Abstract
The IG20 gene undergoes alternative splicing resulting in the differential expression of six putative splice variants. Four of these (IG20pa, MADD, IG20-SV2 and DENN-SV) are expressed in virtually all human tissues. However, investigations examining alternative splicing of the IG20 gene to date have been largely limited to non-neural malignant and non-malignant cells. In this study, we investigated the expression of alternative splice isoforms of the IG20 gene in human neuroblastoma (NB) cells. We found that six IG20 splice variants (IG20-SVs) were expressed in two human NB cell lines (SK-N-SH and SH-SY5Y), highlighted by the expression of two unique splice isoforms, namely KIAA0358 and IG20-SV4. Similarly, we found enriched expression of these two IG20-SVs in human neural tissues derived from cerebral cortex, hippocampus, and, to a lesser extent, spinal cord. Utilizing gain of function studies and siRNA technology, we determined that these “neural-enriched isoforms” exerted significant and contrasting effects on vulnerability to apoptosis in NB cells. Specifically, expression of KIAA0358 exerted a potent anti-apoptotic effect in both the SK-N-SH and SH-SY5Y NB cell lines, while expression of IG20-SV4 had pro-apoptotic effects directly related to the activation of caspase-8 in these cells, which have minimal or absent constitutive caspase-8 expression. These data indicate that the pattern of expression of these neural-enriched IG20-SVs regulates the expression and activation of caspase-8 in certain NB cells, and that manipulation of IG20-SV expression pattern may represent a potent therapeutic strategy in the therapy of neuroblastoma, and perhaps other cancers.
Keywords: Neuroblastoma, IG20, KIAA0358, IG20-SV4, caspase-8, apoptosis
Introduction
Neuroblastoma (NB) is one of the most frequently occurring solid tumors in children, particularly in the first year of life, when it accounts for 50% of all tumors (1). Although improvement in outcome has been observed in small, well-defined subsets of patients over the past several years, the outcome for patients with a high-risk clinical phenotype has not improved, with long-term survival less than 40%(2). A characteristic feature of NB is its remarkable clinical and biological heterogeneity(3). While advanced stage NB in older children typically responds poorly to aggressive chemotherapy regimens, certain tumors in patients below one year of age may spontaneously regress or differentiate into benign ganglioneuromas (2, 3). This spontaneous regression likely represents the activation of an apoptotic and/or differentiation pathway, and the prognosis in NB patients may be related to the level of expression of molecules involved in the regulation of apoptosis.
The IG20 (insulinoma-glucagonoma) gene has been implicated in cancer cell survival and apoptosis(4–10), neurotransmission (11, 12) and neurodegeneration(13). We have previously shown that various splice isoforms of the IG20 gene (IG20-SVs), including IG20pa, MADD/DENN, and DENN-SV, act as negative or positive regulators of apoptosis, and their levels of expression can profoundly affect cell survival in non-neural cells (6–10). IG20-SVs are believed to act, in part, by modulating inflammatory and apoptotic signaling pathways, effects mediated through interactions with tumor necrosis factor receptor 1 (TNFR1). TNFα interacts with TNFR1 to trigger pro-inflammatory actions through various stress-activated protein kinases (SAPKs), such as c-Jun N-terminal kinase (JNK) and p38 mitogen-activated protein kinase (p38 MAPK) (14, 15). IG20 interacts strongly with TNFR1, and all putative IG20-SVs contain the death domain homology region (DDHR) required for this binding. We have shown that expression of MADD/DENN is required and sufficient for cancer cell survival in non-neuronal cancer cells, and mediates its effects by acting as a negative regulator of caspase-8 activation (16). The over-expression of IG20pa, on the other hand, results in enhanced apoptosis and activation of caspase-8 through enhanced DISC formation (9). The caspase-8 (CASP8) gene encodes a key enzyme at the top of the apoptotic cascade. In NB cell lines and tumor samples, CgG methylation of CASP8 at the 5′ end has been associated with inactivation of the gene(17), and recent hypotheses have proposed that CASP8 may act as an NB tumor-suppressor gene (17–20). Furthermore, NB cell lines that do not express caspase-8 are resistant to TRAIL-induced apoptosis(19), and suppression of caspase-8 expression has been shown to occur during establishment of NB metastases in vivo (21).
In this study, we demonstrate the preferential expression of two unique splice isoforms (KIAA0358, IG20-SV4) of the IG20 gene in selected nervous system tissue and in two NB cell lines known to be deficient in the expression of caspase-8. Through gain-of-function studies and using siRNA technology, we show that the expression of IG20-SV4 enhances cellular apoptosis and leads to the expression and activation of caspase-8 in SK-N-SH and SH-SY5Y NB cells, thereby sensitizing these cells to the pro-apoptotic effects of TNFα. In contrast, expression of KIAA0358 effectively renders cells resistant to apoptosis, even when IG20-SV4 is co-expressed, and down-modulation of this isoform causes markedly enhanced apoptotic cell death and activation of caspase-8.
Materials and methods
Cell culture
SK-N-SH, SH-SY5Y, and SK-N-BE(2)-C human neuroblastoma cell lines were purchased from ATCC and cultured according their instructions. Briefly, SK-N-SH cells were cultured in Dulbecco’s modified Eagle’s medium (Invitrogen, CA, USA) supplemented with 10% fetal bovine serum, 0.1 mM non-essential amino acids, 1.5g/L sodium bicarbonate, 1.0 mM sodium pyruvate, and 100 units of penicillin/ml, and 100 μg of streptomycin/ml. SH-SY5Y and BE(2)-C cells were cultured in a 1:1 mixture of Eagle’s minimum essential medium with non-essential amino acids and Ham’s F12 medium (Invitrogen, CA, USA) supplemented with 10% fetal bovine serum and 100 units of penicillin and 100 μg of streptomycin/ml. The cell lines were maintained at 37 °C in a humidified chamber with 5% CO2.
Design of small inhibitory RNAs
The siRNAs utilized in this study are shown in Figure 2A and Supplementary Figure 5. The siRNAs targeting exons13L, 16E, and 15 (“Mid”) and the SCR (negative control) are identical to those previously described (10). The siRNA targeting exon34 was designed using OligoEngine Workstation 2 and purchased from OligoEngine, Inc. (Seattle, WA). These siRNAs were screened in SK-N-SH cells and the most efficient were used to construct the 34E-shRNA lentivirus.
Figure 2. IG20-SVs and down modulation effect of exon-specific siRNAs directed against specific isoforms on endogenous IG20-SVs in SK-N-SH cells.
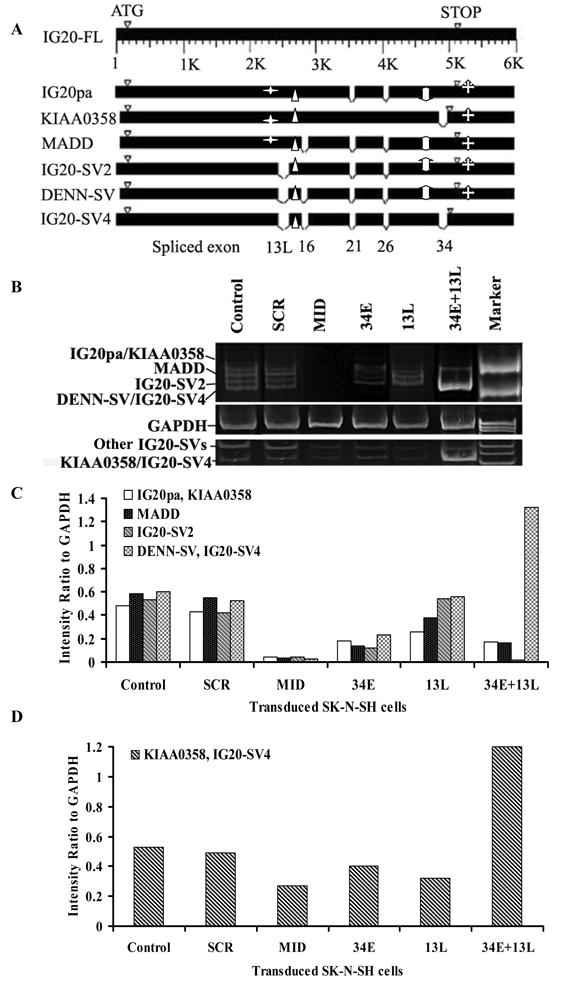
A. Shows human IG20-SVs generated by alternative mRNA splicing. Solid bars represent regions of complete cDNA sequence homology between variants. Empty areas indicate spliced exons 13L, 16, 21, 26 and 34, which when spliced in different combinations can give rise to the six IG20-SVs. B. Effect of down modulation of endogenous IG20-SVs by exon-specific siRNAs in SK-N-SH cells. One microgram total RNA obtained from GFP-positive SK-N-SH cells obtained by fluorescence-activated cell sorting (FACS) at 5 days post-transduction was used for reverse transcription–polymerase chain reaction. The products were separated on a 5% PAGE. Amplification of IG20-SVs using F1-B2 primers (upper panel) and F4824-B5092 (lower panel) is shown. C. Quantification of relative intensities of bands from panel B (upper panel) using ImageJ. D. Quantification of relative intensities of bands from panel B (lower panel) using ImageJ.
Plasmid construction
The siRNAs were cloned into the pSUPER vector using BgI II and HindIII sites (22) to generate pSup-34 plasmids. The shRNA cassettes (including the H1 RNA promoter and the shRNA) were excised from pSup-34 using XbaI and ClaI sites and ligated into the pNL-SIN-CMV-GFP vector to generate 34E lentivirus constructs. The pcTat, pcRev and pHIT/G were gifts from Dr. B.R. Cullen and Dr. T.J. Hope. The YFP-IG20pa plasmid (16) was used as a backbone to subclone YFP-KIAA0358 from the corresponding pBKRSV plasmid (6) using the BstZ 171 and BsiWI sites. The YFP-KIAA0358 and YFPIG20-SV4 mid-sh-RNA resistant mutant constructs were generated using the Quickchange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) according to the manufacturer’s protocol. Briefly, the primers 5′-CGGAACCACAGTACAAGCTTTAGCCTCTCAAACCTCA CACTGCC-3′ (forward) and 5′-GGCAGTGTGAGGTTTGAGAGGCTAAAGCTTGTACTGTGGTT CCG-3′ (reverse) were used to insert four silent mutations (bold and underlined lettering) in the cDNAs without affecting the amino-acid sequence. Hind III restriction sites in the mutants, generated due to base substitutions, were used to identify positive clones that were further confirmed by sequencing. The caspase-8 promoter luciferase vector was constructed by PCR amplification of a 1.2kb fragment from pBLCAT-Casp8 vector, and cloning into promega pGL4.17 luciferase vector at KpnI and XhoI site. The pBLCAT3 vector contain fragment −1161/+16 of caspase-8 promoter was gift from Dr. Silvano Ferrini’s lab(23),
Preparation of Lentivirus stocks
Lentivirus stocks were prepared as described previously (22). Briefly, subconfluent 293FT cells grown in 100mm plates were co-transfected with 10.8 mg of lentivirus vector, 0.6 mg pcRev, 0.6 mg of pcTat and 0.3 mg of pHIT/G using calcium phosphate. Culture medium was replaced after 16 h, and the supernatant was harvested at 40 h and filtered using a 0.45 mm filter. The optimal viral titer for each cell type was determined as the least amount of viral supernatant required to transduce at least 50% of target cells without apparent cytotoxicity.
RNA preparation
Total RNA extracted from human cerebral cortex, hippocampus, cerebellum, and human thyroid, skeletal muscle, lung and liver were purchased from BD Clontech (MountainView, CA, USA). Total RNA extracted from primary NB was a gift from Dr. Jill Lahti’s lab of St. Jude’s Children’s Research Hospital. For testing the efficiency of down-modulation of IG20 splice variants by different siRNAs, the transduced GFP positive SK-N-SH cells were sorted on the MoFlo™ High-Performance Cell Sorter (Dako Denmark, Glostrup, Denmark). Total RNA was extracted from 1×106 GFP-positive NB cells and other described cell lines using Trizol reagent (Invitrogen Life Technologies, Carlsbad, CA, USA).
Reverse transcription–polymerase chain reaction
We used 1 μg of RNA for reverse transcription-polymerase chain reaction (RT-PCR) using the Super-Script III One-Step RT-PCR system (Invitrogen Life Technologies, Carlsbad, CA, USA). Briefly, the cDNAs were synthesized at 50 °C for 30 minutes followed by incubation at 94 °C for 2 minutes. Subsequently, 30 cycles of PCR were carried out with denaturation at 94 °C for 50 seconds, annealing at 55 °C for 50 seconds and extension at 72 °C for variable time periods (as described below); followed by a final incubation at 72°C for 7 min. For amplifying exons 13L and 16, F-1 and B-1 primer pairs (5′-CGG GAC TCT GAC TCC GAA CCT AC-3′ and 5′-GCG GTT CAG CTT GCT CAG GAC-3′, respectively) were used, with 1 minute extension time. For amplifying exon 34, F4824 and B5092 primer pairs (5′ CTG CAG GTG ACC CTG GAA GGG ATC 3′ and 5′ TGT ACC CGG GTC AGC TAG AGA CAG GCC 3′, respectively) were used, with 30 second extension time as described previously(8). The sequence of GAPDH has been previously published(9). The PCR products were then separated on a 5% polyacrylamide gel.
Cell proliferation assay
Cell proliferation assays were performed according to the Vybrant MTT cell proliferation assay kit (V-13154, Molecular Probes, Invitrogen, CA, USA) instructions. Briefly, twenty-four-hour post-transduction, 1×104 sorted GFP-positive SK-N-SH cells were plated onto 96-well plates. Every other day, cells were washed with PBS and labeled with 10 μL of 12mM stock solution MTT in each well, incubated at 37 °C for 4 hours, washed with PBS. 50 μL of DMSO was added to each well and mixed thoroughly with a pipette, and absorbance was recorded at 540 nm.
CFSE dilution assay
Twenty-four hours post-transduction, 1 × 105 SK-N-SH cells were stained with 2 mM SNARF-1carboxylic acid, acetate, succinimidyl ester (S-22801, Molecular Probes, Invitrogen, CA, USA) for 15 minutes at 37 °C. Cells were washed and either used immediately for FACS analysis, or plated into six-well plates. Every other day, cells were collected, washed and CFSE dilution, as an indicator of cell division, was determined in GFP-positive cells by FACS analysis at excitation/emission = 480/640nm.
DiIC staining
SK-N-SH (1.5×105) cells were plated into six-well plates. Twenty-four hours later, cells were treated with different shRNA-expressing lentiviruses for 4 hours, washed and replenished with fresh warm medium immediately, and then every other day. At five days, the transduced cells were trypsinized with 0.05% trypsin, 0.53mM EDTA and suspended in1mL warm PBS. Then, 5μL of 10μM DiIC (Molecular Probes, Invitrogen, Carlsbad, California) was added and the cells were incubated at 37 °C, 5% CO2 for 20 min. Cells were washed once by adding 2mL of warm PBS, and resuspended in 500μL of PBS. DiIC stained cells were analyzed on CyAn™ ADP Flow Cytometer (Dako Denmark, Glostrup, Denmark). Only GFP positive cells were gated and analyzed.
Apoptosis assay
Annexin V-phycoerythrin/7-amino-actinomycin D labeling was done according to the manufacturer’s instructions (BD PharMingen) and samples were analyzed by flow cytometry. NB (1.5×105) cells were plated into six-well plates. Twenty-four hours later, cells were treated with different shRNA-expressing lentiviruses for 4 h, washed and replenished with fresh warm medium immediately, and then every other day. At five days, the transduced cells were trypsinized and washed twice with cold PBS and then resuspended in 1X assay binding buffer. Annexin V-phycoerythrin/7-amino-actinomycin D labeling was performed at room temperature for 15 minutes before analysis by flow cytometry (BD FACScan). Only GFP positive cells were gated and analyzed.
Caspase-8 inhibition
At 3 days post-transduction with different shRNAs, SK-N-SH cells were treated with 40 μM and 80μM of Z-IETD-FMK (BD PharMingen) for an additional two days, or with 10 μg/ml cycoheximide (Sigma) for an additional day. Collected cells were either subjected to Annexin V-PE/7-AAD staining followed by FACS or western blot analysis to determine active caspases.
Western Blot Analysis
Different shRNA-expressing, lentivirus-transduced NB cells were trypsinized and washed with phosphate-buffered saline and lysed at 0 °C for 30 min in a lysis buffer (20 mM Hepes, pH 7.4, 2 mM EGTA, 420mM NaCL, 1% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 10 μ/ml aprotinin, using a 1 mM Na3VO4, and 5 mM NaF). The protein content was determined dye-binding microassay (Bio-Rad), and after boiling the samples for 2 min in a 1X SDS protein sample buffer, 20 μg of protein per lane was loaded and separated on 10% SDS-polyacrylamide gel. The proteins were blotted onto Hybond ECL membranes (Amersham Biosciences). After electroblotting, the membranes were blocked with Tris-buffered saline with Tween-20 (TBST 10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 0.1% Tween-20) containing 5% milk, and were incubated with antibodies diluted in a 5% BSA TBST buffer that can detect cleaved caspase-8 (Santa Cruz, C-20), caspase-9(Cell signaling,), and full length caspase-3 (R & D system, 84803) overnight. The primary antibody dilutions were those recommended by the manufacturer. The membranes were then washed, incubated with the appropriate secondary antibodies (1:5,000) in a blocking buffer for 1 h, and repeatedly washed. Proteins were detected using an enhanced chemiluminescence plus western blotting detection system (Amersham, UK). The anti-GAPDH-HRP (abcam) antibodies were used as loading controls.
Transient transfections and luciferase assays
We seeded 1.5 ×105 SK-N-SH cells per well in 12-well plates and cotransfected them with either 1.6 μg of pEYFP-C1 or pEYFP-IG20-SV4, 1 μg of pGL4.17 (a promoterless control) or 1 μg of pGL4.17-caspase-8 promoter. 20ng of pSV40-Renilla luciferase vector was cotransfected as a normalizing control. Transfections were carried out in triplicate. After 48 h of incubation, cells were collected and analyzed for luciferase activity with the Dual-Luciferase Reporter Assay System (Promega).
Dominant-negative FADD (pcDNA-DN-FADD) or control vector (pcDNA3.1) were transfected (5ug each) into 6×106 SK-N-SH cells, and distributed into 6-well plate. To increase the transfection efficiency of DN-FADD, nucleofection® from Amaxa biosystems was used. . After 24 hours culture, the cells were either transduced with SCR or 34+13L sh-RNA. At 3 days post-transducion, the cells were treated or un-treated with 10ng/ml TNFα for 48 hrs. The cells were trypsinized and stained with Annexin V-PE/7-AAD for FACS analysis. Only GFP positive cells were gated and analyzed.
Statistical analysis
All results are expressed as mean ± SE. Student’s t test was used to determine P values using Microsoft Excel Software (version 2003).
Results
Expression of IG20 splice variants in neuroblastoma cell lines and nervous system tissues
To begin to understand the potential relevance of IG20 alternative splicing in the control of apoptosis in NB cells, we first tested the constitutive expression patterns of IG20-SVs in several NB cell culture lines. We used RNA extracted from the SK-N-SH, SH-SY5Y, and SK-N-BE(2)-C human NB cell lines and performed RT-PCR using multiple sets of IG20-specific primers as described in the Materials and Methods section. Figure 1 shows the expression pattern of IG20-SVs in the tested tissues and cell lines.
Figure 1. Expression of IG20 splice isoforms in human NB cell lines, primary NB tumor lines, and various human tissues.
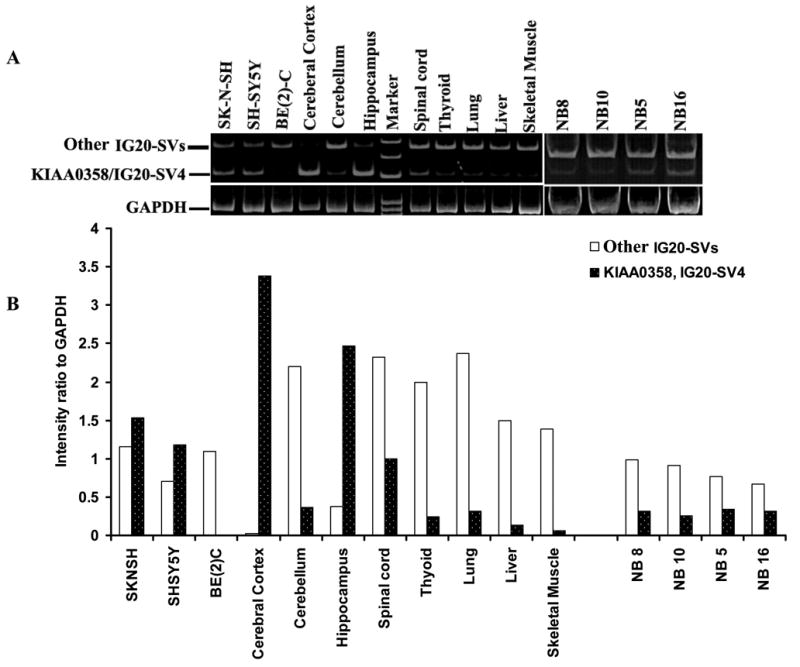
1 μg of total RNA was used for reverse transcription-polymerase chain reaction (RT-PCR) using the Super-Script III OneStep RT-PCR system (Invitrogen Life Technologies, Carlsbad, CA, USA). A. Shows amplification of exon 34 region of IG20-SVs using F4824 and B5092 primers. B. Shows quantification of relative intensities of bands in relation to the housekeeping gene GAPDH from panel A using ImageJ (http://rsb.info.nih.gov/ij/).
Although only one representative sample for each tissue type is shown, we used RNAs from multiple samples of each tissue type to validate the RT-PCR results. In agreement with previous studies(6, 7, 16), we confirmed that different IG20 splice variants are expressed in different patterns and levels in various human tissues. In addition, we found that two isoforms, KIAA0358 and IG20-SV4, which are not significantly expressed in non-neural tissues, are highly expressed in two of the three human NB cell lines (SK-N-SH and SH-SY5Y) tested, and in human cerebral cortex, hippocampus, and, to a lesser extent, spinal cord (Figure 1). In addition, these two isoforms were expressed in both caspase 8-expressing (NB5, NB16) and caspase 8-deficient (NB8, NB10) primary NB tumor lines. The levels of expression of KIAA-0358 and IG20-SV4 did not correlate with constitutive expression of caspase-8 in these cells.
Small inhibitory RNAs effectively down-modulate expression of endogenous IG20-SVs in neuroblastoma cells
To analyze the effects of IG20-SVs on NB cell survival and apoptosis, we designed small inhibitory RNAs (siRNAs) to selectively down-modulate specific IG20-SVs as shown in Figure 2A and Supplementary Figure 5. We had previously identified the most effective siRNAs targeting all isoforms and targeting exons 13L in studies using Hela cells and PA-1 cells (16, 22, 24). We similarly screened several siRNAs targeting exon 34 and chose the most effective for use in this study. We cloned each siRNA in lentiviral vectors to allow for stable expression of the siRNAs that could be detected through GFP expression.
The targeted exons and resulting down-modulated IG20 isoforms for each siRNA used in this study are summarized in Figure 2A and Table 1. We cloned shRNAs into a self-inactivating lentivirus vector (pNL-SIN-GFP) (25) and generated 13L, Mid-, 34E and SCR (negative control shRNA) constructs. Utilizing GFP, this enabled us to monitor expression of double copy cassettes likely resulting in enhanced silencing (25). The transduction efficiency was greater than 50% as determined by GFP expression. For testing the down-modulation efficiency, total RNA from transduced and GFP-positive SK-N-SH cells was used for RT-PCR. The results are shown in Figures 2B – 2D. As expected, SK-N-SH cells expressing Mid-shRNA showed decreased expression levels of all IG20- SVs relative to control (SCR). 13L-shRNA caused down-modulation of IG20pa, MADD, and KIAA0358. 34E-shRNA caused down-modulation of IG20pa, MADD, IG20-SV2, and DENN-SV; and 34E+13L-shRNA caused down-modulation of all of these IG20-SVs with the addition of KIAA0358. Interestingly, when all isoforms except IG20-SV4 were down-modulated, expression of this sole isoform appeared to be increased at five days post-transduction (Figure 2B, C, D).
Down-modulation of KIAA0358 in neuroblastoma cells leads to spontaneous apoptosis, but has no apparent effect on cellular proliferation
Down-modulation of IG20-SVs has no effect on cellular proliferation of NB cells
In order to assess the influence of IG20-SVs on NB cell growth and proliferation, various shRNA-expressing viable cells were counted using a MTT assay and CFSE dilution. Relative to controls, a significant decrease in the numbers of viable cells expressing Mid-, 34E, 13L and 34E + 13L shRNA was observed (Supplemental Figure 1). However, there was no difference in CFSE dilution (SNARF-1 carboxylic acid, acetate, succinimidyl ester) over time amongst the SCR control, Mid-, 34 and 13L and 34+13L -shRNA-treated cells suggesting that the differences in cell numbers were not due to decreased cellular proliferation (Supplemental Figure 1B). Further, shRNA-treated cells failed to show significant differences in cell cycle progression (data not shown). Together, these results indicated that manipulation of the expression patterns of IG20-SVs had little or no effect on cell proliferation or cell cycle progression.
Down-modulation of KIAA0358 induces apoptosis in SK-N-SH NB cells
Since there is no single methods that can conclusively demonstrate cellular apoptosis, we determined spontaneous cell death using both mitochondrial membrane potential DiIC staining (Figure 3A and 3B) and Annexin V-PE/7-AAD staining (Figure 3C) to assure the reliability of our findings. We observed that down-modulation of all IG20-SVs with Mid-shRNA resulted in a significant increase in spontaneous apoptosis. Down-modulation of IG20pa, MADD, KIAA0358 (by targeting exon 13L) and down-modulation of all isoforms with the exception of IG20-SV4 (by targeting exons 13L and 34E) also resulted in significantly increased spontaneous apoptosis. These results were consistently observed using both methods of apoptosis determination and after repeating all experiments a minimum of three times. This suggested to us that certain IG20-SVs may act as pro-survival factors since their knock down resulted in spontaneous apoptosis. The most likely candidates for this pro-survival function were MADD/DENN and KIAA0358 based on the pro-apoptotic results of down-modulation of these two IG20-SVs. Interestingly, the selective expression of KIAA0358 and IG20-SV4 in the absence of other isoforms (targeting exon 34) resulted in markedly reduced apoptotosis. This finding strongly indicated that expression of KIAA0358 had a pronounced anti-apoptotic effect, since expression of IG20-SV4 alone (in the absence of all other isoforms including KIAA0358) resulted in very high levels of spontaneous apoptosis (Figure 3A – 3C), which were suppressed by DN-FADD overexpression (Figure 4B).
Figure 3.
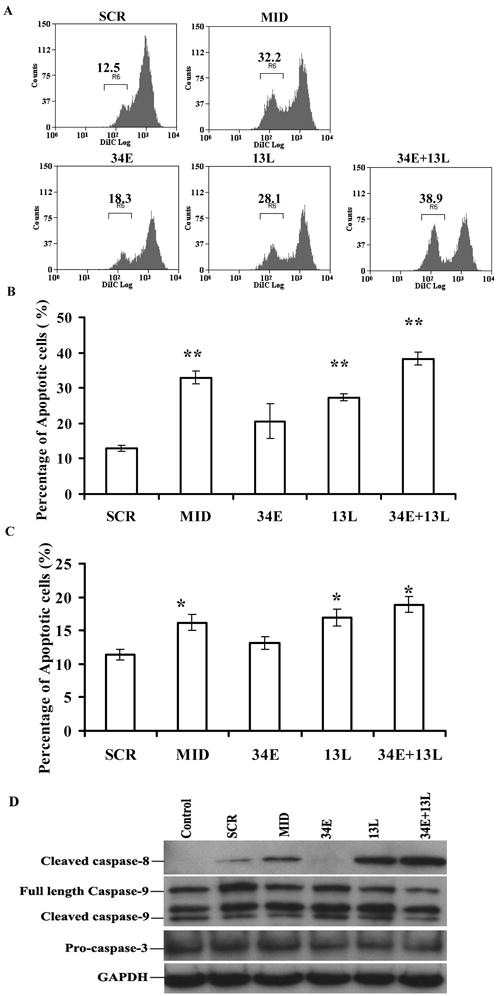
Apoptotic effects and caspase-8 activity with down modulation of IG20-SVs in SK-N-SH cells. A. Representative data showing mitochondrial depolarization as determined by DilC staining. Five days post-transduction, SK-N-SH cells were collected and one-third cells of the collected cells were stained with 50 nM of DiIC, Loss of staining (as a marker of mitochondrial depolarization) was detected by FACS analysis. Percentage of apoptotic cells are indicated on the histograms. B. Summary of the results showing percentages of cells with increased mitochondrial depolarization as measured by DiIC staining from three independent experiments. The P-value was **P<0.01, for test groups vs SCR. C. Summary of results showing percentage of cells with increased apoptosis as determined by Annexin V-PE/7-AAD staining. Another one-third of collected cells as described in A were stained with Annexin V-PE/7-AAD and detected by FACS. The P-value was *P <0.05 for test groups vs SCR. The data were gated from GFP-positive cells only. D. Caspase-8 activity in NB cells transduced with siRNAs. The final one-third of cells from A were lysed and subjected to western blot analysis for caspase-8, caspase-9, and caspase-3. The data shown are representative of three separate experiments.
Figure 4. Effects of TNF-α treatment on apoptosis of siRNA-transduced SK-N-SH cells.
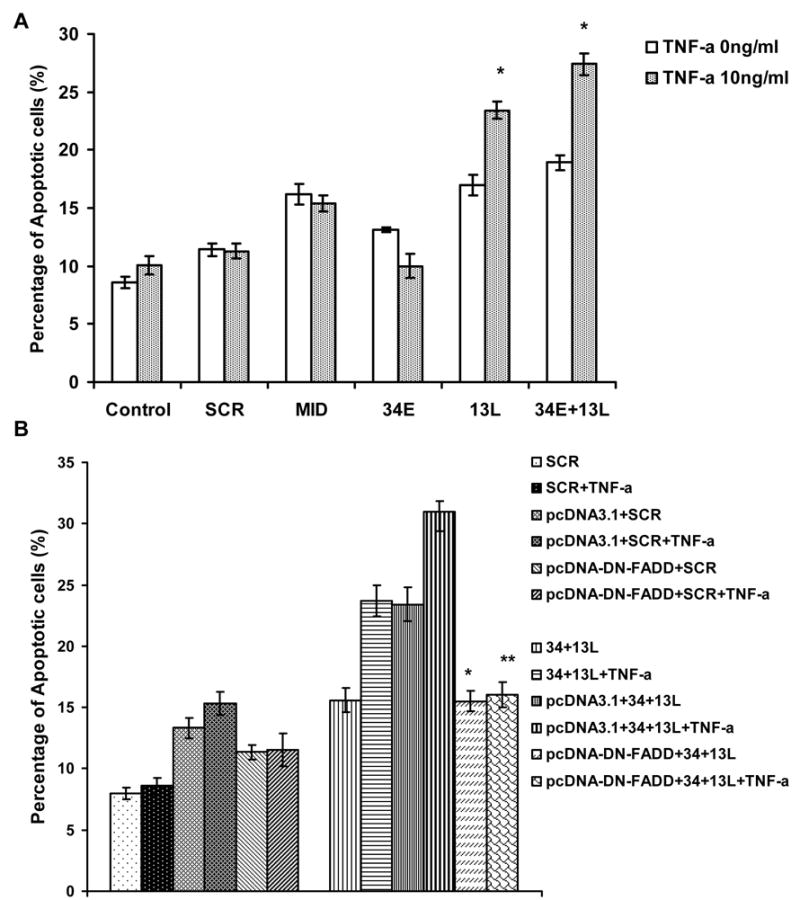
Three days post-transduction, SK-N-SH cells were treated with 10ng/ml TNF-alpha for 2 days, and cells were collected and stained with Annexin V-PE/7-AAD. A. Summarized results showing percentage of cells with increased apoptosis from three independent experiments. The P-value was *P<0.05 for TNF-α treated cells vs untreated cells. B. Summarized results showing percentage of apoptosis in transduced NB cells, cells treated or un-treated with 10ng/ml TNFα after transfection with pcDNA or pcDNA-DN-FADD. Results are from three independent experiments. The P-value was *P<0.05, **P<0.01 for pcDNA-DN-FADD transfected cells vs pcDNA3.1 transfected cells. The data were collected from GFP-positive cells only.
Enhanced apoptosis in SK-N-SH cells depleted of KIAA0358 is due to expression and activation of caspase-8
In order to identify the mechanism of enhanced apoptosis induced by IG20-SV down-modulation, we examined whether specific caspases were activated in transduced SK-N-SH cells. Cells depleted of KIAA0358 (Mid, 13L and 13L + 34E cells) showed enhanced expression of cleaved caspase-8. There was accompanying evidence for processing of caspase 3 (slightly reduced expression of pro-caspase-3), but no change in caspase-9 (Figure 3D).
Manipulation of IG20-SVs in other NB cell lines (SH-SY5Y and SK-N-BE(2)-C)
Similarly, SH-SY5Y cells transduced with 13L and 13L+34E siRNAs showed enhanced apoptosis associated with prominent expression and activation of caspase-8 (Supplemental figure 2). Since we determined that SK-N-BE(2)-C cells did not express KIAA0358 and IG20-SV4, we obviously did not expect that the siRNAs targeting these isoforms would be relevant in this cell line. Instead, we over-expressed IG20-SV4 and KIAA0358 in SK-N-BE(2)-C cells and examined the effect on caspase-8 activation. Introduction of these isoforms had no effect on expression or activation of caspase-8 (Supplemental Figure 3) which was expressed at very low baseline levels in these cells, consistent with previous reports(26, 27).
Treatment with TNF-α enhances apoptosis in NB cells expressing IG20-SV4 in a FADD-dependent manner, but does not attenuate the anti-apoptotic effect of KIAA0358
As a binding partner for the tumor necrosis factor receptor 1 (TNFR1), the IG20 gene promotes both pro-apoptotic and anti-apoptotic signals in Hela cells (6,7). Therefore, we tested the apoptotic effect of TNF-α on SK-N-SH cells. Treatment with TNFα enhanced apoptosis in cells transduced with shRNAs targeting the 13L exon and the combination of exons 13L and 34E (Figure 4A). This induced sensitization to TNFα was significantly suppressed by DN-FADD over-expression (Figure 4B). However, cells transduced with shRNA targeting exon 34 that did not alter endogenous expression of KIAA0358 and IG20-SV4, continued to be resistant to apoptosis even after TNF-α treatment (Figure 4A).
Over-expression of KIAA0358 can rescue SK-N-SH cells from spontaneous apoptosis induced by down-modulation of all IG20-SVs by dampening caspase-8 activation
We created silent mutations in cDNAs encoding KIAA0358 at sites corresponding to the 5th, 7th, 11th and 14th nucleotides of the Mid-shRNA target sequence. These mutations neither affected the amino-acid sequence nor protein expression. We generated SK-N-SH cells stably expressing YFP-KIAA0358-Mut. We confirmed that the Mid-shRNA was unable to down-modulate YFP-KIAA0358-Mut, but effectively down-modulated expression of all endogenous IG20-SVs (Figure 5A). Interestingly, expression of this KIAA0358 mutant was sufficient to rescue SK-N-SH cells from spontaneous apoptosis caused by Mid-shRNA transduction (Figure 5B), confirming the anti-apoptotic properties of KIAA0358. These pro-survival effects were associated with nearly complete dampening of caspase-8 activation (Figure 5C).
Figure 5. Expression of KIAA0358 in isolation can prevent apoptosis and suppress caspase-8 activity in SK-N-SH cells.
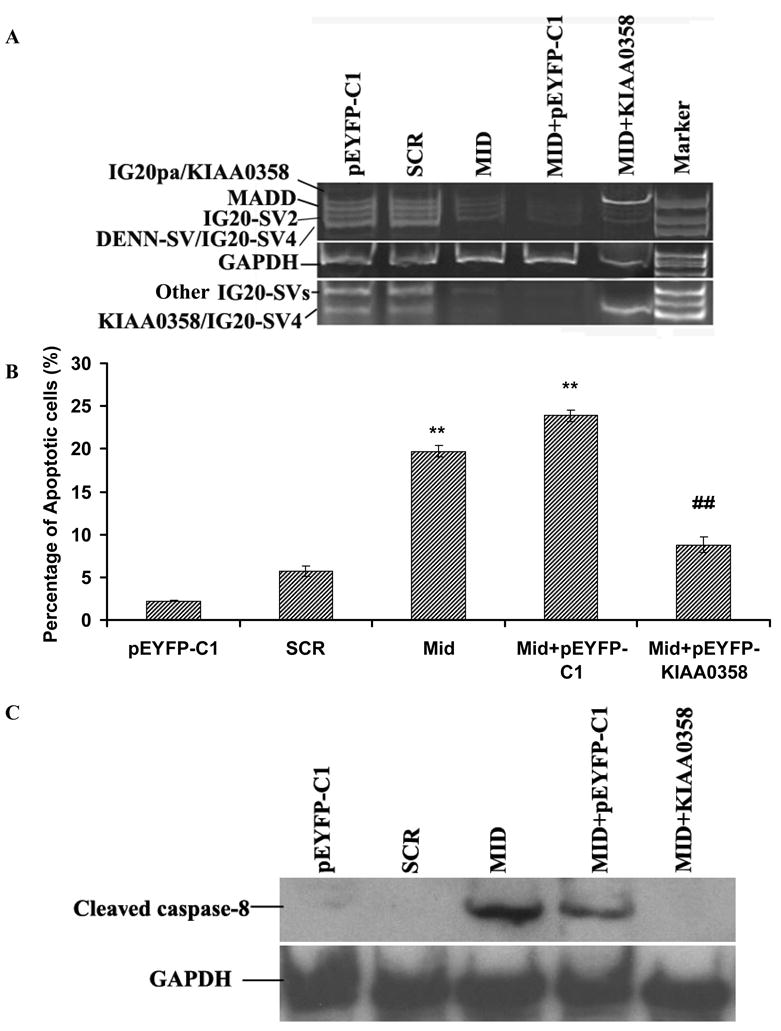
A. RT-PCR of IG20-SVs from stable cells expressing control vector (pEYFP-C1) or YFP-KIAA0358-Mut and infected with Mid-shRNA for five days. B. Mitochondrial depolarization assay. SK-N-SH cells were stained with DiIC to determine spontaneous apoptosis. Data shown are representative of three independent experiments (**P<0.01 vs SCR, ## P<0.01 vs Mid+pEYFP-C1). The data were collected from YFP and GFP double-positive cells only. C. Western blot showing caspase-8 activity. Cell lysates were subjected to western blot analysis of caspase-8. The data shown are representative of three individual experiments.
Down-modulation of KIAA0358 and selective expression of IG20-SV4 modulates expression of caspase-8 in caspase-8-deficient SK-N-SH cells
To determine whether the increased apoptosis induced utilizing 34 + 13L shRNA was due to modulation of the expression of caspase-8, we measured the expression of caspase-8 transcripts in SK-N-SH cells treated with the different combinations of siRNAs. We found that SK-N-SH cells in which all isoforms were down-modulated leaving expression of IG20-SV4 unperturbed (13L + 34E), expressed increased levels of caspase-8 mRNA compared to control cells (Supplemental figure 4). To confirm that the increased expression of caspase-8 was due to induction of gene expression, we exposed the cells to 10 μg/mL cycloheximide as an inhibitor of new protein synthesis. This inhibited the expression of caspase-8 protein (Figure 6A) suggesting that the effects of IG20-SV manipulation were mediated at the level of CASP8 gene expression. This result was further confirmed by using a luciferase assay, in which overexpression of IG20-SV4 caused a significant (4-fold) increase in activation of the CASP8 promoter compared to control or pEFYP-c1 (empty vector) (Figure 6B).
Figure 6. Down modulation of KIAA0358 or selective expression of IG20-SV4 enhances apoptosis through expression/activation of caspase-8 in SK-N-SH cells.
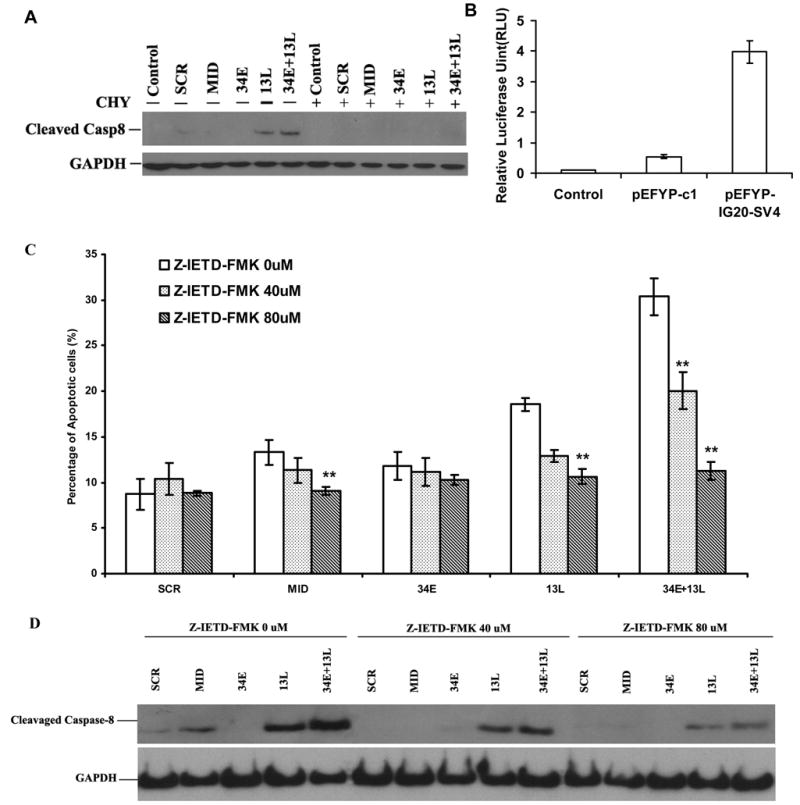
A. Effects of cycloheximide on expression/activation of caspase-8. Three days post-transduction with shRNA-expressing virus, SK-N-SH cells were treated with 10 μg/ml cycloheximde (a protein synthesis inhibitor) for two days. Whole cell lysates were subjected to western blot analysis. B. Caspase-8 reporter assay. SK-N-SH cells were cotransfected with pGL4.17-caspase-8 promoter vector, pSV40-Renilla luciferase vector and pEYFP-C1/or pEYFP-IG20-SV4 using Lipofectamine2000, 48 hrs later, cells were collected and analyzed for luciferase activity with the Dual-Luciferase Reporter Assay System (Promega). C and D. Effects of caspase-8 inhibition. Three days post-transduction with shRNA-expressing virus, SK-N-SH cells were treated with 40 μM and 80μM of Z-IETD-FMK (a caspase 8 inhibitor) for two days. Collected cells were either subjected to Annexin V-PE/7-AAD stain for FACS analysis (C) or western blot analysis (D). C. Percentage apoptosis in cells transduced with different shRNAs in the presence or absence of the caspase inhibitor. The P-value was ** P<0.01 for Z-IETD-FMK treated vs untreated. D. Western blot showing inhibitory effect of Z-IETD-FMK on caspase-8 activity. Representative data are from three independent experiments.
Inhibition of caspase-8 effectively decreases apoptosis in 13L- and (34E+13L)-transduced SK-N-SH cells in dose dependent manner
We then pre-treated the cells with the specific caspase-8 inhibitor, Z-IETD-FMK (40 μM and 80 μM) which significantly attenuated the apoptotic effect caused by down-modulation of KIAA0358 in a dose-dependent fashion (Fig. 6C). The inhibitory effect of Z-IETD-FMK on caspase-8 expression was confirmed by western blot analysis (Fig. 6D). Inhibition of caspase 8 did not significantly affect apoptosis in cells treated with shRNA targeting 34E (Figure 6C).
Discussion
Previously reported cDNAs designated MADD and DENN share the same location on human chromosome 11p11 as IG20 which comprises 36 exons (4,5,14,15). Other proteins with high homology and almost identical death domains have been discovered and include rat Rab3-GEP, a GDP/GTP exchange protein (28, 29), and KIAA0358, a human brain protein of unknown function(30). Since the publishing of these reports, it has become clear that IG20, MADD, DENN and KIAA0358 are different isoforms of the same gene(6) that stem from alternative splicing of exons 13L, 16, 21, 26 and 34. In fact, a total of seven putative IG20-SVs have been identified, namely, IG20pa, MADD, DENN-SV, IG20-SV2, KIAA0358, IG20-SV4, and IG20-FL (6, 7). IG20-SVs can differentially regulate cell proliferation and death in non-neuronal cell lines (4–10).
In the current study, we first observed that IG20-SVs are highly expressed in select nervous system tissues and discovered that KIAA0358 and IG20-SV4, which are not highly expressed in non-neural cells, were significantly expressed in cerebral cortex, hippocampus, and to a lesser extent, spinal cord. We therefore designated IG20-SV4 and KIAA0358 as “neural-enriched” IG20-SVs. These neural-enriched isoforms were also found to be expressed in two NB cell lines (SK-N-SH, and SH-SY5Y) known to be deficient in caspase-8 expression, but not in the SK-N-BE(2) NB cell line which is known to express caspase-8. There was relatively little mRNA expression of neural-enriched IG20-SVs in human cerebellum or skeletal muscle. The differential presence of these neural-specific IG20-SVs is consistent with current evidence of tissue specific differences in alternative splicing of pre-mRNAs (31, 32).
To investigate the physiological relevance of the expression of the neural-enriched IG20-SVs in NB cells, we down-modulated select combinations of IG20-SVs using siRNAs in SK-N-SH and SH-SY5Y NB cells. We found that down-modulation of MADD/DENN using shRNA targeting exon 13L enhanced spontaneous apoptosis (SK-N-SH and SH-SY5Y) and TNF-α-induced apoptosis (SK-N-SH). It is important to understand that the 13L siRNA will also down-modulate KIAA0358 expression. As expected, down-modulation of all IG20-SVs also resulted in enhanced apoptosis of NB cells in SK-N-SH cells, although not significantly in SH-SY5Y cells. However, selective down-modulation of IG20pa, MADD, IG20-SV2, and DENN-SV, allowing for unaltered endogenous expression of IG20-SV4 and KIAA0358, resulted in markedly enhanced cellular survival in both NB cell lines. In contrast, knock-down of all splice isoforms except for IG20-SV4 caused a significant enhancement of apoptosis in both SK-N-SH and SH-SY5Y cells. These results suggested that KIAA0358 exerts a predominant suppressive effect on IG20-SV4 in certain NB cells.
Since previous studies have shown that MADD/DENN acts as a negative regulator of caspase-8 activation in non-neural cells(33), we hypothesized that the mechanism responsible for enhanced apoptosis in NB cells mediated by manipulation of IG20-SV4 and KIAA0358 expression involves activation of caspase-8. While the expression of KIAA0358 and IG20-SV4 in selected primary NB tumor lines did not suggest a reciprocal relationship between IG20-SVs and caspase-8, we hypothesized that these IG20-SVs may be involved in the regulation of caspase-8 activation in NB cells.
We first noted that caspase-8 expression was increased in cells in which KIAA0358 was down-modulated (treated with 13L and 34E+13 siRNAs, and, to a lesser extent, in cells in which all IG20-SVs were knocked down). When transduced SK-N-SH cells were treated with cycloheximide, the induced caspase-8 was inhibited, consistent with it being newly synthesized protein, and suggesting that the pattern of IG20-SV4 and KIAA0358 expression may be involved in the regulation of CASP8 gene expression. This was confirmed by showing the effect of IG20-SV4 on activation of the CASP8 promoter utilizing a luciferase assay. The marked activation of the CASP8 promoter by IG20-SV4 is direct evidence that IG20-SVs may exert their effects through regulation of CASP8 gene expression. As expected, inhibition of caspase-8 protected cells from undergoing apoptosis only when KIAA0358 was down-modulated, i.e., utilizing 13L, 34E+13L and mid siRNAs.
This argued strongly that the mechanism of enhanced apoptosis in these cells was related to caspase-8 expression and activation. Furthermore, the selective expression of IG20-SV4 sensitized NB cells to the pro-apoptotic effects of TNFα, and this sensitization was suppressed by DN-FADD, offering further support for the mechanistic role of caspase-8 in enhancement of both spontaneous and TNFα-induced apoptosis mediated by selective overexpression of IG20-SV4. Whether these effects occur at the level of the death-inducing signaling complex (DISC) or possibly after dissociation from TNFR1, i.e., complex II (34), will be explored in future studies.
While levels of apoptosis and caspase-8 activation were very high in NB cells in which all IG20-SVs except IG20-SV4 were down-modulated, selective expression of KIAA0358 in the presence of IG20-SV4 (or in the setting of down-modulation of all other isoforms) effectively prevented apoptosis and caspase-8 expression, suggesting that KIAA0358 may have a dominant-negative effect on IG20-SV4. To further confirm the pro-survival effects of KIAA0358 on NB cell survival, we generated SK-N-SH cells stably expressing a mutant KIAA0358 which contained silent mutations that did not affect protein expression but prevented down-modulation of KIAA0358 by mid-shRNA, the cell was transduced with MID-shRNA for 5 days. As expected, SK-N-SH cell lines expressing this KIAA0358 mutant were largely resistant to apoptosis compared to control cells treated with mid-shRNA. This effect was accompanied by a nearly complete dampening of caspase-8 activation. While the effects of manipulation of neural IG20-SVs were similar in the SK-N-SH and SH-SY5Y cell-lines (both deficient in caspase-8), we observed no effect of introduction of either IG20-SV4 or KIAA0358 on caspase-8 expression in the SK-N-BE(2)-C cell line which has constitutive expression of caspase-8, suggesting that the effects of IG20-SV expression are likely tissue/cell dependent (33).
Silencing of the CASP8 gene may play a role in NB tumor progression by the induction of tumor cell resistance to apoptosis induced by cytotoxic agents, or by death-inducing ligands, such as TNF-Alpha or TRAIL (17–19, 35, 36). Induction of caspase-8 expression by gene transfer in NB cells has been shown to confer sensitivity to death-receptor ligands and to their combinations with cytotoxic agents (23, 37). Further, interferon-γ can sensitize neoplastic cells to apoptosis through up-regulation of caspase-8 (18), and an interferon-sensitive response element (ISRE) in the caspase-8 promoter may play a role in this IFN-γ-driven regulation of caspase-8 expression in cancer cells(23). The regulation of caspase-8 expression likely involves other complex interactions involving the CASP8 gene. Our results suggest that the expression of IG20-SVs may play a role in determining caspase-8 expression/activation and susceptibility to apoptosis in NB cells.
In summary, we show for the first time that pro-apoptotic signaling caused by down-modulation of KIAA0358 or overexpression of IG20-SV4 effectively induces spontaneous apoptosis and sensitization to TNFα-induced apoptosis through expression and activation of caspase-8 in NB cells known to be deficient in caspase-8. Furthermore, enhanced expression of IG20-SV4 alone can overcome the transcriptional inhibition of the CASP8 gene, and upregulate its expression, while KIAA0358 acts as a negative regulator of caspase-8 expression and activation in these cells. Further study examining the precise mode of action of IG20-SVs in modulating expression of caspase-8 in NB and other cancer cells may elucidate novel targets that can be manipulated to enhance apoptosis (both spontaneous and in response to cytotoxic drugs) in cancer cells, and is currently under investigation.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Jill Lahti of the St. Jude’s Children’s Research Hospital for providing RNA from primary neuroblastoma tumor lines, and Dr. Silvano Ferrini for the generous gift of the pBLCAT-Casp8 plasmid, Dr Prasad Kanteti for critical review of the manuscript, Dr. Karen L. Hagen for assistance with flow cytometry, and Mrs. Lixia Qian for assistance in plasmid construction. This work was supported in part by grant R01 CA107506 to BSP from the National Institutes of Health, Bethesda, Maryland.
Abbreviations
- IG-20
Insulinoma Glucagonoma clone-20
- NB
Neuroblastoma
- MADD
Map-kinase Activating Death Domain containing protein
- DENN
Differentially Expressed in Normal and Neoplastic tissues
- SVs
Splice Variants
- TNFR1
Tumor Necrosis Factor Receptor 1
- YFP
Yellow Fluorescent Protein
- GFP
Green Fluorescent Protein
References
- 1.Brodeur G, Mris JM. Neuroblastoma. In: Pizzo PA, Poplack DG, editors. Principles and practice of pediatric oncology. 5. Philadelphia: JB Lippincott; 2006. pp. 933–70. [Google Scholar]
- 2.Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- 3.Brodeur GM. Neuroblastoma: biological insights into a clinical enigma. Nat Rev Cancer. 2003;3:203–16. doi: 10.1038/nrc1014. [DOI] [PubMed] [Google Scholar]
- 4.Chow VT, Lee SS. DENN, a novel human gene differentially expressed in normal and neoplastic cells. DNA Seq. 1996;6:263–73. doi: 10.3109/10425179609020873. [DOI] [PubMed] [Google Scholar]
- 5.Chow VT, Lim KM, Lim D. The human DENN gene: genomic organization, alternative splicing, and localization to chromosome 11p11.21–p11.22. Genome. 1998;41:543–552. doi: 10.1139/g98-050. [DOI] [PubMed] [Google Scholar]
- 6.Al-Zoubi AM, Efimova EV, Kaithamana S, et al. Contrasting effects of IG20 and its splice isoforms, MADD and DENN-SV, on tumor necrosis factor alpha-induced apoptosis and activation of caspase-8 and -3. J Biol Chem. 2001;276:47202–11. doi: 10.1074/jbc.M104835200. [DOI] [PubMed] [Google Scholar]
- 7.Efimova E, Martinez O, Lokshin A, Arima T, Prabhakar BS. IG20, a MADD splice variant, increases cell susceptibility to gamma-irradiation and induces soluble mediators that suppress tumor cell growth. Cancer Res. 2003;63:8768–76. [PubMed] [Google Scholar]
- 8.Efimova EV, Al-Zoubi AM, Martinez O, et al. IG20, in contrast to DENN-SV, (MADD splice variants) suppresses tumor cell survival, and enhances their susceptibility to apoptosis and cancer drugs. Oncogene. 2004;23:1076–87. doi: 10.1038/sj.onc.1207210. [DOI] [PubMed] [Google Scholar]
- 9.Ramaswamy M, Efimova EV, Martinez O, Mulherkar NU, Singh SP, Prabhakar BS. IG20 (MADD splice variant-5), a proapoptotic protein, interacts with DR4/DR5 and enhances TRAIL-induced apoptosis by increasing recruitment of FADD and caspase-8 to the DISC. Oncogene. 2004;23:6083–94. doi: 10.1038/sj.onc.1207804. [DOI] [PubMed] [Google Scholar]
- 10.Mulherkar N, Ramaswamy M, Mordi DC, Prabhakar BS. MADD/DENN splice variant of the IG20 gene is necessary and sufficient for cancer cell survival. Oncogene. 2006;25:6252–61. doi: 10.1038/sj.onc.1209650. [DOI] [PubMed] [Google Scholar]
- 11.Yamaguchi K, Tanaka M, Mizoguchi A, et al. A GDP/GTP exchange protein for the Rab3 small G protein family up-regulates a postdocking step of synaptic exocytosis in central synapses. Proc Natl Acad Sci USA. 2002;99:14536–41. doi: 10.1073/pnas.212511399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tanaka M, Miyoshi J, Ishizaki H, et al. Role of Rab3 GDP/GTP exchange protein in synaptic vesicle trafficking at the mouse neuromuscular junction. Mol Biol Cell. 2001;12:1421–30. doi: 10.1091/mbc.12.5.1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Del Villar K, Miller CA. Down-regulation of DENN/MADD, a TNF receptor binding protein, correlates with neuronal cell death in Alzheimer’s disease brain and hippocampal neurons. Proc Natl Acad Sci USA. 2004;101:4210–15. doi: 10.1073/pnas.0307349101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schievella AR, Chen JH, Graham JR, Lin LL. MADD, a novel death domain protein that interacts with the type 1 tumor necrosis factor receptor and activates mitogen-activated protein kinase. J Biol Chem. 1997;272:12069–75. doi: 10.1074/jbc.272.18.12069. [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Zhou L, Miller CA. A splicing variant of a death domain protein that is regulated by a mitogen-activated kinase is a substrate for c-Jun N-terminal kinase in the human central nervous system. Proc Natl Acad Sci USA. 1998;95:2586–91. doi: 10.1073/pnas.95.5.2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mulherkar N, Prasad KV, Prabhakar BS. MADD/DENN splice variant of the IG20 gene is a negative regulator of caspase-8 activation. Knockdown enhances TRAIL-induced apoptosis of cancer cells. J Biol Chem. 2007;282:11715–21. doi: 10.1074/jbc.M701085200. [DOI] [PubMed] [Google Scholar]
- 17.Teitz T, Wei T, Valentine MB, et al. Caspase 8 is deleted or silenced preferentially in childhood neuroblastomas with amplification of MYCN. Nat Med. 2000;6:529–35. doi: 10.1038/75007. [DOI] [PubMed] [Google Scholar]
- 18.Fulda S, Debatin KM. IFNgamma sensitizes for apoptosis by upregulating caspase-8 expression through the Stat1 pathway. Oncogene. 2002;21:2295–308. doi: 10.1038/sj.onc.1205255. [DOI] [PubMed] [Google Scholar]
- 19.Hopkins-Donaldson S, Bodmer JL, Bourloud KB, Brognara CB, Tschopp J, Gross N. Loss of caspase-8 expression in highly malignant human neuroblastoma cells correlates with resistance to tumor necrosis factor-related apoptosis-inducing ligand-induced apoptosis. Cancer Res. 2000;60:4315–19. [PubMed] [Google Scholar]
- 20.Teitz T, Lahti JM, Kidd VJ. Aggressive childhood neuroblastomas do not express caspase-8: an important component of programmed cell death. J Mol Med. 2001;79:428–36. doi: 10.1007/s001090100233. [DOI] [PubMed] [Google Scholar]
- 21.Stupack DG, Teitz T, Potter MD, et al. Potentiation of neuroblastoma metastasis by loss of caspase-8. Nature. 2006;439:95–9. doi: 10.1038/nature04323. [DOI] [PubMed] [Google Scholar]
- 22.Lee MT, Coburn GA, McClure MO, Cullen BR. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J Virol. 2003;77:11964–72. doi: 10.1128/JVI.77.22.11964-11972.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.De Ambrosis A, Casciano I, Croce M, et al. An interferon-sensitive response element is involved in constitutive caspase-8 gene expression in neuroblastoma cells. Int J Cancer. 2007;120:39–47. doi: 10.1002/ijc.22173. [DOI] [PubMed] [Google Scholar]
- 24.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–3. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 25.Cullen LM, Arndt GM. Genome-wide screening for gene function using RNAi in mammalian cells. Immunol Cell Biol. 2005;83:217–23. doi: 10.1111/j.1440-1711.2005.01332.x. [DOI] [PubMed] [Google Scholar]
- 26.Fulda S, Kufer MU, Meyer E, van Valen F, Dockhorn-Dworniczak B, Debatin KM. Sensitization for death receptor- or drug-induced apoptosis by re-expression of caspase-8 through demethylation or gene transfer. Oncogene. 2001;20:5865–77. doi: 10.1038/sj.onc.1204750. [DOI] [PubMed] [Google Scholar]
- 27.Banelli B, Casciano I, Croce M, et al. Expression and methylation of CASP8 in neuroblastoma: identification of a promoter region. Nat Med. 2002;8:1333–5. doi: 10.1038/nm1202-1333. [DOI] [PubMed] [Google Scholar]
- 28.Coppola T, Perret-Menoud V, Gattesco S, et al. The death domain of Rab3 guanine nucleotide exchange protein in GDP/GTP exchange activity in living cells. Biochem J. 2002;362:273–9. doi: 10.1042/0264-6021:3620273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sakisaka T, Takai Y. Purification and properties of Rab3 GEP (DENN/MADD) Methods Enzymol. 2005;403:254–61. doi: 10.1016/S0076-6879(05)03021-1. [DOI] [PubMed] [Google Scholar]
- 30.Nagase T, Ishikawa K, Nakajima D, et al. Prediction of the coding sequences of unidentified human genes. VII. The complete sequences of 100 new cDNA clones from brain which can code for large proteins in vitro. DNA Res. 1997;4:141–50. doi: 10.1093/dnares/4.2.141. [DOI] [PubMed] [Google Scholar]
- 31.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 32.Licatalosi DD, Darnell RB. Splicing regulation in neurologic disease. Neuron. 2006;52:93–101. doi: 10.1016/j.neuron.2006.09.017. [DOI] [PubMed] [Google Scholar]
- 33.Mulherkar N, Prasad KVS, Prabhakar BS. Differential effects of the IG20 gene splice variants in cancer cell survival. In: Venables JP, editor. Alternative splicing in cancer. Kerala, India: Transworld Research Network; 2006. pp. 123–39. [Google Scholar]
- 34.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–90. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 35.Prabhakar BS, Mulherkar N, Prasad KV. Role of IG20 splice variants in TRAIL resistance. Clin Cancer Res. 2008;14:347–51. doi: 10.1158/1078-0432.CCR-07-0493. [DOI] [PubMed] [Google Scholar]
- 36.Yang X, Merchant MS, Romero ME, et al. Induction of caspase 8 by interferon gamma renders some neuroblastoma (NB) cells sensitive to tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) but reveals that a lack of membrane TR1/TR2 also contributes to TRAIL resistance in NB. Cancer Res. 2003;63:1122–29. [PubMed] [Google Scholar]
- 37.Muhlethaler-Mottet A, Bourloud KB, Auderset K, Joseph JM, Gross N. Drug-mediated sensitization to TRAIL-induced apoptosis in caspase-8-complemented neuroblastoma cells proceeds via activation of intrinsic and extrinsic pathways and caspase-dependent cleavage of XIAP, Bcl-xL and RIP. Oncogene. 2004;23:5415–25. doi: 10.1038/sj.onc.1207704. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


