Abstract
A female patient is described with clinical symptoms of both microphthalmia with linear skin defects (MLS or MIDAS) and dental enamel defects, having an appearance compatible with X-linked amelogenesis imperfecta (XAI). Genomic DNA was purified from the patient's blood and semiquantitative multiplex PCR revealed a deletion encompassing the amelogenin gene (AMELX). Because MLS is also localized to Xp22, genomic DNA was subjected to array comparative genomic hybridization, and a large heterozygous deletion was identified. Histopathology of one primary and one permanent molar tooth showed abnormalities in the dental enamel layer, and a third tooth had unusually high microhardness measurements, possibly due to its ultrastructural anomalies as seen by scanning electron microscopy. This is the first report of a patient with both of these rare conditions, and the first description of the phenotype resulting from a deletion encompassing the entire AMELX gene. More than 50 additional genes were monosomic in this patient.
Keywords: MLS syndrome, amelogenesis imperfecta, X-chromosomal deletion, array CGH, mineral microhardness
Introduction
The microphthalmia with linear skin defects syndrome (MLS; OMIM309801) is a rare and sporadic X-linked condition that is usually lethal in males. Females with this clinically complex condition are characterized by irregular linear areas of erythematous skin hypoplasia on the head and neck, microphthalmia, corneal opacities and orbital cysts [Temple et al., 1990], but can have additional symptoms affecting brain and heart [Morleo et al., 2005]. The variability in severity between affected females has been proposed to be due to the pattern of X-inactivation in this region of the X chromosome, the extent of the region of monosomy and the nature of the chromosomal anomaly [Van den Veyver, 2002; Kayserili et al., 2001]. The condition has also been referred to as Gazali-Temple syndrome [Al-Gazali et al., 1990], or as MIDAS (microphthalmia, dermal aplasia and sclerocornea) [Happle et al., 1993].
A minimal critical region for MLS of 610 kb at Xp22.2 was identified by cytogenetic and breakpoint analyses of affected and unaffected individuals [Wapenaar et al., 1994]. Most MLS patients have segmental monosomy in this region, which includes genes MID1, HCCS and the 3′ region of ARHGAP6 [Schaefer et al., 1996; 1997]. Recently, MLS patients with normal karyotype but short deletions or point mutations in the HCCS gene, which encodes a mitochondrial protein involved in oxidative phosphorylation and apoptotic cell death, have been reported [Wimplinger et al., 2006]. Mutations in HCCS explain both male lethality and the clinical presentation in females because of the role of the HCCS protein in housekeeping functions.
ARHGAP6 includes AMELX entirely located within intron 1 in the opposite orientation [Prakash et al., 2000]. Mutations within AMELX lead to a group of dental enamel defects referred to as X-linked amelogenesis imperfecta (XAI; MIM300391) in which patients have either hypoplastic enamel or enamel with various mineral defects, or both [Wright, 2006].
An 18-year-old female patient is described who presented with eye and skin defects, characteristic of MLS syndrome, and vertical bands of normal and deeply pitted enamel, characteristic of XAI. In addition, she had short stature, skeletal deformities, delayed bone age and polycystic ovary syndrome, findings that were seen in patients with deletions of the Short Stature Homeobox (SHOX) [Rao et al., 1997], as well as learning impairment and autistic tendencies, findings that were seen in a patient with deletion of VCX3A (also called VCX-A) and NLGN4X [Chocholska et al., 2006]. Both parents and her older sister appeared to be clinically normal. Because the patient's clinical appearance was hypothesized to be due to a deletion of contiguous genes, a DNA analysis was undertaken including the Xp22 region of her X chromosome, and three teeth extracted because of pain were evaluated for histological or structural defects.
Materials and Methods
Clinical and histopathological examination of teeth
This study was approved by the Royal Liverpool and Broadgreen Hospitals NHS Trust and Huntingdon Research Ethics Committee, and informed consent was obtained. For dental histopathology, one extracted permanent molar and one primary molar were bisected with one half used for decalcified sections and the other half for ground sections.
Enamel mineral microdensity, microhardness and ultrastructure
A primary molar from the patient was embedded in methyl methacrylate. Mesio-distal sections were polished to a thickness of approximately 100 μm, etched for 15 sec with 35% phosphoric acid and washed in distilled water. Mineral density across the full thickness of enamel at 6 equi-distant sites was determined by quantitative transverse microradiography (TMR) based upon a modification of the technique routinely used to quantify mineral loss in dental decay [Angmar et al., 1963; DeJosselin De Jong et al., 1987]. The microradiographs were analyzed using customized TMRW software (version 20.0.27.16, 2000; Inspektor Research Systems, Amsterdam, The Netherlands). Microhardness analysis of the enamel was by conventional testing [Koulourides et al., 1961] determined from 500 μm sections using a Struers Duramin testing rig (Struers Ltd., Solihull, West Midlands, UK) equipped with a Knoop diamond tip. A load of 100 g was applied at 6 different sites for 15 sec and the analysis carried out using Duramin™ 5 software. All measurements were compared to an age-matched unaffected control tooth of the same type. For ultrastructural analysis, one of the sections was mounted on an aluminum stub and sputter coated with gold before imaging in a JEOL 35 scanning electron microscope fitted with the Deben “Genie” upgrade (Deben Engineering, Debenham, UK).
Primer design and multiplex PCR
To investigate the hypothesis that the patient has an X-chromosomal deletion, primers for PCR amplification (Table SI) were designed using the human genome reference sequence (University of California Santa Cruz [UCSC] version HG 18; National Center for Biotechnology Information [NCBI] Build 36) and MacVector or DS Gene software (Accelrys, Burlington, MA). Genomic DNA was prepared from blood lymphocytes by three-fold phenol/chloroform/isoamyl alcohol extraction, followed by precipitation with ethanol. Semi-quantitative multiplex PCR was performed with the patient's DNA using the Multiplex PCR Kit (Qiagen, Valencia, CA) as described [Woodward et al., 2005]. Two primer pairs for regions flanking AMELX were multiplexed with primer pairs for two regions of PLP1 on Xq, a region of DMD on Xp, a region of CFTR on chromosome 7, and a region of SRY on chromosome Y. One member of each primer pair was labeled with TET.
Detection and quantitation of products were done using capillary electrophoresis on an ABI Prism 310 Genetic Analyzer with Genescan software (Applied Biosystems, Foster City, CA). Three normal female control DNA samples were included in each experiment along with patient DNA. Average results of the normal control DNA samples were used to calculate normalized ratios. If a region in or near AMELX was deleted on one allele in the female patient, the expected normalized ratios of the region to the DMD control, to the PLP1 control and to the autosomal CFTR control were 0.5. If the region was not deleted, the expected ratios were 1. No amplification was expected with the primers for the SRY gene since all DNA samples were from females.
Array comparative genomic hybridization
High resolution fine-tiling array comparative genomic hybridization (aCGH) using the HG18 CHRX FT array with median probe spacing of 340 bp (aCGH service, NimbleGen Systems, Inc., Madison, WI) was used for identification of the extent of the deletion. Patient and normal female DNAs were differentially labeled, mixed and hybridized to the array chip. Two-color array data signal intensities were normalized to one another using qspline normalization and expressed as log2 ratio [Workman et al., 2002]. DNA segmentation analysis for copy number was performed using CGH-segMNT analysis (NimbleGen). Genes in the deleted region were identified using the human genome reference sequence hg18, Build 36, release 49 (Ensembl) [Hubbard et al., 2007].
X-inactivation analysis
The X-inactivation pattern was evaluated by examining the methylation pattern at the AR (androgen receptor) locus [Allen et al., 1992]. DNA aliquots of 250 ng were incubated in a total volume of 12.5 μl with no enzyme, DdeI only, HpaII only, or DdeI and HpaII for 18 h at 37 °C (New England Biolabs) using the manufacturer's instructions. Digestion with DdeI can prevent falsely skewed X-inactivation patterns by allowing equal amplification of methylated and unmethylated DNA [van Dijk et al., 2002]. Subsequently, regions of the AR and JARID1C (formerly SMCX) loci were amplified by PCR in separate 25 μl reactions with 2.5 μl of the digested DNA by adding Taq Buffer (5× Taq Buffer is 83 mM Tris-HCl [pH 8.8], 850 mg/mL BSA, 83 mM (NH4)2SO4, 33.5 mM MgCl2, 34 mM EDTA, and 50mM β-mercaptoethanol), 5% dimethyl sulfoxide, 12.5 pmol of each primer, 1.5 mM dNTPs, and 0.625 U AmpliTaq (Applied Biosystems). The JARID1C locus, which escapes X-inactivtion, was used as a control for complete digestion by HpaII. The conditions were as follows: melting at 94 °C for 5 min; 25 cycles at 94 °C for 30 s, 55 °C for 30 s, and 65 °C for 1 min; and a final extension at 65 °C for 6 min. Primers were AR F: 5′-CCCCAGGCACCCAGAGGC and AR R-TET: GAGAACCATCCTCACCCTGC, SMCX F: 5′-ACCCATACCAGCCTATTTAGC and SMCX R-TET: 5′-GCCTTCGCCACCACAGTTAC. PCR products were analyzed and quantification was performed using an ABI 310 (Genetic Analyzer with Genotyper software (Applied Biosystems).
PCR products obtained before HpaII digestion were from both the active and inactive X chromosomes and those obtained after HpaII digestion were from the inactive X chromosomes only.
DNA sequence analysis
Genomic DNA was subjected to PCR amplification and sequence analysis either directly or after cloning into the TA vector pCR4-TOPO (Invitrogen, Carlsbad, CA) using primers that are specific for coding exons as described previously [Collier et al., 1997].
Results
Clinical characterization
The patient was a 20-year-old female, born after a normal pregnancy at 41 weeks, and weighing 2636 g. At birth, no limb abnormalities were noted, but she had microphthalmia of the right eye and corneal opacity of the left. There were ‘weeping raw’ skin areas on her face and neck.
By 3 years of age, delays in her development were noted and her height was below the 3rd centile. Her right eye had a perforated cornea and the prolapsed iris was enucleated, but her left eye was noted to have only a small corneal opacity, a clear lens and normal intraocular pressure. The skin lesions were described as eczematous in appearance.
At 12 years of age, her medical reports note short stature with height below the 3rd centile, a bone age of 1.7 years lower than her chronological age, learning difficulties and autistic tendencies.
By 14 years of age, she still had short stature, which had not responded to growth hormone, and she was diagnosed with thoracic scoliosis. Although she still had learning difficulties, improvements in comprehension and speech were noted. Following laser treatment of the skin, the lesions had improved but residual scarring of the face and neck was present (Fig. 1A). Menarche, pubic hair and breast stage development were all within normal limits. She had polycystic ovary syndrome, with hirsutism, irregular periods, a reverse FSH/LH and high testosterone level.
Fig. 1.
Clinical appearance of patient. A. Lateral view of face and neck showing erythematous skin lesions and scarring. B. Anterior view of permanent dentition, showing vertical bands of normal and abnormal enamel which vary between different teeth.
At 20 years, she is 142 cm tall. The scoliosis is stable and no surgery is planned. The vision in her left eye is stable, and she has no hearing problems. Her learning difficulty is moderate and her autistic tendencies, e.g. obsessive behaviors, routines, lack of empathy, continue. The symptoms of polycystic ovary syndrome are controlled by medication.
At the first dental examination by AHB at 12 years of age, the permanent upper left lateral incisor was absent and the permanent upper right lateral incisor was microdont. Generalized enamel defects, which were predominantly hypoplasia with a vertical presentation, were present, but there were also several opacities (Fig. 1B). A striking finding was the asymmetry of the hypoplastic defects between the left and right sides, as shown for example between the lower canine teeth (Fig. 1B). Non-carious enamel breakdown was apparent on three of the permanent molars on the right side. Radiographs showed areas of enamel with normal thickness and radiodensity while other areas of enamel were deficient. The molar teeth were taurodont.
Measurements of physical properties of the enamel layer and ultrastructural appearance
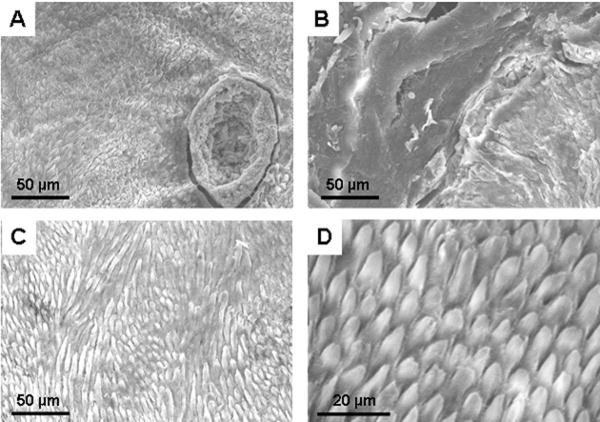
Quantitative transverse microradiography was used to determine mineral density for both enamel and dentine; both were within the normal range for primary teeth (Table I). Dentine microhardness was also within the normal range, but the enamel was 91% harder than that of similar control teeth (Table I). Only one tooth was available for these analyses but the results are included here as the findings regarding microhardness were startlingly different than the expected norm. Scanning electron microscopy revealed areas of pitting (Fig. 2A) and disturbed enamel architecture (Fig. 2B) interrupted by areas of what appeared to be normal prismatic structure (Fig. 2C). A normal control is shown (Fig. 2D) for comparison.
Table I. Measurements of enamel and dentine mineral density and microhardness.
| Affected Tooth: Enamel | Control Tooth: Enamel | Affected Tooth: Dentine | Control Tooth: Dentine | |
|---|---|---|---|---|
| Mineral Density (Vol %) | 86.0 ± 7.6 | 83.0 ± 1.0 | 43.3 ± 5.1 | 34.7 ± 3.1 |
| Microhardness (kH) | 607.2 ± 28.7 | 317.4 ± 15.2 | 177.3 ±17.3 | 207.9 ±10.3 |
Measurements for an affected compared to a matched control tooth. Measurements were made at 6 different locations across sections of both tooth types, showing means and SDs.
Fig. 2.
Scanning electron micrographs. A,B. Sections taken from an affected primary tooth revealed areas with deep pitting (A) and disturbed enamel architecture with loss of typical prismatic structure (B). C. Other areas within the same section revealed more normal prismatic appearance. D. These more normal areas in (C) were comparable to that seen in the same anatomical areas of matched control teeth.
Identification of the deletion
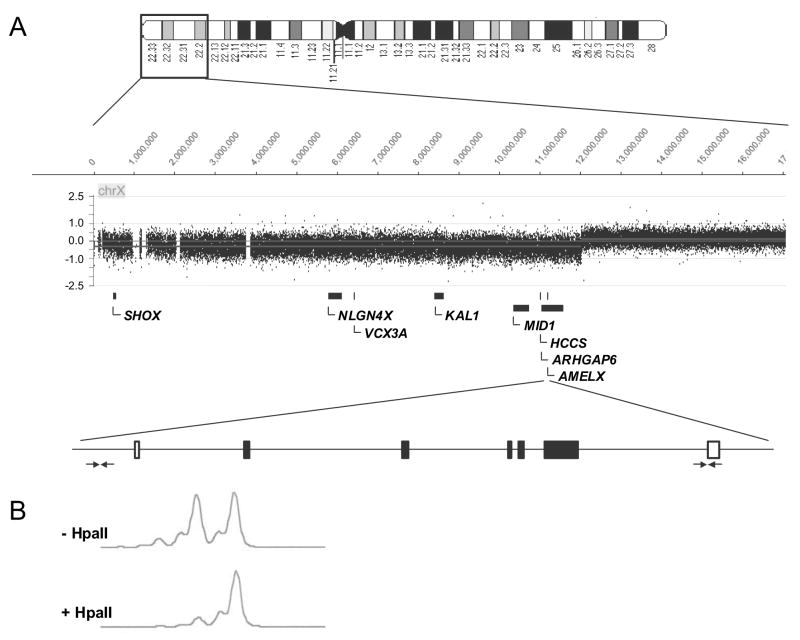
Semi-quantitative multiplex PCR experiments using primers listed in Table SI revealed that the ratio of amplification from each of the AMELX primer pairs to each of the other primer pairs was half that seen with the average of three normal control females (Table II), indicating that the patient had a deletion on one X chromosome extending at least from bases 11221896 to 11229693. This includes the 5′ end of AMELX to exon 7, and a region of intron 1 of ARHGAP6 (Fig. 3A).
Table II. Analysis of copy number by semi-quantitative multiplex PCR.
| Peak Areas | ||||||
|---|---|---|---|---|---|---|
| Primer Pairs | Patient | NL1 | NL2 | NL3 | NL Ave | |
| G1092/G1093 | 14579 | 53407 | 27775 | 36077 | 39086 | |
| G1094/G1095 | 13800 | 52028 | 26539 | 33456 | 37341 | |
| PLPpromF6&R5 | 32971 | 62074 | 30738 | 40346 | 44386 | |
| PLP 5F&5R | 28508 | 54494 | 26510 | 34826 | 38610 | |
| Hdys23F&R | 33700 | 64955 | 32346 | 41251 | 46184 | |
| HCFTR13F&13R | 23695 | 46612 | 22356 | 32011 | 33660 | |
| Ratios of Peak Areas | ||||||
| G1092/G1093 | G1094/G1095 | PLPpromF6&R5 | PLP 5F&5R | Hdys23F&R | HCFTR13F&13R | |
| G1092/G1093 | 1.0 | 1.0 | 0.5 | 0.5 | 0.5 | 0.5 |
| G1094/G1095 | 1.0 | 0.5 | 0.5 | 0.5 | 0.5 | |
| PLPpromF6&R5 | 1.0 | 1.1 | 1.0 | 1.1 | ||
| PLP 5F&5R | 1.0 | 1.0 | 1.0 | |||
| Hdys23F&R | 1.0 | 1.0 | ||||
| HCFTR13F&13R | 1.0 | |||||
Ratios of peak areas are calculated by the following formula for each region in the matrix against each other region using the patient data and the average data from 3 normal females: (Patient region a ÷ region b) ÷ (NL Average region a ÷ region b)
Fig. 3.
Molecular analyses of the patient's DNA. A. Semi-quantitative multiplex PCR using the AMELX primers indicated by horizontal arrows and aCGH revealed that the patient has a heterozygous deletion from Xpter to Xp22.2 and totaling 12 Mb. More than 50 genes were deleted in the patient (SII). Only those mentioned in the text are shown. B. Restriction enzyme digestion with the HpaII enzyme that is sensitive to methylation revealed skewed X-inactivation in the patient's peripheral blood lymphocytes.
Genomic DNA analysis by aCHG (NimbleGen) confirmed that the patient has an X-chromosome terminal deletion extending from Xpter to Xp22.2 approximately at base 12,024,000 (Fig. 3A). Deleted genes and associated diseases are listed in Table SII, including genes responsible for Kallman Syndrome and Opitz G/BBB as well as genes linked to autism, mental retardation and chondrodysplasia. No changes within coding regions of the AMELX gene on the remaining X chromosome were detected by DNA sequence analysis.
X-chromosome inactivation studies
We examined the X-inactivation status in the peripheral blood cells of the patient by examining methylation at the AR locus. X-chromosome inactivation (XCI) was skewed in the patient at a ratio of 15% to 85% (Fig. 3B). The direction of skewing was not determined.
Discussion
Understanding the underlying cause of MLS has presented a challenge as affected patients have rare sporadic mutations, and affected individuals from a large kindred are not available for confirmation by linkage analysis. Difficulties in distinguishing MLS from Aicardi and Goltz syndromes were earlier thought to be due to variable patterns of X-inactivation in females, but several clinical differences prompted the suggestion that these syndromes have different genetic causes [Lindsay et al., 1994; Van den Veyver, 2002]. At this time, it is known that focal dermal dysplasia or Goltz syndrome is caused by PORCN mutations [Wang et al., 2007], but gene defects responsible for Aicardi syndrome have not yet been established.
MLS was localized to a minimal critical region on Xp22, and candidate genes have included MID1, ARHGAP6 and HCCS [Van den Veyver, 2002]. MID1 mutations are now associated with Opitz G/BBB syndrome [Quaderi et al., 1997]. The conserved murine Arhgap6 gene encodes a Rho GTPase activating protein, which has GAP activity for RhoA [Prakash et al., 2000]. Mice with a large engineered deletion in the minimal critical region including these genes were able to survive when they expressed a human HCCS transgene, thereby linking HCCS to the cause of male lethality in MLS [Prakash et al., 2002]. HCCS encodes a mitochondrial holocytochrome c-type synthetase, and catalyzes a reaction leading to mature cytochrome c in the mitochondrial respiratory chain [Schaefer et al., 1996]. The HCCS protein is also implicated in regulation of decisions between apoptosis and necrosis [Prakash et al., 2002]. HCCS mutations have been identified in several patients with MLS and in patients with severe eye defects [Wimplinger et al., 2006; 2007], and it is now assumed that defects in oxidative phosphorylation and apoptosis plus skewed X-inactivation contribute to the observed MLS phenotypic heterogeneity.
The AMELX gene, located within an intron of ARHGAP6, encodes amelogenin proteins, which represent 90% of the enamel organic matrix [Termine et al., 1980]. Deletions or point mutations within AMELX lead to varied dental enamel defects, known collectively as X-linked amelogenesis imperfecta or XAI [Wright et al., 2003]. Random X-inactivation is thought to lead to the vertical ridges in incisor teeth of female XAI patients [Sauk et al., 1972; Collier et al., 1997], where adjacent groups of ameloblast cells would produce enamel in which either the normal or the mutated AMELX gene is active. The abnormal enamel in these patients may be defective in thickness and/or in mineralization and may retain excess organic material.
In the only other report of a large deletion in the AMELX gene causing XAI, a 5 kb deletion involving five of the seven exons resulted in a phenotype of hypomineralized (softer than normal) enamel in males, while females had vertically arranged ridges of normal and abnormal appearing enamel [Lagerstrom et al., 1991; Lagerstrom-Fermer et al., 1993]. The female patient reported here also had vertical ridges but with either normal enamel or enamel reduced in thickness, so that the affected enamel was principally hypoplastic (Figs. 1 and 2) with some hypomature areas seen as opacities. Female mice generated to have a heterozygous Amelx deletion also develop abnormal vertical ridges in their enamel [Gibson et al., 2005].
In the current study, the primary molar tooth subjected to microhardness testing had areas of deep pitting in the enamel, increased microhardness, and regions of abnormal architecture. The increase in enamel microhardness was remarkable given that mature enamel is the most highly mineralized and hardest of all of the skeletal tissues. However, increased mineral hardness has also been measured in transgenic mice when the ameloblasts over-expressed certain dentin matrix proteins [White et al., 2007]. The mechanical properties of enamel are related to both its high degree of mineralization and its characteristic structure. It is possible that the increased hardness seen in this tooth was therefore related to its highly abnormal structural organization. Normal prismatic structure was also observed in certain regions of the enamel, presumably related to the macroscopic observations reported above, where bands of “normal” enamel are seen and the “normal” AMELX gene is expressed. Apoptosis of 25-50% of the enamel producing ameloblasts normally occurs during development in rat teeth [Smith and Warshawsky, 1977], and therefore a defect in the apoptotic pathway due to an HCCS mutation could be a contributing factor to an enamel defect in this patient. The loss of other genes in the deleted region of the X chromosome may also contribute to alterations in enamel structure or to changes in normal gene expression in enamel-forming cells.
Although several syndromes are known to affect both eyes and teeth, this is the first report of a patient with both MLS and AI, and other symptoms associated with deletions of this region in females, including short stature and autistic tendencies. Our patient has a skewed X-inactivation pattern in peripheral blood, as has been reported for other females with X-linked diseases caused by genomic copy number changes [Wells et al., 1991; Woodward et al., 2000; [Megarbane et al., 2002]. Presumably, this is secondary skewing that occurred in many tissues of the patient due to death of cells in which the normal X chromosome was inactivated and it is probably a skewed pattern that allows the patient to survive when deletion of so many genes would be lethal in a male. Our patient has linear defects of the skin and teeth. Although not formally tested, it has been hypothesized that the linear defects could be due to a mosaic pattern of XCI early in tissue development, with regions skewed toward the normal X alternating with regions skewed toward the abnormal X [Morleo and Franco, 2008]. Further, ameloblasts die after tooth formation, but in skin, the “sick” cells in which the normal X chromosome is inactivated may die and be replaced by cells in which the abnormal X is inactivated over time [Morleo, 2008]. Indeed, the skin lesions of MLS patients are replaced by hyper-pigmented areas as they age.
In many MLS cases, the length of the X-chromosomal deletion does not correlate with severity of clinical phenotype [see review Morleo and Franco, 2008]. Skewed XCI has been reported in most MLS cases studied, generally done by analysis of patient blood cells. It is likely that XCI will prove to vary between tissues in these patients, providing a mechanism to explain heterogeneity of skin and eye defects. Phenotypic heterogeneity between teeth in a single dentition or between affected members of a kindred with XAI has been repeatedly observed, and is also expected to be due to variations in patterns of XCI. Management of both MLS and AI involves treatment of the symptoms [Seow and Amaratunge, 1998], which for MLS can change significantly over time.
Supplementary Material
Acknowledgments
This work was supported by Nemours and NCRR grant P20RR020173 (GMH), Wellcome Trust research grant 075945/Z/04/Z (AB and JK) and NIDCR grant DE011089 (CWG). We thank S. Labadessa and Dr. S. Myers for technical assistance. We express our gratitude to the family who participated in this study.
References
- Al-Gazali LI, Mueller RF, Caine A, Antoniou A, McCartney A, Fitchett M, Dennis NR. Two 46,XX,t(X;Y) females with linear skin defects and congenital microphthalmia: a new syndrome at Xp22.3. J Med Genet. 1990;27:59–63. doi: 10.1136/jmg.27.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RC, Zoghbi HY, Moseley AB, Rosenblatt HM, Belmont JW. Methylation of HpaII and HhaI sites near the polymorphic CAG repeat in the human androgen-receptor gene correlates with X chromosome inactivation. Amer J Hum Genet. 1992;51:1229–1239. [PMC free article] [PubMed] [Google Scholar]
- Angmar B, Carlstrom D, Glas JE. Studies on the ultrastructure of dental enamel. IV. The mineralization of normal human enamel. J Ultrastruct Res. 1963;8:12–23. doi: 10.1016/s0022-5320(63)80017-9. [DOI] [PubMed] [Google Scholar]
- Chocholska S, Rossier E, Barbi G, Kehrer-Sawatzki H. Molecular cytogenetic analysis of a familial interstitial deletion Xp22.2-22.3 with a highly variable phenotype in female carriers. Am J Med Genet A. 2006;140:604–610. doi: 10.1002/ajmg.a.31145. [DOI] [PubMed] [Google Scholar]
- Collier PM, Sauk JJ, Rosenbloom J, Yuan ZA, Gibson CW. An amelogenin gene defect associated with human X-linked amelogenesis imperfecta. Arch Oral Biol. 1997;42:235–242. doi: 10.1016/s0003-9969(96)00099-4. [DOI] [PubMed] [Google Scholar]
- De Josselin De Jong E, van der Linden AH, Ten Bosch JJ. Longitudinal microradiography: a non-destructive automated quantitative method to follow mineral changes in mineralised tissue slices. Phys Med Biol. 1987;32:1209–1220. doi: 10.1088/0031-9155/32/10/001. [DOI] [PubMed] [Google Scholar]
- Gibson CW, Kulkarni AB, Wright JT. The use of animal models to explore amelogenin variants in amelogenesis imperfecta. Cells Tissues Organs. 2005;181:196–201. doi: 10.1159/000091381. [DOI] [PubMed] [Google Scholar]
- Happle R, Daniels O, Koopman RJ. MIDAS syndrome (microphthalmia, dermal aplasia, and sclerocornea): an X-linked phenotype distinct from Goltz syndrome. Am J Med Genet. 1993;47:710–713. doi: 10.1002/ajmg.1320470525. [DOI] [PubMed] [Google Scholar]
- Hubbard TJP, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, Down T, Dyer SC, Fitzgerald S, Fernandez-Banet J, Graf S, Haider S, Hammond M, Herrero J, Holland R, Howe K, Howe K, Johnson N, Kahari A, Keefe D, Kokocinski F, Kulesha E, Lawson D, Longden I, Melsopp C, Megy K, Meidl P, Ouverdin B, Parker A, Prlic A, Rice S, Rios D, Schuster M, Sealy I, Severin J, Slater G, Smedley D, Spudich G, Trevanion S, Vilella A, Vogel J, White S, Wood M, Cox T, Curwen V, Durbin R, Fernandez-Suarez XM, Flicek P, Kasprzyk A, Proctor G, Searle S, Smith J, Ureta-Vidal A, Birney E. Ensembl 2007. Nucleic Acids Res. 35(Database issue):D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayserili H, Cox TC, Cox LL, Basaran S, Kilic G, Ballabio A, Yuksel-Apak M. Molecular characterisation of a new case of microphthalmia with linear skin defects (MLS) J Med Genet. 2001;38:411–417. doi: 10.1136/jmg.38.6.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koulourides T, Cueto H, Pigman W. Rehardening of softened enamel surfaces of human teeth by solutions of calcium phosphates. Nature. 1961;189:226–227. doi: 10.1038/189226a0. [DOI] [PubMed] [Google Scholar]
- Lagerstrom M, Dahl N, Nakahori Y, Nakagome Y, Backman B, Landegren U, Pettersson U. A deletion in the amelogenin gene (AMG) causes X-liked amelogenesis imperfecta. Genomics. 1991;10:971–975. doi: 10.1016/0888-7543(91)90187-j. [DOI] [PubMed] [Google Scholar]
- Lagerstrom-Fermer M, Pettersson U, Landegren U. Molecular basis and consequences of a deletion in the amelogenin gene, analyzed by capture PCR. Genomics. 1993;17:89–92. doi: 10.1006/geno.1993.1287. [DOI] [PubMed] [Google Scholar]
- Lindsay EA, Grillo A, Ferrero GB, Roth EJ, Magenis E, Grompe M, Hulten M, Gould C, Baldini A, Zoghbi HY, Ballabio A. Microphthalmia with linear skin defects (MLS) syndrome: clinical, cytogenetic, and molecular characterization. Am J Med Genet. 1994;49:229–234. doi: 10.1002/ajmg.1320490214. [DOI] [PubMed] [Google Scholar]
- Megarbane A, Vabres P, Slaba S, Smahi A, Loeys B, Okais N. Linear and whorled nevoid hypermalenosis with bilateral giant cerebral aneurysms. Am J Med Genet. 2002;112:95–98. doi: 10.1002/ajmg.10666. [DOI] [PubMed] [Google Scholar]
- Morleo M, Pramparo T, Perone L, Gregato G, Le Caignec C, Mueller RF, Ogata T, Raas-Rothschild A, de Blois MC, Wilson LC, Zaidman G, Zuffardi O, Ballabio A, Franco B. Microphthalmia with linear skin defects (MLS) syndrome: clinical, cytogenetic and molecular characterization of 11 cases. Am J Med Genet. 2005;137A:190–198. doi: 10.1002/ajmg.a.30864. [DOI] [PubMed] [Google Scholar]
- Morleo M, Franco B. Dosage compensation of the mammalian X chromosome influences the phenotypic variability of X-linked dominant male-lethal disorders. J Med Genet. 2008;45:401–408. doi: 10.1136/jmg.2008.058305. [DOI] [PubMed] [Google Scholar]
- Prakash SK, Paylor R, Jenna S, Lamarche-Vane N, Armstrong DL, Xu B, Mancini MA, Zoghbi HY. Functional analysis of ARHGAP6: a novel GTPase-activating protein for RhoA. Hum Mol Genet. 2000;9:477–488. doi: 10.1093/hmg/9.4.477. [DOI] [PubMed] [Google Scholar]
- Prakash SK, Cormier TA, McCall AE, Garcia JJ, Sierra R, Haupt B, Zoghbi HY, Van den Veyver IB. Loss of holocytochrome c-type synthetase causes the male lethality of X-linked dominant microphthalmia with linear skin defects (MLS) syndrome. Hum Mol Genet. 2002;11:3237–3248. doi: 10.1093/hmg/11.25.3237. [DOI] [PubMed] [Google Scholar]
- Quaderi NA, Schweiger S, Gaudenz K, Franco B, Rugarli EI, Berger W, Feldman GJ, Volta M, Andolfi G, Gilgenkranz S, Marion RW, Hennekam RCM, Opitz JM, Muenke M, Ropers HH, Ballabio A. Optiz G/BBB syndrome, a defect of midline development, is due to mutations in a new RING finger gene on Xp22. Nat Genet. 1997;17:285–291. doi: 10.1038/ng1197-285. [DOI] [PubMed] [Google Scholar]
- Rao E, Weiss B, Fukami M, Rump A, Niesier B, Mertz A, Muroya K, Binder G, Kirsch S, Winkelmann M, Nordsiek G, Heinrich U, Breuning MH, Ranke MB, Rosenthal A, Ogata T, Rappoid GA. Pseudoautosomal deletions encompassing a novel homeobox gene cause growth failure in idiopathic short stature and Turner syndrome. Nat Genet. 1997;16:54–63. doi: 10.1038/ng0597-54. [DOI] [PubMed] [Google Scholar]
- Sauk JJ, Jr, Lyon HW, Witkop CJ., Jr Electron optic microanalysis of two gene products in enamel of females heterozygous for X-linked hypomaturation amelogenesis imperfecta. Am J Hum Genet. 1972;24:267–276. [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Ballabio A, Zoghbi HY. Cloning and characterization of a putative human holocytochrome c-type synthetase gene (HCCS) isolated from the critical region for microphthalmia with linear skin defects (MLS) Genomics. 1996;34:166–172. doi: 10.1006/geno.1996.0261. [DOI] [PubMed] [Google Scholar]
- Schaefer L, Prakash S, Zoghbi HY. Cloning and characterization of a novel rho-type GTPase-activating protein gene (ARHGAP6) from the critical region for microphthalmia with linear skin defects. Genomics. 1997;46:268–277. doi: 10.1006/geno.1997.5040. [DOI] [PubMed] [Google Scholar]
- Seow WK, Amaratunge A. The effects of acid-etching on enamel from different clinical variants of amelogenesis imperfecta: an SEM study. Pediatr Dent. 1998;20:37–42. [PubMed] [Google Scholar]
- Smith CE, Warshawsky H. Quantitative analysis of cell turnover in the enamel organ of the rat incisor. Evidence for ameloblast death immediately after enamel matrix secretion. Anat Rec. 1977;187:63–98. doi: 10.1002/ar.1091870106. [DOI] [PubMed] [Google Scholar]
- Temple IK, Hurst JA, Hing S, Butler L, Baraitser M. De novo deletion of Xp22.2-pter in a female with linear skin lesions on the face and neck, microphthalmia, and anterior chamber eye anomalies. J Med Genet. 1990;27:56–58. doi: 10.1136/jmg.27.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Termine JD, Belcourt AB, Christner PJ, Conn KM, Nylen MU. Properties of dissociatively extracted fetal tooth matrix proteins. I. Principal molecular species in developing bovine enamel. J Biol Chem. 1980;255:9760–9768. [PubMed] [Google Scholar]
- Van den Veyver IB. Microphthalmia with linear skin defects (MLS), Aicardi, and Goltz symdromes: are they related X-linked dominant male-lethal disorders? Cytogenet Genome Res. 2002;99:289–296. doi: 10.1159/000071606. [DOI] [PubMed] [Google Scholar]
- van Dijk JP, Heuver L, Stevens-Linders E, Jansen JH, Mensink EJ, Raymakers RA, de Witte T. Acquired skewing of Lyonization remains stable for a prolonged period in healthy blood donors. Laukemia. 2002;16:362–367. doi: 10.1038/sj.leu.2402379. [DOI] [PubMed] [Google Scholar]
- Wang X, Sutton R, Peraza-Llanes JO, Yu Z, Rosetta R, Kou YC, Eble TN, Patel A, Thaller C, Fang P, Van den Veyver IB. Mutations in X-linked PORCN, a putative regulator of Wnt signaling, cause focal dermal hypoplasia. Nature Genetics. 2007;39:836–838. doi: 10.1038/ng2057. [DOI] [PubMed] [Google Scholar]
- Wapenaar MC, Schiaffino MV, Bassi MT, Schaefer L, Chinault AC, Zoghbi HY, Ballabio A. A YAC-based binning strategy facilitating the rapid assembly of cosmid contigs: 1.6 Mb of overlapping cosmids in Xp22. Hum Mol Genet. 1994;3:1155–1161. doi: 10.1093/hmg/3.7.1155. [DOI] [PubMed] [Google Scholar]
- Wells S, Mould S, Robins D, Robinson D, Jacobs P. Molecular and cytogenetic analysis of a familial microdeletion of Xq. J Med Genet. 1991;28:163–166. doi: 10.1136/jmg.28.3.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White SN, Paine ML, Ngan AY, Miklus VG, Luo W, Wang HJ, Snead ML. Ectopic expression of dentin sialoprotein during amelogenesis hardens bulk enamel. J Biol Chem. 2007;282:5340–5345. doi: 10.1074/jbc.M604814200. [DOI] [PubMed] [Google Scholar]
- Wimplinger I, Morleo M, Rosenberger G, Iaconis D, Orth U, Meinecke P, Lerer I, Ballabio A, Gal A, Franco B, Kutsche K. Mutations of the mitochondrial holocytochrome c-type synthase in X-linked dominant microphthalmia with linear skin defects syndrome. Am J Hum Genet. 2006;79:878–889. doi: 10.1086/508474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimplinger I, Shaw GM, Kutsche K. HCCS loss-of-function missense mutation in a female with bilateral microphthalmia and sclerocornea: a novel gene for severe ocular malformations? Molecular Vision. 2007;13:1475–1482. [PubMed] [Google Scholar]
- Woodward K, Kirtland K, Dlouhy S, Raskind W, Bird T, Malcolm S, Abeliovich D. X inactivation phenotype in carriers of Pelizaeus-Merzbacher disease: skewed in carriers of a duplication and random in carriers of point mutations. Eur J Hum Genet. 2000;8:449–454. doi: 10.1038/sj.ejhg.5200480. [DOI] [PubMed] [Google Scholar]
- Woodward KJ, Cundall M, Sperle K, Sistermans EA, Ross M, Howell G, Gribble SM, Burford DC, Carter NP, Hobson DL, Garbern JY, Kamholz J, Heng H, Hodes ME, Malcolm S, Hobson GM. Heterogeneous duplications in patients with Pelizaeus-Merzbacher disease suggest a mechanism of coupled homologous and nonhomologous recombination. Am J Hum Genet. 2005;77:966–987. doi: 10.1086/498048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Workman C, Jensen LJ, Jarmer H, Berka R, Gautier L, Nielser HB, Saxild HH, Nielsen C, Brunak S, Knudsen S. A new non-linear normalization method for reducing variability in DNA microarray experiments. Genome Biol. 2002;3:research0048.1–0048.16. doi: 10.1186/gb-2002-3-9-research0048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright JT, Hart PS, Aldred MJ, Seow K, Crawford PJ, Hong PS, Gibson CW, Hart TC. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res. 2003;44 1:72–78. [PubMed] [Google Scholar]
- Wright JT. The molecular etiologies and associated phenotypes of amelogenesis imperfecta. Am J Med Genet A. 2006;140:2547–2555. doi: 10.1002/ajmg.a.31358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.