Abstract
Charcot-Marie-Tooth (CMT) disease or hereditary motor and sensory neuropathy (HMSN) is a genetically heterogeneous group of conditions that affect the peripheral nervous system. The disease is characterized by degeneration or abnormal development of peripheral nerves and exhibits a range of patterns of genetic transmission. In the majority of cases, CMT first appears in infancy, and its manifestations include clumsiness of gait, predominantly distal muscular atrophy of the limbs, and deformity of the feet in the form of foot drop. It can be classified according to the pattern of transmission (autosomal dominant, autosomal recessive, or X linked), according to electrophysiological findings (demyelinating or axonal), or according to the causative mutant gene. The classification of CMT is complex and undergoes constant revision as new genes and mutations are discovered. In this paper, we review the most efficient diagnostic algorithms for the molecular diagnosis of CMT, which are based on clinical and electrophysiological data.
1. Introduction
Hereditary neuropathies are a genetically heterogeneous group of diseases that affect the peripheral nervous system. The most common form is the hereditary motor and sensory neuropathy (HMSN), also called Charcot-Marie-Tooth disease (CMT). Almost 120 years have elapsed since the first contemporary description of the same familial neurological syndrome, “peroneal muscular atrophy” (Charcot and Marie, 1886; Tooth, 1886). Dyck and Lambert (1968) were the first to observe, using electrophysiological studies, a marked nerve conduction slowing in some families, whereas conduction was preserved in others. These authors contributed to a rational classification of the complex peroneal muscular atrophies according to inheritance pattern and clinical, electrophysiological, and pathological features. They introduced the term “hereditary motor and sensory neuropathy” and classified the autosomal-dominant form with low conduction velocities as HMSN type I (or CMT 1) and the autosomal-dominant form with preserved conduction velocities as HMSN type II (or CMT 2). Harding and Thomas (1980) observed that the motor nerve conduction velocities (NCV) showed a bimodal distribution and set a median NCV of 38 m/s as an arbitrary but rational threshold between the demyelinating HMSN type I (motor NCV <38 m/s) and the axonal HMSN type II (motor NCV >38 m/s).
2. Classification
The most commonly used classification combines clinical findings with the inheritance pattern (autosomal dominant, autosomal recessive, or X linked) and either electrophysiological or anatomical pathology findings (axonal or demyelinating). The demyelinating forms are characterized by predominantly affecting the sheath of myelin that surrounds the axon of peripheral nerves. The main function of myelin is to increase NCV. Therefore, the most notable electrophysiological finding in this form of the disorder is a low NCV. The axonal forms of the disease are caused by specific effects on the axon itself; neurophysiological studies reveal conserved NCVs, while the amplitude of the motor and sensory potentials (which are a reliable indication of axon conservation) are greatly reduced. Recently, disease variants have been described that combine effects on both the myelin and the axon and that, therefore, have intermediate conduction speeds. The transmission of many of these new variants is X linked [1].
The combination of these data allows the identification of several main categories of CMT. The majority of cases can be classified as CMT1 (i.e., demyelinating forms with low NCVs and autosomal-dominant transmission), CMT2 (i.e., axonal form, usually showing a dominant inheritance pattern), or CMT4 (i.e., recessive and severe forms). Each of these types can be further divided into subtypes, depending on the underlying causative molecular defect (Table 1). However, it should be noted that there is not a good genotype—phenotype correlation and that great variability exists, both within and between families, regarding the degree of clinical expression [2]. With the advent of genetic testing, nerve biopsies are reserved for patients for whom genetic testing did not lead to a molecular diagnosis, for patients with atypical presentation, or for patients with suspected inflammatory neuropathy. Recently, peripheral nerve, magnetic resonance studies and skin biopsy have emerged as potential diagnostic aids for certain types of hereditary neuropathies [3].
Table 1.
Summary of CMT classification, including clinical data (such as electrophysiological results) and molecular findings.
| Dominant forms | |||
|---|---|---|---|
| CMT 1 (demyelinating) | |||
| Type | Gene | Inheritance | Locus |
| CMT1A: | PMP-22 | Dominant/sporadic | 17p11 |
| CMT1B: | P0 protein | Dominant | 1q22 |
| CMT1C: | LITAF | Dominant | 16p13 |
| CMT1D: | EGR2 | Dominant | 10q21 |
| CMT1E: | P0 protein | Dominant | 1q22 |
| CMT1F: | Neurofilament | Dominant/sporadic | 8p21 |
| Light chain | |||
| CMT 2 (axonal) | |||
| Type | Gene | Inheritance | Locus |
| CMT 2A1: | KIF1B | Dominant | 1p36 |
| CMT 2A2: | MFN2 | Dominant | 1p36 |
| CMT 2B: | RAB7 | Dominant | 3q13-q22 |
| CMT 2C: | Dominant | 12q23-q24 | |
| CMT 2D: | GARS | Dominant | 7p15 |
| CMT 2E: | Neurofilament light chain | Dominant | 8p21 |
| CMT 2F: | HSPB1 | Dominant/recessive | 7q11-q21 |
| CMT 2G: | Dominant | 12q12 | |
| CMT 2I: | P0 | Dominant | 1q22 |
| CMT 2J: | P0 | Dominant | 1q22 |
| CMT 2L: | HSPB8 | Dominant | 12q24 |
| AR-CMT2A | Lamin A/C | Recessive | 1q21.2 |
| AR-CMT2B | Med25 | Recessive | 19q13.3 |
| CMT 2K: | GDAP1 | Recessive/dominant | 8q21 |
| Intermediate NCV | |||
| Type | Gene | Inheritance | Locus |
| Connexin-32 | GJB1 | X-linked | Xq13 |
| CMT DIA | Dominant | 10q24 | |
| CMT DIB | DNM2 | Dominant | 19p12 |
| CMT DIC | Tyrosyl-tRNA synthetase | Dominant | 1p34 |
| CMT DI3 | P0 | Dominant | 1q22 |
| CMT 2E: | Neurofilament light chain | Dominant | 8p21 |
| CMT 4 | |||
| Type | Gene | Inheritance | Locus |
| CMT 4A: | GDAP1 | Recessive | 8q21 |
| CMT 4B1: | MTMR2 | Recessive | 11q23 |
| CMT 4B2: | SBF2 | Recessive | 11p15 |
| CMT 4C: | SH3TC2 (KIAA1985) | Recessive | 5q32 |
| CMT 4D (Lom): | NDRG1 | Recessive | 8q24 |
| CMT 4E: | EGR2 | Recessive/dominant | 10q21 |
| CMT 4F: | Periaxin | Recessive | 19q13 |
| HMSN-Russe | (4G) | Recessive | 10q23 |
| CMT 4H: | FGD4 | Recessive | 12q12 |
3. Epidemiology of CMT
The prevalence of CMT ranges from 10 to 30 per 100,000, depending on the geographical region of origin [4, 5]. Two extensive studies in the literature indicate that the most common subtypes are CMT1 (demyelinating with autosomal-dominant transmission) and CMT2 (axonal and usually dominant). In Western countries, the most common cause of CMT1 is CMT1A, which results from the duplication of a genomic fragment that encompasses the PMP22 gene on chromosome 17. It would appear that CMT1B is more common in Japan than in Western countries [6, 7]. In the recent years, all the published studies identify CMT2A as the most common cause of CMT2 (which would account for 10–33% of cases) [8].
4. CMT1
This form is characterized by autosomal-dominant transmission and NCVs in the demyelinating range, as indicated by electrophysiological studies, that is, NCVs <38 m/s (the normal NCV being >50 m/s). The degree of reduction in NCV does not correlate with the clinical severity of the disease, and low NCVs can even be detected in asymptomatic individuals and as early as one year of age [9].
Nerve biopsy, which is not routinely carried out, reveals a loss of large myelinated fibers, demyelination and remyelination, and an “onion bulb” appearance.
CMT1 can be divided into CMT1 A, B, C, D, E, or F, depending on the underlying causative molecular defect (Table 1).
4.1. CMT1A
CMT1A is the most common cause of CMT1 in Western countries. In the majority of cases, it is caused by the duplication of a 1.5 Mb fragment on chromosome 17p.11 that includes the PMP22 gene. The PMP22 protein is predominantly expressed in Schwann cells and can be found in compact myelin. It has a structural function (myelination) and regulates cell growth. Overexpression of PMP22 reduces the proliferation of Schwann cells. This trait is dominant; in 89% of cases, it is of paternal origin, while in 11% of cases it is of maternal origin [2, 9]. It is stimated that about 5–24% of the the duplication mutation may ocurr de novo thus, the absence of family history does not preclude genetic testing.
Molecular studies of PMP22 are carried out using multiplex ligation-dependent probe amplification (MLPA), in which all 38 fragments of the gene are amplified at once (nine within the different exons of the PMP22 gene, seven in the region that encompasses the duplication, two outside the duplicated region, and the rest in different chromosomes, as controls to calculate gene dosage). Once this Polimerase Chain Reaction has been performed, sequence analysis is carried out using an ABI Prism sequencer. To complete the study, seven molecular markers or microsatellites present in the 1.5 Mb duplicated region are analyzed. Microsatellites are repetitions of 2 to 6 nucleotides that exhibit great variability, which renders them very polymorphic. They are normally found in noncoding regions of the genome. The microsatellites considered here are D17S839, D17S921, D17S1356, D17S1357, D17S1358, D17S955, and D17S261. They are amplified using PCR and are then analyzed using the ABI Prism sequencer.
Point mutations of this gene have also been described as a cause of CMT1A, although not very frequently (2–5% of CMT1A cases) [7]. It is estimated that about one third of the point mutations may ocurr de novo. The molecular analysis of these cases should be performed by direct sequencing of each of the coding fragments and exons of the gene.
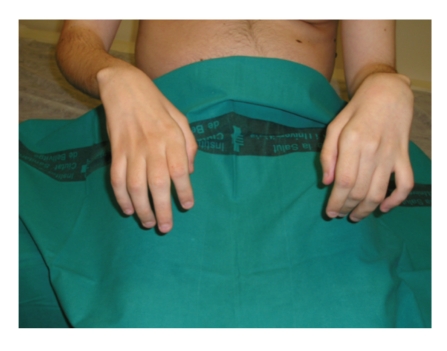
Symptoms usually first appear in infancy and include clumsiness of gait and deformity of the feet. Patients may suffer from cramps, which are heightened after physical exercise. Positive sensory alterations are extremely rare and represent a departure feature from acquired polyneuropathies. Foot drop is evident on physical examination (Figure 1) and so are retraction of the Achilles tendon and the presence of an abundance of corns. Over time, these symptoms are compounded by weakness and atrophy of the intrinsic foot and peroneal muscles, which cause “steppage” gait. The hands can also be affected, with the presence of claw hand in extreme cases. Generalized areflexia is an early and very frequent finding. Some patients present with scoliosis (10%) and nerve hypertrophy (25–66%). Extensive electrophysiological studies reported in the literature reveal that NCVs are always in the demyelinating range and remain constant throughout the evolution of the disease. This confirms that the determination of motor NCV of the median nerve is an excellent marker for genetically determined neuropathies. Conduction blocks are rare in CMT1A, as are temporal dispersion phenomena, as opposed to what is observed in acquired demyelinating neuropathies or CMT1 Types B and C. There may also be signs of secondary axonal degeneration [7].
Figure 1.
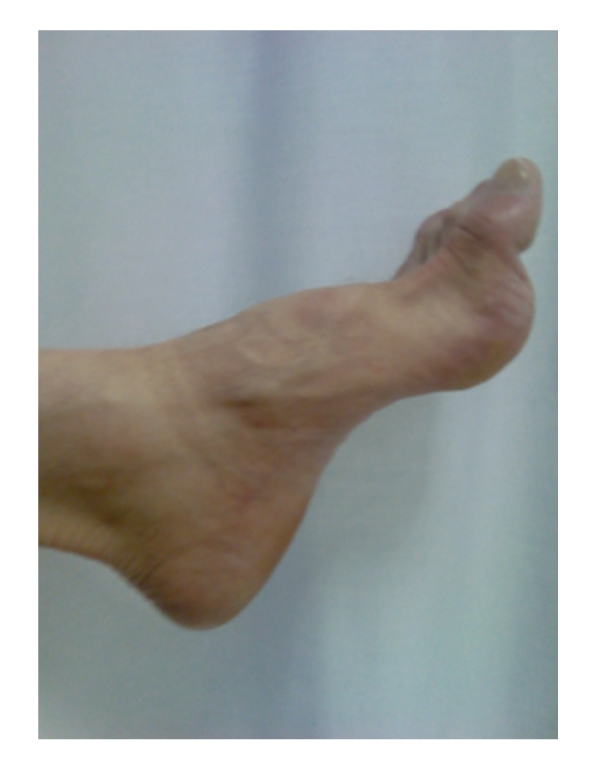
Foot drop in a patient with CMT1 caused by duplication of the genomic fragment that encompasses PMP22 (CMT1A).
Hereditary Neuropathy with Liability to Pressure Palsies (HNPP).
In the majority of cases (85%), HNPP is caused by the deletion of a 1.5 Mb fragment on chromosome 17p.11, which includes the PMP22 gene. Therefore, the deleted region is identical to the region duplicated in CMT1A. Nonsense mutations, frameshifts with premature termination, missense mutations, and splice site changes have been reported in the remaining 15% of cases. The clinical phenotype of these patients is characterized by recurrent nerve dysfunction at compression sites. Asymmetric palsies can occur after relatively minor compression or trauma. With ageing, these patients can have a significant clinical overlap with CMT1, as the repeated injuries to the nerve can prevent full reversal. Electrophysiological findings include mildly slowed NCV and increased motor latencies and conduction blocks [3, 10–12].
4.2. CMT1B
This form of the disease is caused by mutations in the myelin zero protein (MZP), which is a 28 kDa protein comprising 219 amino acids. MZP is found exclusively in Schwann cells and is the most abundant protein in the myelin of peripheral nerve tissues, whereas it is not found in the myelin of the central nervous system. It is indispensable for the normal structure and function of myelin. The gene that encodes MZP has six exons and is located on chromosome 1q22. In the majority of cases, the disease is caused by point mutations, particularly in exons 2 and 3, which correspond to the immunoglobulin-like extracellular domain [8–10].
In CMT1B, the onset of symptoms may be delayed, movement of the pupils may be affected, and deafness may be present. Electrophysiological studies reveal that patients can be segregated into two groups, according to NCV: one with NCVs <38 m/s and the other with NCVs above this threshold (i.e., in the axonal range). These findings render the classification of CMT even more difficult, as mutations in the same gene can cause either CMT1 (demyelinating), CMT2 (axonal), CMT with intermediate NCV (CMTDI3) or CMT associated with deafness (CMT1E) [6]. Nerve conduction blocks have also been described in CMT1B [13].
4.3. Other Types of CMT1
Mutations in the LITAF and ERG2 genes cause CMT1C and D, respectively. In CMT1C, electrophysiological findings include the presence of temporal dispersion and nerve conduction blocks. The molecular study of both genes is carried out by direct sequencing to search for point mutations in coding regions [6].
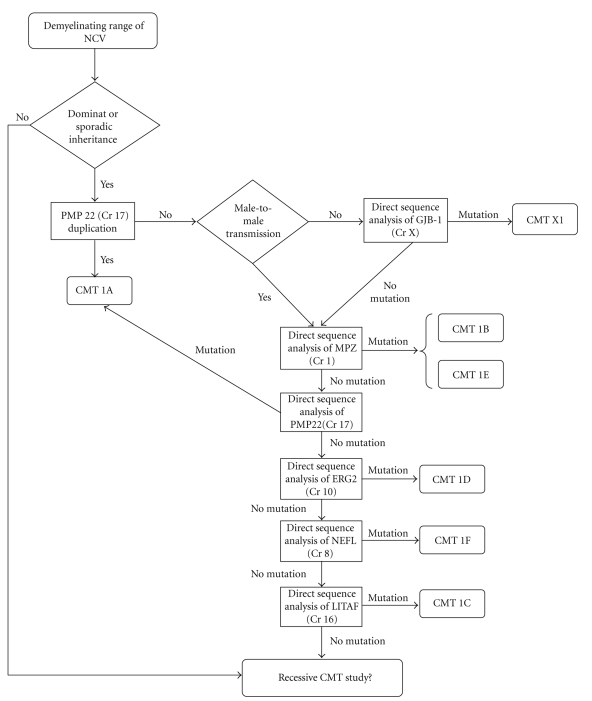
5. Diagnostic Protocol for CMT1
The diagnostic protocol for demyelinating and dominant CMT (CMT1), which has been adapted from England et al. [14, 15], is summarized in Figure 2. Given that the data available suggest that duplication of PMP22 is the most frequent cause of CMT1 and that mutation of PMP22 is the most common cause of sporadic CMT1 [6, 16], it is proposed that the study of demyelinating CMT should begin by checking for duplication of the genomic fragment encompassing PMP22. Once this duplication has been ruled out, in sporadic cases or in cases where there is no male-to-male transmission, researchers should check for point mutations in the GJB1 gene (which can be used to diagnose up to 12% of cases). If there are no changes in GJB1, MPZ and then PMP22 should be analyzed for the presence of point mutations. In the remaining cases, one should look for alterations in genes that are known to cause CMT at a lower frequency: EGR2, NEFL, and LITAF.
Figure 2.
Diagnostic protocol for demyelinating CMT (adapted from England et al.) [10, 11].
6. CMT2
The most common form of CMT2 is CMT2A [17–19]. It is caused by mutations in MFN2, which codes for the mitofusin 2 protein. Mitofusin 2 is a GTPase that is involved in the fusion of mitochondria. It is ubiquitously expressed and is present in the spinal cord, muscle, heart, and peripheral nerves. Intracellularly, it is attached to the outer mitochondrial membrane via its C-terminal domain. It also has an N-terminal GTPase domain, which is located in the cytoplasm. This domain can form a complex with another mitochondrial protein, mitofusin 1. Mitofusin 2 regulates the architecture of the mitochondrial network via mitochondrial fusion, in a stage downstream of mitofusin 1 [19–24].
The majority of MFN2 mutations described in the literature are of the missense type, although stop mutations have also been described. These mutations have been found in cytoplasmic domains, within or immediately upstream of the GTPase domain, within two coiled-coil domains, or in association with the functioning or mitochondrial targeting of mitofusin 2. De novo mutations have also been found occasionally. Although most cases are autosomal dominant, homozygous or compound heterozygous mutations that produce a more severe form of the disease have been described. The molecular study of CMT2 is carried out by direct sequencing of the 19 exons of MFN2 [18–20].
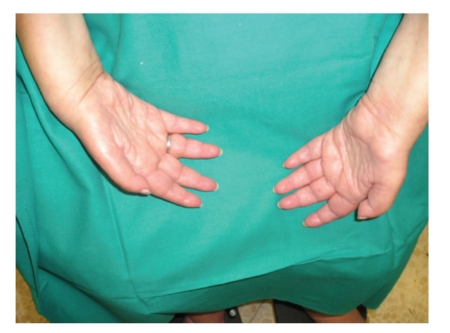
The CMT2 phenotype is highly heterogeneous, with variable penetrance, and there are mutations associated with both early- and late-onset forms of the disease. Some frequent clinical associations have been described, such as the presence of optical atrophy and tremors, migraine, and effects on the central nervous system. In general, we expect to find greater atrophy of the limbs (Figure 3) and an absence of nerve hypertrophy on palpation [23, 25].
Figure 3.
Severe atrophy of the intrinsic hand muscles of a patient with CMT2 who carries a mutation in MFN2 (CMT2A).
Other less common CMT2 subtypes with unusual characteristics may be encountered. CMT2B presents with ulcerative-mutilating phenomena, CMT2C exhibits paralysis of the vocal chords and affects the diaphragm, CMT2D has a greater effect on the intrinsic muscles of the hand, and CMT2J is associated with hearing impairment, alterations of the pupils, and severe sensory disruption. Subtypes 2I and 2J are caused by mutations in the MPZ gene and exhibit conserved NCVs; that is, in the axonal range. Therefore, defects in the production of MPZ can lead to both Type 1 and Type 2 CMT. Cases of CMT caused by mutations in MPZ that result in NCVs in the intermediate range have also been reported [8, 9, 16].
The majority of cases of CMT2 show an autosomal-dominant inheritance pattern. However, some of the CMT2 causative genes display recessive inheritance (RAB7 and LMNA, among others), but these are limited to a few families.
An initial electrophysiological study is necessary to distinguish CMT2 from CMT1.
NCVs are normal in CMT2, but the amplitudes of the motor and sensory potentials are severely reduced. It differs from distal spinal muscular atrophy in that the latter presents normal sensory neurography. The anatomical pathology points to axonal degeneration with relative preservation of myelin.
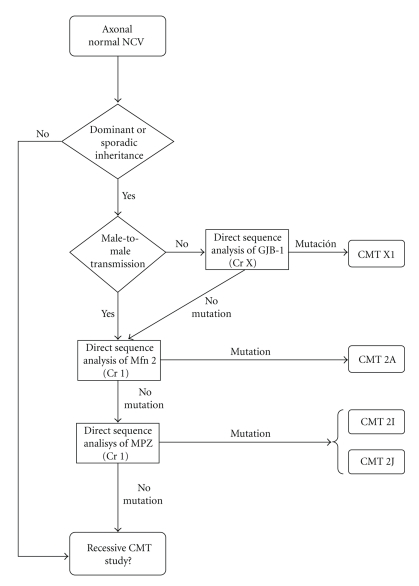
7. Diagnostic Protocol for CMT2
It is proposed to start the study of axonal CMT by checking for point mutations of MFN2 (Figure 4, adapted from England et al. [14, 15]). This is probably the most common cause of CMT2 and is responsible for up to 33% of cases in families where the inheritance pattern is known to be dominant. The data currently available suggest that variations in the GJB1 gene may explain up to 12% of CMT (both axonal and demyelinating). Hence, the presence of mutations in this gene should be ruled out in cases of axonal CMT that have no evidence of mutations in MFN2 and that are sporadic or have no known male-to-male transmission. After this, and in selected cases, the study should continue by checking for mutations in the genes that code for MPZ and NEFL (if dominant) or for GDAP1 (if sporadic or recessive). Other genes that could be studied in selected cases are RAB7, GARS, and HSPB1 [13, 24–31].
Figure 4.
Diagnostic protocol for axonal CMT; adapted from England JD et al. [10, 11].
8. CMT4
CMT4A is the most common form of CMT4 and is caused by mutations in the GDAP1 gene. It is a severe motor and sensory neuropathy with early onset (Figure 5). Neurophysiological studies reveal NCVs in both the axonal and demyelinating range. CMT4A can be associated with paresis of the vocal cords [32, 33].
Figure 5.
Very severe atrophy in the arms of a patient carrying a homozygous mutation in the GDAP1 gene (CMT4).
CMT4B1 is caused by mutations in the gene encoding the myotubularin-related protein 2 (MTMR2). Disease onset is in early infancy. Facial, bulbar, and diaphragmatic involvement has been reported in some families [34–37].
The CMT4B2 locus was mapped to chromosome 11p15 in Tunisian families [38].
CMT4C was mapped to chromosome 5q23-q33 [39] and was recently associated with mutations in the KIAA1985 gene [40]. The hallmark of CMT4C is the presence of early and severe scoliosis, which is reportedly the presenting sign in the majority of patients [41].
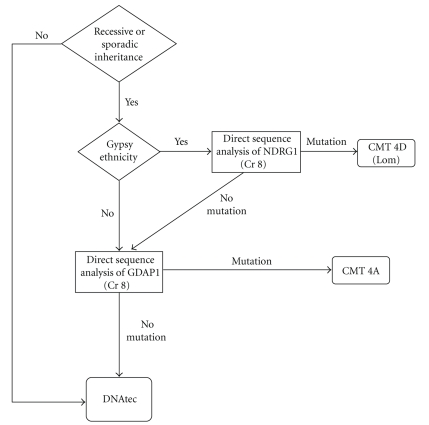
Recessive CMT caused by mutations in NDRG1 is particularly common among the Gypsy population. This form is classified as CMT4D (or Type “Lom”; Lom is the region of Bulgaria where this mutation originated). In our diagnostic protocol (Figure 6), we suggest studying this gene first if the subject is of Gypsy ethnicity; if no mutation is found or the subject is from a different ethnic group, then GDAP1 should be analyzed [14, 15]. The other CMT related to the Gypsy population is the CMT Russe type. This entity has been described in Bulgarian, Romanian, and Spanish Gypsies [42, 43] and was mapped to chromosome 10q23. It is characterized by an age of onset ranging between 8 to 16 years and progression to severe distal weakness. NCVs are moderately reduced [41].
Figure 6.
Diagnostic protocol of CMT with a suspected recessive inheritance pattern (adapted from England et al.) [10, 11].
CMT4E and CMT4F are associated with mutations in the EGR2 and periaxin (PRX) genes, respectively. Finally, CMT4H was recently mapped to a new locus on chromosome 12p11.21-q13.11 [44] (Table 1).
9. CMT with Intermediate NCV
DI-CMT Type A has been mapped to chromosome 10q24.1-q25.1 in an Italian family. It has dominant inheritance, and the age of onset is in the first or second decade of life. It is characterized by distal weakness, mild sensory loss, absent tendon reflexes, and slow progression. Many of these patients never use a wheelchair. Nerve conduction studies reveal NCVs ranging between 25 and 45 m/s (i.e., in the intermediate range). Loss of large myelinated axons and occasional onion bulbs are found in nerve biopsies.
DI-CMT Type B is related to mutations in the dynamin 2 (DNM2) gene located on chromosome 19p12-p13.2. Dominant inheritance of the trait has been described in Australian, Belgian, and North-American families. This disease is allelic with centronuclear myopathy. The clinical onset of disease occurs in the first or second decade of life, and patients present with a slow evolution. Electrodiagnostic studies reveal NCVs in the intermediate range (24–54 m/s).
DI-CMT Type C is associated with mutations in the tyrosyl-tRNA synthetase (YARS) gene on chromosome 1p34-p35. Two DI-CMT Type C families have been reported, one from Midwestern USA and one from Bulgaria. Because of slow disease progression over decades, many patients never use a wheelchair. Electrodiagnostic studies revealed NCVs in the intermediate range (30–50 m/s). Other CMT disorders with intermediate NCV have been reported in association with mutations in MPZ (CMT-DI3), in the neurofilament light chain protein (NEFL) gene, and in GJB-1 (CMT X) [3].
10. CMTX
The most common form of CMTX is dominant, X linked, and caused by point mutations in the GJB1 gene, which is located in chromosome Xq13.1. Many different mutations have been described that are normally located on the cell surface domains of the protein (i.e., extracellular domains). At present, mutation of GJB1 is the second most common cause of CMT in our setting [6, 7, 27, 45–49]. GJB1 encodes connexin 32, which acts as a gap junction at the level of compact myelin.
Boys experience symptoms in the first decade of life and are seriously affected. In women, the disease follows a less severe course, with onset at 20–30 years of age and a mild presentation of the disease.
The slowing of nerve conduction is proportional to clinical severity. Unlike CMT1, there are more myelinated fibers, more regenerating fibers, and fewer “onion bulbs”, as assessed by anatomical pathology.
Characteristically, electrophysiological studies reveal NCVs in the intermediate range; that is, between the demyelinating range of CMT1 and the axonal range of CMT2.
Although rare, there are two recessive X-linked forms of CMT, which are associated with deafness, mental retardation, and encephalomyelitis [48, 50].
11. Evolution and Treatment of CMT
The disease follows a slow but progressive course. At present, there is no etiological treatment; however, a prospective European multicentre study is under way to assess the use of high doses of ascorbic acid in patients with CMT1A. Ascorbic acid has been associated with improved clinical evolution in mice. It is particularly important that patients with CMT avoid other known risk factors for the development of neuropathies, such as the use of neurotoxins (e.g., drugs and alcohol). The literature describes transitory clinical improvements in CMT1B after treatment with corticoids [4].
12. Conclusions
CMT is a disease that is highly heterogeneous, both clinically and genetically. Clinical and electrophysiological data are indispensable for performing efficient molecular diagnoses of this entity. It is crucial to know which subtype affects each patient in order to provide good clinical treatment, effective diagnosis of family members, and valid prognosis.
Acknowledgment
The first two authors (Isabel Banchs and Carlos Casasnovas) have contributed at the same level.
References
- 1.Dyck P, Chance P, Lebo R, et al. Hereditary motor and sensory neuropaties. In: Dick P, editor. Peripheral Neuropathy. 3rd edition. Philadelphia, Pa, USA: W.B. Saunders; 1993. pp. 1094–1136. [Google Scholar]
- 2.Sevilla T, Vilchez JJ. Diferentes fenotipos del síndrome de Charcot-Marie-Tooth causados por mutaciones del mismo gen: siguen siendo útiles los criterios de clasificación clásicos? Neurologia. 2004;19(5):264–271. [PubMed] [Google Scholar]
- 3.Szigeti K, Lupski JR. Charcot-Marie-Tooth disease. European Journal of Human Genetics. 2009;17(6):703–710. doi: 10.1038/ejhg.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berciano J, Combarros O. Hereditary neuropathies. Current Opinion in Neurology. 2003;16(5):613–622. doi: 10.1097/01.wco.0000093105.34793.dd. [DOI] [PubMed] [Google Scholar]
- 5.Emery AEH. Population frequencies of inherited neuromuscular diseases—a world survey. Neuromuscular Disorders. 1991;1(1):19–29. doi: 10.1016/0960-8966(91)90039-u. [DOI] [PubMed] [Google Scholar]
- 6.Kurihara S, Adachi Y, Wada K, Awaki E, Harada H, Nakashima K. An epidemiological genetic study of Charcot-Marie-Tooth disease in Western Japan. Neuroepidemiology. 2002;21(5):246–250. doi: 10.1159/000065643. [DOI] [PubMed] [Google Scholar]
- 7.Hattori N, Yamamoto M, Yoshihara T, et al. Demyelinating and axonal features of Charcot-Marie-Tooth disease with mutations of myelin-related proteins (PMP22, MPZ and Cx32): a clinicopathological study of 205 Japanese patients. Brain. 2003;126(1):134–151. doi: 10.1093/brain/awg012. [DOI] [PubMed] [Google Scholar]
- 8.Shy ME, Jáni A, Krajewski K, et al. Phenotypic clustering in MPZ mutations. Brain. 2004;127(2):371–384. doi: 10.1093/brain/awh048. [DOI] [PubMed] [Google Scholar]
- 9.Laurà M, Milani M, Morbin M, et al. Rapid progression of late onset axonal Charcot-Marie-Tooth disease associated with a novel MPZ mutation in the extracellular domain. Journal of Neurology, Neurosurgery and Psychiatry. 2007;78(11):1263–1266. doi: 10.1136/jnnp.2006.112276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Meuleman J, Pou-Serradell A, Löfgren A, et al. A novel 3′-splice site mutation in peripheral myelin protein 22 causing hereditary neuropathy with liability to pressure palsies. Neuromuscular Disorders. 2001;11(4):400–403. doi: 10.1016/s0960-8966(00)00214-5. [DOI] [PubMed] [Google Scholar]
- 11.Shy ME, Scavina MT, Clark A, et al. T118M PMP22 mutation causes partial loss of function and HNPP-like neuropathy. Annals of Neurology. 2006;59(2):358–364. doi: 10.1002/ana.20777. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Krajewski K, Shy ME, Lewis RA. Hereditary neuropathy with liability to pressure palsy: the electrophysiology fits the name. Neurology. 2002;58(12):1769–1773. doi: 10.1212/wnl.58.12.1769. [DOI] [PubMed] [Google Scholar]
- 13.Bort S, Nelis E, Timmerman V, et al. Mutational analysis of the MPZ, PMP22 and Cx32 genes in patients of Spanish ancestry with Charcot-Marie-Tooth disease and hereditary neuropathy with liability to pressure palsies. Human Genetics. 1997;99(6):746–754. doi: 10.1007/s004390050442. [DOI] [PubMed] [Google Scholar]
- 14.England JD, Gronseth GS, Franklin G, et al. Practice Parameter: evaluation of distal symmetric polyneuropathy: role of laboratory and genetic testing (an evidence-based review). Report of the American Academy of Neurology, American Association of Neuromuscular and Electrodiagnostic Medicine, and American Academy of Physical Medicine and Rehabilitation. Neurology. 2009;72(2):185–192. doi: 10.1212/01.wnl.0000336370.51010.a1. [DOI] [PubMed] [Google Scholar]
- 15.England JD, Gronseth GS, Franklin G, et al. Evaluation of distal symmetric polyneuropathy: the role of laboratory and genetic testing (an evidence-based review) Muscle and Nerve. 2009;39(1):116–125. doi: 10.1002/mus.21226. [DOI] [PubMed] [Google Scholar]
- 16.Street VA, Meekins G, Lipe HP, et al. Charcot-Marie-Tooth neuropathy: clinical phenotypes of four novel mutations in the MPZ and Cx 32 genes. Neuromuscular Disorders. 2002;12(7-8):643–650. doi: 10.1016/s0960-8966(02)00021-4. [DOI] [PubMed] [Google Scholar]
- 17.Züchner S, Mersiyanova IV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nature Genetics. 2004;36(5):449–451. doi: 10.1038/ng1341. [DOI] [PubMed] [Google Scholar]
- 18.Sołtysińska E, Kabzińska D, Kochański A. Mutations in the mitofusin 2 gene are the most common cause of Charcot-Marie-Tooth type 2 disease. Neurologia i Neurochirurgia Polska. 2007;41(4):350–354. [PubMed] [Google Scholar]
- 19.Amiott EA, Lott P, Soto J, et al. Mitochondrial fusion and function in Charcot-Marie-Tooth type 2A patient fibroblasts with mitofusin 2 mutations. Experimental Neurology. 2008;211(1):115–127. doi: 10.1016/j.expneurol.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Loiseau D, Chevrollier A, Verny C, et al. Mitochondrial coupling defect in Charcot-Marie-Tooth type 2A disease. Annals of Neurology. 2007;61(4):315–323. doi: 10.1002/ana.21086. [DOI] [PubMed] [Google Scholar]
- 21.Ishihara N, Eura Y, Mihara K. Mitofusin 1 and 2 play distinct roles in mitochondrial fusion reactions via GTPase activity. Journal of Cell Science. 2004;117(26):6535–6546. doi: 10.1242/jcs.01565. [DOI] [PubMed] [Google Scholar]
- 22.Züchner S, Vance JM. Molecular genetics of autosomal-dominant axonal Charcot-Marie-Tooth disease. Neuromolecular Medicine. 2006;8(1-2):63–74. doi: 10.1385/nmm:8:1-2:63. [DOI] [PubMed] [Google Scholar]
- 23.Verhoeven K, Claeys KG, Züchner S, et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129(8):2093–2102. doi: 10.1093/brain/awl126. [DOI] [PubMed] [Google Scholar]
- 24.Züchner S, De Jonghe P, Jordanova A, et al. Axonal neuropathy with optic atrophy is caused by mutations in mitofusin 2. Annals of Neurology. 2006;59(2):276–281. doi: 10.1002/ana.20797. [DOI] [PubMed] [Google Scholar]
- 25.Banchs I, Casasnovas C, Montero J, Martínez-Matos JA, Volpini V. Two Spanish families with Charcot-Marie-Tooth type 2A: clinical, electrophysiological and molecular findings. Neuromuscular Disorders. 2008;18(12):974–978. doi: 10.1016/j.nmd.2008.09.006. [DOI] [PubMed] [Google Scholar]
- 26.Hoogendijk JE, Hensels GW, Gabreëls-Festen AA, et al. De-novo mutation in hereditary motor and sensory neuropathy type I. The Lancet. 1992;339(8801):1081–1082. doi: 10.1016/0140-6736(92)90668-s. [DOI] [PubMed] [Google Scholar]
- 27.Janssen EA, Kemp S, Hensels GW, et al. Connexin32 gene mutations in X-linked dominant Charcot-Marie-Tooth disease (CMTX1) Human Genetics. 1997;99(4):501–505. doi: 10.1007/s004390050396. [DOI] [PubMed] [Google Scholar]
- 28.Silander K, Meretoja P, Juvonen V, et al. Spectrum of mutations in Finnish patients with Charcot-Marie-Tooth disease and related neuropathies. Human Mutation. 1998;12(1):59–68. doi: 10.1002/(SICI)1098-1004(1998)12:1<59::AID-HUMU9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 29.Nelis E, Van Broeckhoven C, De Jonghe P, et al. Estimation of the mutation frequencies in Charcot-Marie-Tooth disease type 1 and hereditary neuropathy with liability to pressure palsies: a European collaborative study. European Journal of Human Genetics. 1996;4(1):25–33. doi: 10.1159/000472166. [DOI] [PubMed] [Google Scholar]
- 30.Nicholson GA. Mutation testing in Charcot-Marie-Tooth neuropathy. Annals of the New York Academy of Sciences. 1999;883:383–388. [PubMed] [Google Scholar]
- 31.Choi BO, Lee MS, Shin SH, et al. Mutational analysis of PMP22, MPZ, GJB1, EGR2 and NEFL in Korean Charcot-Marie-Tooth neuropathy patients. Human Mutation. 2004;24(2):185–186. doi: 10.1002/humu.9261. [DOI] [PubMed] [Google Scholar]
- 32.Cuesta A, Pedrola L, Sevilla T, et al. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nature Genetics. 2002;30(1):22–25. doi: 10.1038/ng798. [DOI] [PubMed] [Google Scholar]
- 33.Sevilla T, Cuesta A, Chumillas MJ, et al. Clinical, electrophysiological and morphological findings of Charcot-Marie-Tooth neuropathy with vocal cord palsy and mutations in the GDAP1 gene. Brain. 2003;126(9):2023–2033. doi: 10.1093/brain/awg202. [DOI] [PubMed] [Google Scholar]
- 34.Bolino A, Muglia M, Conforti FL, et al. Charcot-Marie-Tooth type 4B is caused by mutations in the gene encoding myotubularin-related protein-2. Nature Genetics. 2000;25(1):17–19. doi: 10.1038/75542. [DOI] [PubMed] [Google Scholar]
- 35.Bolino A, Levy ER, Muglia M, et al. Genetic refinement and physical mapping of the CMT4B gene on chromosome 11q22. Genomics. 2000;63(2):271–278. doi: 10.1006/geno.1999.6088. [DOI] [PubMed] [Google Scholar]
- 36.Houlden H, King RH, Hashemi-Nejad A, et al. A novel TRK A (NTRK1) mutation associated with hereditary sensory and autonomic neuropathy type V. Annals of Neurology. 2001;49(4):521–525. [PubMed] [Google Scholar]
- 37.Verny C, Ravisé N, Leutenegger A-L, et al. Coincidence of two genetic forms of Charcot-Marie-Tooth disease in a single family. Neurology. 2004;63(8):1527–1529. doi: 10.1212/01.wnl.0000142082.65144.ee. [DOI] [PubMed] [Google Scholar]
- 38.Othmane KB, Johnson E, Menold M, et al. Identification of a new locus for autosomal recessive Charcot-Marie- Tooth disease with focally folded myelin on chromosome 11p15. Genomics. 1999;62(3):344–349. doi: 10.1006/geno.1999.6028. [DOI] [PubMed] [Google Scholar]
- 39.LeGuern E, Guilbot A, Kessali M, et al. Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23-q33. Human Molecular Genetics. 1996;5(10):1685–1688. doi: 10.1093/hmg/5.10.1685. [DOI] [PubMed] [Google Scholar]
- 40.Senderek J, Bergmann C, Stendel C, et al. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. American Journal of Human Genetics. 2003;73(5):1106–1119. doi: 10.1086/379525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dubourg O, Azzedine H, Verny C, et al. Autosomal-recessive forms of demyelinating Charcot-Marie-Tooth disease. Neuromolecular Medicine. 2006;8(1-2):75–85. doi: 10.1385/nmm:8:1-2:75. [DOI] [PubMed] [Google Scholar]
- 42.Rogers T, Chandler D, Angelicheva D, et al. A novel locus for autosomal recessive peripheral neuropathy in the EGR2 region on 10q23. American Journal of Human Genetics. 2000;67(3):664–671. doi: 10.1086/303053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thomas PK, Kalaydjieva L, Youl B, et al. Hereditary motor and sensory neuropathy-russe: new autosomal recessive neuropathy in Balkan gypsies. Annals of Neurology. 2001;50(4):452–457. doi: 10.1002/ana.1137. [DOI] [PubMed] [Google Scholar]
- 44.De Sandre-Giovannoli A, Delague V, Hamadouche T, et al. Homozygosity mapping of autosomal recessive demyelinating Charcot-Marie-Tooth neuropathy (CMT4H) to a novel locus on chromosome 12p11.21-q13.11. Journal of Medical Genetics. 2005;42(3):260–265. doi: 10.1136/jmg.2004.024364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Colomer-Oferil J. Aspectos clínicos y abordaje diagnóstico y terapéutico de las neuropatías hereditarias sensitivo-motoras. Revista de Neurologia. 2002;35(3):239–245. [PubMed] [Google Scholar]
- 46.Berciano J, Combarros O, Calleja J, et al. The application of nerve conduction and clinical studies to genetic counseling in hereditary motor and sensory neuropathy type I. Muscle and Nerve. 1989;12(4):302–306. doi: 10.1002/mus.880120408. [DOI] [PubMed] [Google Scholar]
- 47.Lupski JR, de Oca-Luna RM, Slaugenhaupt S, et al. DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell. 1991;66(2):219–232. doi: 10.1016/0092-8674(91)90613-4. [DOI] [PubMed] [Google Scholar]
- 48.Casasnovas C, Banchs I, Corral J, Martínez-Matos JA, Volpini V. Clinical and molecular analysis of X-linked Charcot-Marie-Tooth disease type 1 in Spanish population. Clinical Genetics. 2006;70(6):516–523. doi: 10.1111/j.1399-0004.2006.00724.x. [DOI] [PubMed] [Google Scholar]
- 49.Taylor RA, Simon EM, Marks HG, Scherer SS. The CNS phenotype of X-linked Charcot-Marie-Tooth disease: more than a peripheral problem. Neurology. 2003;61(11):1475–1478. doi: 10.1212/01.wnl.0000095960.48964.25. [DOI] [PubMed] [Google Scholar]
- 50.Bergoffen J, Scherer SS, Wang S, et al. Connexin mutations in X-linked Charcot-Marie-Tooth disease. Science. 1993;262(5142):2039–2042. doi: 10.1126/science.8266101. [DOI] [PubMed] [Google Scholar]