Abstract
During cell proliferation, DNA damage inflicted by intrinsic or extrinsic genotoxic stresses impose a thereat to DNA replication. The stability of the DNA replication forks that encounter DNA damage is crucial for genomic integrity. Both the ATR-regulated checkpoint pathway and the translesion DNA synthesis mediated by the ubiquitinated PCNA are important for continuous replication of damaged DNA. We have recently shown that Chk1, a key effector kinase of ATR in checkpoint response, is required for efficient PCNA ubiquitination after DNA damage. Surprisingly, the ubiquitination of PCNA is independent of ATR, but regulated by Claspin, a replication protein that mediates the activation of Chk1 by ATR. Like Claspin, Timeless and Rad17, two other Chk1 regulators at stressed replication forks, are also implicated in PCNA ubiquitination. These findings suggest that while ATR signaling and PCNA ubiquitination are two independent processes, they are mediated by a common group of proteins including Chk1 and it regulators at replication forks. Furthermore, these data raise the possibility that Chk1 and its regulators may constitute a functional module at replication forks to enable multiple stress responses.
Keywords: DNA damage, checkpoint, replication fork, PCNA, ATR, Chk1, ubiquitination
Introduction
The stability of DNA replication forks is crucial for genomic stability and cell survival. When DNA replication forks encounter DNA damage in the genome, they rely on several signaling and DNA repair/recombination mechanisms to overcome the impediment. Among these mechanisms are the DNA damage-signaling pathway regulated by the Ataxia Telangiectasia-mutated and Rad3-related (ATR) kinase, which is often referred to as the ATR checkpoint 1, 2, and the process of translesion DNA synthesis (TLS) mediated by the ubiquitinated form of PCNA 3. Both the ATR pathway and TLS are important for continuous replication of damaged DNA and avoidance of fork collapse. The ATR pathway is critical for stabilizing stressed replication forks, whereas TLS allows replication forks to progress through certain types of DNA lesions.
The activation of the ATR pathway is regulated by specific DNA structures and DNA damage sensor proteins at stressed DNA replication forks 2. Several components of normal replication forks are known to play important roles in mediating the signaling from ATR to its effector kinase Chk1. Like the signaling of ATR pathway, TLS is also induced by DNA replication stress. PCNA, a key structural and functional component of replication forks, is ubiquitinated in response to replication interference. The mono-ubiquitination of PCNA facilitates its association with several translesion DNA polymerases and may have additional functions 4–6. Recent studies by others and us have revealed that ATR activation and PCNA ubiquitination are two independent processes but, surprisingly, are regulated by a common group of proteins at stressed DNA replication forks 7. These studies have shed new lights on how multiple protective mechanisms are orchestrated by specific proteins and protein-DNA structures at stressed replication forks.
PCNA ubiquitination in human cells
In response to DNA damage, PCNA is either mono- or poly-ubiquitinated in yeast. In human cells, PCNA is predominantly mono-ubiquitinated after treatment with ultraviolet light (UV) or hydroxyurea (HU), an inhibitor of DNA replication. The mono-ubiquitination of PCNA is primarily mediated by the E2-E3 complex composed of Rad6 and Rad184, 5. In cells, PCNA is ubiquitinated on chromatin after DNA damage. In an in vitro assay using purified proteins, PCNA is ubiquitinated by Rad6-Rad18 only when it is loaded onto DNA by RFC 8. These results suggest that the loading of PCNA onto DNA is a prerequisite for its mono-ubiquitination. In response to UV damage, Rad18 and PCNA colocalize in nuclear foci 5, suggesting that the accumulation of Rad18 at stressed replication forks may be an important step for the induction of PCNA mono-ubiquitination.
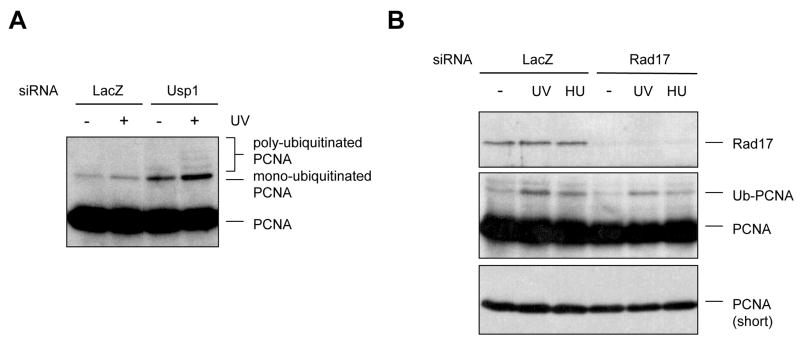
The levels of PCNA mono-ubiquitination are also regulated by the de-ubiquitinase Usp19. Depletion of Usp1 in human cells led to accumulation of mono-ubiquitinated PCNA even in the absence of DNA damage (Fig. 1A), suggesting that PCNA mono-ubiquitination may occur during the normal cell cycle, and that the steady-state levels of PCNA mono-ubiquitination are determined by the balance between Rad18 and Usp1. Mono-ubiquitinated PCNA has been detected in Xenopus extracts during unperturbed DNA replication 10. Depletion of Usp1 also elevated the levels of ubiquitinated PCNA after DNA damage (Fig. 1A), suggesting that Usp1 counteracts Rad18 at stressed forks. Moreover, poly-ubiquitinated PCNA became readily detectable when Usp1 was removed (Fig. 1A), suggesting that Usp1 limits the poly-ubiquitination of PCNA in human cells. It has been shown that Usp1 is degraded after UV damage through autocleavage 9, providing another possible mechanism for the damage induction of PCNA ubiquitination.
Figure 1.
Regulation of PCNA ubiquitination by Usp1 and Rad17. (A) Depletion of Usp1 leads to elevated levels of ubiquitinated PCNA in undamaged and damaged cells. HeLa cells transfected with Usp1 siRNA or control siRNA were treated with UV or left untreated. The unmodified and ubiquitinated PCNA was detected with an anti-PCNA antibody. (B) Depletion of Rad17 leads to reduced levels of ubiquitinated PCNA in UV- and HU-treated cells. Cells transfected with Rad17 siRNA or control siRNA were treated with UV and HU as indicated, and were analyzed by immunoblotting as above.
Independent ATR activation and PCNA ubiquitination
The potential link between the ATR checkpoint and PCNA ubiquitination was first assessed in yeast. In fission yeast, neither Rad3 nor Tel1, the respective homologous of ATR and ATM (a kinase with overlapping functions with ATR in DNA damage signaling), is required for damage-induced PCNA ubiquitination 11. Furthermore, elimination of the ubiquitination site of PCNA did not affect the phosphorylation of a Rad3 substrate, suggesting that PCNA ubiquitination is not needed for Rad3 activation 11. In Xenopus extracts, inhibition or depletion of the ATR-ATRIP complex did not affect PCNA ubiquitination 12. Consistent with these findings, we found that neither ATR nor ATM is required for UV- or HU-induced PCNA ubiquitination 2. Together, these results strongly suggest that ATR activation and PCNA ubiquitination are two independent processes from yeast to humans.
Parallels between the ATR pathway and PCNA ubiquitination
1. RPA and ssDNA
While ATR signaling and PCNA ubiquitination are independent of each other, they are induced by a similar spectrum of DNA damage and replication stress, suggesting that they may share common mechanisms for activation. In response to UV-induced DNA damage or replication inhibitors such as HU and aphidicolin, DNA synthesis on the leading and/or lagging strands of replication forks is interrupted, and the coordination between DNA helicase and DNA polymerases at replication forks is compromised 13. These changes at stressed replication forks lead to accumulation of increased amounts of single-stranded DNA (ssDNA) at or behind the forks 14, 15. The ssDNA induced by replication stress is rapidly coated by RPA, resulting in the RPA-ssDNA complex. RPA-ssDNA is directly recognized by ATRIP, the regulatory partner of ATR, thereby providing a landing pad for the ATR-ATRIP complex at stressed forks or ssDNA gaps 16, 17. Like ATR activation, PCNA ubiquitination is also induced by uncoupling DNA helicase and polymerase in Xenopus extracts 12. Rad18 is capable of binding ssDNA under mild conditions in vitro through its SAP domain 18. However, depletion of RPA from human cells compromises PCNA ubiquitination 6, 19, suggesting that RPA contributes to Rad18 recruitment in vivo. Remarkably, RPA directly associates with Rad18 and stimulates its binding to ssDNA, providing a key mechanism by which Rad18 is recruited to stressed forks 20. Thus, ATR and Rad18, the two central players for the ATR signaling pathway and the ubiquitination of PCNA, respectively, are both recruited by the same protein-DNA structure at stressed forks, RPA-ssDNA.
2. Rad17
During the process of ATR activation, RPA-ssDNA not only recruits the ATR-ATRIP kinase complex, but also its key regulators and substrates. One of the regulators of ATR is the RFC-like Rad17 complex 21, 22. Once recruited to stressed forks by RPA-ssDNA, the Rad17 complex recognizes the junctions between single- and double-stranded DNA and loads the PCNA-like 9–1–1 (Rad9-Rad1-Hus1) complex onto DNA. The 9–1–1 complex interacts with TopBP1, a protein that directly stimulates the ATR-ATRIP kinase 23, providing a means for ATR activation at stressed forks. We found that the levels of ubiquitinated PCNA in cells treated Rad17 siRNA were reduced compared to those in control cells, suggesting that Rad17 and perhaps 9–1–1 contribute to PCNA ubiquitination (Fig. 1B). Interestingly, it was recently shown that the yeast homologue of human Rad9, like PCNA, is ubiquitinated by Rad6-Rad1824. These findings suggest that the Rad17 and 9–1–1 complexes may function in parallel with the RFC and PCNA complexes to recruit Rad6-Rad18 to stressed forks. Given that the loading of 9–1–1 by Rad17 is greatly stimulated by DNA damage, one interesting possibility is that 9–1–1 plays a role in recruiting Rad18 or delivering Rad18 to PCNA. Further studies are needed to assess this model.
3. Chk1 and Claspin
Chk1 is a key effector kinase of ATR that is required for stabilizing stressed replication forks. The phosphorylation of Chk1 by ATR not only requires the recruitment and activation of the ATR kinase, but also a number of replication proteins including Claspin, Timeless, and Tipin. Using siRNA to deplete Chk1 from human cells, we found that Chk1 is required for efficient PCNA ubiquitination 7. Surprisingly, the compromised PCNA ubiquitination in Chk1-depleted cells was partially rescued by a kinase-defective Chk1 mutant, suggesting that the kinase activity of Chk1 is not essential for its function in PCNA ubiquitination. This novel function of Chk1 is at least in part attributed to its role in stabilizing the replication protein Claspin. Depletion of Claspin also leads to compromised PCNA ubiquitination in human cells, whereas overexpression of Claspin partially rescues PCNA ubiquitination in Chk1-depleted cells. Depletion of Timeless, a Claspin-associated protein, also results in reduced PCNA ubiquitination.
How do Chk1, Claspin, and Timeless contribute to PCNA ubiquitination? We found that depletion of Claspin from human cells significantly reduced the amounts of Rad18 on chromatin 7. In addition, we showed that both Claspin and Timeless associated with PCNA in human cells. The stability of Chk1 and Claspin relies on each other even in the absence of DNA damage 7, suggesting that they exist in a complex. It was recently reported that Chk1 interacts with PCNA through a PCNA-interacting-protein (PIP) box, and that the Chk1-PCNA interaction is important for both DNA replication and checkpoint response 25. These findings suggest that Chk1, Claspin, and Timeless may associate with PCNA as a complex and facilitate the recruitment of Rad18 to PCNA. Furthermore, Tipin, a protein stably associated with Timeless, has been shown to bind RPA 26, 27. This Tipin-RPA-ssDNA interaction may allow the complex above to contribute to the recruitment of Rad18 to RPA-ssDNA.
The functions of Chk1, Claspin, and Timesless in PCNA ubiquitination could also be explained by their roles in configuring replication forks. Mrc1 and Tof1, the budding yeast homologues of Claspin and Timeless, respectively, are important for maintaining stressed replication forks at the sites of DNA synthesis 28. In the absence of Mrc1 or Tof1, replication forks disengage from nascent DNA strands when stressed by HU. If such an event occurs at sites of DNA lesion, the separation of replication proteins from the termini of impeded DNA strands would present a problem for TLS, and may affect the positioning of PCNA or the recruitment of Rad18. Mrc1 is also known to interact with the MCM helicase, Cdc45, and DNA polymerase ε (Pol ε), the polymerase functions on the leading strand of replication forks 29. The lack of Mrc1 may compromise the coordinated movement of helicase and polymerase, or the coordination between the proteins on leading and lagging strands. These structural changes in replication forks may in turn affect the interaction between PCNA and its ubiquitination ligase. These possibilities remain to be assessed by further studies.
A multi-functional module at replication forks
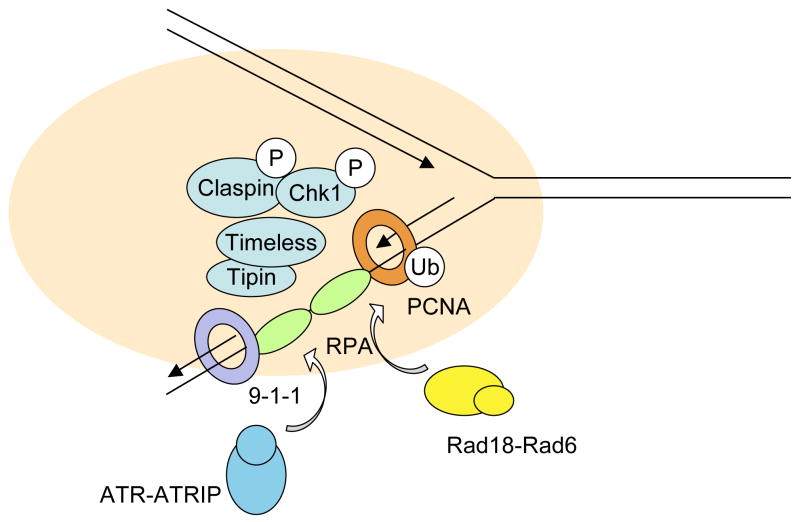
The involvement of RPA, Rad17, Claspin, Timeless, and Chk1 in PCNA ubiquitination has revealed that these proteins play multiple roles in different stress responses at replication forks. Among these proteins, RPA and Rad17 recognize specific DNA structures induced at stressed replication forks. Both RPA-ssDNA and the DNA-bound 9–1–1 complexes loaded by Rad17 are important protein-DNA structures upon which the checkpoint-signaling complex is assembled 2. Claspin, Timeless, and Chk1 function downstream of RPA and Rad17 in checkpoint signaling, suggesting that they may be part of the signaling complex organized by RPA-ssDNA, Rad17, and 9–1–130. The interactions of Claspin, Timeless, and Chk1 with PCNA and their functions in PCNA ubiquitination suggest that PCNA may also contribute to the organization of this complex (Fig. 2). Furthermore, the interaction between Chk1 and PCNA, the role of Chk1 in stabilizing Claspin, and the involvement of Chk1 in fork progression in unperturbed cells have linked Chk1 to normal DNA replication 7, 25, 31, 32.
Figure 2.
A model for a multi-functional module at DNA replication forks. Chk1, Claspin, Timeless, and Tipin may constitute a functional module at replication forks. Through its interactions with RPA-ssDNA, 9–1–1, and PCNA, this module may play dual roles in ATR checkpoint signaling and PCNA ubiquitination.
Yeast genetic studies have strongly suggested that Mrc1, Tof1, and Csm3 (the yeast homologue of Tipin) constitute a multi-functional module at replication forks 33. These proteins are not only involved in checkpoint signaling, but also in replication fork progression, stable replication pausing, and sister chromatid cohesion 28, 34–38. While these proteins clearly interact with each other, some functional differences between Mrc1 and Tof1 have been reported 37, 39, 40, suggesting that this module is not a static complex. Like their yeast homologues, human Claspin, Timeless, and Tipin have also been implicated in checkpoint signaling and fork progression 26, 41, 42. Our finding that Claspin and Timeless contribute to PCNA ubiquitination independently of ATR suggests that similar multi-functional modules may exist in both yeast and human cells. In human cells, the stability of Clapsin and Chk1 is dependent on each other, thus linking Chk1 to this module 7. Likewise, Timeless and Tipin rely on each other to be stable 7, 42, 43. These findings suggest that the human module may be consisted of two complexes: Claspin-Chk1 and Timeless-Tipin. The presence of this pre-assembled module at replication forks may have important functions during normal replication, and enable the forks to response swiftly to different stresses (Fig. 2). Further analyses of normal and stressed DNA replication forks both in vivo and in vitro are needed to test this model.
Acknowledgments
The Zou laboratory is supported by grants from the NIH (GM076388), Susan G. Komen for the Cure, and the Ellison Medical Foundation.
References
- 1.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat Rev Mol Cell Biol. 2008;9:616–27. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zou L. Single- and double-stranded DNA: building a trigger of ATR-mediated DNA damage response. Genes Dev. 2007;21:879–85. doi: 10.1101/gad.1550307. [DOI] [PubMed] [Google Scholar]
- 3.Lehmann AR. Translesion synthesis in mammalian cells. Exp Cell Res. 2006;312:2673–6. doi: 10.1016/j.yexcr.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Kannouche PL, Wing J, Lehmann AR. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol Cell. 2004;14:491–500. doi: 10.1016/s1097-2765(04)00259-x. [DOI] [PubMed] [Google Scholar]
- 5.Watanabe K, et al. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. Embo J. 2004;23:3886–96. doi: 10.1038/sj.emboj.7600383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bi X, et al. Rad18 regulates DNA polymerase kappa and is required for recovery from S-phase checkpoint-mediated arrest. Mol Cell Biol. 2006;26:3527–40. doi: 10.1128/MCB.26.9.3527-3540.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang XH, Shiotani B, Classon M, Zou L. Chk1 and Claspin potentiate PCNA ubiquitination. Genes Dev. 2008;22:1147–52. doi: 10.1101/gad.1632808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garg P, Burgers PM. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc Natl Acad Sci U S A. 2005;102:18361–6. doi: 10.1073/pnas.0505949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang TT, et al. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat Cell Biol. 2006;8:339–47. doi: 10.1038/ncb1378. [DOI] [PubMed] [Google Scholar]
- 10.Leach CA, Michael WM. Ubiquitin/SUMO modification of PCNA promotes replication fork progression in Xenopus laevis egg extracts. J Cell Biol. 2005;171:947–54. doi: 10.1083/jcb.200508100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frampton J, et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol Biol Cell. 2006;17:2976–85. doi: 10.1091/mbc.E05-11-1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang DJ, Lupardus PJ, Cimprich KA. Monoubiquitination of proliferating cell nuclear antigen induced by stalled replication requires uncoupling of DNA polymerase and mini-chromosome maintenance helicase activities. J Biol Chem. 2006;281:32081–8. doi: 10.1074/jbc.M606799200. [DOI] [PubMed] [Google Scholar]
- 13.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–52. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sogo JM, Lopes M, Foiani M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science. 2002;297:599–602. doi: 10.1126/science.1074023. [DOI] [PubMed] [Google Scholar]
- 15.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 16.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–8. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 17.Namiki Y, Zou L. ATRIP associates with replication protein A-coated ssDNA through multiple interactions. Proc Natl Acad Sci U S A. 2006;103:580–5. doi: 10.1073/pnas.0510223103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tsuji Y, et al. Recognition of forked and single-stranded DNA structures by human RAD18 complexed with RAD6B protein triggers its recruitment to stalled replication forks. Genes Cells. 2008;13:343–54. doi: 10.1111/j.1365-2443.2008.01176.x. [DOI] [PubMed] [Google Scholar]
- 19.Niimi A, et al. Regulation of proliferating cell nuclear antigen ubiquitination in mammalian cells. Proc Natl Acad Sci U S A. 2008;105:16125–30. doi: 10.1073/pnas.0802727105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davies AA, Huttner D, Daigaku Y, Chen S, Ulrich HD. Activation of ubiquitin-dependent DNA damage bypass is mediated by replication protein a. Mol Cell. 2008;29:625–36. doi: 10.1016/j.molcel.2007.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zou L, Liu D, Elledge SJ. Replication protein A-mediated recruitment and activation of Rad17 complexes. Proc Natl Acad Sci U S A. 2003;100:13827–32. doi: 10.1073/pnas.2336100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zou L, Cortez D, Elledge SJ. Regulation of ATR substrate selection by Rad17-dependent loading of Rad9 complexes onto chromatin. Genes Dev. 2002;16:198–208. doi: 10.1101/gad.950302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kumagai A, Lee J, Yoo HY, Dunphy WG. TopBP1 activates the ATR-ATRIP complex. Cell. 2006;124:943–55. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 24.Fu Y, et al. Rad6-Rad18 mediates a eukaryotic SOS response by ubiquitinating the 9–1–1 checkpoint clamp. Cell. 2008;133:601–11. doi: 10.1016/j.cell.2008.02.050. [DOI] [PubMed] [Google Scholar]
- 25.Scorah J, et al. A conserved proliferating cell nuclear antigen-interacting protein sequence in Chk1 is required for checkpoint function. J Biol Chem. 2008;283:17250–9. doi: 10.1074/jbc.M800369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gotter AL, Suppa C, Emanuel BS. Mammalian TIMELESS and Tipin are evolutionarily conserved replication fork-associated factors. J Mol Biol. 2007;366:36–52. doi: 10.1016/j.jmb.2006.10.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Unsal-Kacmaz K, et al. The human Tim/Tipin complex coordinates an Intra-S checkpoint response to UV that slows replication fork displacement. Mol Cell Biol. 2007;27:3131–42. doi: 10.1128/MCB.02190-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–83. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 29.Lou H, et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Mol Cell. 2008;32:106–17. doi: 10.1016/j.molcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang X, et al. Rad17 phosphorylation is required for claspin recruitment and Chk1 activation in response to replication stress. Mol Cell. 2006;23:331–41. doi: 10.1016/j.molcel.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 31.Chini CC, Wood J, Chen J. Chk1 is required to maintain claspin stability. Oncogene. 2006;25:4165–71. doi: 10.1038/sj.onc.1209447. [DOI] [PubMed] [Google Scholar]
- 32.Petermann E, et al. Chk1 requirement for high global rates of replication fork progression during normal vertebrate S phase. Mol Cell Biol. 2006;26:3319–26. doi: 10.1128/MCB.26.8.3319-3326.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tong AH, et al. Global mapping of the yeast genetic interaction network. Science. 2004;303:808–13. doi: 10.1126/science.1091317. [DOI] [PubMed] [Google Scholar]
- 34.Osborn AJ, Elledge SJ. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 2003;17:1755–67. doi: 10.1101/gad.1098303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xu H, Boone C, Klein HL. Mrc1 is required for sister chromatid cohesion to aid in recombination repair of spontaneous damage. Mol Cell Biol. 2004;24:7082–90. doi: 10.1128/MCB.24.16.7082-7090.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tourriere H, Versini G, Cordon-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 37.Calzada A, Hodgson B, Kanemaki M, Bueno A, Labib K. Molecular anatomy and regulation of a stable replisome at a paused eukaryotic DNA replication fork. Genes Dev. 2005;19:1905–19. doi: 10.1101/gad.337205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Szyjka SJ, Viggiani CJ, Aparicio OM. Mrc1 is required for normal progression of replication forks throughout chromatin in S. cerevisiae. Mol Cell. 2005;19:691–7. doi: 10.1016/j.molcel.2005.06.037. [DOI] [PubMed] [Google Scholar]
- 39.Hodgson B, Calzada A, Labib K. Mrc1 and Tof1 regulate DNA replication forks in different ways during normal S phase. Mol Biol Cell. 2007;18:3894–902. doi: 10.1091/mbc.E07-05-0500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mohanty BK, Bairwa NK, Bastia D. The Tof1p-Csm3p protein complex counteracts the Rrm3p helicase to control replication termination of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2006;103:897–902. doi: 10.1073/pnas.0506540103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petermann E, Helleday T, Caldecott KW. Claspin promotes normal replication fork rates in human cells. Mol Biol Cell. 2008;19:2373–8. doi: 10.1091/mbc.E07-10-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yoshizawa-Sugata N, Masai H. Human Tim/Timeless-interacting protein, Tipin, is required for efficient progression of S phase and DNA replication checkpoint. J Biol Chem. 2007;282:2729–40. doi: 10.1074/jbc.M605596200. [DOI] [PubMed] [Google Scholar]
- 43.Chou DM, Elledge SJ. Tipin and Timeless form a mutually protective complex required for genotoxic stress resistance and checkpoint function. Proc Natl Acad Sci U S A. 2006;103:18143–7. doi: 10.1073/pnas.0609251103. [DOI] [PMC free article] [PubMed] [Google Scholar]