.1 | 22 ± 1.5 | 11 ± 1.4 |
| NOR-VHHn03-VHHn41+ | |||
| VHHn27 | 7 ± 2 | 39 ± 0.5 | 5.9 ± 1.3 |
nc: not calculable.
Crystallization and X-ray diffraction
Conditions giving rise to diffracting crystals were not found with NOR alone, at least in our hands. On the basis of the experience of other laboratories and confident that complex formation with antibody fragments may facilitate the crystallization of membrane proteins,32,36–38 we systematically used NOR-VHH binary, ternary, and quaternary complexes for crystallization trials. For each complex, excepted for the ternary NOR-VHHn03 + 41, we have obtained crystals, in general after 2 weeks. Average resolution of the crystals was between 12 and 6 Å. A preliminary data set was collected at 7 Å resolution with NOR-VHHn02 crystals at the Swiss Light Source Synchrotron Radiation Facility at the Paul Scherrer Institut (SLS, Switzerland) beam line X10SA (PXII). Crystals belong to space group P21 with cell dimensions a = 85.0 Å, b = 135.3 Å, c = 137.2Å, and β = 95.6°. The Vm values and water percentages calculated for the asymmetric unit are of 4.6 Å2/Da = 73%, 3.1 Å2/Da = 60%, and 2.3 Å2/Da = 47%, for 2, 3, and 4 molecules, respectively.
Enzymatic activity
Reduced Ps. nautica cytochrome c552 or ascorbate/PMS are efficient electron donors for NOR (Timóteo CG, et al., in preparation), and were used to evaluate the activity of NOR in complex with VHHs in comparison to the activity of NOR alone. All complexes are active with ascorbate/PMS, although at different level (see Fig. 6); NOR complexed with VHHn02 and VHHn03 is inactive with reduced cytochrome c552 but active with ascorbate/PMS. The binding of VHHn27 partially affects the kinetic behavior of NOR with c552 as electron donor, while VHHn41 scarcely modifies activity in the presence of either electron donor system.
Figure 6.
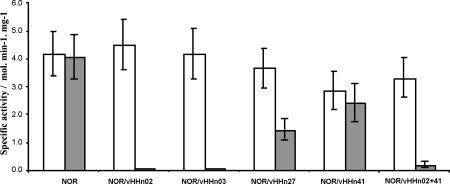
Specific activity plot of different NOR-VHHn complexes using two electron donating systems (white—ascorbate/PMS; grey—reduced Ps. nautica cytochrome c552).
Discussion
We report for the first time camelid immunization and VHHs selection against an integral membrane protein. The VHHs obtained by this means can be used, as Swiss knives, in a variety of situations: difficult protein purification, functional studies, epitope mapping, crystallization.
Using VHHs, we could purify NOR readily in two chromatography steps and reduce the purification time to two days. A similar approach was used for the first time to purify COX with antibody fragments.39 In our previous protocol, three chromatography steps were necessary and the overall process lasted for 5 days. As membrane proteins are often fragile and detergents expensive, a shorter and faster purification may be crucial for obtaining a stable preparation. The idea of using VHHs for protein purification was also proposed recently in the case of soluble proteins.7 As often observed with the VHHs domains, five of the six antigen-specific binders selected by panning could be expressed in E. coli readily and in quantities sufficient for BIAcore binding experiments, crystallization and enzymatic assays. Four of these VHHs have excellent Kd values, ranging from 1.3 to 9.2 nM, while a fifth has a Kd value of 45 nM. The affinity values class these VHHs among the best raised against soluble proteins.4,12–22 The best binder, VHHn03 forms a strong complex with NOR (1.3 nM). When the complex NOR-VHHn03 was passed over the four other VHHs attached to the BIAcore chip, two of them (VHHn01 and 02) did not interact with the complex, indicating that they share the same epitope as VHHn03. In contrast, the two remaining VHHs (VHHn027 and 41) interact with the complex, demonstrating that these three binders have distinct epitopes. It should be noticed that VHHn41 Kd value does not change significantly when binding to NOR alone or to NOR-VHHn03 complex, indicating that VHHn41 should bind to an epitope quite distinct from that of VHHn03. In contrast, when VHHn27 binds to the NOR-VHHn03 complex, the Kd value decreases fourfold as compared to the binding to NOR alone. We propose, therefore, that VHHn27 binds to an epitope distinct but close enough to that of VHHn03, and that the association of VHHn03 to NOR may induce a conformational change favorable to the further binding of VHHn27. The ternary complex NOR-VHHn03-VHHn41 is still able to bind to the VHHn27, demonstrating that VHHn41 and VHHn27 have also distinct epitopes. The Kd of VHHn27 with NOR-VHHn03-VHHn41 is slightly lower (1.7-fold) than with the binary complex, suggesting some limited effect (conformational or steric) of VHHn41 on the subsequent binding by VHHn27.
It is interesting to know the location of the four VHH binders epitopes on NOR. To this end, we measured the electron transfer activity of NOR with ascorbate/PMS or cytochrome c552 as electron donors in presence of four VHHs. When the bulky dimeric cytochrome c55240 is used as electron donor, electron transfer is abolished by VHHn02 and VHHn03. This means that the epitope common to both VHHs coincides with the reduced cytochrome c552 binding site on NorC subunit, and that these VHHs prevent cytochrome c552 to transfer electrons to NOR. In contrast, when the small molecules of ascorbate and PMS are used in the experiments, electron transfer is not altered by any of the VHHs, meaning that they can sneak into the cytochrome domain close to the heme iron. Alternatively, ascorbate and PMS may bypass the cytochrome c552 electron delivery pathway and deliver electrons to the active site through a different mechanism that is not blocked by the VHHs. Torndycroft et al.41 also consider this possibility based on the information that ascorbate/PMS can also effectively deliver electrons to other classes of NORs which lack the c subunit.42
In contrast to VHHn02 and VHHn03, VHHn27 alters partially (by ∼60%) the electron transfer, which suggests that they interact partially with reduced cytochrome c552. BIAcore experiments have confirmed that VHHn27 binding site is different to that common to VHHn02 and VHHn03, the presence of which abolish completely cytochrome c552 binding. Therefore VHHn27 should not interact at all with the VHHn02 or VHHn03, but should interact partially with the large dimeric cytochrome c552 that binds at the same place as VHHn02 or VHHn03.
In conclusion, neither of the small VHHn02 nor VHHn03 molecules interfere with the VHHn27 binding site, while the large dimeric cytochrome c552 does. The binding of VHHn41 does not alter significantly the electron transfer by reduced cytochrome c552. Considering the respective sizes of VHHs and cytochrome c552 and that of the NorC subunit (as estimated from COX in Fig. 1), it is very likely that two or even three VHHs, such as VHHn02, VHHn27, and VHHn41, that we found able to co-crystallize with NOR, can bind simultaneously on NorC subunit (Fig. 1, right). The VHHn41, however, might also bind to a completely different epitope, on the cytochrome c small subunit side, or even internally.
It is well-established that binders favor protein crystallization, particularly that of integral membrane proteins.32,38 In this later case, they may help by providing nondetergent covered protein surfaces, favorable to tight packing, or by stabilizing the flexible membrane protein in a given conformation.38 This has been illustrated with COX, a topological relative of NOR,32 with the β2-adrenergic receptor38 and in several other cases. Here we have shown that all our complexes of VHHs with NOR crystallize readily, while NOR alone does not, despite several years of intensive efforts. However, most crystals diffract to 6 Å at best. We expect that further optimizations will make it possible to get crystals diffracting at higher resolution.
Material and Methods
Materials
Detergents (Anatrace), S-sec-phenyl-ethyl-alcohol (Sigma), DEAE-Biogel A gel ion exchange (Bio-rad), Macroprep Ceramic hydroxyapatite type I and Bio-Scale CHT20-I column (Ceramic hydroxyapatite type I) (BioRad), Ni-NTA column, 6xHis ladder and penta-His-HRP conjugate (Qiagen), ECL chemiluminescent detection system, Superdex 75 column, His-trap crude FF column (GE Healthcare), PVDF membranes (Roche), bicinchoninic acid (BCA) protein assay kit (Pierce), Amicon-Ultra cutoff 10–50 kDa concentrators (Millipore).
Ps. nautica NOR (NORn) production
Ps. nautica cells were grown as described by Prudêncio et al.43 Ps. nautica medium culture contained: 11.7 g/L NaCl, 12.3 g/L MgSO47H2O, 0.75 g/L KCl, 6.05 g/L Tris, 3 g/L NH4Cl, 2 mL sodium lactate 60%, 1 g/L Yeast-Extract, 1 g/L KNO3, 2 mg/L FeSO47H2O, 74.4 mg/L K2HPO4, 1.47 g/L CaCl22H2O, 1 ml/L Starkey oligo-elements stock solution pH7.5. 1000× oligo-elements stock solution: 263 mM MgO, 20 mM CaCO3, 22.3 mM FeSO47H2O, 5 mM ZnSO47H2O, 3.6 mM MnSO4H2O, 0.1 mM CuSO45H2O, 3.2 mM CoSO47H2O, 0.97 mM BO3H3, 0.8 mM Mo7(NH4)6(O2)44H2O, 0.14 mM Ni(NO3)26H2O, 0.12 mM Na2SeO3. A volume of 200 ml of medium, inoculated with a 20 mL Ps. nautica overnight culture, were first grown in bottle under aerobic conditions at 30°C. The 200 ml culture was used to inoculate 2 L of medium and let grown 24 hours under anaerobic conditions to reach an OD600 of 0.8. This 2 L culture was used to inoculate 18 L of medium and let grown 5 hours under aerobic conditions (air supply of 0.2 volume per volume per minute) and 19 hours under anaerobic conditions to reach an OD600 of 0.8. Volume of 200 L of medium in a 300 L working volume fermentor (CHEMAP) was finally inoculated with this 18 L culture and grown again at 30°C 5 hours under aerobic conditions (air supply of 0.2 v/v per minute) and 19 hours under anaerobic conditions. Cells were harvested by centrifugation at 14000g at 4°C under N2 atmosphere (100 L/h). Resulted ∼250 g wet pellet was resuspended in ∼1 L 10 mM Tris pH 7.2, disintegrated (1.8 Kbars) and stored aliquoted at −20°C.
Ps. nautica NOR purification
NOR was purified as described by Timóteo C. G. et al. (manuscript in preparation): 250 mL of defrost Ps nautica fermentor pellet (corresponding to 50 L of culture) were diluted two times in 50 mM Tris pH8.0 at 4°C and ultracentrifuged 1H30 at 40,000 rpm, 4°C. Pellets were resuspended in ∼500 mL 50 mM Tris pH 8.0, 100 mM KCl with a Potter-S homogenizer (Braun). Protein concentration was quantified following the BCA method using bovine serum albumin as a standard and brought to a concentration of 10 mg/ml. Resuspended pellets were sonicated, washed two times in 50 mM Tris pH 8.0 by ultracentrifugation 1H30 at 40,000 rpm, 4°C, aliquoted in 3 × 250 mL at ∼10 mg/ml and stored at −80°C. Solubilization of membranes from 250 mL defrost sample was done by addition of 0.6% (w/v) n-dodecyl-β-D-maltopyranoside (DDM) and 0.01% (v/v) S-sec-phenyl-ethyl alcohol (phenyl ethanol) 15 min at 4°C under agitation and membrane proteins were recovered in supernatant after ultracentrifugation 1H30 at 40,000 rpm, 4°C. Sample was applied on DEAE column equilibrated in 50 mM Tris pH 8.0, 0.02% DDM, 0.01% phenyl ethanol. After washing the column, elution of NOR protein by a linear gradient from 0 to 500 mM NaCl was followed spectrophotometrically at 410 and 278 nm absorbance. Eluate was buffer exchanged and concentrated in 10 mM K phosphate pH 7.0, 400 mM NaCl, 0.02% DDM, 0.01%, phenyl ethanol (buffer A), aliquoted in 3 × 20 mL and stored at −80°C. A 20 mL defrost sample was brought to 0.1% DDM and centrifuged 15 min at 7000g before being applied on a Macroprep hydroxyapatite column equilibrated in buffer A. A gradient from 0 to 100% of buffer B (1.5M K phosphate pH 7.0, 0.02% DDM, 0.01% phenyl ethanol) was applied to the column and eluted fractions were pooled, buffer exchanged and concentrated in buffer A. Pool was brought again to 0.1% DDM and centrifuged 15 min at 7000g before being applied on a Bio-Scale hydroxyapatite column equilibrated in buffer A. A gradient from 0 to 100% of buffer B was applied to the column. Eluted fractions were pooled, buffer exchanged and concentrated in 100 mM K phosphate pH 7.0, 0.02% DDM, 0.01% phenyl ethanol. Purified NOR concentration was determined spectrophotometrically from absorbance at 410 nm.
Dromedary immunization, VHH library construction, and binders selection
A dromedary was injected subcutaneously on days 0, 7, 14, 21, 28, 35 with about 500 μg (per injection) of purified NOR in Gerbu adjuvant. On day 39, anticoagulated blood was collected from the vein for the preparation of lymphocytes.
The lymphocytes from the ‘immune’ blood were purified on Lymphoprep. The total RNA from the peripheral blood lymphocytes was extracted and used as template for first strand cDNA synthesis with oligo (dT) primer. Using this cDNA, the VHH encoding sequences were amplified by PCR according to Saerens et al.,44 digested with PstI and NotI, and cloned into the PstI and NotI sites of the phage-display phagemid vector pHEN4.45 A VHH library of about 4 × 107 independent transformants was obtained. About 82% of these transformants harbored the vector with an insert corresponding to the expected VHH gene size. Three consecutive rounds of phage display and panning (1011 phages per well of microtiter plate) were performed on solid-phase coated NOR (100 μg/mL, 10 μg/well). The enrichment for antigen-specific phages within the pools at each round of panning was assessed by polyclonal phage ELISA. A clear enrichment was seen after the second and third round of panning. Totally 96 individual colonies (48 from 2nd round and 48 from third round) were randomly selected and analyzed by ELISA for the presence of antigen specific VHHs in their periplasmic extracts. The VHH sequences from the ELISA-positive colonies were subject to RFLP analysis using HinfI restriction enzyme and nucleotide sequencing. Seven different nanobodies were identified.
VHHs expression and purification
VHH genes of the binders were sequenced and subcloned in pHEN6 vector in fusion with pelB sequence on N-ter and six histidine tag on C-ter.46 Escherichia coli WK6 cells transformed with the appropriate vector were grown at 37°C in Terrific Broth medium containing 100 μg/mL ampicillin and 0.1% glucose until optical density reached 0.8. Expression was induced by the addition of 1 mM IPTG and growth was continued for an additional 16 hours at 28°C. Periplasmic fraction containing the VHH fragments was prepared.47 The His-tail-containing fusion protein was purified by immobilized metal affinity chromatography on a 5 mL Ni-NTA column in 50 mM Na/K phosphate pH 8.0, 300 mM NaCl, 10% glycerol. Eluted fractions in 250 mM imidazole were concentrated on a Amicon-Ultra 10 kDa cutoff concentrator prior to be loaded on a HiLoad 10/30 Superdex75 PG gel filtration column in PBS. Protein concentration of the VHHs was determined by UV spectrometry from absorbance at 278 nm, using their calculated extinction coefficient.
NOR-VHHs complexes purification
NOR–VHH complexes were prepared following two different procedures.
Purified NOR in 100 mM KPB (potassium phosphate buffer) pH 7.0, 0.02% DDM, 0.01%, phenyl ethanol (PE) was incubated with pure VHH protein in PBS pH 7.2, for 1 h on ice, in a 1:1.2 molar ratio. NOR-VHH complex was purified and separated from free antibody excess by gel filtration chromatography on a Superdex75 column in 100 mM KPB pH 7.0, 0.005% (w/v) DDM, 0.01% (v/v) PE.
Solubilized protein from membrane fraction (see NOR purification (Timóteo CG, et al. in preparation) was dialyzed against 100 mM KPB pH 7.0, 0.02% DDM, 0.01% PE and incubated with pure VHH (in the same buffer), in 1:1.2 ratio for 1 h. The mixture was applied onto a His-trap FF crude column (GE Healthcare) equilibrated in the same buffer. The column was washed with 60 mM imidazole and eluted with 250 mM imidazole in the same buffer. Eluted fractions were pooled and concentrated. When necessary the complex was mixed with one or two additional distinct VHH. NOR-VHH complex was separated from free antibody excess by gel filtration chromatography as described in 1. NOR-VHH concentration was determined using NOR extinction coefficients (ɛ411 nm = 295 mM−1 cm−1) (Timóteo CG, et al. in preparation) and purity was evaluated by SDS-PAGE.
NOR-VHHs complexes characterization
Western blot
Proteins were transferred from a 15% SDS-PAGE gel to a Hybond PVDF membrane and blocked with 5% nonfat dried milk in 20 mM Tris-HCl pH 7.6, 150 mM NaCl, 0.1% Tween 20 at 4°C. The membrane was then treated with anti His-HRP primary antibodies. Visualization of VHH-specific bands was done by ECL chemiluminiscence.
Mass spectrometry
Mass spectrometry was performed on a matrix-assisted laser desorption/ionization time of flight (MALDI-TOF) mass spectrometer (Bruker Autoflex, Bruker Daltonics, Bremen, Germany) according to standard procedures, slightly modified for membrane proteins.
MALS/UV/RI (Wyatt)
Size exclusion chromatography was performed on an Alliance 2695 HPLC system (Waters) using a Silica Gel KW804 column (Shodex) eluted with 50 mM Tris-HCl pH 8, 300 mM NaCl, 0.05% DDM at a flow rate of 0.5 mL/min. 30 μL sample between 0.5 and 1 mg/mL was routinely applied on the column. Detection was performed using a triple-angle light scattering detector (Mini-DAWN™ TREOS, Wyatt technology, Sanata Barbara, USA). Molecular weight determination was performed by ASTRA V software (Wyatt technology).
SPR affinity measurements
Kinetic analysis of NOR-VHHs interactions were performed on BIAcore 1000 using CM5 (carboxymethylated dextran) sensor chip coated with 500–900 resonance units of individual purified VHH fragments immobilized by amine coupling. VHHs non-specific for NOR were used as control. For the kinetic constants determination, diluted solutions of NOR or NOR in complex with VHHs (0.3–40 nM in 10 mM Hepes pH 7.2, 150 mM NaCl, 2 mM EDTA, 0.005% surfactant P20) were passed over the flow-cell with the specific VHHs and on the control flow-cell. Binding traces were recorded for at least five concentrations of analyte, in triplicate. Each binding-regeneration cycle was performed at room temperature with a constant flow rate of 40 μL/min. In each cycle, buffer (50 μL) was injected first to stabilize the baseline, the analyte (80 μL) was then injected and the spontaneous dissociation was followed for 6 min; regeneration of the binding surface was achieved by injection of 5 μL of ethylenglycol and/or 0.5% SDS and further washing with the running buffer for 6 min. Curves obtained after subtraction of the reference and the buffer signals were fitted to a (1:1) Langmuir binding model and a (1:1) binding with mass transfer with BIAevaluation (BIAcore). Better results, in terms of chi2 and standard error of the calculated values, were obtained with the first model.
Crystallization and X-ray diffraction
Initial crystallization screening of NOR-VHHs complexes were performed with commercial kits MemStart, MemSys, MemGold I and II, MemPlus (Molecular Dimensions Ltd), MB I and II (QIAGE). The nanodrops experiments were carried out with a Cartesian HoneyBee X8 robot, using sitting-drops in Greiner plates (100–200-300) nL of protein mixed with 100 nL of well solution). In the first round of crystallization trials, the concentration of binary (NOR-VHHn02, NOR-VHHn03, NOR-VHHn27 and NOR-VHHn41), ternary (NOR-VHHn03 + 27 and NOR-VHHn03 + 41) and quaternary (NOR-VHHn03 + 27 + 41) complex varied between 3 and 12 mg/mL in the purification buffer 100 mM potassium phosphate pH 7.0, 0.01% phenyl ethanol, 0.005% DDM.
Kinetic activity assays
NO reductase activity measurements were performed as described by Timóteo CG, et al. (manuscript in preparation). Purified NOR was incubated for 30 min on ice with each or mixtures of VHHs in equimolar ratio (1:1). After incubation, the complexes were assayed for NO reductase activity with ascorbate/PMS or reduced Ps. nautica cytochrome c552 as electron donors.
Acknowledgments
The technical assistance of Céline Huyghe is acknowledged, and that of Marielle Bauzan (IBSM, Marseille) who performed Ps. nautica cells production. We thank Giuliano Sciara and David Veesler for their help with the Wyatt instrument, Kieron Brown for first crystallization of NOR, Laurent Gautier and Christophe Quetard for their help in BIAcore experiments.
Glossary
Abbreviations:
- cAb
camel antibody
- CDR
complementary determining regions
- DDM
n-dodecyl-ß-D-maltopyranoside
- IPTG
isopropyl-(-D-thiogalactopyranoside
- NOR
Nitric Oxide Reductase consists in a complex of NorB and NorC
- PBS
phosphate saline buffer
- Ps. nautica
Pseudomonas nautica
- SPR
Surface Plasmon Resonance
- vH
variable heavy chain domain of antibody
- VHH
variable heavy chain domain of camel heavy chain antibody.
References
- 1.Hamers-Casterman C, Atarhouch T, Muyldermans S, Robinson G, Hamers C, Songa EB, Bendahman N, Hamers R. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 2.Muyldermans S, Cambillau C, Wyns L. Recognition of antigens by single-domain antibody fragments: the superfluous luxury of paired domains. Trends Biochem Sci. 2001;26:230–235. doi: 10.1016/s0968-0004(01)01790-x. [DOI] [PubMed] [Google Scholar]
- 3.Dumoulin M, Last AM, Desmyter A, Decanniere K, Canet D, Larsson G, Spencer A, Archer DB, Sasse J, Muyldermans S, Wyns L, Redfield C, Matagne A, Robinson CV, Dobson CM. A camelid antibody fragment inhibits the formation of amyloid fibrils by human lysozyme. Nature. 2003;424:783–788. doi: 10.1038/nature01870. [DOI] [PubMed] [Google Scholar]
- 4.Dolk E, van der Vaart M, Lutje Hulsik D, Vriend G, de Haard H, Spinelli S, Cambillau C, Frenken L, Verrips T. Isolation of llama antibody fragments for prevention of dandruff by phage display in shampoo. Appl Environ Microbiol. 2005a;71:442–450. doi: 10.1128/AEM.71.1.442-450.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison RA, Hasson SS, Harmsen M, Laing GD, Conrath K, Theakston RD. Neutralisation of venom-induced haemorrhage by IgG from camels and llamas immunised with viper venom and also by endogenous, non-IgG components in camelid sera. Toxicon. 2005;47:364–368. doi: 10.1016/j.toxicon.2005.10.017. [DOI] [PubMed] [Google Scholar]
- 6.Saerens D, Frederix F, Reekmans G, Conrath K, Jans K, Brys L, Huang L, Bosmans E, Maes G, Borghs G, Muyldermans S. Engineering camel single-domain antibodies and immobilization chemistry for human prostate-specific antigen sensing. Anal Chem. 2005;77:7547–7555. doi: 10.1021/ac051092j. [DOI] [PubMed] [Google Scholar]
- 7.Rothbauer U, Zolghadr K, Muyldermans S, Schepers A, Cardoso MC, Leonhardt H. A versatile nanotrap for biochemical and functional studies with fluorescent fusion proteins. Mol Cell Proteomics. 2008;7:282–289. doi: 10.1074/mcp.M700342-MCP200. [DOI] [PubMed] [Google Scholar]
- 8.Conrath KE, Lauwereys M, Galleni M, Matagne A, Frere JM, Kinne J, Wyns L, Muyldermans S. Beta-lactamase inhibitors derived from single-domain antibody fragments elicited in the camelidae. Antimicrob Agents Chemother. 2001;45:2807–2812. doi: 10.1128/AAC.45.10.2807-2812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cortez-Retamozo V, Lauwereys M, Hassanzadeh Gh G, Gobert M, Conrath K, Muyldermans S, De Baetselier P, Revets H. Efficient tumor targeting by single-domain antibody fragments of camels. Int J Cancer. 2002;98:456–462. doi: 10.1002/ijc.10212. [DOI] [PubMed] [Google Scholar]
- 10.Aires da Silva F, Santa-Marta M, Freitas-Vieira A, Mascarenhas P, Barahona I, Moniz-Pereira J, Gabuzda D, Goncalves J. Camelized rabbit-derived VH single-domain intrabodies against Vif strongly neutralize HIV-1 infectivity. J Mol Biol. 2004;340:525–542. doi: 10.1016/j.jmb.2004.04.062. [DOI] [PubMed] [Google Scholar]
- 11.Harmsen MM, van Solt CB, van Zijderveld-van Bemmel AM, Niewold TA, van Zijderveld FG. Selection and optimization of proteolytically stable llama single-domain antibody fragments for oral immunotherapy. Appl Microbiol Biotechnol. 2006;72:544–551. doi: 10.1007/s00253-005-0300-7. [DOI] [PubMed] [Google Scholar]
- 12.Desmyter A, Transue TR, Ghahroudi MA, Thi MH, Poortmans F, Hamers R, Muyldermans S, Wyns L. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 13.Spinelli S, Frenken L, Bourgeois D, de Ron L, Bos W, Verrips T, Anguille C, Cambillau C, Tegoni M. The crystal structure of a llama heavy chain variable domain. Nat Struct Biol. 1996;3:752–757. doi: 10.1038/nsb0996-752. [DOI] [PubMed] [Google Scholar]
- 14.Decanniere K, Desmyter A, Lauwereys M, Ghahroudi MA, Muyldermans S, Wyns L. A single-domain antibody fragment in complex with RNase A: non-canonical loop structures and nanomolar affinity using two CDR loops. Structure. 1999;7:361–370. doi: 10.1016/s0969-2126(99)80049-5. [DOI] [PubMed] [Google Scholar]
- 15.Decanniere K, Transue TR, Desmyter A, Maes D, Muyldermans S, Wyns L. Degenerate interfaces in antigen-antibody complexes. J Mol Biol. 2001;313:473–478. doi: 10.1006/jmbi.2001.5075. [DOI] [PubMed] [Google Scholar]
- 16.Desmyter A, Decanniere K, Muyldermans S, Wyns L. Antigen specificity and high affinity binding provided by one single loop of a camel single-domain antibody. J Biol Chem. 2001;276:26285–26290. doi: 10.1074/jbc.M102107200. [DOI] [PubMed] [Google Scholar]
- 17.Desmyter A, Spinelli S, Payan F, Lauwereys M, Wyns L, Muyldermans S, Cambillau C. Three camelid VHH domains in complex with porcine pancreatic alpha-amylase. Inhibition and versatility of binding topology. J Biol Chem. 2002;277:23645–23650. doi: 10.1074/jbc.M202327200. [DOI] [PubMed] [Google Scholar]
- 18.Renisio JG, Perez J, Czisch M, Guenneugues M, Bornet O, Frenken L, Cambillau C, Darbon H. Solution structure and backbone dynamics of an antigen-free heavy chain variable domain (VHH) from Llama. Proteins. 2002;47:546–555. doi: 10.1002/prot.10096. [DOI] [PubMed] [Google Scholar]
- 19.Loris R, Marianovsky I, Lah J, Laeremans T, Engelberg-Kulka H, Glaser G, Muyldermans S, Wyns L. Crystal structure of the intrinsically flexible addiction antidote MazE. J Biol Chem. 2003;278:28252–28257. doi: 10.1074/jbc.M302336200. [DOI] [PubMed] [Google Scholar]
- 20.Spinelli S, Desmyter A, Frenken L, Verrips T, Tegoni M, Cambillau C. Domain swapping of a llama VHH domain builds a crystal-wide beta-sheet structure. FEBS Lett. 2004;564:35–40. doi: 10.1016/S0014-5793(04)00304-7. [DOI] [PubMed] [Google Scholar]
- 21.Dolk E, van Vliet C, Perez JM, Vriend G, Darbon H, Ferrat G, Cambillau C, Frenken LG, Verrips T. Induced refolding of a temperature denatured llama heavy-chain antibody fragment by its antigen. Proteins. 2005;59:555–564. doi: 10.1002/prot.20378. [DOI] [PubMed] [Google Scholar]
- 22.Spinelli S, Desmyter A, Verrips CT, de Haard HJ, Moineau S, Cambillau C. Lactococcal bacteriophage p2 receptor-binding protein structure suggests a common ancestor gene with bacterial and mammalian viruses. Nat Struct Mol Biol. 2006;13:85–89. doi: 10.1038/nsmb1029. [DOI] [PubMed] [Google Scholar]
- 23.Spinelli S, Frenken LG, Hermans P, Verrips T, Brown K, Tegoni M, Cambillau C. Camelid heavy-chain variable domains provide efficient combining sites to haptens. Biochemistry. 2000;39:1217–1222. doi: 10.1021/bi991830w. [DOI] [PubMed] [Google Scholar]
- 24.Spinelli S, Tegoni M, Frenken L, van Vliet C, Cambillau C. Lateral recognition of a dye hapten by a llama VHH domain. J Mol Biol. 2001;311:123–129. doi: 10.1006/jmbi.2001.4856. [DOI] [PubMed] [Google Scholar]
- 25.Einsle O, Kroneck PM. Structural basis of denitrification. Biol Chem. 2004;385:875–883. doi: 10.1515/BC.2004.115. [DOI] [PubMed] [Google Scholar]
- 26.Fulop V, Moir JW, Ferguson SJ, Hajdu J. The anatomy of a bifunctional enzyme: structural basis for reduction of oxygen to water and synthesis of nitric oxide by cytochrome cd1. Cell. 1995;81:369–377. doi: 10.1016/0092-8674(95)90390-9. [DOI] [PubMed] [Google Scholar]
- 27.Nurizzo D, Cutruzzola F, Arese M, Bourgeois D, Brunori M, Cambillau C, Tegoni M. Conformational changes occurring upon reduction and NO binding in nitrite reductase from Pseudomonas aeruginosa. Biochemistry. 1998;37:13987–13996. doi: 10.1021/bi981348y. [DOI] [PubMed] [Google Scholar]
- 28.Brown K, Djinovic-Carugo K, Haltia T, Cabrito I, Saraste M, Moura JJG, Moura I, Tegoni M, Cambillau C. Revisiting the catalytic CuZ cluster of nitrous oxide (N2O) reductase - Evidence of a bridging inorganic sulfur. J Biol Chem. 2000;275:41133–41136. doi: 10.1074/jbc.M008617200. [DOI] [PubMed] [Google Scholar]
- 29.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NC. Insights into the respiratory electron transfer pathway from the structure of nitrate reductase A. Nat Struct Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 30.Haltia T, Brown K, Tegoni M, Cambillau C, Saraste M, Mattila K, Djinovic-Carugo K. Crystal structure of nitrous oxide reductase from Paracoccus denitrificans at 1.6 angstrom resolution. Biochem J. 2003;369:77–88. doi: 10.1042/BJ20020782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jormakka M, Richardson D, Byrne B, Iwata S. Architecture of NarGH reveals a structural classification of Mo-bisMGD enzymes. Structure. 2004;12:95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 32.Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- 33.Forte E, Urbani A, Saraste M, Sarti P, Brunori M, Giuffre A. The cytochrome cbb3 from Pseudomonas stutzeri displays nitric oxide reductase activity. Eur J Biochem. 2001;268:6486–6491. doi: 10.1046/j.0014-2956.2001.02597.x. [DOI] [PubMed] [Google Scholar]
- 34.Kabat EA, Wu TT, Perry HM, Gottesmann KS, Foeller C. NH Publication no. 91-3242. 5th Ed. U.S. Department of Health and Human Services; 1991. [Google Scholar]
- 35.Fan SF, Kao CY. Three-dimensional structure of an Fv from a human IgM immunoglobulin. Eur J Pharmacol. 1992;229:259–263. [Google Scholar]
- 36.Hunte C, Michel H. Crystallisation of membrane proteins mediated by antibody fragments. Curr Opin Struct Biol. 2002;12:503–508. doi: 10.1016/s0959-440x(02)00354-8. [DOI] [PubMed] [Google Scholar]
- 37.Rothlisberger D, Pos KM, Pluckthun A. An antibody library for stabilizing and crystallizing membrane proteins selecting binders to the citrate carrier CitS. FEBS Lett. 2004;564:340–348. doi: 10.1016/S0014-5793(04)00359-X. [DOI] [PubMed] [Google Scholar]
- 38.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, Schertler GF, Weis WI, Kobilka BK. Crystal structure of the human beta2 adrenergic G-protein-coupled receptor. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 39.Kleymann G, Ostermeier C, Ludwig B, Skerra A, Michel H. Engineered Fv fragments as a tool for the one-step purification of integral multisubunit membrane protein complexes. Biotechnology (N Y) 1995;13:155–160. doi: 10.1038/nbt0295-155. [DOI] [PubMed] [Google Scholar]
- 40.Brown K, Nurizzo D, Besson S, Shepard W, Moura J, Moura I, Tegoni M, Cambillau C. MAD structure of Pseudomonas nautica dimeric cytochrome c(552) mimicks the c(4) dihemic cytochrome domain association. J Mol Biol. 1999;289:1017–1028. doi: 10.1006/jmbi.1999.2838. [DOI] [PubMed] [Google Scholar]
- 41.Thorndycroft FH, Butland G, Richardson DJ, Watmough NJ. A new assay for nitric oxide reductase reveals two conserved glutamate residues form the entrance to a proton-conducting channel in the bacterial enzyme. Biochem J. 2007;401:111–119. doi: 10.1042/BJ20060856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cramm R, Pohlmann A, Friedrich B. Purification and characterization of the single-component nitric oxide reductase from Ralstonia eutropha H16. FEBS Lett. 1999;460:6–10. doi: 10.1016/s0014-5793(99)01315-0. [DOI] [PubMed] [Google Scholar]
- 43.Prudêncio M, Pereira AS, Tavares P, Besson S, Cabrito I, Brown K, Samyn B, Devreese B, Van Beeumen J, Rusnak F, Fauque G, Moura JJ, Tegoni M, Cambillau C, Moura I. Purification, characterization, and preliminary crystallographic study of copper-containing nitrous oxide reductase from Pseudomonas nautica 617. Biochemistry. 2000;39:3899–3907. doi: 10.1021/bi9926328. [DOI] [PubMed] [Google Scholar]
- 44.Saerens D, Kinne J, Bosmans E, Wernery U, Muyldermans S, Conrath K. Single domain antibodies derived from dromedary lymph node and peripheral blood lymphocytes sensing conformational variants of prostate-specific antigen. J Biol Chem. 2004;279:51965–51972. doi: 10.1074/jbc.M409292200. [DOI] [PubMed] [Google Scholar]
- 45.Arbabi Ghahroudi M, Desmyter A, Wyns L, Hamers R, Muyldermans S. Selection and identification of single domain antibody fragments from camel heavy-chain antibodies. FEBS Lett. 1997;414:521–526. doi: 10.1016/s0014-5793(97)01062-4. [DOI] [PubMed] [Google Scholar]
- 46.Lauwereys M, Arbabi Ghahroudi M, Desmyter A, Kinne J, Holzer W, De Genst E, Wyns L, Muyldermans S. Potent enzyme inhibitors derived from dromedary heavy-chain antibodies. EMBO J. 1998;17:3512–3520. doi: 10.1093/emboj/17.13.3512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skerra A, Pluckthun A. Assembly of a functional immunoglobulin Fv fragment in Escherichia coli. Science. 1988;240:1038–1041. doi: 10.1126/science.3285470. [DOI] [PubMed] [Google Scholar]


