Abstract
Many functionally important membrane proteins are cleaved within their transmembrane helices to become activated. This unusual reaction is catalyzed by a group of highly specialized and membrane-bound proteases. Here I briefly summarize current knowledge about their structure and mechanism, with a focus on the rhomboid family. It has now become clear that rhomboid protease can cleave substrates not only within transmembrane domains, but also in the solvent-exposed juxtamembrane region. This dual specificity seems possible because the protease active site is positioned in a shallow pocket that can directly open to aqueous solution through the movement of a flexible capping loop. The narrow membrane-spanning region of the protease suggests a possible mechanism for accessing scissile bonds that are located near the end of substrate transmembrane helices. Similar principles may apply to the metalloprotease family, where a crystal structure has also become available. Although how the GxGD proteases work it is still less clear, recent results indicate that presenilin also appears to clip substrate from the end of transmembrane helices.
Keywords: rhomboid, site-2 protease, presenilin, intramembrane proteolysis, membrane structure
1. Introduction
Many proteins are synthesized initially as inactive precursors that are later converted to functional molecules through proteolysis. In the past ten years or so, we have observed an important addition to this general theme: it is now evident that proteolytic modifications can occur not only in soluble regions of the protein, but also in the transmembrane (TM) domain of many integral membrane proteins. This novel reaction, or intramembrane proteolysis, is involved in many crucial cellular and developmental processes (see reviews in ref. 1–6), and mutations that interfere with its mechanism are known to cause human disease (for example, see ref. 7, 8).
This chapter is concerned primarily with aspects of the biochemical mechanism that are unique to intramembrane proteolysis. Much of the discussion will be on the rhomboid protease [9, 10]. Rhomboid was first described in Drosophila as a key regulator of the epidermal growth factor receptor signaling pathway [11]. This function is due to its proteolytic activity towards several membrane-bound growth factors, which is required for their release from signal sending cells [12]. Homologous proteases, identifiable through sequence similarities, have been found in bacteria, archaea, other eukaryotic organisms and mitochondria. These proteases have completely different biological functions, some of which will only be briefly mentioned here. Interested readers can find more detailed and thorough accounts of the functions of rhomboid protease in a number of previously published reviews [5, 13, 14].
The hydrophobic interior of cell membrane, typically 30 Å wide [15], is packed with the hydrocarbon tails of membrane lipid, and is generally devoid of water molecules. On both sides of this 30 Å wide space are the so-called interfacial regions, which are rich in lipid head groups, and contain water and other ions of the aqueous medium [16]. To traverse the membrane (its hydrophobic core), a polypeptide usually contains 20 consecutive hydrophobic residues that fold into a continuous α-helix (in a regular helix, each residue rises by about 1.5 Å along the helical axis). Peptide bonds inside TM helices are highly resistant to proteolysis: to become cleavable, the helix has to unfold, which would expose its polar backbone (in aqueous solution, this is less of a problem because the backbone can form hydrogen bonds with water); the proteolytic reaction itself also requires water. Figure 1 summarizes two ideas that have been raised to explain how intramembrane proteolysis can occur. According to one hypothesis, part of substrate TM helix moves first out of the membrane, and then gets cleaved (for example, see ref. 17, 18). According to the other hypothesis, substrate enters the membrane-embedded protease active site laterally: since the active site is hydrophilic, it must be initially closed on the side, and some sort of gating mechanism would be required to allow substrate access (for example, see discussion in ref. 19). Both models have received some experimental support, but recent development, especially in the area of structural biology, has started to indicate that at least some proteases may work in accordance with the first model.
Fig. 1.
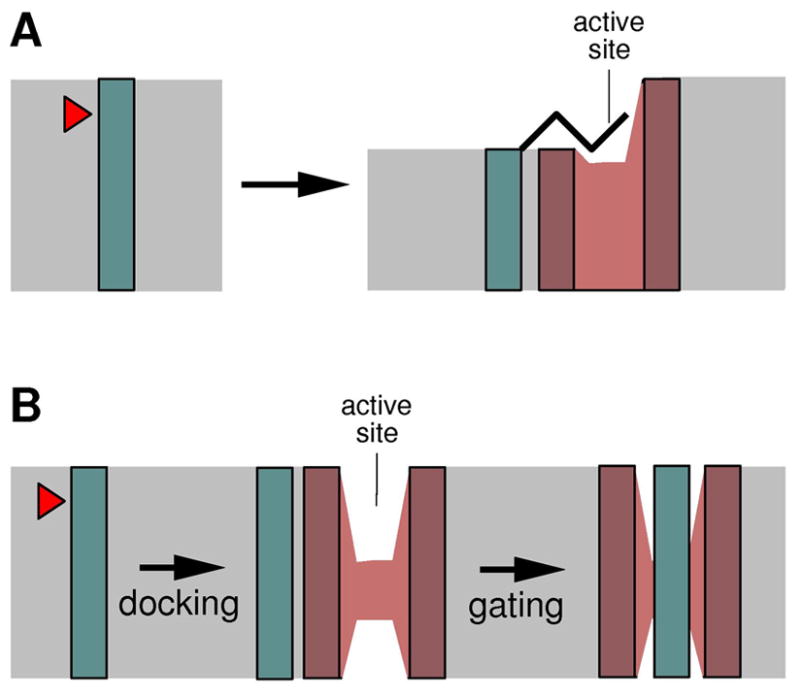
Hypothetical models to explain how peptide bonds in TM helices can be cleaved by the membrane protease. Blue box, substrate TM helix; red, protease (a cut-open view); grey, membrane. The red arrow head marks the scissile bond. (A) A portion of substrate TM helix partitions out of the membrane and unfolds before entering the active site. This may be facilitated by changes in the bilayer structure. (B) Substrate enters the protease through a gate that laterally opens inside the membrane.
2. The rhomboid family of serine proteases
Rhomboid was initially described in drosophila as a gene required for pattern formation in the ventral ectoderm, in which mutation caused a rhomboid-shaped head skeleton [20]. This phenotype is due to the role of fly rhomboid in epidermal growth factor receptor signaling [21]. Using site-directed mutagenesis and class-specific protease inhibitors, Urban et al. showed that drosophila rhomboid-1 was directly responsible for cleaving, thereby activating, membrane-anchored growth factor spitz [22]. Their proposal that rhomboid-1, as well as other members of the rhomboid family [23], were novel serine proteases was confirmed by later biochemical experiments using purified protein [24–26].
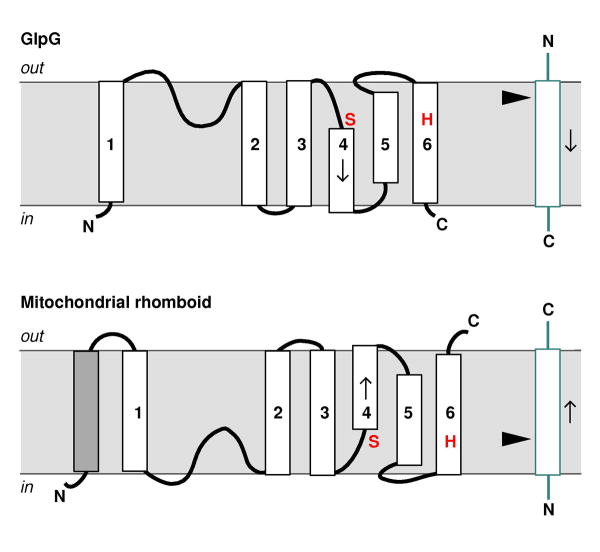
Rhomboids are integral membrane proteins [27]. The structure of E. coli GlpG is the simplest within the family. Its predicted 6-TM-domain topology [28], shown in Fig. 2, has been confirmed by β-lactamase fusion experiment [26], and by crystallographic analysis of the detergent-solubilized protein [29–32]. Most eukaryotic rhomboids, including the fly rhomboid-1 mentioned above, have an extra TM domain after the 6-TM-domain core, which brings the carboxyl terminus of the protein to the luminal side of the membrane [33, 34]. The mitochondrial rhomboids have an extra TM domain on the amino terminal side, and the rest of the protein appears to adopt a different membrane orientation (Fig. 2): their active sites now face the matrix side of the membrane (equivalent to the cytosolic side) [33, 34]. The proteolytic activity of mitochondrial rhomboids towards type-II (Nin-Cout) membrane protein substrates, instead of the type-I (Nout-Cin) substrates that other rhomboids cleave, may be related to this flip of enzyme structure.
Fig. 2.
The membrane topology models for bacterial GlpG, mitochondrial rhomboid and their TM substrates (blue box). The arrow head points at the cleavage site. Arrows indicate the direction of the polypeptide chain. The catalytically essential serine and histidine are shown in red. The TM domains (white box) within the core region are sequentially labeled from 1 to 6. Mitochondrial rhomboid has an additional TM domain (grey box).
The catalytically essential serine and histidine of rhomboid (Ser-201, His-254 in E. coli GlpG), previously identified by mutagenesis [23, 24, 26], are separately located in TM domains S4 and S6. In the folded structure, these two residues are near each other and form a hydrogen bond [29], which probably enables the histidine to activate the serine for direct nucleophilic attack on substrate, in a fashion similar to other serine proteases such as chymotrypsin. At this time, however, a direct proof of the proposed catalytic mechanism remains lacking. For example, it was noticed that, among the many serine protease inhibitors, rhomboid was only universally sensitive to 3,4-dichloroisocoumarin (DCI) [22, 24, 25]. DCI is supposed to covalently modify the catalytic serine nucleophile [35, 36]. There is yet no report, however, of this covalent adduct formation, and a previous attempt to soak DCI into preformed GlpG crystals has failed to generate enzyme:inhibitor complex [37].
2.1. Rhomboids are intramembrane proteases
The initial evidence came from an analysis of the gel mobility of rhomboid-1-released growth factor fragment in cultured cells [22], which roughly mapped the proteolytic site to a small region within the predicted TM domain of spitz near its amino terminus (bold in Fig. 3A). Swapping this region (“substrate motif”) into other membrane proteins rendered them susceptible to rhomboid-1 cleavage [38]. Baker et al. have used mass spectrometry to try to determine the exact site of cleavage by E. coli GlpG [39], using an artificial substrate bearing the spitz motif, and making the assumptions that bacterial rhomboid cut at an identical site [23], and that cleavage specificity was not affected by fusing spitz with another TM sequence, or by conducting the reaction in detergent solutions. Based on this study, it was suggested that spitz was cleaved at two adjacent sites (after Ala-141 and Gly-143 respectively) within the substrate motif.
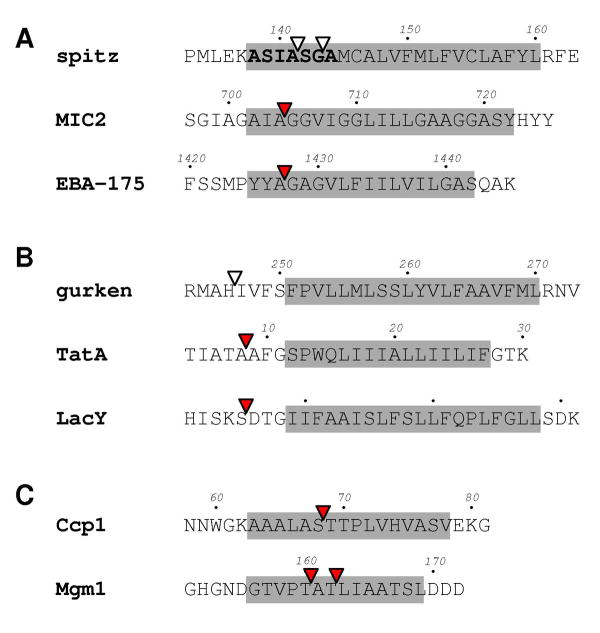
Fig. 3.
Rhomboid substrates with known cleavage sites (red arrow head). The cleavage sites in spitz and gurken, indicated by white arrow heads, were derived from reactions catalyzed by bacterial rhomboid proteases [39, 45]. The sequence around the second TM domain of LacY is listed here because it can be efficiently cleaved by E. coli GlpG, and has been used as an artificial substrate. The membrane-spanning region for each sequence is highlighted by the grey box. The “substrate motif” in spitz is shown in bold. (A) Substrates undergoing proteolysis within TM domains; (B) substrates cleaved in the juxtamembrane region; (C) mitochondrial rhomboid substrates.
A second line of independent evidence came from studies of the shedding of apicomplexan parasite surface adhesion molecules: the ectodomain of Toxoplasma gondii adhesin MIC2, a type-I membrane protein, is shed during host cell invasion; by directly purifying the processed adhesin from the parasite, and by mass spectroscopic analysis, it was discovered that the shedding resulted from proteolysis within the TM domain of MIC2 (Fig. 3A) [40, 41]. The location of the cleavage site towards the extracellular side of the membrane, like that in spitz, suggested that a rhomboid protease might be involved, and Brossier et al. subsequently showed that parasite rhomboid TgROM5 could provide this proteolytic activity [42]. The adhesin EBP-175 of Plasmodium falciparum also undergoes ectodomain shedding, in a reaction likely catalyzed by plasmodium rhomboid pfROM4 [43, 44]. O’Donnell et al. have determined the carboxyl terminus of the released EBP-175 by mass spectroscopy, which confirmed that pfROM4 also cleaved within the TM region of the adhesin (Fig. 3A) [43].
2.2. Features of rhomboid active site
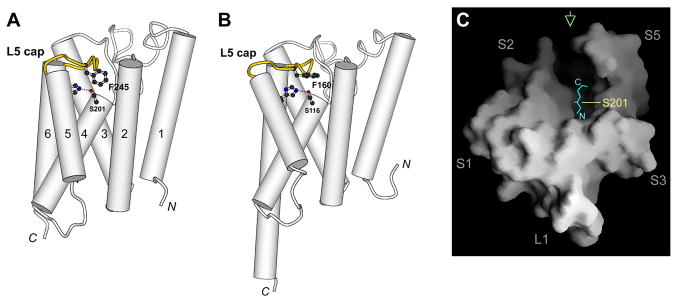
The structure of bacterial GlpG, which has been solved by x-ray crystallography [29–32], is representative of the family (Fig. 4A, B). The active site of GlpG is found at the bottom of a shallow pocket that faces the extracellular side of the membrane. Immediately above the catalytic dyad is a loop structure that we previously called L5 cap (highlighted in yellow in Fig. 4A, B), which lies roughly parallel to the membrane plane [29, 37]. A side chain from the loop (Phe-245) drops down and seals a gap between two TM helices (S2 and S5). L5 cap in this conformation, which has been seen twice (once in the E. coli GlpG structure [29], and once in the H. influenzae structure [32]), would sterically hinder substrate access to Ser-201. The facts that the loops in the two bacterial GlpGs have different amino acid sequences, and that their crystals were obtained from totally different detergent and solution conditions, suggest that a closed cap likely reflects the conformation of the enzyme in its native membrane environment.
Fig. 4.
L5 cap (yellow) blocks access to the Ser-His dyad (stick model and dashed line). (A) E. coli GlpG (PDB entry 2ic8; ref. 29). (B) H. influenzae GlpG (PDB entry 2nr9; ref. 32). (C) A surface representation of GlpG, viewed from periplasm onto the membrane plane, with L5 cap removed to reveal the substrate binding site. A di-peptide (cyan) is manually put above the catalytic serine to illustrate the size of the groove. Green arrow indicates the direction from which substrate binds to the protease. Panel (C) is taken from ref. 37 and is copyrighted by The National Academy of Sciences of the USA.
L5 cap appears to be intrinsically flexible. Under some crystallization or soaking conditions, the loop can adopt different conformations, or become disordered in the crystal [30, 31, 37]. Maegawa et al. found that cysteines introduced into the active site of GlpG could react with fairly bulky alkylating reagents [45]. Their observation suggests that, in the membrane-bound state, L5 cap sometimes can spontaneously “open” even while substrate is absent. The movement of L5 cap alone will generate a wide open groove on the periplasmic surface of the membrane protein with the catalytic dyad at its bottom, creating the opportunity for an extended, and probably solvent-exposed, peptide to lie across it to become cleaved (Fig. 4C).
2.3. Cleavage sites outside the membrane
Given that almost all the initially identified rhomboid substrates were found to be cleaved within their TM domains, and that rhomboid active site has now been confirmed to lie inside membranes, and that intramembrane proteases were generally thought to function only inside lipid bilayers, the idea that rhomboid might also cleave solvent-exposed peptide bonds has been greeted with suspicion. Despite the concerns, however, now there have been a number of reports indicating rhomboid’s ability to cleave substrates within their juxtamembrane regions.
Drosophila growth factor gurken, like spitz, also requires rhomboid cleavage for activation [46]. The rhomboid cleavage site lies within a long stretch of hydrophobic sequence in gurken (after His-246 [45]), which contains the single TM domain of the protein (Fig. 3B) [24]. The exact amino terminus of the TM helix and whether the helix includes the scissile bond were initially unclear: the helix was thought to begin at Met-244 [24], but this would make it 27aa long (assuming the helix ends at Leu-270), and include a polar residue (His-246). Maegawa et al. later studied the questionable amino terminal region using single cysteine mutants: they found that cysteines introduced to positions 247 and 250, both lying between the scissile bond and the rest of the hydrophobic sequence, reacted readily with AMS, a charged alkylating reagent that could not penetrate into membranes [45], which suggested that the cleavage site in gurken either resided outside the membrane, or could partition readily out of the bacterial membrane.
Providencia stuartii rhomboid protease AarA removes an 8aa leader sequence from TatA, the channel component of the twin-arginine translocase system, and enables the channel to secrete a quorum-sensing molecule [47, 48]. All bacterial TatA proteins have a single TM helix near the amino terminus that is required for channel activity [49]. Sequence alignment of P. stuartii TatA with homologous TatAs from other species that do not have the 8aa extension (and thus do not require rhomboid for activation), indicates that the bond cleaved by AarA is not located within the common TM region (Fig. 3B).
While attempting to identify new substrates, Maegawa et al. discovered by chance that an artificial fusion protein bearing the sequence around the second TM domain of LacY was cleavable by E. coli GlpG in intact bacterial cells [26]. After purifying the carboxyl terminal fragment and sequencing it, they found that a Ser-Asp bond was cleaved (Fig. 3B). The Ser-Asp bond, located on the amino terminal side of a type-I (Nout-Cin) TM helix like those in gurken and TatA, is obviously not buried inside the membrane. Therefore, the finding by Maegawa et al. most convincingly demonstrated that a well established intramembrane protease in its native membrane environment had the potential to efficiently cleave a juxtamembrane bond. Indeed, recent work from Freeman’s group has also shown that at least one mammalian rhomboid can cleave a bond that is even further away from the TM domain (Freeman, personal communications), suggesting that this activity is not uniquely due to the bacterial protease or bacterial membrane.
2.4. Membrane compression around the protease
Besides its functional significance, the dual specificity of rhomboid proteases towards both buried and exposed peptide bonds has important mechanistic implications. We first consider how E. coli GlpG cleaves the exposed Ser-Asp bond in LacY. One possibility is that the TM helix may tilt to bring the scissile bond into the membrane so that it can be cleaved “properly” like the others. This seems unlikely because Maegawa et al. found that insertion of up to seven residues between the Ser-Asp bond and the TM region had little effect on proteolysis [45]. The charge of Asp on the P1′ location also argues strongly against any model where this residue has to enter the membrane first. Therefore, it appears almost certain that the scissile bond in LacY must enter the protease directly from solution: for example, the membrane-integrated substrate may approach the protease from the direction of TM domains S2 and S5, and bend into the active site to position the juxtamembrane Ser-Asp bond over the catalytic serine for proteolysis (Fig. 4C) [37].
The ability of the protease active site to directly open to aqueous solution [37], and receive peptide substrate through this opening, was unexpected. An important question now is whether or not this mode of substrate entry applies also to buried bonds, which are initially in TM helices. We have speculated that they may enter the protease through a similar route by first moving partially out of the membrane [18]. The crystal structure of GlpG appears to contain features that are supportive of this idea.
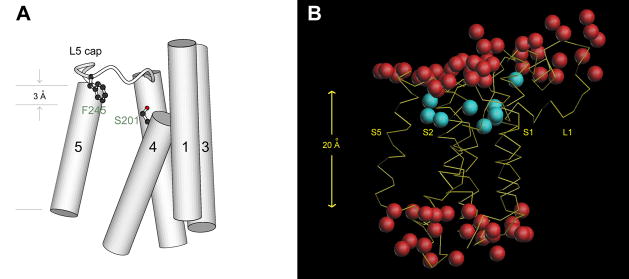
The location and geometry of the catalytic serine provide a clue about how substrate is finally bound to the active site (Fig. 5A). The crystal structure of E. coli GlpG has now been refined to a very high resolution (1.7 Å), at which many water and detergent molecules bound to the membrane protein can be easily resolved [18]. This has led to a better estimation of the upper boundary of the membrane, and thus the depth of the catalytic serine: it now seems that the hydroxyl oxygen of Ser-201, which is supposed to form a transient covalent bond with the substrate, is positioned only 3 Å below the membrane surface. The up-pointing configuration of Ser-201, and the locations of His-254 and other potentially important residues (for example, His-150 and Asn-154, which may form the oxyanion hole) [31, 32, 37], suggest that substrate is bound on top of the serine, which would position it flat on the surface of the membrane. Therefore, at its final destination, the substrate has already moved partially out of the membrane.
Fig. 5.
Features of the high-resolution crystal structure of E. coli GlpG. (A) The catalytic serine is located 3 Å below the membrane surface, and points up. (B) The α-carbon trace of the protein backbone is shown in yellow. Waters bound inside the protein are shown in cyan. The externally bound waters, observed in the 1.9 Å resolution crystal structure (PDB entry 3b45) and shown in red, are found in two layers, which roughly correspond to the boundaries of the membrane.
Another important feature of the GlpG structure is about the width of its hydrophobic belt. This is the region of the protein that directly contacts the hydrocarbon groups of the lipid, and its dimension should match the thickness of the membrane. Based on the high resolution crystal structures, it has now been estimated that the hydrophobic dimension around GlpG, in the direction of TM helix S5 at least where substrate comes in to bind, is quite narrow (about 20 Å; Fig. 5B) [18], suggesting that membrane bilayer has to significantly compress there (membranes frequently deform around membrane proteins to match their thickness; for example, see reviews in ref. 50, 51). This is unlikely to be unique for the bacterial protease because a structure-based alignment of diverse rhomboid sequences has shown that most, if not all, have a short hydrophobic S5 [29]. It is conceivable that this membrane compression may destabilize the ends of single TM helices that come into contact with the protease, including those that bear cleavable sites near their amino termini (Fig. 3A). The protease could have acquired this structural feature through evolution to facilitate the presentation of membrane-embedded bonds in the substrate.
2.5. A lateral gate?
The mechanism of the protease has been compared by some researchers to that of the protein-conducting channel SecY [30], where movements of channel TM helices create a lateral opening inside the membrane through which substrate TM helices can pass [52].
Along this line of reasoning, it was proposed that substrate entry into rhomboid might be gated by a lateral movement of TM helix S5 [30]. This hypothesis originated from the observation that in one crystal form of E. coli GlpG, one S5 helix had tilted 35 degrees, which created a large opening on the side of the protein. The tilted S5 conformation is however unusual because it is different not only from those observed in three other crystal forms of GlpG [29, 31, 32], but also from that of a second GlpG molecule in the same crystal [30]. The questionable S5 helix makes extensive crystal packing interactions, which could have influenced its conformation. The so-called “open state” is also intriguing because there is no substrate present in the crystal to trigger the conformational change in S5 helix.
A considerable amount of enzymatic data had been later generated to try to confirm the gating model [39]. In one experiment, disulfide bridges were engineered between TM helices S2 and S5 to lock the gate in a closed state: the oxidized enzyme, in which the disulfide bond had presumably formed, indeed appeared less active than the reduced enzyme. Nevertheless, some activity was retained in the oxidized enzyme: if a large movement of S5 is required to open an essential gate as proposed [30], one might expect that closing it should prevent substrate entry, and thus completely eliminate activity.
2.6. Substrate specificity
Generally, how well the side chains of a polypeptide substrate fit in the binding site of the protease influences its cleavability. Akiyama et al. performed an elegant screen for sequences that can be efficiently cleaved by E. coli GlpG by randomizing the P1 and P1′ residues of LacY (the P1 residue is the first residue on the amino side of the scissile bond; P1′ is on the carboxyl side) [53]. Their results seemed to suggest that this bacterial rhomboid had a strong preference for small side chains at P1 (Ser, Ala or Gly) and a carboxylate group at P1′ (Asp or Glu). This is somewhat in contrast with earlier views that intramembrane proteases in general might not be sequence specific.
Unlike the exposed Ser-Asp bond in LacY, cleavage sites that are initially buried inside membranes pose an additional challenge for being in α-helices, which are resistant to proteolysis due to steric hindrance. Because of this, it has been hypothesized that rhomboid substrates, like those for signal peptide peptidase [54], carry helix-breaking residues near their cleavage site [38]. Urban et al. identified a 7aa region near the top of spitz TM domain as the determinant for cleavability (“substrate motif”; Fig. 3A), and showed that swapping only two residues of the motif (Gly-143, Ala-144) into TGFα, a protein normally not cleaved by rhomboid-1, rendered it fully cleavable (introducing Gly-143 alone generated some cleavage) [38]. This seemed to indicate that Gly-143 was important, but how a single glycine residue could function as a helix-breaker is not clear: in soluble proteins, glycine is rarely found in α-helices [55]; but in the non-polar environment of the membrane, glycine has a fairly high helical propensity (ranked number 6 of the 20 amino acids), and is abundantly found in TM helices [56].
The hypothesis we developed earlier allows us to make a different prediction: we have postulated that buried cleavage site need to move partially out of the membrane (and unfold) before entering the protease [18]; according to this model, TM helices with a less hydrophobic amino terminus are more likely to become cleavable by rhomboid protease because the energy cost for moving the cleavage sites into water would be smaller. This prediction can be partly tested by the sequence of spitz. In Fig. 6, the transfer free energy, cost for moving an amino acid from a membrane-spanning α-helix into water [57], is plotted for each residue around the TM region of spitz (note that this is not a typical hydropathy plot, which represents the energy for transferring a peptide segment of usually 20 residues). Most residues in the “substrate motif” have transfer energies around 1 kcal/mol (average 1.4 kcal/mol), whereas those in the rest of the TM domain have higher transfer energies (average 2.6 kcal/mol). A typical single pass TM helix of 20aa has a total transfer energy around 48 kcal/mol [58], which means that each single residue by average should contribute about 2.4 kcal/mol. Comparing these numbers, one may conclude, at least in principle, that the “substrate motif” of spitz could represent a region of the TM helix that is only weakly inserted in the membrane, and have an intrinsic potential to partition into water. Terminally located glycines, like Gly-143 of spitz, and those in MIC2 and EBA-175 (Fig. 3A), may be indicative of a cleavable TM sequence, because the transfer free energy of glycine is only 1 kcal/mol.
Fig. 6.
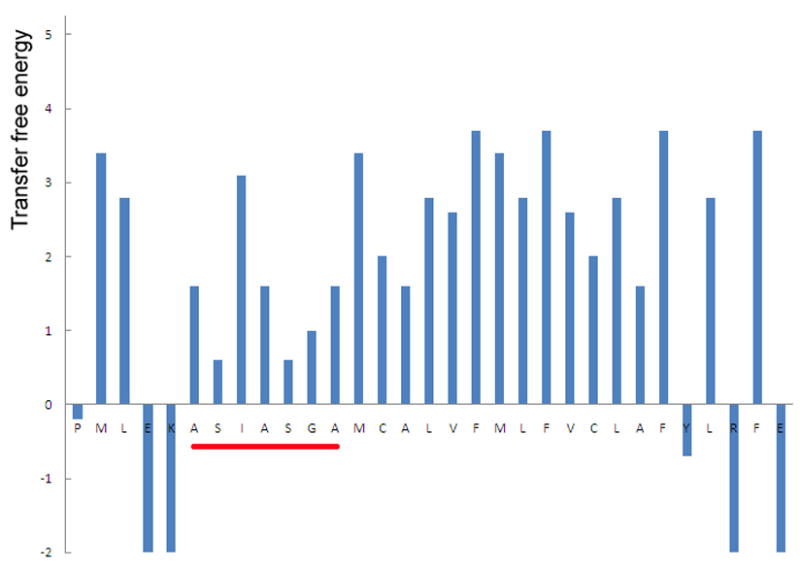
The transfer free energy (Y-axis, in kcal/mol [57]) for each residue around the TM domain of spitz. The “substrate motif” is underlined in red.
In the non-polar environment of the membrane, there is a strong correlation between the hydrophobicity of an amino acid and its helical propensity [56]: insertion of peptides into membrane is mainly determined by the hydrophobicity of amino acid side chains (for example, see ref. 59); once inserted, they almost invariably adopt a helical conformation to satisfy the need for hydrogen bonding within its main chain. In a polar environment, however, the formation and breaking of α-helices are more subtly influenced by many other factors (for example, see ref. 60). Therefore, the two types of ideas summarized above may not be mutually exclusive at a more fundamental level.
Besides regions that immediately flank the cleavage site, other parts of the substrate have also been shown to affect proteolysis, but through what mechanisms it is presently unknown. For example, Akiyama et al. found that a Gln-Pro sequence in the middle of LacY TM domain influence the cleavage efficiency of LacY by E. coli GlpG greatly (Fig. 3B) [53]. The glutamine and proline could be substituted by other helix-destabilizing residues, but it is unclear how helix destabilization at a distal location could influence proteolysis of a scissile bond that is already exposed and not part of the TM helix. In another example, Lohi et al. found that the cytosolic domain of thrombomodulin, a potential TM substrate for mammalian rhomboid RHBDL2, was critical for its proteolysis by RHBDL2 [61].
2.7. The mitochondrial rhomboid
The mitochondrial rhomboids deserve a separate mention not only because their catalytic domains adopt a different membrane orientation (Fig. 2B) [62], but also because of some of the unique features in their substrates [63–66]. Saccharomyces cerevisiae mitochondrial rhomboid Pcp1/Rbd1 has two known substrates: Ccp1, a mitochondrial cytochrome c peroxidase [63]; and Mgm1, a dynamin-like GTPase [64, 65]. Both Ccp1 and Mgm1 span the mitochondrial inner membrane once (with a type-II Nin-Cout orientation); their enzymatic domains are on the carboxyl terminal side of the TM region, and after cleavage by the protease, the carboxyl half of the molecule is released into solution. This is different from some of the examples cited above, e.g., TatA, where the carboxyl terminal fragment remains embedded in the membrane. Sequencing of the processed proteins has allowed mapping of the potential cleavage sites, which for both Ccp1 and Mgm1, reside in a short stretch of moderately hydrophobic sequence (Fig. 3C) [65, 67, 68]. Again common in both proteins these short hydrophobic sequences are preceded by another hydrophobic segment on the amino terminal side that could also serve as the TM domain, which if so, would leave the short sequence in the solution and on the wrong side of the membrane; to position the rhomboid cleavage site properly in the membrane for Pcp1/Rbd1 cleavage, it appears that other protein machineries are required to pull the substrate from the amino end (matrix side of the membrane) to dislocate the first hydrophobic segment [69, 70]. The model we developed earlier for other rhomboid substrates can not easily explain the processing of Ccp1 or Mgm1 by Pcp1/Rbd1: for example, if the scissile bond needs to move partially out of the membrane (into mitochondrial matrix), the remaining hydrophobic sequence would become even shorter to be able to span the membrane bilayer in a helical conformation. How the short hydrophobic sequence in Ccp1 or Mgm1, both containing a helix-breaking proline, initially interacts with membrane is also unclear.
3. The metalloproteases
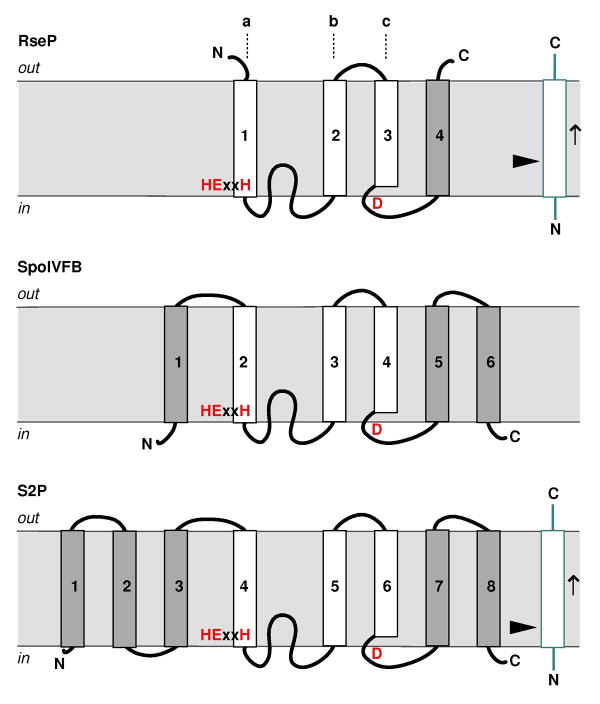
The membrane metalloprotease is another large enzyme family whose members have been found in all kingdoms of life [71, 72]. Mammalian site-2 protease (S2P) is the first known intramembrane protease [71]. It cleaves inside the TM region of sterol regulatory element binding protein (SREBP), and releases the transcription factor domain of SREBP from the membrane [73]. S2P also cleaves transcription factor ATF6, and plays a role in endoplasmic reticulum stress response [74]. Besides S2P, a number of bacterial metalloproteases have been well characterized both in function and in mechanism: the B. subtilis protease SpoIVFB, for example, was found to be responsible for removing a hydrophobic segment from transcription factor pro-σK, which converted it to the mature and active σK during sporulation [75, 76]; E. coli RseP (YaeL) is known to cleave RseA, a membrane protein that binds and inactivates transcription factor σE, in response to misfolded protein in the envelope [77, 78].
All membrane metalloproteases contain at least four hydrophobic and potentially membrane-spanning segments [72]: the first three are conserved, and each individually forms a single TM domain (a–c); the last hydrophobic segment, depending on protease, folds into either one or two TM helices [79, 80]. Figure 7 illustrates the membrane topologies for RseP, SpoIVFB and S2P, representatives of the three known structural subclasses within the family [72]. RseP is the simplest in structure, and contains only four TM domains (the common three plus the last hydrophobic segment that forms a single TM domain) [79]. SpoIVFB has an additional TM domain on the amino terminal side, and its last hydrophobic segment appears to fold into two TM helices, bringing the total number of TM domains to six [80]. The structure of S2P is the most complex, and least certain. Since its conserved core of three TM domains most likely adopts the same membrane orientation as those in RseP and SpoIVFB, and its carboxyl terminus is in the cytoplasm [81], the last hydrophobic segment of the protein, which is 28aa long and has a helix-breaking asparagine near the middle, must have also folded into two TM helices like that of SpoIVFB (Fig. 7). Towards its amino terminus, which is also in the cytoplasm [81], S2P has either one or three additional TM domains. In all three structural subclasses, between the first conserved TM domain (a) and the second (b), there is another hydrophobic region that forms a reentrant loop; the metal chelating motifs are located within the first (a) and third segments (c) of the conserved three, and face the cytosolic side of the membrane [72].
Fig. 7.
The membrane topology models for RseP, SpoIVFB and S2P (substrates shown in blue). The arrow head points at the cleavage site; the arrow indicates the direction of the polypeptide; the metal binding motifs are shown in red. The conserved three TM domains (a–c) are represented by white boxes. An 8-TM model of S2P is shown here (see text). Note that the figure is not in scale; the various “loops” that connect the TM domains may fold into α-helices (or even continuous with the TM helix) or β-strands in the real structure.
S2P cleaves a buried Leu-Cys bond close to the boundary with cytoplasm [82, 83]. RseP also cleaves type II (Nin-Cout) TM helices [84]. The bond in pro-σK that is cleaved by SpoIVFB, however, does not appear to be in a TM helix: the 20aa pro-sequence removed by the protease, which is responsible for membrane association, contains two charged residues near the middle (a lysine and a glutamate), and it has been shown that pro-σK could be partially solubilized from membrane by salts [85, 86]. Since the catalytic motifs in SpoIVFB are similarly located in the conserved and hydrophobic regions of the protein sequence, its ability to cleave a bond not in TM helices seems to suggest that the entrance to metalloprotease active site can not be positioned inside the membrane.
The crystal structure of Y392_METJA, a putative archaea metalloprotease from Methanocaldococcus jannaschii, was recently solved [87]. This represents another major achievement in structural biology, and will certainly stimulate further research into the detailed mechanism for the metalloproteases. The biological function of Y392 is not yet known, but its sequence is most similar to that of SpoIVFB, and the protein falls into the 6-TM-domain structural subclass (Fig. 7) [72, 80]. The sequence similarity, however, does not rule out the possibility that Y392 may normally cleave inside TM helices, but there is biochemical evidence suggesting that at least detergent-solubilized Y392 can cleave a protein substrate that does not have any TM helices [87].
The structure of Y392 is composed mainly of six α-helices (α1–α6; α4 has a break in the middle). The zinc atom is bound near the middle of the protein, about 14 Å from the cytosolic ends of the helices (α2–α6), which has led to the suggestion that the active site is deeply buried inside the membrane [87]. This, however, may not be correct because a significant portion of those five helices (α2–α6), from the point of the bound zinc towards cytosol, does not appear to be embedded in the membrane: a number of charged residues are exposed on the helices (red and blue in Fig. 8); between the two halves of α4, there is a conserved loop whose main chain is also exposed and needs to form hydrogen bond with water (yellow in Fig. 8). The loop contributes an aspartyl residue that binds to the zinc, and based on its position, it seems that the active site of the protease has to be very close to membrane surface (Fig. 8).
Fig. 8.
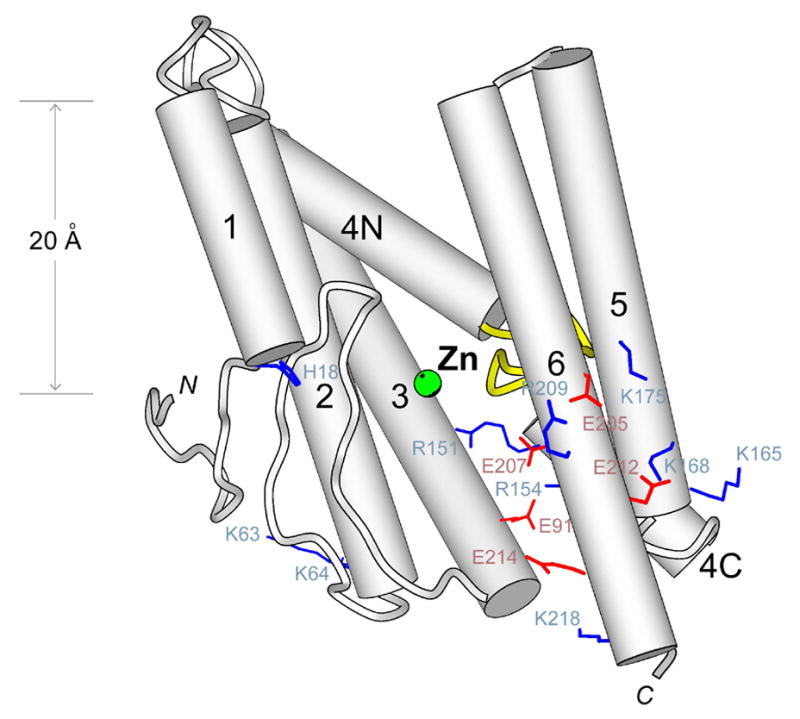
The crystal structure of Y392_METJA [87]. The bound zinc is shown in green. The possible boundaries of the membrane are indicated on the left.
After establishing the correct boundary with cytoplasm, it also seems that the membrane bilayer around Y392 can not be much thicker than 20 Å (Fig. 8). Besides the crystal structure, one may get an independent confirmation of this by examining the sequence segment that forms the last two TM domains of Y392: the hydrophobic segment, which is flanked by many charged residues, is 29aa long, and each TM domain has only 14 residues (the two TM domains are separated by an asparagine, which forms an N-cap to the second TM helix). Therefore, no matter how the two TM domains are inserted in the membrane, which is currently uncertain (because there are two independent copies of Y392 in the same crystal, and they are different in structure, especially around α5 and α6), they can only span a distance of around 20 Å. This does not appear to be unique to the SpoIVFB structural subclass: the last hydrophobic segment in human S2P is also short (28aa), and appears to have folded into two TM domains.
Based on differences between the two Y392 structures, it has been suggested that α1 and α6 may function as a lateral gate, the opening of which would allow substrate access to the active site [87, 88]. The proposal has not been tested by experiment. It should be noted, however, that this gating mechanism, correct or not, may not be generally relevant because RseP and other members in the 4-TM-domain structural subclass lack both helices (Fig. 7).
From the brief review provided here, we see at least three striking parallels between the rhomboid and metallo-proteases. First, within each family, some members specialize in cleaving bonds that are not in TM helices despite the conservation of the core structure of the enzyme. Secondly, their active sites are physically located very close to membrane surface. Thirdly, proteases in both families appear to cause significant membrane compression around the enzyme. These similarities suggest that the mechanism we proposed for the rhomboid protease may also apply to the metalloproteases.
4. The GxGD aspartyl proteases
Major members in this enzyme family include presenilin [89], signal peptide peptidase (SPP) [90], type IV prepilin peptidase (TFPP) and the related preflagellin peptidase [91, 92]. Steiner et al. were the first to notice the similarity between presenilin and the bacterial peptidase (TFPP) [93]: as a group, they are different from classic aspartyl proteases such as pepsin, which has two copies of the Asp-Thr-Gly (DTG) sequence motif, each contributing an aspartyl residue to the active site [94]. The pair of catalytic aspartates in presenilin and SPP is found in the middle of two hydrophobic and membrane-spanning regions of the protein: the first aspartate is preceded by a highly preferred tyrosine (YD), and the second follows a preferred leucine and two conserved glycines (LGxGD). TFPP has similar sequence motifs: its first aspartate usually follows an isoleucine (ID), and the second is part of a GxGD sequence.
Ponting et al. performed a multiple sequence alignment of presenilin and its homologs (some later identified as SPP and SPP-like proteases) [95], which showed that the carboxyl terminal halves of presenilin and SPP, including the two catalytic aspartates, were the most conserved. Narayanan et al. further demonstrated that this conserved region of four TM domains in SPP, when expressed alone, was proteolytically active [96]. The 4-TM-domain core structure, common among presenilin, SPP and their homologs (except for membrane orientation), may also be present in the prokaryotic proteases: TFPP and preflagellin peptidase have additional TM domains inserted between the four TM segments, but the number of inserted domains is always even so that the directionalities of the common four are maintained.
Unlike presenilin and SPP, TFPP does not cleave inside TM helices. The bacterial peptidase removes a short and positively charged leader sequence from pilin or pilin-like proteins by cleaving after an invariant glycine (for a review, see ref. 97). Five residues downstream of the glycine, before a type-II membrane anchor, there is a highly conserved glutamate, the charge of which would prevent the peptidase cleavage site from being buried inside the membrane.
Because of its link to Alzheimer’s disease, presenilin has been drawing the most attention [7, 8]. It now seems that many membrane proteins are potentially cleavable by this versatile protease; among them, amyloid precursor protein (APP) and notch receptor, validated by genetic analysis [98–101], are still the best known and characterized. From early on, it was realized that both APP and notch could be cleaved at multiple sites, either near the center of the TM domain or close to the border with cytoplasm [102, 103]. Using high-resolution gel electrophoresis, Qi-Takakara et al. carefully analyzed the size of APP fragments generated by presenilin cleavage [104]. They discovered a full range of cleavage products, mainly Aβ46, Aβ43 and Aβ40, all differing by three residues at the carboxyl terminus (there is another minor set from Aβ48 to Aβ39, which is not discussed here, but may be relevant to Alzherimer’s disease). Intriguingly, there appears to be an order by which these fragments are generated (APP needs to be processed initially by β-secretase, which produces a 99aa carboxyl terminal fragment C99 or Aβ99, before presenilin can cleave it): first, Aβ99 is converted to Aβ49 in a reaction that also generates Aβ50–99, which is detectable as a major soluble species inside the cell (APP intracellular domain or AICD); Aβ49 is quickly turned over to Aβ46 (which may explain why Aβ49 level is so low); Aβ46 is then shortened to Aβ43, and finally to Aβ40. This order, supported by more studies [105–109], provides a link between the deep and shallow cleavage sites, but also raises the question of why APP has to be cleaved in such an order. One possibility is that each time only a small segment of the TM domain (for the intermediate species, this includes the neo-carboxyl terminus that carries a negative charge) can partition out of the membrane for proteolysis by presenilin.
Presenilin is part of a larger protein complex (γ-secretase) that includes three other membrane protein components (for example, see ref. 110). The complex has been studied by electron microscopy through single particle averaging [111, 112], but the achieved resolutions were still too low to reveal the fold of the protein, or to identify the active site. The active site of presenilin has been independently probed in two studies that analyzed the accessibility of single cysteines introduced into it by mutagenesis [113, 114]. Both studies showed that the two catalytic aspartates were in an aqueous cavity that was continuous with cytoplasm. However, the lack of a high-resolution crystal structure in this family has hindered the type of mechanistic insights that we are beginning to gain for the other two membrane protease families.
5. Concluding remarks
How to unfold a TM helix to make it cleavable is a unique challenge that intramembrane proteases face. The problem could be solved in theory by creating an aqueous channel inside the membrane-embedded protease into which substrate TM helix can enter, but recently solved crystal structures, representing two of three known membrane protease families, have revealed no such channels; instead, both proteases, totally unrelated in fold or catalytic mechanism, appear to have narrow membrane-spanning regions. This common feature suggests a different type of mechanism to achieve a similar goal: in the thinner membrane around the protease, part of the substrate that bears the cleavage site, which is often near the end of TM helices, can partition into aqueous solution (or into protease active site), unfold and become cleaved, whereas the rest of the TM helix is still of sufficient length to span the bilayer (Fig. 1A). Whether this represents a universal principle awaits experimental test, and requires analysis of more crystal structures, especially one from the GxGD family.
Acknowledgments
Some of the ideas presented here emerged from discussions with Y. Akiyama and M. Freeman. Research in the author’s laboratory has been supported by Yale University, Ellison Medical Foundation, Neuroscience Education and Research Foundation, and NIH.
Abbreviations
- aa
amino acids
- APP
amyloid precursor protein
- DCI
3,4-dichloroisocoumarin
- S2P
site-2 protease
- SPP
signal peptide peptidase
- SREBP
sterol regulatory element binding protein
- TFPP
type 4 prepilin peptidase
- TM
transmembrane
References
- 1.Brown MS, Ye J, Rawson RB, Goldstein JL. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell. 2000;100:391–8. doi: 10.1016/s0092-8674(00)80675-3. [DOI] [PubMed] [Google Scholar]
- 2.Kroos L, Yu YT. Regulation of sigma factor activity during Bacillus subtilis development. Curr Opin Microbiol. 2000;3:553–60. doi: 10.1016/s1369-5274(00)00140-5. [DOI] [PubMed] [Google Scholar]
- 3.Weihofen A, Martoglio B. Intramembrane-cleaving proteases: controlled liberation of proteins and bioactive peptides. Trends Cell Biol. 2003;13:71–8. doi: 10.1016/s0962-8924(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 4.Alba BM, Gross CA. Regulation of the Escherichia coli sigma-dependent envelope stress response. Mol Microbiol. 2004;52:613–9. doi: 10.1111/j.1365-2958.2003.03982.x. [DOI] [PubMed] [Google Scholar]
- 5.Freeman M. Proteolysis within the membrane: rhomboids revealed. Nat Rev Mol Cell Biol. 2004;5:188–97. doi: 10.1038/nrm1334. [DOI] [PubMed] [Google Scholar]
- 6.Wolfe MS, Kopan R. Intramembrane proteolysis: theme and variations. Science. 2004;305:1119–23. doi: 10.1126/science.1096187. [DOI] [PubMed] [Google Scholar]
- 7.Haass C, Steiner H. Alzheimer disease gamma-secretase: a complex story of GxGD-type presenilin proteases. Trends Cell Biol. 2002;12:556–62. doi: 10.1016/s0962-8924(02)02394-2. [DOI] [PubMed] [Google Scholar]
- 8.Selkoe DJ, Wolfe MS. Presenilin: running with scissors in the membrane. Cell. 2007;131:215–21. doi: 10.1016/j.cell.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 9.Ha Y. Structural principles of intramembrane proteases. Curr Opin Struct Biol. 2007;17:405–11. doi: 10.1016/j.sbi.2007.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lemberg MK, Freeman M. Cutting proteins within lipid bilayers: rhomboid structure and mechanism. Mol Cell. 2007;28:930–40. doi: 10.1016/j.molcel.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 11.Wasserman JD, Urban S, Freeman M. A family of rhomboid-like genes: Drosophila rhomboid-1 and roughoid/rhomboid-3 cooperate to activate EGF receptor signaling. Genes Dev. 2000;14:1651–63. [PMC free article] [PubMed] [Google Scholar]
- 12.Lee JR, Urban S, Garvey CF, Freeman M. Regulated intracellular ligand transport and proteolysis control EGF signal activation in Drosophila. Cell. 2001;107:161–71. doi: 10.1016/s0092-8674(01)00526-8. [DOI] [PubMed] [Google Scholar]
- 13.Urban S. Rhomboid proteins: conserved membrane proteases with divergent biological functions. Genes Dev. 2006;20:3054–68. doi: 10.1101/gad.1488606. [DOI] [PubMed] [Google Scholar]
- 14.Pellegrini L, Scorrano L. A cut short to death: Parl and Opa1 in the regulation of mitochondrial morphology and apoptosis. Cell Death Differ. 2007;14:1275–84. doi: 10.1038/sj.cdd.4402145. [DOI] [PubMed] [Google Scholar]
- 15.Bretscher MS, Munro S. Cholesterol and the Golgi apparatus. Science. 1993;261:1280–1. doi: 10.1126/science.8362242. [DOI] [PubMed] [Google Scholar]
- 16.Gawrisch K, Gaede HC, Mihailescu M, White SH. Hydration of POPC bilayers studied by 1H-PFG-MAS-NOESY and neutron diffraction. Eur Biophys J. 2007;36:281–91. doi: 10.1007/s00249-007-0142-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ye J, Davé UP, Grishin NV, Goldstein JL, Brown MS. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc Natl Acad Sci USA. 2000;97:5123–8. doi: 10.1073/pnas.97.10.5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang Y, Maegawa S, Akiyama Y, Ha Y. The role of L1 loop in the mechanism of rhomboid intramembrane protease GlpG. J Mol Biol. 2007;374:1104–13. doi: 10.1016/j.jmb.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe MS. The gamma-secretase complex: membrane-embedded proteolytic ensemble. Biochemistry. 2006;45:7931–9. doi: 10.1021/bi060799c. [DOI] [PubMed] [Google Scholar]
- 20.Mayer U, Nüsslein-Volhard C. A group of genes required for pattern formation in the ventral ectoderm of the Drosophila embryo. Genes Dev. 1988;2:1496–511. doi: 10.1101/gad.2.11.1496. [DOI] [PubMed] [Google Scholar]
- 21.Wasserman JD, Freeman M. Control of EGF receptor activation in Drosophila. Trends Cell Biol. 1997;7:431–6. doi: 10.1016/S0962-8924(97)01143-4. [DOI] [PubMed] [Google Scholar]
- 22.Urban S, Lee JR, Freeman M. Drosophila rhomboid-1 defines a family of putative intramembrane serine proteases. Cell. 2001;107:173–82. doi: 10.1016/s0092-8674(01)00525-6. [DOI] [PubMed] [Google Scholar]
- 23.Urban S, Schlieper D, Freeman M. Conservation of intramembrane proteolytic activity and substrate specificity in prokaryotic and eukaryotic rhomboids. Curr Biol. 2002;12:1507–12. doi: 10.1016/s0960-9822(02)01092-8. [DOI] [PubMed] [Google Scholar]
- 24.Lemberg MK, Menendez J, Misik A, Garcia M, Koth CM, Freeman M. Mechanism of intramembrane proteolysis investigated with purified rhomboid proteases. EMBO J. 2005;24:464–72. doi: 10.1038/sj.emboj.7600537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Urban S, Wolfe MS. Reconstitution of intramembrane proteolysis in vitro reveals that pure rhomboid is sufficient for catalysis and specificity. Proc Natl Acad Sci U S A. 2005;102:1883–8. doi: 10.1073/pnas.0408306102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maegawa S, Ito K, Akiyama Y. Proteolytic action of GlpG, a rhomboid protease in the Escherichia coli cytoplasmic membrane. Biochemistry. 2005;44:13543–52. doi: 10.1021/bi051363k. [DOI] [PubMed] [Google Scholar]
- 27.Bier E, Jan LY, Jan YN. Rhomboid, a gene required for dorsoventral axis establishment and peripheral nervous system development in Drosophila melanogaster. Genes Dev. 1990;4:190–203. doi: 10.1101/gad.4.2.190. [DOI] [PubMed] [Google Scholar]
- 28.Daley DO, Rapp M, Granseth E, Melén K, Drew D, von Heijne G. Global topology analysis of the Escherichia coli inner membrane proteome. Science. 2005;308:1321–3. doi: 10.1126/science.1109730. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Zhang Y, Ha Y. Crystal structure of a rhomboid family intramembrane protease. Nature. 2006;444:179–83. doi: 10.1038/nature05255. [DOI] [PubMed] [Google Scholar]
- 30.Wu Z, Yan N, Feng L, Oberstein A, Yan H, Baker RP, Gu L, Jeffrey PD, Urban S, Shi Y. Structural analysis of a rhomboid family intramembrane protease reveals a gating mechanism for substrate entry. Nat Struct Mol Biol. 2006;13:1084–91. doi: 10.1038/nsmb1179. [DOI] [PubMed] [Google Scholar]
- 31.Ben-Shem A, Fass D, Bibi E. Structural basis for intramembrane proteolysis by rhomboid serine proteases. Proc Natl Acad Sci USA. 2007;104:462–6. doi: 10.1073/pnas.0609773104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemieux MJ, Fischer SJ, Cherney MM, Bateman KS, James MN. The crystal structure of the rhomboid peptidase from Haemophilus influenzae provides insight into intramembrane proteolysis. Proc Natl Acad Sci USA. 2007;104:750–4. doi: 10.1073/pnas.0609981104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koonin EV, Makarova KS, Rogozin IB, Davidovic L, Letellier MC, Pellegrini L. The rhomboids: a nearly ubiquitous family of intramembrane serine proteases that probably evolved by multiple ancient horizontal gene transfers. Genome Biol. 2003;4:R19. doi: 10.1186/gb-2003-4-3-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemberg MK, Freeman M. Functional and evolutionary implications of enhanced genomic analysis of rhomboid intramembrane proteases. Genome Res. 2007;17:1634–46. doi: 10.1101/gr.6425307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harper JW, Hemmi K, Powers JC. Reaction of serine proteases with substituted isocoumarins: discovery of 3,4-dichloroisocoumarin, a new general mechanism based serine protease inhibitor. Biochemistry. 1985;24:1831–41. doi: 10.1021/bi00329a005. [DOI] [PubMed] [Google Scholar]
- 36.Cole LB, Kilpatrick JM, Chu N, Babu YS. Structure of 3,4-dichloroisocoumarin-inhibited factor D. Acta Crystallogr D Biol Crystallogr. 1998;54:711–7. doi: 10.1107/s0907444997010457. [DOI] [PubMed] [Google Scholar]
- 37.Wang Y, Ha Y. Open-cap conformation of intramembrane protease GlpG. Proc Natl Acad Sci USA. 2007;104:2098–102. doi: 10.1073/pnas.0611080104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urban S, Freeman M. Substrate specificity of rhomboid intramembrane proteases is governed by helix-breaking residues in the substrate transmembrane domain. Mol Cell. 2003;11:1425–34. doi: 10.1016/s1097-2765(03)00181-3. [DOI] [PubMed] [Google Scholar]
- 39.Baker RP, Young K, Feng L, Shi Y, Urban S. Enzymatic analysis of a rhomboid intramembrane protease implicates transmembrane helix 5 as the lateral substrate gate. Proc Natl Acad Sci U S A. 2007;104:8257–62. doi: 10.1073/pnas.0700814104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opitz C, Di Cristina M, Reiss M, Ruppert T, Crisanti A, Soldati D. Intramembrane cleavage of microneme proteins at the surface of the apicomplexan parasite Toxoplasma gondii. EMBO J. 2002;21:1577–85. doi: 10.1093/emboj/21.7.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou XW, Blackman MJ, Howell SA, Carruthers VB. Proteomic analysis of cleavage events reveals a dynamic two-step mechanism for proteolysis of a key parasite adhesive complex. Mol Cell Proteomics. 2004;3:565–76. doi: 10.1074/mcp.M300123-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Brossier F, Jewett TJ, Sibley LD, Urban S. A spatially localized rhomboid protease cleaves cell surface adhesins essential for invasion by Toxoplasma. Proc Natl Acad Sci U S A. 2005;102:4146–51. doi: 10.1073/pnas.0407918102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.O’Donnell RA, Hackett F, Howell SA, Treeck M, Struck N, Krnajski Z, Withers-Martinez C, Gilberger TW, Blackman MJ. Intramembrane proteolysis mediates shedding of a key adhesin during erythrocyte invasion by the malaria parasite. J Cell Biol. 2006;174:1023–33. doi: 10.1083/jcb.200604136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baker RP, Wijetilaka R, Urban S. Two Plasmodium rhomboid proteases preferentially cleave different adhesins implicated in all invasive stages of malaria. PLoS Pathog. 2006;2:e113. doi: 10.1371/journal.ppat.0020113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maegawa S, Koide K, Ito K, Akiyama Y. The intramembrane active site of GlpG, an E. coli rhomboid protease, is accessible to water and hydrolyses an extramembrane peptide bond of substrates. Mol Microbiol. 2007;64:435–47. doi: 10.1111/j.1365-2958.2007.05679.x. [DOI] [PubMed] [Google Scholar]
- 46.Urban S, Lee JR, Freeman M. A family of Rhomboid intramembrane proteases activates all Drosophila membrane-tethered EGF ligands. EMBO J. 2002;21:4277–86. doi: 10.1093/emboj/cdf434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallio M, Sturgill G, Rather P, Kylsten P. A conserved mechanism for extracellular signaling in eukaryotes and prokaryotes. Proc Natl Acad Sci U S A. 2002;99:12208–13. doi: 10.1073/pnas.192138799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stevenson LG, Strisovsky K, Clemmer KM, Bhatt S, Freeman M, Rather PN. Rhomboid protease AarA mediates quorum-sensing in Providencia stuartii by activating TatA of the twin-arginine translocase. Proc Natl Acad Sci U S A. 2007;104:1003–8. doi: 10.1073/pnas.0608140104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee PA, Buchanan G, Stanley NR, Berks BC, Palmer T. Truncation analysis of TatA and TatB defines the minimal functional units required for protein translocation. J Bacteriol. 2002;184:5871–9. doi: 10.1128/JB.184.21.5871-5879.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelman DM. Membranes are more mosaic than fluid. Nature. 2005;438:578–80. doi: 10.1038/nature04394. [DOI] [PubMed] [Google Scholar]
- 51.Killian JA, Nyholm TK. Peptides in lipid bilayers: the power of simple models. Curr Opin Struct Biol. 2006;16:473–9. doi: 10.1016/j.sbi.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 52.Van den Berg B, Clemons WM, Jr, Collinson I, Modis Y, Hartmann E, Harrison SC, Rapoport TA. X-ray structure of a protein-conducting channel. Nature. 2004;427:36–44. doi: 10.1038/nature02218. [DOI] [PubMed] [Google Scholar]
- 53.Akiyama Y, Maegawa S. Sequence features of substrates required for cleavage by GlpG, an Escherichia coli rhomboid protease. Mol Microbiol. 2007;64:1028–37. doi: 10.1111/j.1365-2958.2007.05715.x. [DOI] [PubMed] [Google Scholar]
- 54.Lemberg MK, Martoglio B. Requirements for signal peptide peptidase-catalyzed intramembrane proteolysis. Mol Cell. 2002;10:735–44. doi: 10.1016/s1097-2765(02)00655-x. [DOI] [PubMed] [Google Scholar]
- 55.Chou PY, Fasman GD. Conformational parameters for amino acids in helical, beta-sheet, and random coil regions calculated from proteins. Biochemistry. 1974;13:211–22. doi: 10.1021/bi00699a001. [DOI] [PubMed] [Google Scholar]
- 56.Liu LP, Deber CM. Uncoupling hydrophobicity and helicity in transmembrane segments. Alpha-helical propensities of the amino acids in non-polar environments. J Biol Chem. 1998;273:23645–8. doi: 10.1074/jbc.273.37.23645. [DOI] [PubMed] [Google Scholar]
- 57.Engelman DM, Steitz TA, Goldman A. Identifying nonpolar transbilayer helices in amino acid sequences of membrane proteins. Annu Rev Biophys Biophys Chem. 1986;15:321–53. doi: 10.1146/annurev.bb.15.060186.001541. [DOI] [PubMed] [Google Scholar]
- 58.Arkin IT, Brunger AT. Statistical analysis of predicted transmembrane alpha-helices. Biochim Biophys Acta. 1998;1429:113–28. doi: 10.1016/s0167-4838(98)00225-8. [DOI] [PubMed] [Google Scholar]
- 59.Hessa T, Kim H, Bihlmaier K, Lundin C, Boekel J, Andersson H, Nilsson I, White SH, von Heijne G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature. 2005;433:377–81. doi: 10.1038/nature03216. [DOI] [PubMed] [Google Scholar]
- 60.Shi Z, Olson CA, Bell AJ, Jr, Kallenbach NR. Stabilization of alpha-helix structure by polar side-chain interactions: complex salt bridges, cation-pi interactions, and C-H em leader O H-bonds. Biopolymers. 2001;60:366–80. doi: 10.1002/1097-0282(2001)60:5<366::AID-BIP10177>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 61.Lohi O, Urban S, Freeman M. Diverse substrate recognition mechanisms for rhomboids; thrombomodulin is cleaved by Mammalian rhomboids. Curr Biol. 2004;14:236–41. doi: 10.1016/j.cub.2004.01.025. [DOI] [PubMed] [Google Scholar]
- 62.Jeyaraju DV, Xu L, Letellier MC, Bandaru S, Zunino R, Berg EA, McBride HM, Pellegrini L. Phosphorylation and cleavage of presenilin-associated rhomboid-like protein (PARL) promotes changes in mitochondrial morphology. Proc Natl Acad Sci U S A. 2006;103:18562–7. doi: 10.1073/pnas.0604983103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Esser K, Tursun B, Ingenhoven M, Michaelis G, Pratje E. A novel two-step mechanism for removal of a mitochondrial signal sequence involves the mAAA complex and the putative rhomboid protease Pcp1. J Mol Biol. 2002;323:835–43. doi: 10.1016/s0022-2836(02)01000-8. [DOI] [PubMed] [Google Scholar]
- 64.McQuibban GA, Saurya S, Freeman M. Mitochondrial membrane remodelling regulated by a conserved rhomboid protease. Nature. 2003;423:537–41. doi: 10.1038/nature01633. [DOI] [PubMed] [Google Scholar]
- 65.Herlan M, Vogel F, Bornhovd C, Neupert W, Reichert AS. Processing of Mgm1 by the rhomboid-type protease Pcp1 is required for maintenance of mitochondrial morphology and of mitochondrial DNA. J Biol Chem. 2003;278:27781–8. doi: 10.1074/jbc.M211311200. [DOI] [PubMed] [Google Scholar]
- 66.Cipolat S, Rudka T, Hartmann D, Costa V, Serneels L, Craessaerts K, et al. Mitochondrial rhomboid PARL regulates cytochrome c release during apoptosis via OPA1-dependent cristae remodeling. Cell. 2006;126:163–75. doi: 10.1016/j.cell.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 67.Kaput J, Goltz S, Blobel G. Nucleotide sequence of the yeast nuclear gene for cytochrome c peroxidase precursor. Functional implications of the pre sequence for protein transport into mitochondria. J Biol Chem. 1982;257:15054–8. [PubMed] [Google Scholar]
- 68.Takio K, Titani K, Ericsson LH, Yonetani T. Primary structure of yeast cytochrome c peroxidase. II. The complete amino acid sequence. Arch Biochem Biophys. 1980;203:615–29. doi: 10.1016/0003-9861(80)90219-2. [DOI] [PubMed] [Google Scholar]
- 69.Herlan M, Bornhövd C, Hell K, Neupert W, Reichert AS. Alternative topogenesis of Mgm1 and mitochondrial morphology depend on ATP and a functional import motor. J Cell Biol. 2004;165:167–73. doi: 10.1083/jcb.200403022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tatsuta T, Augustin S, Nolden M, Friedrichs B, Langer T. m-AAA protease-driven membrane dislocation allows intramembrane cleavage by rhomboid in mitochondria. EMBO J. 2007;26:325–35. doi: 10.1038/sj.emboj.7601514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rawson RB, Zelenski NG, Nijhawan D, Ye J, Sakai J, Hasan MT, Chang TY, Brown MS, Goldstein JL. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol Cell. 1997;1:47–57. doi: 10.1016/s1097-2765(00)80006-4. [DOI] [PubMed] [Google Scholar]
- 72.Lewis AP, Thomas PJ. A novel clan of zinc metallopeptidases with possible intramembrane cleavage properties. Protein Sci. 1999;8:439–42. doi: 10.1110/ps.8.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown MS, Goldstein JL. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 1997;89:331–40. doi: 10.1016/s0092-8674(00)80213-5. [DOI] [PubMed] [Google Scholar]
- 74.Ye J, Rawson RB, Komuro R, Chen X, Davé UP, Prywes R, Brown MS, Goldstein JL. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–64. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 75.Rudner DZ, Fawcett P, Losick R. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc Natl Acad Sci U S A. 1999;96:14765–70. doi: 10.1073/pnas.96.26.14765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yu YT, Kroos L. Evidence that SpoIVFB is a novel type of membrane metalloprotease governing intercompartmental communication during Bacillus subtilis sporulation. J Bacteriol. 2000;182:3305–9. doi: 10.1128/jb.182.11.3305-3309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kanehara K, Ito K, Akiyama Y. YaeL (EcfE) activates the sigma(E) pathway of stress response through a site-2 cleavage of anti-sigma(E), RseA. Genes Dev. 2002;16:2147–55. doi: 10.1101/gad.1002302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Alba BM, Leeds JA, Onufryk C, Lu CZ, Gross CA. DegS and YaeL participate sequentially in the cleavage of RseA to activate the sigma(E)-dependent extracytoplasmic stress response. Genes Dev. 2002;16:2156–68. doi: 10.1101/gad.1008902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kanehara K, Akiyama Y, Ito K. Characterization of the yaeL gene product and its S2P-protease motifs in Escherichia coli. Gene. 2001;281:71–9. doi: 10.1016/s0378-1119(01)00823-x. [DOI] [PubMed] [Google Scholar]
- 80.Green DH, Cutting SM. Membrane topology of the Bacillus subtilis pro-sigma(K) processing complex. J Bacteriol. 2000;182:278–85. doi: 10.1128/jb.182.2.278-285.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Zelenski NG, Rawson RB, Brown MS, Goldstein JL. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J Biol Chem. 1999;274:21973–80. doi: 10.1074/jbc.274.31.21973. [DOI] [PubMed] [Google Scholar]
- 82.Sakai J, Duncan EA, Rawson RB, Hua X, Brown MS, Goldstein JL. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 1996;85:1037–46. doi: 10.1016/s0092-8674(00)81304-5. [DOI] [PubMed] [Google Scholar]
- 83.Duncan EA, Davé UP, Sakai J, Goldstein JL, Brown MS. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J Biol Chem. 1998;273:17801–9. doi: 10.1074/jbc.273.28.17801. [DOI] [PubMed] [Google Scholar]
- 84.Akiyama Y, Kanehara K, Ito K. RseP (YaeL), an Escherichia coli RIP protease, cleaves transmembrane sequences. EMBO J. 2004;23:4434–42. doi: 10.1038/sj.emboj.7600449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kroos L, Kunkel B, Losick R. Switch protein alters specificity of RNA polymerase containing a compartment-specific sigma factor. Science. 1989;243:526–9. doi: 10.1126/science.2492118. [DOI] [PubMed] [Google Scholar]
- 86.Zhang B, Hofmeister A, Kroos L. The prosequence of pro-sigmaK promotes membrane association and inhibits RNA polymerase core binding. J Bacteriol. 1998;180:2434–41. doi: 10.1128/jb.180.9.2434-2441.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Feng L, Yan H, Wu Z, Yan N, Wang Z, Jeffrey PD, Shi Y. Structure of a site-2 protease family intramembrane metalloprotease. Science. 2007;318:1608–12. doi: 10.1126/science.1150755. [DOI] [PubMed] [Google Scholar]
- 88.Urban S, Shi Y. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr Opin Struct Biol. 2008 doi: 10.1016/j.sbi.2008.03.005. in the press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wolfe MS, Xia W, Ostaszewski BL, Diehl TS, Kimberly WT, Selkoe DJ. Two transmembrane aspartates in presenilin-1 required for presenilin endoproteolysis and gamma-secretase activity. Nature. 1999;398:513–7. doi: 10.1038/19077. [DOI] [PubMed] [Google Scholar]
- 90.Weihofen A, Binns K, Lemberg MK, Ashman K, Martoglio B. Identification of signal peptide peptidase, a presenilin-type aspartic protease. Science. 2002;296:2215–8. doi: 10.1126/science.1070925. [DOI] [PubMed] [Google Scholar]
- 91.LaPointe CF, Taylor RK. The type 4 prepilin peptidases comprise a novel family of aspartic acid proteases. J Biol Chem. 2000;275:1502–10. doi: 10.1074/jbc.275.2.1502. [DOI] [PubMed] [Google Scholar]
- 92.Bardy SL, Jarrell KF. Cleavage of preflagellins by an aspartic acid signal peptidase is essential for flagellation in the archaeon Methanococcus voltae. Mol Microbiol. 2003;50:1339–47. doi: 10.1046/j.1365-2958.2003.03758.x. [DOI] [PubMed] [Google Scholar]
- 93.Steiner H, Kostka M, Romig H, Basset G, Pesold B, Hardy J, Capell A, Meyn L, Grim ML, Baumeister R, Fechteler K, Haass C. Glycine 384 is required for presenilin-1 function and is conserved in bacterial polytopic aspartyl proteases. Nat Cell Biol. 2000;2:848–51. doi: 10.1038/35041097. [DOI] [PubMed] [Google Scholar]
- 94.Rawlings ND, Barrett AJ. Families of aspartic peptidases, and those of unknown catalytic mechanism. Methods Enzymol. 1995;248:105–20. doi: 10.1016/0076-6879(95)48009-9. [DOI] [PubMed] [Google Scholar]
- 95.Ponting CP, Hutton M, Nyborg A, Baker M, Jansen K, Golde TE. Identification of a novel family of presenilin homologues. Hum Mol Genet. 2002;11:1037–44. doi: 10.1093/hmg/11.9.1037. [DOI] [PubMed] [Google Scholar]
- 96.Narayanan S, Sato T, Wolfe MS. A C-terminal region of signal peptide peptidase defines a functional domain for intramembrane aspartic protease catalysis. J Biol Chem. 2007;282:20172–9. doi: 10.1074/jbc.M701536200. [DOI] [PubMed] [Google Scholar]
- 97.Lory S, Strom MS. Structure-function relationship of type-IV prepilin peptidase of Pseudomonas aeruginosa--a review. Gene. 1997;192:117–21. doi: 10.1016/s0378-1119(96)00830-x. [DOI] [PubMed] [Google Scholar]
- 98.De Strooper B, Saftig P, Craessaerts K, Vanderstichele H, Guhde G, Annaert W, et al. Deficiency of presenilin-1 inhibits the normal cleavage of amyloid precursor protein. Nature. 1998;391:387–90. doi: 10.1038/34910. [DOI] [PubMed] [Google Scholar]
- 99.De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–22. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 100.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–5. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 101.Ye Y, Lukinova N, Fortini ME. Neurogenic phenotypes and altered Notch processing in Drosophila Presenilin mutants. Nature. 1999;398:525–9. doi: 10.1038/19096. [DOI] [PubMed] [Google Scholar]
- 102.Weidemann A, Eggert S, Reinhard FB, Vogel M, Paliga K, Baier G, et al. A novel epsilon-cleavage within the transmembrane domain of the Alzheimer amyloid precursor protein demonstrates homology with Notch processing. Biochemistry. 2002;41:2825–35. doi: 10.1021/bi015794o. [DOI] [PubMed] [Google Scholar]
- 103.Okochi M, Steiner H, Fukumori A, Tanii H, Tomita T, Tanaka T, Iwatsubo T, Kudo T, Takeda M, Haass C. Presenilins mediate a dual intramembranous gamma-secretase cleavage of Notch-1. EMBO J. 2002;21:5408–16. doi: 10.1093/emboj/cdf541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Qi-Takahara Y, Morishima-Kawashima M, Tanimura Y, Dolios G, Hirotani N, Horikoshi Y, et al. Longer forms of amyloid beta protein: implications for the mechanism of intramembrane cleavage by gamma-secretase. J Neurosci. 2005;25:436–45. doi: 10.1523/JNEUROSCI.1575-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Sato T, Tanimura Y, Hirotani N, Saido TC, Morishima-Kawashima M, Ihara Y. Blocking the cleavage at midportion between gamma- and epsilon-sites remarkably suppresses the generation of amyloid beta-protein. FEBS Lett. 2005;579:2907–12. doi: 10.1016/j.febslet.2005.04.037. [DOI] [PubMed] [Google Scholar]
- 106.Zhao G, Cui MZ, Mao G, Dong Y, Tan J, Sun L, Xu X. gamma-Cleavage is dependent on zeta-cleavage during the proteolytic processing of amyloid precursor protein within its transmembrane domain. J Biol Chem. 2005;280:37689–97. doi: 10.1074/jbc.M507993200. [DOI] [PubMed] [Google Scholar]
- 107.Yagishita S, Morishima-Kawashima M, Tanimura Y, Ishiura S, Ihara Y. DAPT-induced intracellular accumulations of longer amyloid beta-proteins: further implications for the mechanism of intramembrane cleavage by gamma-secretase. Biochemistry. 2006;45:3952–60. doi: 10.1021/bi0521846. [DOI] [PubMed] [Google Scholar]
- 108.Kakuda N, Funamoto S, Yagishita S, Takami M, Osawa S, Dohmae N, Ihara Y. Equimolar production of amyloid beta-protein and amyloid precursor protein intracellular domain from beta-carboxyl-terminal fragment by gamma-secretase. J Biol Chem. 2006;281:14776–86. doi: 10.1074/jbc.M513453200. [DOI] [PubMed] [Google Scholar]
- 109.Funamoto S, Morishima-Kawashima M, Tanimura Y, Hirotani N, Saido TC, Ihara Y. Truncated carboxyl-terminal fragments of beta-amyloid precursor protein are processed to amyloid beta-proteins 40 and 42. Biochemistry. 2004;43:13532–40. doi: 10.1021/bi049399k. [DOI] [PubMed] [Google Scholar]
- 110.Takasugi N, Tomita T, Hayashi I, Tsuruoka M, Niimura M, Takahashi Y, et al. The role of presenilin cofactors in the gamma-secretase complex. Nature. 2003;422:438–41. doi: 10.1038/nature01506. [DOI] [PubMed] [Google Scholar]
- 111.Lazarov VK, Fraering PC, Ye W, Wolfe MS, Selkoe DJ, Li H. Electron microscopic structure of purified, active gamma-secretase reveals an aqueous intramembrane chamber and two pores. Proc Natl Acad Sci U S A. 2006;103:6889–94. doi: 10.1073/pnas.0602321103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ogura T, Mio K, Hayashi I, Miyashita H, Fukuda R, Kopan R, Kodama T, Hamakubo T, Iwatsubo T, Tomita T, Sato C. Three-dimensional structure of the gamma- secretase complex. Biochem Biophys Res Commun. 2006;343:525–34. doi: 10.1016/j.bbrc.2006.02.158. [DOI] [PubMed] [Google Scholar]
- 113.Tolia A, Chávez-Gutiérrez L, De Strooper B. Contribution of presenilin transmembrane domains 6 and 7 to a water-containing cavity in the gamma-secretase complex. J Biol Chem. 2006;281:27633–42. doi: 10.1074/jbc.M604997200. [DOI] [PubMed] [Google Scholar]
- 114.Sato C, Morohashi Y, Tomita T, Iwatsubo T. Structure of the catalytic pore of gamma-secretase probed by the accessibility of substituted cysteines. J Neurosci. 2006;26:12081–8. doi: 10.1523/JNEUROSCI.3614-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]