Abstract
DNA repair occurs in a chromatin context, and nucleosome remodeling is now recognized as an important regulatory feature by allowing repair factors access to damaged sites. The yeast mating type locus (MAT) has emerged an excellent model to study the role of chromatin remodeling at a well-defined DNA double-strand break (DSB). We discuss methods to study nucleosome dynamics and DSB repair factor recruitment to the MAT locus after a DSB has been formed.
Overview of double-strand break repair
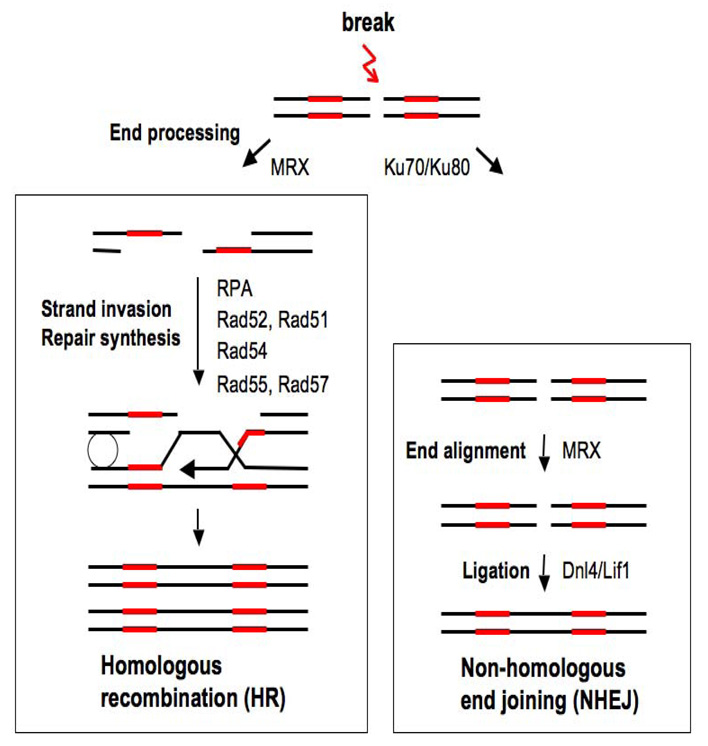
Both exogenous factors such as irradiation and endogenous factors such as stalled replication forks can lead to DSBs in eukaryotic chromosomes. The presence of unrepaired DSBs promotes genome instability, and cells have evolved two major repair pathways to restore intact DNA strands: the nohomolous end-joining (NHEJ) and homologous recombination (HR) pathways (Figure 1). NHEJ involves the direct religation of broken ends, while HR is initiated by 5’ to 3’ DNA resection at ends, utilizing information present on an intact donor chromosome or sister chromatid to repair DNA (1–4) Both pathways employ a set of unique proteins to carry out repair, and the order of protein recruitment and the action of these factors at DSBs have been defined by a combination of biochemical, genetic, molecular, and cytological approaches (5–11). NHEJ can occur throughout the cell cycle, but because strand resection, the essential initial step in HR is promoted in S and G2 phases, NHEJ is the predominant repair pathway in G1 phase and HR is favored in S and G2 phases (3, 12).
Figure 1.
DSB repair takes place in the context of chromatin, which is generally inhibitory to protein-DNA interactions, and over the past several years there have been numerous reports linking chromatin remodeling to the execution of specific steps in DSB repair pathways. Chromatin remodeling encompasses two general classes of factors – those that modify specific histones and those that use ATP hydrolysis to disrupt histone-DNA interactions. Both sets of factors feature prominantly in DSB repair and have been the topic of a number of recent reviews (13–20). Briefly, one of the earliest chromatin-remodeling events at a DSB is the C-terminal phosphorylation of the histone H2A variant H2A.X (H2A in yeast) in a large chromatin domain surrounding the break (10, 21). Other histone modifications also accumulate in the vicinity of DSBs, including H4 acetylation and phosphorylation, and H2A (H2A.X) ubiquitylation (22–29). A third histone modification, H3 lysine 79 methylation, is important for checkpoint signaling in response to a DSB, but its levels are not regulated (30–32). In addition to histone modifications, a number of different ATP-dependent nucleosome remodeling factors also accumulate at DSBs, as primarily identified in yeast. These include the Swi/Snf, RSC, INO80, and SWR1 complexes, but, with the exception of RSC, most of these factors appear at DSBs later than factors that modify histones (19, 27, 33–38). Finally, recent evidence has shown that nucleosomes are displaced from chromatin adjacent to a DSB, with nucleosome reassembly accompanying the completion of DSB repair by HR (39, 40).
The relationship between various chromatin remodeling events and specific steps in DSB repair has been the subject of intense investigation for the past several years. Generally, it is assumed that chromatin remodeling is required for the sequential recruitment or stabilization of repair factors at DSBs as the result of specific alterations of chromatin. Several systems have been developed to assess repair of an induced DSB in vivo, and in this review, we focus on methods to measure chromatin remodeling and repair factor recruitment in the context of DSB repair at the S. cerevisiae MAT locus.
The yeast MAT locus
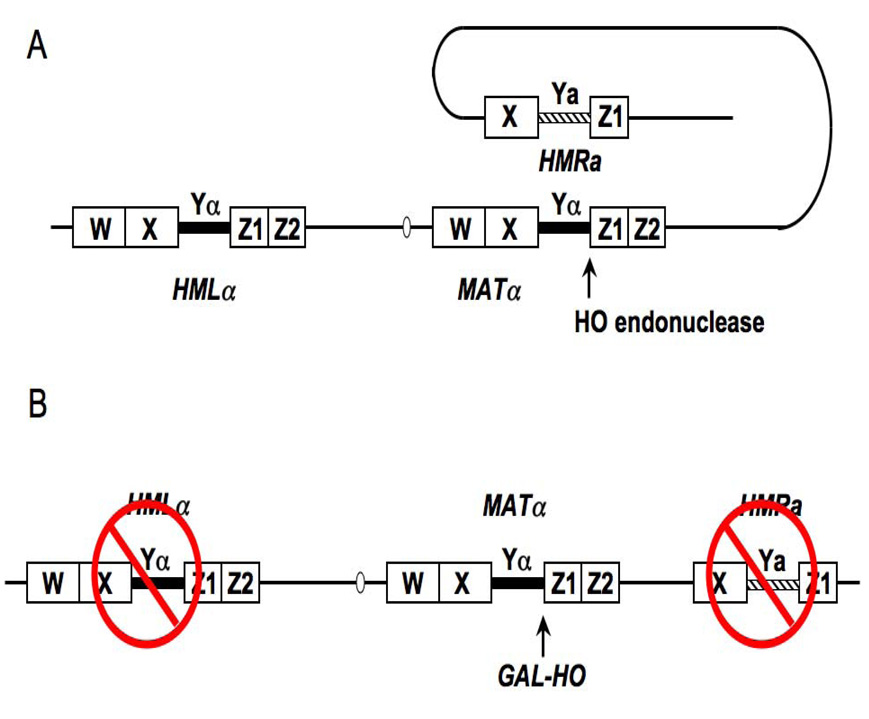
The MAT locus is present on budding yeast chromosome III and is represented by two allelic forms: MAT α and MATa (Figure 2A). These alleles encode regulatory proteins that determine cell type, and thus the MAT locus is transcriptionally active. The MAT locus can interconvert between the two alleles by a specialized form of HR that uses information from one of two silent mating type cassettes at HMRa and HMLα (41) HR is is initiated by a site-specific endonuclease, HO, that makes a DSB at a unique sequence in the MAT locus in the late G1 phase of the cell cycle. Depending on the MAT allele, the DSB will be repaired by gene conversion from either HMRa or HMLα. Through a series of genetic alterations created in the laboratory of James Haber, this system has emerged as an excellent model to study various features of DSB repair (Figure 2B) (42). First, the HO gene has been replaced with a version whose expression can be regulated such that HO can be expressed at all phases of the cell cycle. The most common version is a GAL-HO gene, which can be kept inactive until galactose is added to the medium, and a more recent version contains HO under control of a tetracycline-regulated promoter (35, 42). When HO is expressed, it cleaves its recognition sequences at MAT with almost 100% efficiency. Second, the silent HM donors required for DSB repair by HR have been deleted (Figure 2B). In this configuration, the DSB can only be repaired by NHEJ. However, because GAL-HO can be expressed throughout the cell cycle, strand resection will occur, leading to the recruitment of factors that would normally participate in HR (seeFigure 1). To exclusively study NHEJ, the HO cut can be produced in G1 arrested cells, where strand resection leading to HR is suppressed. Thus, using this system, a researcher can follow the kinetics of chromatin remodeling and recruitment of either NHEJ or HR repair factors to a defined DSB. In the sections below, we discuss methods to follow histone displacement and HR repair factor recruitment to the MAT DSB.
Figure 2.
1. Strains and growth conditions
The parent strain for these studies is JKM179 (MAT α Δho Δhml::ADE1 Δhmr::ADE1 ade1 leu2,3–112 lys5 trp1::hisG ura3–52 ade3::GAL10-HO) (42). Using this strain, appropriate genes can be deleted or their protein products can be tagged with Myc, HA, or Flag epitopes using standard yeast genetics technique (43). Typically, strains will be constructed with one extra chromosomal copy of a Flag tagged H2B or H3 gene to follow histone occupancy around the DSB (39). Cells from relevant strains are grown overnight in a small volume of GLGYP medium (3% glycerol, 2% lactate, 0.05% glucose, 1% yeast extract, 2% peptone, pH 5.5), in which GAL-HO is not expressed. The culture is then diluted into a large volume of GLGYP medium (e.g., 500–1000 ml) and grown to a density of 0.6–0.8 OD600 units per ml, at which time galactose is added to a final concentration of 2% to induce the HO endonuclease. Samples are taken for chromatin analysis as well as for measurement of DNA cleavage and strand resection immediately before and at various times after HO induction as described in more detail in the following sections.
Because DNA repair pathways are greatly influenced by cell cycle stage, it is important to perform these studies in exponentially growing cells, as cells that are predominantly in G1 phase will not undergo the initial steps in HR due to the suppression of DNA strand resection. In addition, deletion of some genes could alter cell cycle distribution. Thus, it is advisable to perform FACS analysis on wild type and mutant strains grown under noninducing conditions to determine cell cycle distributions and thereby eliminate indirect effects on chromatin analyses or factor recruitment.
1.1 Analysis of HO cutting and strand resection
Critical factors to consider when assessing the chromatin changes and patterns of HR factor recruitment that occur in response to the MAT DSB are the efficiency of strand-cleavage by HO and the extent of 5’ to 3’ DNA strand resection at MAT, as these factors can significantly influence data interpretation. Two general methods are used to measure these factors: Southern blot hybridization and qPCR. Here, we present an overview of these methods, and the reader is directed to the appropriate references for detailed protocols.
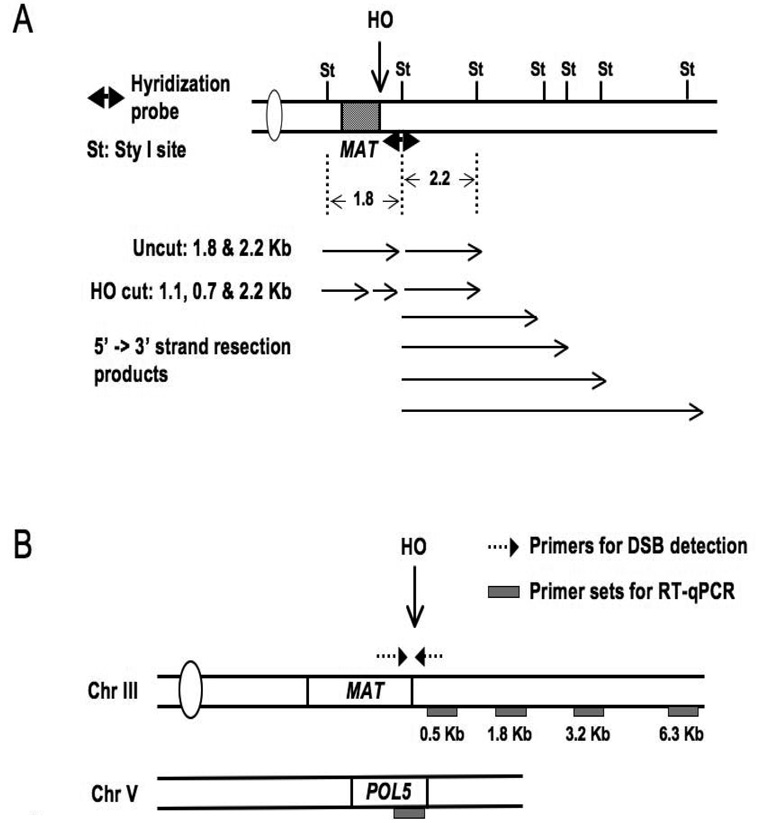
Both methods utilize genomic DNA isolated from unfixed cells before and at various times after HO induction, and qPCR can also be performed on DNA isolated from whole cell extracts as part of the chromatin immunoprecipitation procedure (Section 2) (44). For Southern blot analysis, genomic DNA is isolated and digested with a restriction enzyme such as StyI, whose recognition sequences flank the HO cut site (Figure 3A) (39, 45, 46). The products are separated on either alkaline or non-denaturing agarose gels, and blots are hybridized with a probe that encompasses the StyI site on the right side of the HO cut site. Both undigested MAT sequences and HO-cleaved DNA will be detected by this method. Additionally, the extent of strand resection can be readily assessed, as MAT-distal StyI sites will disappear when single-strand DNA is formed, leading to additional, more slowly migrating bands on the Southern blot. A variation on this method is to use non-denaturing slot-blot hybridization with MAT-specific probes (47). These hybridization methods are suitable for measurement of extended stretches of strand resection and for comparing the kinetics of resection between strains, but they are more visual than quantitative.
Figure 3.
qPCR analysis of non-denatured genomic DNA is used as a quantitative approach to measure both the efficiency of DSB formation as well as the extent of 5’ to 3’ DNA resection. The efficiency of HO cutting can be probed by qPCR simply by using primers that flank the MAT DSB to compare the amplification signal before and after digestion with HO relative to the amplification signal from a control location (Figure 3B) (39). Two general qPCR methods have been employed to measure 5’ to 3’ strand resection. In the first, the difference in qPCR template levels between unresected and resected DNA is measured after digestion with a restriction enzyme that cleaves at defined positions distal to the HO cut site (47). As ssDNA is formed, the restriction enzyme cleavage sites are lost from the amplicons of resected DNA, but not from those of unresected DNA. Thus, after treatment with the restriction enzyme, there will be a reduction in amplifiable DNA from dsDNA templates but not from ssDNA templates. A second qPCR-based method termed QAOS (quantitative amplification of single strand DNA) has been used to measure limited extents of strand resection at MAT (35, 48). In this method, a “tag” single strand oligonucleotide is designed to possess 10–16 nucleotides complementary to a specific region in the developing 3’ ssDNA, plus a second “tag” that does not match sequences in the yeast genome. Annealing this oligonucleotide to nondenatured genomic DNA followed by extension yields a product that can be further amplified and quantitated by qPCR using primers specific to the “tag” and the MAT locus. Because amplification by QAOS depends on the presence of ssDNA with a 3’ overhang, the absence of a PCR signal can be attributed to the absence of resection or to the degradation of the 3’ overhang. Finally, a third PCR- based method, termed LMPCR for ligation mediated PCR, has been used in some instances to determine if resection has been initiated after the HO cut has been formed (11). This method is sensitive enough to detect the resection of as little as a single base at the MAT locus.
2. Chromatin analysis
In this section, we detail the steps to quantitatively measure histone displacement and HR factor recruitment to the MAT DSB by chromatin immunoprecipitation, and then present a general overview of a second method to examine chromatin structure at the MAT locus.
2.1 Chromatin immunoprecipitation (ChIP)
The protocol described in this section was originally developed by Kuo and Allis (49) and has been adapted to follow chromatin remodeling after a DSB has been formed.
2.1.1 Cell harvesting and fixation
Immediately before galactose induction, ~40 OD units of cells (usually 50 ml) are collected for a zero time point and incubated with 1% formaldehyde at room temperature with gentle shaking. Cross-linking with formaldehyde is a time-critical procedure and depends on the lysine content and proximity of these moieties to functional groups on DNA, so the fixation times must be empirically determined for each protein (50). Excessive cross-linking may lead to a reduction in the availability of epitopes for antibody binding, and insufficient cross-linking can increase the likelihood of target migration and reduce the overall yield of DNA template. Because histones are so intimately associated with DNA, a short fixation time of 15 min is sufficient for histone ChIPs. In contrast, a fixation time of 1 hour is required for immunoprecipitation of the single-strand DNA binding proteins RPA and Rad51 that are recruited during early steps in HR. Fixation is stopped by addition of glycine to a final concentration of 0.125M, and incubation is continued for 5 min or for the duration of the experiment.
Galactose is added to the remaining culture at a final concentration of 2% to induce HO, and at 30–60 min intervals after induction, 40 OD units of cells are collected, fixed with formaldehyde for predetermined times, and fixation is stopped by addition of glycine.
The fixed cultures are harvested by centrifugation at 4500 rpm for 2 min in a table top centrifuge, washed twice with ice cold TBS, and the cell pellets are quick frozen in dry ice before storage at −80°C.
2.1.2 Cell lysis
The most common method to lyse cells is by mechanical bead beating. Because every bead-beating device is different, the amount of time required to optimally lyse cells varies. As a rule of thumb, 30–40 OD units of cells are lysed in 0.5 ml lysis buffer in a microfuge tube. For ChIP experiments requiring more than 40 OD units of cells, it is recommended that lysis be performed in multiple tubes. The amount of glass beads should fall 2–3mm below the surface of the suspension in order to obtain efficient lysis. It is also recommended that the protein concentration of the lysate be measured to identify the time at which maximum lysis has occurred.
Each fixed cell pellet is thawed on ice, resuspended in cold 0.5 ml FA lysis buffer containing 1mM PMSF plus 1X protease inhibitor cocktai, and transferred to a prechilled 1.5 ml microfuge tube. Approximately 0.4 gm of acid washed glass beads are then added to the cell suspension.
The cells are lysed at 4°C by vortexing for 15 min at maximum speed using a vortexer fitted with a multitube adaptor.
The whole cell lysate (WCL) is drained into a pre-chilled 15 ml conical tube by puncturing the bottom of the microfuge tube, followed by centrifugation at 1000 rpm for 1 min in a table top centrifuge. Alternatively, the lysate can be removed from the glass beads using a pipetman fitted with a DNA sequencing tip.
2.1.3 Solubilization of chromatin
In most cases, it is beneficial to enrich for cross-linked chromatin in the WCL by a centrifugation step. Eliminating the supernatant, which contains soluble protein, will leave behind most of the crosslinked, chromatin-associated material plus cellular debris. This chromatin-enriched pellet can be resuspended in fresh buffer and solubilized as described below. Including this step can reduce or eliminate background during immunoprecipitation and is recommended when using low affinity antibodies or when the target protein is in low abundance.
Two general methods are employed to fragment chromatin in the WCL or chromatin-enriched fraction: sonication and enzymatic digestion with micrococcal nuclease (MNase). Which method is used will depend upon the type of ChIP experiment being performed and the resolution desired. Here, we describe a method using sonication to solubilize chromatin (51). Sonication creates randomly sized DNA fragments averaging 500–700 base pairs, which would encompass as many as four nucleosomes. While such fragments are sufficient for most ChIP procedures, this size could potentially affect the resolution of ChIP products.
The WCL is spun in a microfuge at 4°C for 10 min at 14,000 rpm to collect the insoluble, chromatin-enriched fraction, and the supernatant is discarded.
The chromatin fraction is resuspended in 0.5 ml FA lysis buffer containing PMSF and 1X protease inhibitors. The lysate is sonicated 6 times for 10 sec each using a Branson Sonifier set at 30% continuous output.
The sonicated lysate is spun in a microfuge at 4°C for 10 min at 14000 rpm and the supernatant is retained. This supernatant contains the solubilized chromatin and is used for all subsequent steps. One OD unit of lysate is set aside to purify input DNA.
2.1.4 Chromatin Immunoprecipitation
The clarified lysate is incubated with appropriate antibodies overnight at 4°C. The amount of lysate used varies with the abundance of the protein being targeted and the affinity of the antibody against the protein. Below, we describe conditions for immunoprecipitation of Flag-tagged H2B or H3 and the HR repair factors RPA and Rad51. Immunoprecipitation conditions for histone modifications and chromatin remodeling factors can be found in relevant publications.
2.1.5 Histone ChIPs
Two and a half OD units of chromatin-enriched lysate are added to a prechilled1.5 ml microfuge tube containing 0.5 ml of cold FA lysis buffer with PMSF and 1X protease inhibitors. Eighty µl of anti-Flag agarose beads are added (Sigma, 50% slurry washed 3X in 125 mM glycine and 1X in FA lysis buffer), and the suspension is rotated overnight at 4°C
The beads are collected by centrifugation at 2000 rpm for 2 min and sequentially washed 2 times each with FA lysis buffer, wash buffer #1, wash buffer #2, and 1X TE.
The beads are suspended in 0.25 ml elution buffer and incubated at 65°C for 15 min to elute the antibody complexes. The eluate is removed to a new 1.5 ml microfuge tube, digested with pronase (2 mg/ml plus 5 mM CaCl) for 2 hr at 42°C, and incubated at 65°C overnight to reverse formaldehyde crosslinks.
DNA from both the retained WCL (input) and immunoprecipitated (IP) samples are purified with a Qiagen PCR purification kit and stored at −20°C.
2.1.6 RPA and Rad51 ChIPs
Twenty OD units are added to 0.5 ml FA lysis buffer containing PMSF and protease inhibitors as described for histone ChIPs. Two microliters of serum containing antibodies against native RPA or Rad51 protein are added and incubation is performed overnight at 4°C.
Eighty microliters of protein A beads equilibrated in FA buffer (Invitrogen, washed 3X with FA buffer plus 0.05% BSA; 1X with FA buffer plus PMSF and protease inhibitors) are added, and incubation is performed for 2 hr at 4°C.
Collection of beads, washes, and DNA elution and purification are performed as outlined in section 2.1.5.
2.2 Quantitation of ChIP DNA and data analysis
2.2.1 Real time PCR (RT-qPCR) analysis
In this section, we discuss methods for quantitation of input and IP DNA using a real time qPCR assay (RT-qPCR) with Sybr Green fluoresecence detection. While free Sybr Green has little fluorescence on its own, the magnitude of its fluorescence increases when bound to double-strand DNA, so as the amount of PCR amplification product increases, the fluorescence signal from Sybr Green also increases. RT-qPCR generates threshold cycle (CT) values, with the threshold cycle representing the amplification cycle at which there is a significant increase in fluorescence signal associated with exponential growth of the PCR product. Thus, the higher the concentration of DNA in the initial sample, the fewer cycles are required to reach exponential accumulation of the PCR product and the lower the CT value. To obtain accurate quantitation of DNA, DNA standards must be included for each set of primers in every PCR run. A large quantity of standard DNA is prepared from cells exactly as described for preparation of input DNAs for ChIP analysis. The standard DNA is 10-fold serially diluted from 1 to 1/10,000 (0.01 ng/µl to 100 ng/µl), RT-qPCR is performed with a given primer pair, and a standard curve is generated from the CT value for each DNA concentration. The amount of input and immunoprecipitated DNA from the experimental samples is then derived from the standard curve. As a rule of thumb, input DNA is diluted 1/100 and IP DNAs are diluted 1/10–1/25 for qPCR analysis. However, If the fluorescent signal from an experimental sample does not fall within the linear range of the standard curve, the sample concentration needs to be adjusted for the qPCR results to be accurate.
2.2.2 Analysis of histone levels at the MAT DSB
Following the induction of the MAT DSB, nucleosomes are gradually displaced around the break site (39). In this section, we outline steps in the analysis of histone levels at the MAT DSB using qPCR on input and IP DNA isolated from strains carrying Flag tagged H2B or H3. qPCR primers are designed to various locations surrounding the DSB site using Primer Express software for analysis of DNA on an ABI real time PCR machine (Figure 3B). Primers to a position on the POL5 gene, where no DSB occurs, are included as a control.
To determine the extent of histone displacement at the MAT DSB, the amount of IP DNA at each MAT location is divided by the amount of IP DNA at POL5 (MAT IP DNA/POL5 IP DNA). The MAT IP DNA/POL5 IP DNA ratio at each time point after HO induction is then normalized to the MAT IP DNA/POL5 DNA IP at zero time, and the normalized values are plotted on a log2 scale to visualize the changes in histone occupancy at MAT over time after DSB formation (39).
2.2.3 Analysis of HR repair factor recruitment to the MAT DSB
For analysis of RPA and Rad51 recruitment to the MAT DSB, DNA is amplified with the same primers described for analysis of histone loss. However, for data analysis, the MAT IP DNA value is divided by the POL5 INPUT DNA value instead of the POL5 IP DNA value. Using POL5 INPUT DNA as denominator avoids artifactual results, because in the case of the repair proteins, the POL5 IP DNA value represents a background signal and experimental variation in this signal can significantly alter the results. For example, if a 2- or 3-fold fluctuation occured in the POL5 IP signal, even if its absolute value was very small, the MAT IP DNA value would be changed by 2- or 3-fold.
2.3. MNase analysis of MAT chromatin structure
In addition to measuring histone occupancy by ChIP, the chromatin structure of MAT can also be assessed by examining the micrococcal (MNase) sensitivity of this locus after induction of the DSB (33, 36, 39). The MAT locus is organized into a highly ordered array of nucleosomes, and changes in this pattern can be easily visualized by Southern blot analysis of DNA purified from nuclei treated with MNase and probed with DNA from the MAT locus (52). In this section, we give a brief overview of this procedure, and the reader is directed to pertinent references for details (39, 53).
To obtain enough material for MNase analysis, generally 2–4 liters of cells are grown in GLGYP medium to mid-log phase; 500–1000 ml are retained as an uninduced control, and galactose is added to 2% to the remaining culture.
At 1, 2, and 3 hrs after induction, 500–1000 ml of culture are removed, the cells are collected by centrifugation, and a crude nuclei preparation is prepared from cells that have been spheroplasted with either Lyticase or Zymolase following published procedures (39, 53). Once prepared, the nuclei are immediately used for MNase digestion.
MNase digestion of the nuclei is performed at 37° using a fixed concentration of MNase and removing samples for DNA isolation over time (0–15 min). The amount of nuclei and concentration of MNase are first empirically determined on a small-scale sample before proceeding with large-scale enzyme digestion.
DNA is purified using proteinase K digestion, followed by phenol-chloroform and EtOH precipitation, and then by RNaseA treatment. The purified DNA is run on a 20–30 cm long 1.25% agaose gel in TBE buffer for best resolution.
DNA is transferred to an appropriate membrane and Southern blot analysis is performed with a labeled 800 bp fragment corresponding to sequences ~200 bp from the right side of the HO cut site (39). In wild type cells, the MNase pattern becomes progressively less organized and fainter with time after HO digestion (39). Mutations in some chromatin remodeling factors prevent this disorganization of MAT chromatin (36, 39).
3. Reagents
ChIP analysis
FA lysis buffer: 50 mM Hepes-KOH pH 7.5, 0.14 M NaCl, 1 mM EDTA, 1% Triton, 0.1% sodium deoxycholate
1X TBS: 0.05 M Tris pH8.0, 0.138 M NaCl, 0.0027 M KCl
1X PI: protease inhibitor cocktail (Sigma cat.#P2714)
Wash buffer #1: 50 mM Hepes-KOH pH 7.5, 0.5 M NaCl, 1 mM EDTA, 1% Triton, 0.1% sodium deoxycholate
Wash buffer #2: 0.25 M LiCl, 1XTE, 0.5% NP-40, 0.5% Sodium deoxycholate
Elution buffer: 1% SDS, 10 mM TrisHCl pH 7.4, 1 mM EDTA
RT-qPCR analysis
Syber Green PCR master mix: Applied Biosystem Inc. cat# 4309155; New England Biolab Cat# F-410; Fermentas cat# K0221
Concluding remarks
Many of the methods originally developed to study chromatin dynamics during transcription have been successfully adapted to study this phenomenon during the repair of DSBs. Extensive parallels exist between the two DNA-mediated processes, and many of the same histone modifications and ATP-dependent nucleososome remodeling factors that contribute to transcriptional regulation also play key roles in the regulation of DSB repair. Progress in understanding the relationship of chromatin remodeling to transcription has come largely from the study of model genes. The yeast MAT system provides a similar model to define the role of chromatin remodeling in DSB repair, and research on this system has greatly advanced our understanding of the range of chromatin remodeling events that participate in the various steps that preserve genome integrity.
Acknowledgments
Research in the authors’ laboratories is supported by grants from the NIH (CA118537; M.A.O.) and the CEA and CNRS (E.M.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Paques F, Haber JE. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cahill D, Connor B, Carney JP. Front Biosci. 2006;11:1958–1976. doi: 10.2741/1938. [DOI] [PubMed] [Google Scholar]
- 3.Shrivastav M, De Haro LP, Nickoloff JA. Cell Res. 2008;18:134–147. doi: 10.1038/cr.2007.111. [DOI] [PubMed] [Google Scholar]
- 4.Sonoda E, Hochegger H, Saberi A, Taniguchi Y, Takeda S. DNA Repair (Amst) 2006;5:1021–1029. doi: 10.1016/j.dnarep.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 5.Krogh BO, Symington LS. Annu Rev Genet. 2004;38:233–271. doi: 10.1146/annurev.genet.38.072902.091500. [DOI] [PubMed] [Google Scholar]
- 6.Lisby M, Barlow JH, Burgess RC, Rothstein R. Cell. 2004;118:699–713. doi: 10.1016/j.cell.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 7.Miyazaki T, Bressan DA, Shinohara M, Haber JE, Shinohara A. Embo J. 2004;23:939–949. doi: 10.1038/sj.emboj.7600091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugawara N, Wang X, Haber JE. Mol Cell. 2003;12:209–219. doi: 10.1016/s1097-2765(03)00269-7. [DOI] [PubMed] [Google Scholar]
- 9.Wolner B, van Komen S, Sung P, Peterson CL. Mol Cell. 2003;12:221–232. doi: 10.1016/s1097-2765(03)00242-9. [DOI] [PubMed] [Google Scholar]
- 10.Shroff R, Arbel-Eden A, Pilch D, Ira G, Bonner WM, Petrini JH, Haber JE, Lichten M. Curr Biol. 2004;14:1703–1711. doi: 10.1016/j.cub.2004.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frank-Vaillant M, Marcand S. Mol Cell. 2002;10:1189–1199. doi: 10.1016/s1097-2765(02)00705-0. [DOI] [PubMed] [Google Scholar]
- 12.Lisby M, Rothstein R. Biochimie. 2005;87:579–589. doi: 10.1016/j.biochi.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 13.Allard S, Masson JY, Cote J. Biochim Biophys Acta. 2004;1677:158–164. doi: 10.1016/j.bbaexp.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 14.Bao Y, Shen X. Curr Opin Genet Dev. 2007;17:126–131. doi: 10.1016/j.gde.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 15.Downs JA, Cote J. Cell Cycle. 2005;4:1373–1376. doi: 10.4161/cc.4.10.2108. [DOI] [PubMed] [Google Scholar]
- 16.Downs JA, Nussenzweig MC, Nussenzweig A. Nature. 2007;447:951–958. doi: 10.1038/nature05980. [DOI] [PubMed] [Google Scholar]
- 17.Moore JD, Krebs JE. Biochem Cell Biol. 2004;82:446–452. doi: 10.1139/o04-034. [DOI] [PubMed] [Google Scholar]
- 18.Morrison AJ, Shen X. Results Probl Cell Differ. 2006;41:109–125. doi: 10.1007/400_008. [DOI] [PubMed] [Google Scholar]
- 19.Osley MA, Shen X. Trends Genet. 2006;22:671–677. doi: 10.1016/j.tig.2006.09.007. [DOI] [PubMed] [Google Scholar]
- 20.Wurtele H, Verreault A. Curr Opin Cell Biol. 2006;18:137–144. doi: 10.1016/j.ceb.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 21.Rogakou EP, Pilch DR, Orr AH, Ivanova VS, Bonner WM. J Biol Chem. 1998;273:5858–5868. doi: 10.1074/jbc.273.10.5858. [DOI] [PubMed] [Google Scholar]
- 22.Bird AW, Yu DY, Pray-Grant MG, Qiu Q, Harmon KE, Megee PC, Grant PA, Smith MM, Christman MF. Nature. 2002;419:411–415. doi: 10.1038/nature01035. [DOI] [PubMed] [Google Scholar]
- 23.Cheung WL, Turner FB, Krishnamoorthy T, Wolner B, Ahn SH, Foley M, Dorsey JA, Peterson CL, Berger SL, Allis CD. Curr Biol. 2005;15:656–660. doi: 10.1016/j.cub.2005.02.049. [DOI] [PubMed] [Google Scholar]
- 24.Murr R, Loizou JI, Yang YG, Cuenin C, Li H, Wang ZQ, Herceg Z. Nat Cell Biol. 2006;8:91–99. doi: 10.1038/ncb1343. [DOI] [PubMed] [Google Scholar]
- 25.Qin S, Parthun MR. Mol Cell Biol. 2006;26:3649–3658. doi: 10.1128/MCB.26.9.3649-3658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Utley RT, Lacoste N, Jobin-Robitaille O, Allard S, Cote J. Mol Cell Biol. 2005;25:8179–8190. doi: 10.1128/MCB.25.18.8179-8190.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Downs JA, Allard S, Jobin-Robitaille O, Javaheri A, Auger A, Bouchard N, Kron SJ, Jackson SP, Cote J. Mol Cell. 2004;16:979–990. doi: 10.1016/j.molcel.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 28.Mailand N, Bekker-Jensen S, Faustrup H, Melander F, Bartek J, Lukas C, Lukas J. Cell. 2007;131:887–900. doi: 10.1016/j.cell.2007.09.040. [DOI] [PubMed] [Google Scholar]
- 29.Huen MS, Grant R, Manke I, Minn K, Yu X, Yaffe MB, Chen J. Cell. 2007;131:901–914. doi: 10.1016/j.cell.2007.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lazzaro F, Sapountzi V, Granata M, Pellicioli A, Vaze M, Haber JE, Plevani P, Lydall D, Muzi-Falconi M. Embo J. 2008;27:1502–1512. doi: 10.1038/emboj.2008.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toh GW, O'Shaughnessy AM, Jimeno S, Dobbie IM, Grenon M, Maffini S, O'Rorke A, Lowndes NF. DNA Repair (Amst) 2006;5:693–703. doi: 10.1016/j.dnarep.2006.03.005. [DOI] [PubMed] [Google Scholar]
- 32.Giannattasio M, Lazzaro F, Plevani P, Muzi-Falconi M. J Biol Chem. 2005;280:9879–9886. doi: 10.1074/jbc.M414453200. [DOI] [PubMed] [Google Scholar]
- 33.Kent NA, Chambers AL, Downs JA. J Biol Chem. 2007;282:27693–27701. doi: 10.1074/jbc.M704707200. [DOI] [PubMed] [Google Scholar]
- 34.Morrison AJ, Highland J, Krogan NJ, Arbel-Eden A, Greenblatt JF, Haber JE, Shen X. Cell. 2004;119:767–775. doi: 10.1016/j.cell.2004.11.037. [DOI] [PubMed] [Google Scholar]
- 35.van Attikum H, Fritsch O, Hohn B, Gasser SM. Cell. 2004;119:777–788. doi: 10.1016/j.cell.2004.11.033. [DOI] [PubMed] [Google Scholar]
- 36.Liang B, Qiu J, Ratnakumar K, Laurent BC. Curr Biol. 2007;17:1432–1437. doi: 10.1016/j.cub.2007.07.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Attikum H, Fritsch O, Gasser SM. Embo J. 2007;26:4113–4125. doi: 10.1038/sj.emboj.7601835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chai B, Huang J, Cairns BR, Laurent BC. Genes Dev. 2005;19:1656–1661. doi: 10.1101/gad.1273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tsukuda T, Fleming AB, Nickoloff JA, Osley MA. Nature. 2005;438:379–383. doi: 10.1038/nature04148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen CC, Carson JJ, Feser J, Tamburini B, Zabaronick S, Linger J, Tyler JK. Cell. 2008;134:231–243. doi: 10.1016/j.cell.2008.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Haber JE. Annu Rev Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 42.Sugawara N, Haber JE. Methods Enzymol. 2006;408:416–429. doi: 10.1016/S0076-6879(06)08026-8. [DOI] [PubMed] [Google Scholar]
- 43.Petracek ME, Longtine MS. Methods Enzymol. 2002;350:445–469. doi: 10.1016/s0076-6879(02)50978-2. [DOI] [PubMed] [Google Scholar]
- 44.Hoffman CS, Winston F. Gene. 1987;57:267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- 45.Lee SE, Moore JK, Holmes A, Umezu K, Kolodner RD, Haber JE. Cell. 1998;94:399–409. doi: 10.1016/s0092-8674(00)81482-8. [DOI] [PubMed] [Google Scholar]
- 46.Clerici M, Mantiero D, Lucchini G, Longhese MP. EMBO Rep. 2006;7:212–218. doi: 10.1038/sj.embor.7400593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zierhut C, Diffley JF. Embo J. 2008;27:1875–1885. doi: 10.1038/emboj.2008.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Booth C, Griffith E, Brady G, Lydall D. Nucleic Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kuo MH, Allis CD. Methods. 1999;19:425–433. doi: 10.1006/meth.1999.0879. [DOI] [PubMed] [Google Scholar]
- 50.Jackson V. Methods. 1999;17:125–139. doi: 10.1006/meth.1998.0724. [DOI] [PubMed] [Google Scholar]
- 51.Orlando V, Strutt H, Paro R. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 52.Weiss K, Simpson RT. Mol Cell Biol. 1998;18:5392–5403. doi: 10.1128/mcb.18.9.5392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fleming AB, Pennings S. Embo J. 2001;20:5219–5231. doi: 10.1093/emboj/20.18.5219. [DOI] [PMC free article] [PubMed] [Google Scholar]