Abstract
Fn14 is the smallest member of the tumor necrosis factor (TNF) receptor superfamily, and specifically binds to its ligand, TWEAK (TNF-like weak inducer of apoptosis), which is a member of the TNF superfamily. The receptor-ligand recognition between Fn14 and TWEAK induces a variety of cellular processes for tissue remodeling and is also involved in the pathogenesis of some human diseases, such as cancer, chronic autoimmune diseases, and acute ischaemic stroke. The extracellular ligand-binding region of Fn14 is composed of 53 amino acid residues and forms a single, cysteine-rich domain (CRD). In this study, we determined the solution structure of the Fn14 CRD (Glu28-Ala70) by heteronuclear NMR, with a 13C-/15N-labeled sample. The tertiary structure of the CRD comprises a β-sheet with two strands, followed by a 310 helix and a C-terminal α-helix, and is stabilized by three disulfide bonds connecting Cys36-Cys49, Cys52-Cys67, and Cys55-Cys64. Comparison of the disulfide bond connectivities and the tertiary structures with those of other CRDs revealed that the Fn14 CRD is similar to the fourth CRD of TNF receptor 1 (A1-C2 module type), but not to the CRD of B-cell maturation antigen and the second CRD of transmembrane activator and CAML (calcium modulator and cyclophilin ligand) interactor (A1-D2 module type). This is the first structural report about the A1-C2 type CRD that could bind to the known target.
Keywords: solution structure, fibroblast growth factor-inducible 14 (Fn14), cysteine-rich domain (CRD)
Introduction
The tumor necrosis factor (TNF) superfamily (TNFSF) and the TNF receptor (TNFR) superfamily (TNFRSF) currently include 19 ligands and 28 receptors, respectively.1,2 The TNF-like weak inducer of apoptosis (TWEAK) has recently been identified as a ligand member of the TNFSF,3,4 and Fn14 (fibroblast growth factor-inducible 14; TWEAK receptor) is considered to be a distant relative of the TNFRSF.5–7 TWEAK and Fn14 specifically bind to each other, with an affinity constant Kd of ∼0.8–2.4 nM.7,8 Conversely, Fn14 is not activated by any other known ligand members in the TNFSF, and TWEAK does not bind to any other known receptor members in the TNFRSF.7 There is a growing body of evidence indicating that the interaction between TWEAK and Fn14 plays a broad spectra of roles in cell proliferation, cell death, apoptosis, angiogenesis, inflammation, and tissue remodeling.9–13
Human Fn14 consists of 129 amino acid residues, and it is the smallest member of the TNFRSF identified so far. The extracellular ligand-binding region of Fn14 is composed of 53 amino acid residues, which form a single cysteine-rich domain (CRD).5,6 Generally, all members of the TNFRSF contain one or more CRDs, and several structural studies on the CRDs of the TNFRSF members have been reported.2,14 For example, an X-ray crystallographic study of TNFR1 revealed the structural features of its four tandemly linked CRDs. Namely, each of the four CRDs in TNFR1 has six conserved Cys residues, forming three disulfide bonds. Based on the amino acid sequence alignment and the structural information, each CRD is considered to be composed of two structural modules. The module types are designated by a letter (A, B, C, and D for modules of known structure) and by a numeral indicating the number of disulfide bonds within the module.2,14,15 Accordingly, the first three CRDs of TNFR1 are designated as A1-B2 modules, and the fourth CRD (TNFR1-d4) is an A1-C2 module. At first, the Fn14 CRD, the BCMA (B-cell maturation antigen) CRD (BCMA-d), and the second CRD of transmembrane activator and CAML (calcium modulator and cyclophilin ligand) interactor (TACI) (TACI-d2) were considered as A1-C2 modules, on the basis of their sequence similarity to TNFR1-d4 (Fig. 1). However, subsequent structural studies of BCMA-d and TACI-d2 revealed that the pattern of their disulfide bond connectivities differs from that observed in TNFR1-d4. Thereafter, BCMA-d and TACI-d2 were classified as the A1-D2 module type.15 On the other hand, mass spectrometry following partial reduction and alkylation recently indicated that the disulfide bond connectivity pattern of the Fn14 CRD is similar to that of the A1-C2 module in TNFR1-d4 (Fig. 1).17
Figure 1.
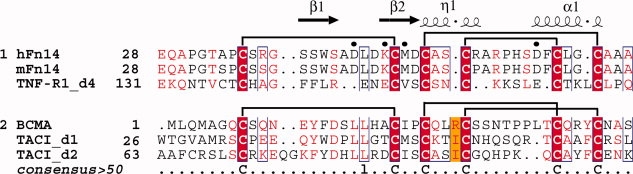
Structure-based multiple sequence alignment of the CRDs of the A1-C2 and A1-D2 modules of the TNFRSF members. Cys residues are highlighted in red and their disulfide bond connectivities are shown above the sequence alignment. Labels and elements of the secondary structure of the Fn14 CRD are reported above the aligned sequences. The crucial residues reported by mutagenesis experiments are marked by dots above the aligned sequence. The sequence alignment output was made with the ESPript program.16
Until now, two types of binding modes between the TNF receptors and the corresponding ligands have been elucidated from complex structures.18 One is observed in the complexes between TNFβ and TNFR119 and between TNF-related apoptosis inducing ligand and death receptor 5.20 The interaction with one ligand molecule is mediated by more than one CRD (of A1-B2 module type) in the receptor molecule. The other binding mode was found in BAFF (B-cell activation factor of the TNF family), complexed with BCMA-d or BAFF receptor15,21 as well as in a proliferation-inducing ligand, complexed with BCMA-d or TACI-d2.22 In these cases, only one CRD in the receptor molecule is responsible for the ligand-receptor recognition in a one-to-one manner, and the complex was formed with 3:3 stoichiometry, as the signaling unit. The CRDs that are involved in this interaction mode differ from the A1-B2 module type; most of them are of the A1-D2 module type. Considering that Fn14 has only one CRD, which is not the A1-B2 module type, the interaction mode between Fn14 and TWEAK is likely to be of the second type despite the lack of structural information for TWEAK and Fn14. Thus, the structural comparison between the Fn14 CRD and the A1-D2 module type CRDs would be interesting for elucidating the specificity between the TNFRSF and the TNFSF.
To gain insight into the structure-function relationship of the Fn14 CRD and its specific recognition of its ligand, TWEAK, we have determined the solution structure of the human Fn14 CRD, which revealed that it forms an A1-C2 module. We discuss the binding mode of the A1-C2 module structure of the Fn14 CRD in comparison with the known A1-D2 module structures in their complexes.
Results and Discussion
Resonance assignments and structure calculation
The assignments of the backbone and the nonlabile side chain resonances for Fn14 CRD were complete, except for the amide proton of Arg38 and all of the side chain protons of Ser37. The imide groups of the asparagine and glutamine residues were assigned, except for Gln21 and Gln44. For all of the Xxx-Pro bonds in Fn14 CRD, the trans conformation was confirmed independently by the intense Xxx (Hα)-Pro (Hδ) sequential nuclear Overhauser enhancement (NOE) spectroscopy (NOESY) cross peaks and by the 13Cβ and 13Cγ chemical shift differences.23 The assignments were confirmed by tracing the dNN, dαN, and dβN connectivities in NOESY spectra.24
Among the 1136 cross peaks identified in the 15N- and 13C-edited 3D NOESY spectra, 99% were assigned by the program CYANA 2.1,25 resulting in a total of 579 nonredundant NOE restraints, including 152 long-range distance restraints for the structure calculation with CYANA 2.1.25 The disulfide bonds Cys36-Cys49, Cys52-Cys67, and Cys55-Cys64, which were already reported,17 were also included in the calculation. The oxidation state of the six Cys residues was confirmed by the chemical shifts of their 13Cβ atoms, which were shifted to ∼40 ppm, implying that the sulfur groups are involved in disulfide bridges.26 In addition, the NOEs between the βHi-S-S-βHj protons were used to confirm the disulfide bond connectivities.
We have determined the solution structure of the human Fn14 CRD, spanning the region (Glu28-Ala70). The structure is well defined and shows excellent agreement with the experimental data. The precision of the structure is characterized by RMSD values to the mean coordinates of 0.11 Å for the backbone and 0.73 Å for all heavy atoms of residues 35–68, excluding the unstructured regions (Glu28-Thr33 and Ala69-Ala70) [Fig. 2(A)]. The NMR assembly of the 20 best conformers and the lowest target function structure are presented in Figure 2. The quality of the structure is also reflected by the fact that less than 1% of the (φ, ψ) backbone torsion angle pairs in the structured region were found in the generously allowed and disallowed regions of the Ramachandran plot, according to the program PROCHECK-NMR.27 The statistics regarding the quality and precision of the best 20 conformers that represent the solution structure of the human Fn14 CRD are summarized in Table I.
Figure 2.
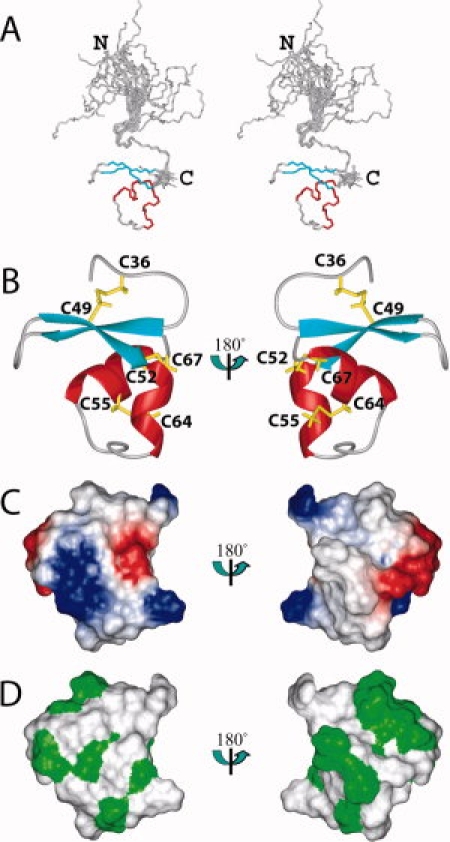
Solution structure of the CRD of human Fn14. (A) Stereo-view of the best 20 structures of the CRD (residues Glu28-Ala70), calculated by CYANA2.1. The helices, the β-sheet, and the loop regions are shown in red, cyan, and gray, respectively. (B) Ribbon presentation of the lowest energy structure of the Fn14 CRD (core part: residues Ala34- Ala69). The colors for the CRD are the same as in A. The side chains of the disulfide bonds are shown in yellow. (C) Electrostatic surface presentation of the CRD domain. The blue and red colors represent positive and negative electrostatic surface potential, respectively. (D) Hydrophobic surface presentation of the CRD domain. The ribbon diagram, electrostatic potential, and hydrophobic surface presentations have the same orientation.
Table I.
Statistics of the 20 Final Solution Structures of the CRD from Human Fn14
| Conformational restraints | |
| Distance restraints | |
| Total NOE | 579 |
| Intraresidue | 133 |
| Sequential (|i − j| = 1) | 161 |
| Medium-range (1 < |i − j| < 5) | 133 |
| Long-range (|i − j ≥ 5) | 152 |
| Disulfide bond restraints (upper limit/lower limit) | 9/9 |
| Hydrogen bond restraints (upper limit/lower limit) | 4/4 |
| φ/ψ dihedral angle restraints (TALOS) | 19/19 |
| Structure statistics | |
| CYANA target function (Å2) | 0.05 |
| Residual distance restraint violations | |
| Maximum (Å) | 0.04 |
| Residual dihedral angle restraint violations | |
| Maximum (°) | 0.09 |
| Ramachandran plot statistics for residues 34–69 (%) | |
| Residues in most favored regions | 66.9 |
| Residues in additional allowed regions | 33.0 |
| Residues in generously allowed regions | 0.1 |
| Residues in disallowed regions | 0.0 |
| Average pairwise RMSD for residues 34–69 (Å) | |
| Backbone | 0.11 |
| Heavy atoms | 0.73 |
Overall structure of the Fn14 CRD
The core part of the CRD (Ala34-Ala69) is well converged, and henceforth, we will discuss the core part as the Fn14 CRD, unless otherwise emphasized [Fig. 2(A)]. The tertiary structure of the Fn14 CRD is a small, compact, global fold with three disulfide bonds, and with the connectivity of Cys36-Cys49, Cys52-Cys67, and Cys55-Cys64 [Fig. 2(B)]. In addition, the electrostatic potential surface and the hydrophobic areas of the Fn14 CRD are represented in Figure 2(C,D).
The Fn14 CRD contains two modules. One is a typical A1 module, which is composed of three β-strands. The second and third strands (Ser40-Ser43 and Lys48-Asp51) are connected by a Type I reverse turn and adopt a β-hairpin structure. The first disulfide bond connectivity, from the first to the third strand, which is well conserved in the A1 modules, was also identified. The region immediately following the A1 module corresponds to the C2 module in TNFR1-d4, and contains a short 310 helix (Cys52-Arg56), followed by a loop region and a C-terminal α-helix (Asp62-Cys67). This region has the second and third disulfide bond connectivities (Cys52-Cys67 and Cys55-Cys64). The loop between the two helices is also well converged [Fig. 2(A)]. Consequently, the Fn14 CRD adopts the A1-C2 module structure that was reported for TNFR1-d4.28 However, as no interaction partner has been reported for TNFR1-d4,19 this is the first structural report about the A1-C2 type module, which has the known ligand.
They share the same pattern of disulfide bond connectivities and have similar tertiary structures [Fig. 3(A,B)]. In the case of the Fn14 CRD, Phe63, which is located on the C-terminal α-helix in the C2 module, plays an important role in the formation of the β-sheet structure of the A1 module [Fig. 3(E)], whereas in TNFR1-d4, the corresponding position is occupied by Phe144, which is within the β-sheet structure [Fig. 3(F)]. Therefore, the interaction between the A1 and C2 modules in the Fn14 CRD is more intimate than that observed in TNFR1-d4 [Fig. 3(A,B,D–F,H)].
Figure 3.
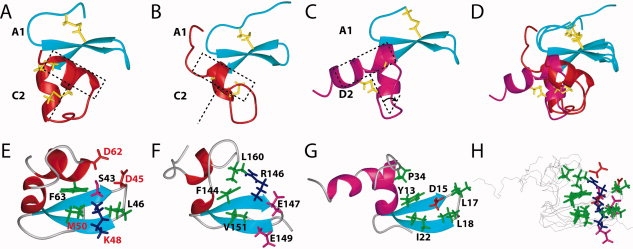
Comparison of the A1-C2 and A1-D2 module structures. (A) Ribbon diagram of the A1-C2 module structure of the Fn14 CRD, with disulfide bonds in yellow. (B) Ribbon diagram of the A1-C2 module structure of the TNFR1-d4, with disulfide bonds in yellow. In A and B, the A1 and C2 modules are shown in cyan and red, respectively. (C) Ribbon diagram of the A1-D2 module structure of the BCMA CRD, with disulfide bonds in yellow. The A1 and C2 modules are colored cyan and magenta, respectively. In A, B, and C, the change of the C-terminal α-helix location between the A1-C2 and A1-D2 module structures approximately corresponds to a 60° rotation, and the orientations of Fn14 CRD, TNFR1-d4, and BCMA-d are the same as those in the ribbon diagram on the right in Figure 2(B). (D) Superposition of the ribbon structures in A, B, and C. (E, F, and G) Ribbon diagrams with selected side chains of the CRD module structures of Fn14 (E), TNFR1 (F), and BCMA (G). The positively charged, negatively charged, hydrophobic, and other residues are colored blue, red, green, and magenta, respectively. The crucial residues (D45, K48, M50, and D62) in the human Fn14 CRD, identified by the mutagenesis study,8 are shown with red annotations. (H) Superposition of the structures in E, F, and G. Backbones are depicted by black lines, and side chains are shown by stick models, colored as in E, F, and G. In (E–H), the orientations correspond to a −120° rotation around the x axis from the right ribbon diagram in Figure 2(B).
Comparison with the A1-D2 module structure
The crystal structures of BCMA-d and TACI-d2 revealed that both of their CRDs have the A1-D2 module structure15,22 although they had been predicted as A1-C2 modules, based on the sequence alignment (Fig. 1). In both module structures, six Cys residues are involved in disulfide bond formation. Between the C2 and D2 modules, the pairs of Cys residues in the second and third disulfide bond connectivities are exchanged (Fig. 1). Namely, in the C2 module, the third and sixth Cys residues are linked in the second disulfide bond, and the fourth and fifth Cys residues form the third disulfide bond (referred to as C3-C6 and C4-C5, respectively). However, in the D2 module, they are linked as C3-C5 and C4-C6 (Fig. 1). Disulfide bonds are considered to play important roles in the folding of CRDs for their functions. Thus, we compared the structure of the Fn14 CRD with that of the A1-D2 module.
The tertiary structure of BCMA-d is shown as a representative of the A1-D2 module type in Figure 3(C,G) and was compared with that of the Fn14 CRD. The structures of the A1 modules in Fn14 CRD, TNFR1-d4, and BCMA-d are quite similar and superimposed very well with each other [blue regions in Fig. 3(A–D)]. Among them, the A1 module of the Fn14 CRD is closer to that of BCMA-d. For example, the length of the hairpin connecting the second and third strands is the same in Fn14 CRD and BCMA-d, but it is longer than that of the TNFR1-d4 (Fig. 1). In the known complex structures of the A1-D2 modules (BCMA-d and TACA-d2), the tip of the hairpin structures fit well with the binding sites on the ligand molecules.15,22 This implies that the hairpin of the Fn14 CRD is also important for its function, although different types of amino acid residues are located at the tip of the hairpin structures (Leu-Leu for BCMA-d and Asp-Leu for the Fn14 CRD) (Fig. 1).
In spite of the different disulfide bond connectivities between Fn14 CRD and BMCA-d, the C2 module of the Fn14 CRD and the D2 module of the BMCA-d share the same secondary structure elements, that is, there are two helical segments linked by a loop. The key difference between the tertiary structures of the CRDs of the A1-C2 module type and those of the A1-D2 module type is the spatial location of the loop just after the first helix and the following C-terminal α-helix, relative to the β-sheet of the A1 module. In the cases of BCMA-d and TACI-d2, the A1-D2 module structure provides a “saddle-like” binding site for the ligand, BAFF.15 Therefore, space is formed between the β-sheet in the A1 module and the loop region in the D2 module, to accommodate the loops from BAFF. Especially, an exposed Asp residue (corresponding to Asp15 in BCMA-d and Asp80 in TACI-d2) from the β-sheet is involved in the formation of a salt bridge with the counterpart Arg residue in the ligand molecule. Furthermore, hydrophobic residues surround the Asp-Arg pair of residues to strengthen the electrostatic interaction in both cases [BCMA example in Fig. 3(G)].15,22 In the A1-C2 module of the Fn14 CRD, however, the C-terminal α-helix is rotated, with respect to the C-terminal α-helix of BCMA-d, by about 60° around a point at the end of the first helix in the D2 module [Fig. 3(A–D)]. Consequently, the C-terminal α-helix in the C2 module moves toward the space under the β-sheet of the A1 module, changing the architecture of the binding site. Thus, the recognition mode of the Fn14 CRD for its ligand TWEAK is considered to be different from that of the A1-D2 module.
Ligand binding by the Fn14 CRD
A previous mutagenesis study based on the murine Fn14 CRD showed that Asp45, Lys48, Met50, and Asp62 are crucial for the interaction with TWEAK.8 By mapping these residues on our Fn14 CRD solution structure, we found that Asp45, Lys48, Met50, and Asp62 are located in the same area [Fig. 3(E)]. Especially, the different location of the C-terminal α-helix of the Fn14 CRD, when compared with that in the A1-D2 module, enables Asp62 to be involved in the charged patch [Fig. 3(H)]. A comparison with the complex structures of BCMA-d and TACI-d215,22 revealed that the charged patch area on the Fn14 CRD corresponds to the binding surfaces of BCMA-d and TACI-d2. However, we found a distinct difference between the binding regions of the A1-C2 module in Fn14 CRD and the A1-D2 module in BCMA-d. In the former, the positively charged residue Lys48 is surrounded by hydrophobic residues (Leu46, Met50) and two acidic residues (Asp45, Asp62) [Fig. 3(E,H)]. In the latter, the negatively charged residue Asp15 is surrounded by hydrophobic residues (Leu17, Ile22) [Fig. 3(G,H)].
The structural information described above will provide the basis for pharmaceutical research targeting the Fn14-TWEAK system.
Materials and Methods
NMR sample preparation
For the protein production, we utilized in vitro cell-free and in vivo protein production systems. In the case of the in vitro cell-free system, the DNA fragment encoding the Fn14 CRD (Glu28-Ala70) (SwissProt accession no. Q9NP84) was amplified by PCR from the human full-length cDNA clone. This DNA fragment was cloned into the expression vector pCR2.1 (Invitrogen), as a fusion with an N-terminal native His affinity tag and a TEV protease cleavage site. The 15N- and 13C-labeled fusion protein was synthesized by the cell-free protein expression system.29–31 The lysate was clarified by centrifugation at 16,000g for 20 min and filtration with a 0.45 mm membrane (Millipore). The clarified lysate was applied to a 5-mL His Trap column (GE Healthcare Biosciences), which was eluted with an imidazole gradient from 20 to 500 mM, and the tag was removed by an incubation with TEV protease for 1 h at 30°C. The cleaved His tag was segregated from the cleaved protein by rechromatography, using a His-trap nickel column. The sample was left at 4°C for several days to allow the formation of disulfide bonds before the NMR measurements. For the structure determination, around 1.1 mM of a uniformly 13C-/15N-labeled NMR sample was prepared in 20 mM d6-Tris-HCl buffer, containing 100 mM NaCl, and 0.02% (w/v) NaN3, pH 7.0, in 90% 1H2O/10% 2H2O, in a Shigemi NMR tube.
NMR spectroscopy
All NMR experiments were carried out at 298 K on Bruker AVANCE 600 and 800 MHz spectrometers with xyz gradients, using the uniformly 13C-/15N-labeled sample. The 1H, 15N, and 13C chemical shifts were assigned with a series of standard triple resonance experiments.32 3D 15N- and 13C-edited NOESY-heteronuclear single quantum coherence spectra were recorded with a mixing time of 80 ms, for collecting NOE restraints. All NMR data were processed with the NMRPipe program.33 The programs NMRView34 and Kujira35 were used for NMR spectra analysis.
Structure calculations
Distance restraints were obtained from 15N- and 13C-edited NOESY spectra and were used for the structure calculation. The NOESY peak lists were generated by automatic peak picking with manual checking, and the peak volumes were integrated by the automation function of NMRView.34 The three-dimensional structure was determined by combined automated NOESY cross peak assignment36 and structure calculation with torsion angle dynamics,37 implemented in the CYANA program.25 Dihedral angle restraints for φ and ψ were obtained from the main chain and 13Cβ chemical shift values, using the program TALOS.38 Upper limit distance restraints (2.25–2.35 Å) and lower limit distance restraints (1.95–2.05 Å) were used for the three disulfide bonds. The quality of the solution structure was evaluated using PROCHECK-NMR.27 Structural figures were prepared with the MOLMOL program.39
Protein data bank accession number
The solution structure of Fn14 CRD, represented by the 20 conformers with the lowest CYANA target function values, has been deposited in the Protein Data Bank (PDB entry 2RPJ).
Acknowledgments
The authors are grateful to Takushi Harada, Yasuko Tomo, Masaomi Ikari, Kazuharu Hanada, and Yukiko Fujikura for the sample preparation.
Glossary
Abbreviations:
- APRIL
a proliferation-inducing ligand
- BAFF
B-cell activation factor of the TNF family
- BAFFR
BAFF receptor
- BCMA
B-cell maturation antigen
- DR5
death receptor 5
- HSQC
heteronuclear single quantum coherence
- NOE
nuclear Overhauser enhancement
- NOESY
NOE spectroscopy
- TACI
transmembrane activator and CAML (calcium modulator and cyclophilin ligand) interactor
- TNF
tumor necrosis factor
- TNFR1
tumor necrosis factor receptor 1
- TNFRSF
TNF receptor superfamily
- TNFSF
TNF superfamily
- TRAIL
tumor necrosis factor-related apoptosis inducing ligand
- TWEAK
TNF-like weak inducer of apoptosis
- TWEAKR
TWEAK receptor.
References
- 1.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 2.Bodmer JL, Schneider P, Tschopp J. The molecular architecture of the TNF superfamily. Trends Biochem Sci. 2002;27:19–26. doi: 10.1016/s0968-0004(01)01995-8. [DOI] [PubMed] [Google Scholar]
- 3.Chicheportiche Y, Bourdon PR, Xu H, Hsu YM, Scott H, Hession C, Garcia I, Browning JL. TWEAK, a new secreted ligand in the tumor necrosis factor family that weakly induces apoptosis. J Biol Chem. 1997;272:32401–32410. doi: 10.1074/jbc.272.51.32401. [DOI] [PubMed] [Google Scholar]
- 4.Marsters SA, Sheridan JP, Pitti RM, Brush J, Goddard A, Ashkenazi A. Identification of a ligand for the death-domain-containing receptor Apo3. Curr Biol. 1998;8:525–528. doi: 10.1016/s0960-9822(98)70204-0. [DOI] [PubMed] [Google Scholar]
- 5.Meighan-Mantha RL, Hsu DK, Guo Y, Brown SA, Feng SL, Peifley KA, Alberts GF, Copeland NG, Gilbert DJ, Jenkins NA, Richards CM, Winkles JA. The mitogen-inducible Fn14 gene encodes a type I transmembrane protein that modulates fibroblast adhesion and migration. J Biol Chem. 1999;274:33166–33176. doi: 10.1074/jbc.274.46.33166. [DOI] [PubMed] [Google Scholar]
- 6.Feng SL, Guo Y, Factor VM, Thorgeirsson SS, Bell DW, Testa JR, Peifley KA, Winkles JA. The Fn14 immediate-early response gene is induced during liver regeneration and highly expressed in both human and murine hepatocellular carcinomas. Am J Pathol. 2000;156:1253–1261. doi: 10.1016/S0002-9440(10)64996-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wiley SR, Cassiano L, Lofton T, Davis-Smith T, Winkles JA, Lindner V, Liu H, Daniel TO, Smith CA, Fanslow WC. A novel TNF receptor family member binds TWEAK and is implicated in angiogenesis. Immunity. 2001;15:837–846. doi: 10.1016/s1074-7613(01)00232-1. [DOI] [PubMed] [Google Scholar]
- 8.Brown SA, Hanscom HN, Vu H, Brew SA, Winkles JA. TWEAK binding to the Fn14 cysteine-rich domain depends on charged residues located in both the A1 and D2 modules. Biochem J. 2006;397:297–304. doi: 10.1042/BJ20051362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wiley SR, Winkles JA. TWEAK, a member of the TNF superfamily, is a multifunctional cytokine that binds the TweakR/Fn14 receptor. Cytokine Growth Factor Rev. 2003;14:241–249. doi: 10.1016/s1359-6101(03)00019-4. [DOI] [PubMed] [Google Scholar]
- 10.Winkles JA, Tran NL, Berens ME. TWEAK and Fn14: new molecular targets for cancer therapy? Cancer Lett. 2006;235:11–17. doi: 10.1016/j.canlet.2005.03.048. [DOI] [PubMed] [Google Scholar]
- 11.Burkly LC, Michaelson JS, Hahm K, Jakubowski A, Zheng TS. TWEAKing tissue remodeling by a multifunctional cytokine: role of TWEAK/Fn14 pathway in health and disease. Cytokine. 2007;40:1–16. doi: 10.1016/j.cyto.2007.09.007. [DOI] [PubMed] [Google Scholar]
- 12.Winkles JA. The TWEAK-Fn14 cytokine-receptor axis: discovery, biology and therapeutic targeting. Nat Rev Drug Discov. 2008;7:411–425. doi: 10.1038/nrd2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng TS, Burkly LC. No end in site: TWEAK/Fn14 activation and autoimmunity associated- end-organ pathologies. J Leukoc Biol. 2008;84:338–347. doi: 10.1189/jlb.0308165. [DOI] [PubMed] [Google Scholar]
- 14.Naismith JH, Sprang SR. Modularity in the TNF-receptor family. Trends Biochem Sci. 1998;23:74–79. doi: 10.1016/s0968-0004(97)01164-x. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Hong X, Kappler J, Jiang L, Zhang R, Xu L, Pan CH, Martin WE, Murphy RC, Shu HB, Dai S, Zhang G. Ligand-receptor binding revealed by the TNF family member TALL-1. Nature. 2003;423:49–56. doi: 10.1038/nature01543. [DOI] [PubMed] [Google Scholar]
- 16.Gouet P, Courcelle E, Stuart DI, Metoz F. ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics. 1999;15:305–308. doi: 10.1093/bioinformatics/15.4.305. [DOI] [PubMed] [Google Scholar]
- 17.Foley SF, Sun Y, Zheng TS, Wen D. Picomole-level mapping of protein disulfides by mass spectrometry following partial reduction and alkylation. Anal Biochem. 2008;377:95–104. doi: 10.1016/j.ab.2008.02.025. [DOI] [PubMed] [Google Scholar]
- 18.Zhang G. Tumor necrosis factor family ligand-receptor binding. Curr Opin Struct Biol. 2004;14:154–160. doi: 10.1016/j.sbi.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Banner DW, D'Arcy A, Janes W, Gentz R, Schoenfeld HJ, Broger C, Loetscher H, Lesslauer W. Crystal structure of the soluble human 55 kd TNF receptor-human TNF beta complex: implications for TNF receptor activation. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 20.Mongkolsapaya J, Grimes JM, Chen N, Xu XN, Stuart DI, Jones EY, Screaton GR. Structure of the TRAIL-DR5 complex reveals mechanisms conferring specificity in apoptotic initiation. Nat Struct Biol. 1999;6:1048–1053. doi: 10.1038/14935. [DOI] [PubMed] [Google Scholar]
- 21.Kim HM, Yu KS, Lee ME, Shin DR, Kim YS, Paik SG, Yoo OJ, Lee H, Lee JO. Crystal structure of the BAFF-BAFF-R complex and its implications for receptor activation. Nat Struct Biol. 2003;10:342–348. doi: 10.1038/nsb925. [DOI] [PubMed] [Google Scholar]
- 22.Hymowitz SG, Patel DR, Wallweber HJ, Runyon S, Yan M, Yin J, Shriver SK, Gordon NC, Pan B, Skelton NJ, Kelley RF, Starovasnik MA. Structures of APRIL-receptor complexes: like BCMA, TACI employs only a single cysteine-rich domain for high affinity ligand binding. J Biol Chem. 2005;280:7218–7227. doi: 10.1074/jbc.M411714200. [DOI] [PubMed] [Google Scholar]
- 23.Schubert M, Labudde D, Oschkinat H, Schmieder P. A software tool for the prediction of Xaa-Pro peptide bond conformations in proteins based on 13C chemical shift statistics. J Biomol NMR. 2002;24:149–154. doi: 10.1023/a:1020997118364. [DOI] [PubMed] [Google Scholar]
- 24.Wüthrich K. NMR of Proteins and Nucleic Acids. New York: Wiley; 1986. [Google Scholar]
- 25.Güntert P. Automated NMR structure calculation with CYANA. Methods Mol Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 26.Sharma D, Rajarathnam K. 13C NMR chemical shifts can predict disulfide bond formation. J Biomol NMR. 2000;18:165–171. doi: 10.1023/a:1008398416292. [DOI] [PubMed] [Google Scholar]
- 27.Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 28.Naismith JH, Devine TQ, Kohno T, Sprang SR. Structures of the extracellular domain of the type I tumor necrosis factor receptor. Structure. 1996;4:1251–1262. doi: 10.1016/s0969-2126(96)00134-7. [DOI] [PubMed] [Google Scholar]
- 29.Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genomics. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda T, Koshiba S, Tochio N, Seki E, Iwasaki N, Yabuki T, Inoue M, Yokoyama S, Kigawa T. Improving cell-free protein synthesis for stable-isotope labeling. J Biomol NMR. 2007;37:225–229. doi: 10.1007/s10858-006-9127-5. [DOI] [PubMed] [Google Scholar]
- 31.Yabuki T, Motoda Y, Hanada K, Nunokawa E, Saito M, Seki E, Inoue M, Kigawa T, Yokoyama S. A robust two-step PCR method of template DNA production for high-throughput cell-free protein synthesis. J Struct Funct Genomics. 2007;8:173–191. doi: 10.1007/s10969-007-9038-z. [DOI] [PubMed] [Google Scholar]
- 32.Clore GM, Gronenborn AM. New methods of structure refinement for macromolecular structure determination by NMR. Proc Natl Acad Sci USA. 1998;95:5891–5898. doi: 10.1073/pnas.95.11.5891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 34.Johnson BA, Blevins RA. NMR View: a computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 35.Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S. KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high-throughput NMR structure studies. J Biomol NMR. 2007;39:31–52. doi: 10.1007/s10858-007-9175-5. [DOI] [PubMed] [Google Scholar]
- 36.Herrmann T, Güntert P, Wüthrich K. Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol. 2002;319:209–227. doi: 10.1016/s0022-2836(02)00241-3. [DOI] [PubMed] [Google Scholar]
- 37.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 38.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 39.Koradi R, Billeter M, Wüthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graphics. 1996;14:51–55. 29–32. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]


