Abstract
Homoisocitrate dehydrogenase (HIcDH) catalyzes the Mg2+- and K+-dependent oxidative decarboxylation of homoisocitrate to α-ketoadipate using NAD as the oxidant. A recent consideration of the structures of enzymes in the same family as HIcDH, including isopropylmalate and isocitrate dehydrogenases, suggests all of the family members utilize a Lys-Tyr pair to catalyze the acid-base chemistry of the reaction (Aktas, D. F., and Cook, P. F. (2009) Biochemistry submitted.). Multiple sequence alignment indicates the active site Lys-Tyr pair consists of lysine-206 and tyrosine-150. Therefore, the K206M and Y150F mutants of HIcDH were prepared and characterized in order to test the potential roles of these residues as acid-base catalysts. The V/Et values of the K206M and Y150F mutant enzymes at pH 7.5 are decreased by about 2400- and 680-fold, respectively, compared to wild type HIcDH; the Km for HIc does not change significantly. V/Et and V/KMgHIc Et for the K206M mutant enzyme are pH independent below pH 6 and decrease to a constant value above pH 7, while V/KNAD Et is independent over the pH range 6.2 to 9.5. In the case of the Y150F mutant enzyme, V/Et and V/KNAD Et are pH independent above pH 9.5 and decrease to a constant value below pH 8. This behavior can be compared to wild type enzyme, where V/Et decreases at high and low pH giving pKa values of about 6.5 and 9.5. Data were interpreted in terms of a group with a pKa of 6.5 that acts as a general base in the hydride transfer step, and a group with a pKa of 9.5 that acts as a general acid to protonate C3 in the tautomerization reaction (Lin, Y., Volkman, J., Nicholas, K. M., Yamamoto, T., Eguchi, T., Nimmo, S. L., West, A. H., Cook, P. F. (2008) Biochemistry 47, 4169–4180.). Solvent deuterium isotope effects on V and V/KMgHIc were near unity for the K206M mutant enzyme, but about 2.2 for the Y150F mutant enzyme. The dramatic decreases in activity, the measured solvent deuterium isotope effects and changes in the pH dependence of kinetic parameters compared to wild type, are consistent with K206 acting as a general base in the hydride transfer step of the wild type enzyme, but as a general acid in the Y150F mutant enzyme, replacing Y150 in the tautomerization reaction. In addition, Y150 acts as a general acid in the tautomerization reaction of the wild type enzyme, and replaces K206 as the general base in the hydride transfer step of the K206M mutant enzyme.
Homoisocitrate dehydrogenase (3-carboxy-2-hydroxyadipate dehydrogenase, EC 1.1.1.87) (HIcDH)1 catalyzes the conversion of homoisocitrate to α-ketoadipate (α-Ka) and CO2 with NAD as the oxidant (1). The HIcDH reaction is specific to the α-aminoadipate pathway (AAA) for lysine biosynthesis in fungi and higher plants, and is a member of the family of pyridine nucleotide-dependent β–hydroxyacid oxidative decarboxylases (2). The reaction is metal ion-dependent and the enzyme selectively binds the MgHIc chelate complex (3). A steady state random kinetic mechanism has been proposed for HIcDH; a preferred ordered release of CO2, α-Ka, and NADH was proposed (3). The rate of the pathway with NAD adding prior to MgHIc is limited by a conformational change, presumably to close the active site prior to catalysis (3, 4). Addition of MgHIc to enzyme, followed by NAD gives a pathway with oxidative decarboxylation contributing to rate limitation. In addition, it was recently shown that the enzyme requires K+ for optimum binding affinity for NAD at pH values greater than 7 (4). The role of the monovalent cation is likely to neutralize charge as a neutral acid in the dinucleotide binding site becomes unprotonated.
On the basis of the pH dependence of kinetic parameters and isotope effects, a general base, general acid chemical mechanism for the HIcDH was proposed, Scheme 1 (5). A general base, with a pKa of about 6.5, was proposed to accept a proton from the 2-hydroxyl of HIc as the 2-keto-3-carboxyadipate intermediate is formed in the hydride transfer step. The metal ion then acts as a Lewis acid and catalyzes the decarboxylation of the β-ketoacid and the base, now acting as an acid, protonates the keto oxygen as the enol of α-ketoadipate is formed. Enol-keto tautomerization is then catalyzed by the base accepting a proton from the oxygen enol and a second enzyme group, with a pKa of about 9.5, protonating C3 to give the final ketone product. The mechanism is very similar to those proposed for malic enzyme (6), isocitrate dehydrogenase (7, 8), and tatrate dehydrogenase (9).
Scheme 1.
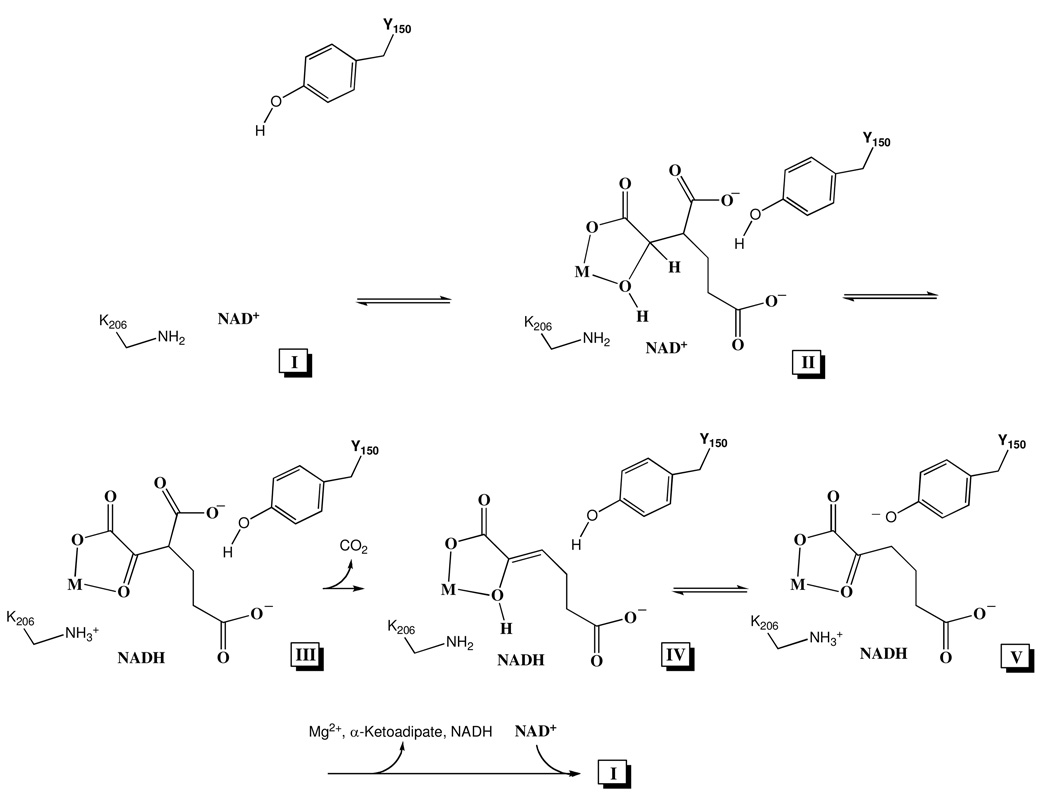
Proposed Chemical Mechanism of Homoisocitrate Dehydrogenase. The enzyme is in an open or partially open form upon binding of NAD (I), and closes upon binding of MgHIc, which brings Y150 into close proximity to HIc (II). Hydride transfer is facilitated by K206 acting as a base catalyst to generate 3-carboxy-α-ketoadipate (III). Decarboxylation occurs with the metal ion acting as a Lewis acid and K206 donating a proton to give the enol (IV). General base (K206), general acid (Y150) catalysis then gives α-ketoadipate (V), and (I) is regenerated upon release of Mg2+ (M), α-ketoadipate, and NADH, and NAD binding. (The acid base chemistry could be catalyzed by K206 and Y150 directly or mediated by a water molecule for one or both residues.)
Although a structure of HIcDH is available, it is for the Thermus thermophilus enzyme and has no ligands bound to the active site (10). The structure of the bacterial HIcDH is superimposable on those of the porcine IcDH (11) and bacterial IPMDH (12), which have ligands bound. Data suggest a conserved active site with a Lys-Tyr pair serving as general base and general acid catalysts. A review of the literature on the metal ion-dependent pyridine nucleotide-linked β-hydroxyacid oxidative decarboxylases has recently been written that suggests a very similar mechanism for this class of enzyme that makes use of very similar catalytic machinery (13). A multiple sequence alignment of HIcDH with sequences of IcDH, IPMDH, and TDH, suggests K206 and Y150 may function as the catalytic groups.
In this paper, site-directed mutagenesis was used to determine whether K206 and Y150 carry out the acid and base catalysis of HIcDH. Thus, K206 was changed to M, and Y150 was changed to F. The resulting mutant enzymes were characterized using initial rate studies and the pH dependence of kinetic parameters and isotope effects. Data are consistent with K206 serving as the catalytic base and Y150 as the general acid in the tautomerization step.
MATERIALS AND METHODS
Chemicals
Potassium acetate (KOAc) was obtained from Sigma. β-NAD and β-NADH were from USB. Hepes, Tris, Bis-Tris, and Ches buffers were from Research Organics, Inc. Deuterium oxide (D2O) (99 atom % D) was purchased from Cambridge Isotope Labotories, Inc. α-Ketoadipate (16), HIc (3), and NADD (17) were prepared according to published procedures.
Enzymes
HIcDH was subjected to site-directed mutagenesis to generate the Y150F and K206M mutant enzymes. The plasmid harboring the HIcDH gene was used as a template for mutagenesis, and the mutant enzymes were prepared using the QuikChange site-directed mutagenesis kit was from Stratagene. Whole gene sequencing was performed for each of the mutations at the Laboratory for Genomics and Bioinformatics of the University of Oklahoma Health Science Center in Oklahoma City. The resulting sequence was compared to that of the wild type HIcDH using BLAST. Successfully mutated plasmids were transformed into BL21 (DE3) competent cells, the expression host. To engineer the Y150F mutant enzyme, the forward and reverse primers are CTGAGGACCTGTTCATTAAAATTG and CAATTTTAATGAACAGGTCCTCAG, respectively, while for the K206M mutant enzyme the forward and reverse primers are GACAGTGACTCATATGTCAAATGTTCTATC and GATAGAACATTTGACATATGAGTCACTGTC, respectively. The mutated codons are shown in bold.
Enzyme Assay
The activity of wt and mutant HIcDHs was measured using a Beckman DU 640 spectrophotometer to monitor the increase or decrease in absorbance at 340 nm as NAD is reduced to NADH. All assays were carried out at 25°C. A unit of enzyme activity is defined as the amount of enzyme catalyzing the production or utilization of 1 µmole of NADH per min at 25°C. Typical assays contained 100 mM buffer, 200 mM KOAc (saturating for both mutant enzymes), 4.2 µM MgHIc and 0.5 mM NAD in a 1 mL volume. The final concentration of HIcDH in the cuvette was 15 nM for wild type enzyme, and about 1 µM for the mutant enzymes. In the case of the mutant enzymes, the total concentration of HIc (HIc plus MgHIc) was at least 10-times greater than the concentration of MgHIc, i.e., HIc was 10 times MgHIc. Thus the lowest ratio of HIctotal/E is 10 for any of the assays carried out.
pH Studies
The pH-dependence of V and V/K for all substrates was measured under conditions in which one substrate concentration was varied with the other maintained at saturation (≥10Km). The pH was maintained using the following buffers at 100 mM concentration: Bis-Tris, 5.0–6.8; Hepes, 6.8–8.2; Ches, 8.2–10.0. Hepes and Ches were titrated to the appropriate pH with KOH, while Bis-Tris was titrated using HCl. Sufficient overlap was obtained upon changing buffers to detect buffer effects; none were observed. The ionic strength of the reaction mixtures over the pH range of our measurements is approximately constant at 0.1. The pH was recorded before and after initial velocity data were measured with observed changes limited to ≤0.1 pH unit. The enzymes were stable when incubated for at least 15 min over the pH range 5.0–10.0. pH-rate profiles were initially evaluated graphically by plotting log V or log(V/K) against pH to determine the proper equation for data fitting and to assess the quality of data.
Solvent Deuterium and Viscosity Effects
Initial velocities were measured in H2O and D2O. For rates measured in D2O, substrates and buffers were first dissolved in a small amount of D2O and lyophilized overnight to remove exchangeable protons. The lyophilized powders were then dissolved in D2O to give the desired concentration, and the pD was adjusted using NaOD. A value of 0.4 was added to the pH meter reading to calculate pD (18). Data were obtained by varying one substrate at a fixed concentration (10Km) of the other one. The isotope effects were obtained by direct comparison of initial rates in H2O and D2O at pH(D) 8.0. Reactions were initiated by adding 5 µL of enzyme solution in H2O, such that the final percentage of D2O was about 99%. The concentration of the enzyme stock solution was adjusted depending on the specific activity.
Initial velocities were determined in H2O at a relative viscosity of 1.24 at pH 8.0 and 25°C; assays contained 9% glycerol (w/v) as the viscosogen, which generates the same relative viscosity as that of 100% D2O at 25°C (19). A standard curve (relative viscosity vs. glycerol) was constructed to determine the amount of glycerol needed for a relative viscosity of 1.24 (20).
Circular Dichroism Spectra
CD spectra were collected on an Aviv 62DS spectropolarimeter at 25°C with a path length of 0.2 cm. Enzyme concentrations of 100 µg/mL were used for far UV CD spectra. The buffer used for all spectra was 10 mM phosphate, pH 7.0 and a buffer blank was subtracted from each spectrum. Spectra were collected with a dwell time of 3 s, and were the average of three spectra. Far-UV spectra were measured from 200–250 nm.
Data Analysis
Initial velocity data were first analyzed graphically using double-reciprocal plots of initial velocities versus substrate concentration and suitable secondary plots. Data were then fitted using the appropriate equation, and the Marquardt-Levenberg algorithm supplied with the EnzFitter program from BIOSOFT, Cambridge, U.K. Kinetic parameters and their corresponding standard errors were estimated using a simple weighting method.
Data obtained as a function of MgHIc at different fixed levels of NAD were fitted using eq 1. Data for solvent deuterium isotope effects on V and V/K were fitted using eq. 2.
| [1] |
| [2] |
In eq. 1 and eq. 2, v and V are initial and maximum velocities, A and B are substrates concentrations, Ka and Kb are Michaelis constants for substrates A and B, respectively. In eq. 1, Kia is the dissociation constant for the EA complex. In eq. 2, Fi is the fraction of D2O in the solvent, and EV/K and EV are the solvent deuterium isotope effects minus 1 on V/K and V, respectively.
Data for pH-rate profiles with a limiting slope of 1 at low pH were fitted using eq. 3, while data for pH-rate profiles that exhibit a partial change at low or high pH were fitted using eq. 4 or eq. 5.
| [3] |
| [4] |
| [5] |
In eq. 3, y is the observed value of V/KMgHIc for the K206M mutant enzyme at any pH, C is the pH-independent value of y, H is the hydrogen ion concentration, and K1 is the acid dissociation constant for an enzyme or substrate functional group that must be unprotonated for optimal binding and/or catalysis. In eq. 4 and eq. 5, y is the observed value of V or V/K, and YL and YH are the pH-independent values of y at low and high pH, respectively. In eq. 4, K2 is the acid dissociation constant for enzyme or substrate functional groups that must be unprotonated for optimal binding and/or catalysis, while in eq. 5, the group must be protonated for optimal binding and/or catalysis. The V/Et and V/KMgHIc Et for the Y150F mutant enzyme decrease from a constant value at low pH to a lower constant value at high pH. Attempts to fit the data to eq. 4 were unsuccessful. The observed pH dependence, Fig. 2, indicates a pKa of ≤6, and a group with a pKa of about 6.5 is observed for the wt enzyme (5). Thus, K2 in eq. 4 was fixed at 10−6 M and data were fitted to generate estimates of YL and YH.
RESULTS
Expression and Purification of Mutant Enzymes
Expression and purification of the Y150F and K206M mutant enzymes was very similar to wild type HIcDH (3). The purity of the wild type and mutant enzymes was >95% on the basis of SDS-PAGE. Far UV CD spectra were very similar for the mutant and wt enzymes suggesting the overall structure of the mutant enzymes is intact and changes are likely restricted to the active site.
Kinetic Parameters of the Y150F and K206M Mutant Enzymes
Initial rates were measured at pH 7.5 as a function of NAD at different fixed concentrations of MgHIc for both mutant enzymes. Double reciprocal plots intersected to the left of the ordinate, consistent with the sequential kinetic mechanism reported previously (3). Kinetic parameters are summarized in Table 1, compared to those obtained for the wt enzyme.
Table 1.
Kinetic Parameters for Wild Type and Mutant Enzymes at pH 7.5.
| Parameter | Wild type | Y150F | K260M |
|---|---|---|---|
| V/Et (s−1) | 13 ± 1 | 0.019 ± 0.001 | 0.0054 ± 0.0005 |
| Fold Decrease | 1 | 680 ± 60 | 2400 ± 290 |
| V/KNAD Et (M−1s−1) | (2.9 ± 0.6) × 104 | (4.2 ± 0.4) × 102 | 6.0 ± 0.6 |
| Fold Decrease | 1 | 70 ± 16 | 4800 ± 1100 |
| V/KMgHIc Et (M−1s−1) | (3.1 ± 0.7) × l06 | (1.8 ± 0.1) × l03 | (1.8 ± 0.6) × 103 |
| Fold Decrease | 1 | 1700 ± 400 | 1700 ± 700 |
| KNAD (mM) | 0.45 ± 0.08 | 0.045 ± 0.004 | 0.84 ± 0.02 |
| Fold Change | 1 | 10 ± 2 (decrease) | 1.9 ± 0.3 (increase) |
| KMgHIc (µM) | 4.2 ± 0.9 | 1.06 ± 0.02 | 3 ± 1 |
| Fold Decrease | 1 | 4.0 ± 0.9 | 1.4 ± 0.6 |
Replacing Y150 with F or K206 with M, results in decreases of 680- and 2400-fold, respectively, in V/Et. KMgHIc is not greatly affected for either of the mutant enzymes, giving decreases in V/KMgHIc Et that reflects the change in V/Et. Although KNAD is increased by only about 2-fold in the case of the K206M mutant enzyme, it is decreased 10-fold for the Y150F mutant enzyme. As a result, the value of V/KNAD Et is decreased by only 70-fold in the case of the Y150F mutant enzyme.
Rescue of Activity by Small Molecules
Addition of small molecules can, in some cases, rescue activity lost when important functional groups are eliminated from an enzyme side chain. Replacement of Y150 with F, results in the elimination of a hydroxyl group, and it is unlikely that addition of small molecules would have an effect. Addition of 10 mM NH4Cl or (NH4)2SO4 had no effect. However, in the case of K206M, addition of the same components resulted in a 30% increase in V/Et.
pH-Rate Profiles
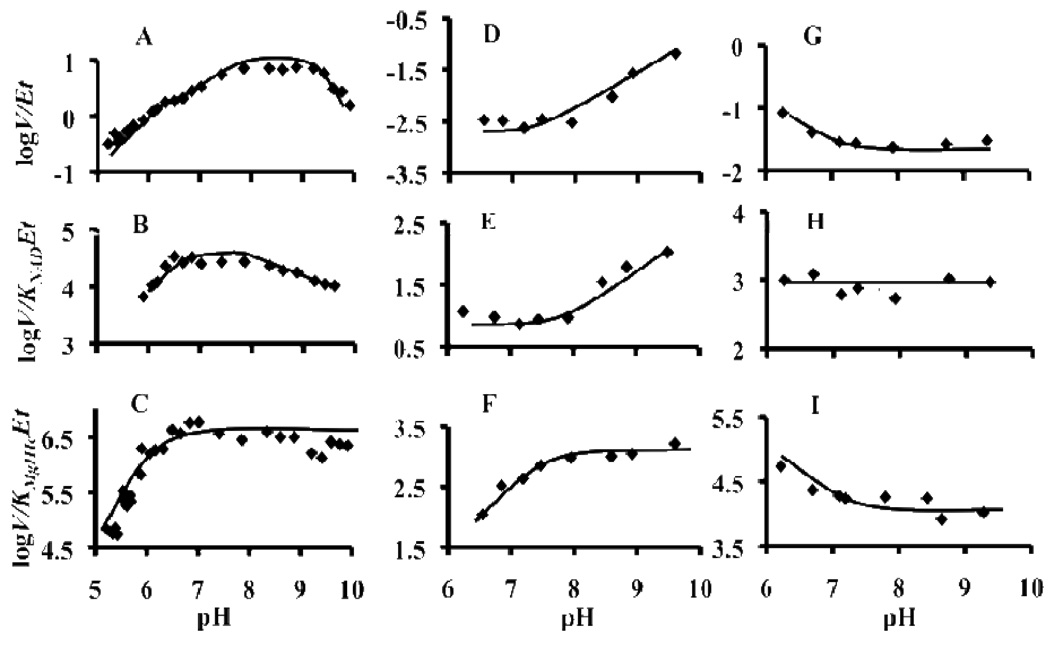
The pH dependence of kinetic parameters obtained previously for the wild type enzyme are reproduced in Figure 1 for comparison to those of the mutant enzymes (5). V/Et gives estimated pKa values of about 7.1 and 9.5. A hollow is observed in the curve as the pH decreases, i.e., the curve exhibits a plateau at about pH 6.5, before it eventually decreases with a limiting slope of 1. V/KNAD Et decreases at low pH, giving a pKa value of about 6.4, while a partial change is observed at high pH giving a pKa value of about 8.3. V/KMgHIc Et decreases at low pH with a limiting slope of +2, with an average pKa value of about 6.0. One of the groups observed in the V/KMgHIc Et pH-rate profile likely reflects the substrate, which has a pKa for its third ionization of 5.8 (5).
Figure 1.
pH Dependence of Kinetic Parameters for Wild Type and Mutant Enzymes. Data for wt enzyme (A–C) are from reference 5 and published with permission of ACS (License number 2121471241144); D-F. K206M mutant enzyme; G-I. Y150F mutant enzyme. Units for V/Et and V/K Et are s−1 and M−1s−1, respectively. All data were obtained at 25°C. Points are the experimentally determined values. The curves are theoretical and based on fits to eq. 5 for D and E, eq. 3 for F, eq. 4 for G and I. The average value is shown for V/KNADEt. Error bars are included in the size of the data point.
The pH dependence of kinetic parameters for the K206M mutant enzyme differ considerably from those of wt and the Y150F mutant enzymes, Figure 2. The V/Et pH-rate profile is pH independent at pH values greater than 9.5, and decreases to a lower constant value below a pKa of 9.6 ± 0.2. The pH independent values of V/Et at high and low pH are 0.13 ± 0.02 s−1 and 0.002 ± 0.001 s−1 (1% and 0.015%, respectively, the V/Et of wt). The V/KNAD Et pH-rate profile is similar to V/Et, and exhibits a pKa of 9.0 ± 0.1, and pH independent values at high and low pH are 140 ± 8 M−1s−1 and 9.0 ± 0.1 M−1s−1 (0.5% and 0.03%, respectively, the values of wt). However, V/KMgHIc Et is pH independent above pH 7.5 and exhibits the requirement for a group that must be unprotonated for optimal activity and/or binding. The observed pKa is 7.3 ± 0.1, and the pH independent value of V/KMgHIc Et is 1120 ± 40 M−1 s−1 (0.04% the value of wt).
The V/Et for the Y150F mutant enzyme decreases from a constant below pH 6 to a lower constant value at high pH, Figure 3. As discussed in Data Analysis above, a pKa of 6.0 was assumed to allow estimates of the low and high pH constant values; the value at low pH is 0.110 ± 0.002 s−1, while that at high pH is 0.026 ± 0.002 s−1 (0.8% and 0.2%, respectively, of the V/Et of wt). The pH dependence of V/KMgHIc Et is similar to that of V/Et, and data were fitted as for V/Et with an assumed pKa of 6.0. pH independent values of about (1.28 ± 0.06) × 105 M−1s−1 and (1.0 ± 0.1) × 104 M−1s−1 are estimated at low and high pH (4% and 3%, respectively, the values of wt). On the other hand, V/KNAD Et is pH independent, and exhibits an average value of about 903 ± 9 M−1s−1 (3% the value of wt). Quantitative anaylsis (see Discussion) of the data will make use of the estimated pH independent values presented above.
Dependence of Kinetic Parameters on Solvent Deuterium and Solvent Viscosity
Finite solvent deuterium isotope effects of about 1.2 and 2.4 are observed on V and V/KMgHIc for Y150F, while equal values of about 2.2 are observed on the two parameters for K206M, Table 2. The wt enzyme exhibits a finite SKIE on V, but an inverse effect on V/KMgHIc that was attributed to the increase in viscosity resulting from 100% D2O. As a result, the effect of 9% glycerol, which gives a viscosity equal to 100% D2O (1.24) on the kinetic parameters of the mutant enzymes was determined. As seen in Table 2, the K206M mutant enzyme exhibits no effect of increased viscosity. The Y150F mutant enzyme, on the other hand, exhibits significant deceases in V and V/KMgHIc, Table 2. Correction of the SKIE for the viscosity effect gives residual effects of about 1 and 1.3 on V and V/KMgHIc.
Table 2.
Effects of Solvent Deuterium and Solvent Viscosity on the HIcDH Mutant Enzymes.
| Enzymes | Fixed substrates | Varied substrate | pH | H2O / D2O | H2O/ 9% glycerol in H2O | ||
|---|---|---|---|---|---|---|---|
| D20(V) | D20(V/K) | n(V) | n(V/K) | ||||
| WTa | NAD (10Km) |
MgHIc | 8.0 | 2.5 ± 0.2 | 1.3 ± 0.2 | 1.5 ± 0.2 | 0.6 ± 0.1 |
| WTa | NAD (10Km) |
Ic | 8.0 | 0.94 ± 0.08 | 0.69 ± 0.08 | 0.66 ± 0.03 | 0.6 ± 0.1 |
| Y150F | NAD (5 mM) |
MgHIc | 8.0 | 1.24 ± 0.06 (0.97 ± 0.06) |
2.4 ± 0.3 (1.3 ± 0.1) |
1.28 ± 0.06 | 1.8 ± 0.2 |
| K206M | NAD (15 mM) |
MgHIc | 8.0 | 2.3 ± 0.4 | 2.1± 0.7 | 1.0 ± 0.1 | 1.0 ± 0.2 |
From Reference 5.
DISCUSSION
Kinetic Mechanism
The large decreases in V/Et observed for the Y150F (500-fold; high pH independent value) and K206M (6500-fold; low pH independent value) mutant enzymes are consistent with the importance of these residues in the catalytic mechanism. Changes in the Km values for reactants are minimal, with the exception of KNAD for the Y150F mutant enzyme, which is decreased by about 10-fold. As a result, V/KNAD Et is decreased by only a factor of 70, while in all other cases the decrease in V/K reflects the decrease in V.
The initial rate of the wild type enzyme reaction is limited by a conformational change triggered by the binding of MgHIc (3). The structural change is likely required to generate the catalytic conformation prior to catalysis (see Correlation of Kinetic and Structural Data below). The primary deuterium isotope effect obtained by direct comparison of initial rates with NADH(D) is within error unity, indicating the net rate constant for the chemical steps is ≥10-fold faster than the rate constant for the conformational change2. Changing Y150 to F and K206 to M likely affects the chemical steps in the oxidative decarboxylation reaction (see Introduction). Thus, the decrease in the net rate constant of the chemical step must be underestimated. In the case of the Y150F mutant enzyme, the decrease must be ≥ 5000-fold (10 × (V/Et)wt/(V/Et)Y150F = 5000), while that for the K206M mutant enzyme must be ≥ 65000-fold (10 × (V/Et)wt/(V/Et)K206M = 65000). The approximately 5 × 103- to 6.5 × 104-fold decreases observed for the two mutant enzymes is consistent with their proposed function as acid-base catalysts. The dramatic decrease in the rate constant of the chemical step(s) of the mutant enzymes compared to those of the wt enzyme indicates the kinetic mechanism will be rapid equilibrium and not steady state. Initial velocity patterns obtained by measuring the initial rate as a function of NAD at different fixed levels of MgHIc (data not shown) are consistent with a random mechanism.3
Isotope effects
As suggested above, K206 and Y150 likely serve as the general base and general acid, respectively, in the oxidative decarboxylation reaction. Primary deuterium isotope effects were measured for wt by direct comparison of initial rates with NADH(D) as the labeled reactant. In the case of the mutant enzymes, the effect could not be measured because of the very low rate in the direction of reductive carboxylation (3). However, solvent deuterium kinetic isotope effects were measured, Table 2. The wt enzyme exhibits a normal solvent isotope effect on V and V/KMgHIc once corrected for solvent viscosity effects. On the basis of primary kinetic deuterium and 13C isotope effects and multiple primary deuterium/solvent deuterium and solvent deuterium/13C effects, data were interpreted in terms of a rate-limiting conformational change prior to the chemical steps with MgHIc limiting, and additionally a contribution to rate-limitation by the tautomerization step with substrates saturating. An inverse solvent kinetic deuterium isotope effect was associated with the conformational change prior to catalysis (5).
The K206M mutant enzyme exhibited a normal isotope effect of about 2.2 on V and V/KMgHIc. The wt enzyme exhibited a finite viscosity effect with the relative viscosity of the medium fixed at that of D2O, 1.24. No viscosity effect was observed for the K206M mutant enzyme. The normal solvent isotope effect is almost certainly a result of the proton transfer from the 2-hydroxyl of HIc as it is oxidized to α-keto-3-carboxyadipate in the hydride transfer step. The hydride transfer step is likely rate determining for the mutant enzyme (see pH-rate studies below), as expected for enzyme without its general base. The normal solvent kinetic deuterium isotope effect is in agreement with a lack of contribution to rate limitation from the conformational change prior to catalysis.
The Y150F mutant enzyme exhibits very small, if any solvent deuterium kinetic isotope effect, once corrected for the significant effect of viscosity, Table 2. The ketonization of the enol of α-ketoadipate is likely the slow step along the reaction pathway of the Y150F mutant, given the absence of the tyrosine hydroxyl, which is believed to serve as the general acid that protonates C3 of the enol. The low value of the solvent isotope effect suggests that if the tautomerization is slow, there must be either an early or late transition state for the proton transfer. Since water likely takes the place of the tyrosine hydroxyl in the ketonization reaction of Y150F, and water is not a good base, the transition state is more likely to be late for the proton transfer. As for the normal K206M mutant enzyme, the solvent kinetic deuterium isotope effect is not inverse, consistent with rate limitation by steps(s) other than the conformational change prior to catalysis.
Interpretation of pH-rate Profiles
Homoisocitrate dehydrogenase from Saccharomyces cerevisiae exhibits a steady state random kinetic mechanism; the predominant pathway is that with addition of MgHIc prior to NAD (3). The dominant species present when V/KMgHIc and V/KNAD are measured are E-NAD and MgHIc, and E:MgHIc and NAD, respectively. The pH dependence under these conditions will reflect group(s) on reactant and/or enzyme that is(are) important for binding and/or catalysis. The pH dependence of V will reflect group(s) on enzyme that is(are) important for catalysis (21).
A general acid-general base mechanism has been previously proposed for the wt enzyme on the basis of the pH-rate profiles shown Figure 1, and isotope effects (5). On the basis of the dramatic decreases in the values of kinetic parameters of the K206M and Y150F mutant enzymes, the slight rescue of K206M by ammonia, and structural data (see Correlation of Kinetic and Structural Data below) data are interpreted in terms of an acid-base mechanism with K206 as the base and Y150 as the acid. For the wt enzyme, a mechanism was proposed in which a general base with a pKa of 6.5–7 acts to accept the proton from the 2-hydroxyl of HIc (or Ic), and a general acid with a pKa of about 9.5 acts to protonate C-3 once decarboxylation has occurred. The pKa of 9.5 is only observed in the V pH-rate profile with MgHIc as a substrate, suggesting the tautomerization of the enol of adipic acid contributed to rate-limitation at saturating reactant concentrations (5). The V/Et pH-rate profiles for the mutant enzymes exhibit pKa values that are very similar to those observed for the wt enzyme, 6 for the Y150F mutant enzyme and 9.5 for the K206M mutant enzyme. However, the pH dependencies are opposite those of the wt enzyme with the Y150F enzyme exhibiting the requirement for a group with a pKa of about 6 that must be protonated for maximum activity and the K206M enzyme exhibiting the requirement for a group with a pKa of 9.5 that must be unprotonated for maximum activity.
The V/Et and V/KNAD Et values of the K206M mutant enzyme decreases below pH 10, giving a pKa value of 9.3, to a lower constant value below pH 8. Overall, this represents an increase by >2 pH units in the intrinsic pKa of 6.5–7 for the base of the wt enzyme (5). Of interest, the pKa is within error identical to that observed on the basic side of the V/Et pH-rate profile for wt enzyme using HIc as the substrate. The latter pKa was attributed to the general acid the protonates the enol of α-ketoadipate in the tautomerization reaction. The pH independent value of V/Et obtained with the group unprotonated is about 0.1 s−1, 90-fold lower than that of wt. It is likely that the group, Y150, which functions as the general acid in the wt enzyme, now serves as a base, either directly or mediated via a water molecule, to accept a proton from the α-hydroxyl of HIc in the overall reaction in the absence of K206. With Y150 acting optimally as a base, at pH values above 9.5, the 90-fold lower value of V/Et compared to wt indicates a single base is not sufficient for the overall reaction. Formally, the reaction requires proton transfer from the α-hydroxyl of HIc to C3 of α-ketoadipate, which could be catalyzed by a single group. However, the geometry of the proton transfer reactions is likely not easily satisfied. The similarity in pH behavior of V/Et and V/KNAD Et suggests the same steps limit the reaction with NAD limiting and with all reactants saturating. On the other hand, V/KMgHIc Et decreases at low pH, giving a pKa of about 7.4. This group is not observed in the pH-rate profile for V, suggesting the presence of group that must be unprotonated for optimal binding of MgHIc. The group is not observed in the pH-rate profiles for the wt or Y150F mutant enzyme. It is thus unmasked once K206 has been eliminated. The identity of the group will have to await further studies.
The pH independent value of V/Et for K206M below pH 8 is about 0.002 s−1, which is 6500-fold lower than the value of V/Et obtained for wt enzyme, and 65-fold lower than the pH independent value below pH 6 of the K206M mutant enzyme (0.13 s−1). At pH 7.5, a SKIE of about 2.3 is measured, suggesting hydride transfer is rate limiting, as suggested above. The ΔΔG‡ estimated from the ratio of (V/Et)wt/(V/Et)K206M is 5.3 kcal/mol, which provides a lower limit to the contribution of K206 to base catalysis.3 The value of 0.002 s−1 is still greater than the value for the un-catalyzed reaction, which would be extremely low for this multistep reaction, and is an indicator that the remaining catalytic functionalities in the active site, not the least of which is the metal ion, together perhaps with active site water, contribute significantly to catalysis.
The V/Et value of the Y150F mutant enzyme decreases above pH 6 (a pKa value of about 6 was assumed, see Methods) to a lower constant value above pH 7. The general shape of the V/KMgHIc Et pH-rate profile is very similar to that of V/Et, while the V/KNAD Et pH-rate profile is pH independent. The general base, presumably K206, now acts as a general acid, taking the place of Y150 in the tautomerization reaction. The pKa of the base decreases from about 6.5–7 in the wt enzyme to about 6 in Y150F. Thus, elimination of Y150 results in a decrease in the pKa, likely as a result of a more hydrophilic environment around the lysine. The pH independence of V/KNAD Et suggests the positive charge on the nicotinamide ring of NAD also affects the pKa of K206. The pH independent value of V/Et obtained with the group protonated in Y150F is ≥0.11 s−1, similar to that of the K206M mutant enzyme with Y150 protonated. It is likely that, in the Y150 mutant enzyme, K206 functions as the general acid and protonates C3 of the enol of α-ketoadipate. With K206 acting optimally as an acid, at pH values below 6, the >90-fold lower value of V/Et compared to wt indicates a single group is not sufficient for the overall reaction, as is true for K206M.
The pH independent value of V/Et for Y150F above pH 7 is 0.026 s−1, which is 500-fold lower than the value of V/Et obtained for wt enzyme, and 4-fold lower than the low pH independent value (0.11 s−1) of the Y150F mutant enzyme. Over this pH range, the SKIE is about 1, indicating a proton is not in flight in the rate-limiting transition state. The ΔΔG‡ estimated from the ratio of (V/Et)wt/(V/Et)Y150F is 3.7 kcal/mol, which provides a lower limit to the contribution of Y150 to base catalysis.4 The value of 0.026 s−1 is still greater than the value for the un-catalyzed reaction, which would be extremely low for this multistep reaction, and is an indicator that the remaining catalytic functionalities in the active site, contribute significantly to catalysis as suggested for K206M.
Another possible explanation of the pH dependence of kinetic parameters of the mutant enzymes is the direct use of H3O+ and OH− in the Y150F and K206M, respectively. This is a possibility in the case of the Y150F mutant enzyme, but is unlikely for the K206M mutant enzyme. V/Et and V/KNADEt clearly approach a limiting value above pH 9, and give a fitted pKa of about 9, very similar to the value observed in V/Et for the wt enzyme (5). Data suggest the use of Y150 as a general base in the K206M-catalyzed reaction, as discussed above. It is thus likely that K206 serves as an acid in the Y150F mutant enzyme. However, even if H3O+ and OH− are used directly, data are consistent with the acid-base catalytic properties of K206 and Y150 in the HIcDH reaction.
Correlation of Kinetic and Structural Data
A recent review of the enzymes in the class of pyridine nucleotide β-hydroxyacid oxidative decarboxylases suggests, on the basis of structure, that all of the enzymes in the class utilize the same catalytic machinery, i.e., a Lys-Tyr pair, to catalyze the reaction. The subclass of enzymes that catalyze the oxidative decarboxylation of R-β-hydroxyacids are dimeric and the active site is comprised of residues from both subunits (13). The structure of the HIcDH from T. thermophilus (10) superimposes on the other members of the subclass quite well (13). The structure of HIcDH, prior to binding substrates is in an open form, and Y150 is distant from the active site lysine (K206); Y150 is contributed by the other subunit in the dimer. However, a conformational change is required to close the site upon substrate binding and this results in Y150 in close proximity to homoisocitrate (10).
Data obtained in these studies are consistent with the proposed acid-base catalytic role of K206 and Y150 in the mechanism of HIcDH, Scheme 1. The open form of the enzyme is shown prior to binding MgHIc with Y150 away from the substrate-binding pocket (I). Binding of MgHIc gives the Michaelis complex with catalytic and binding groups in proper orientation for the hydride transfer step (II). In this complex, K206 and Y150 are neutral; the neutral form of K206 is likely a result of the overall positive nature of the active site (13). Hydride transfer from C3 of HIc to the nicotinamide ring of NAD results in the 3-carboxy-α-ketoadipate intermediate (III). Decarboxylation occurs with the metal ion (M in the figure) acting as a Lewis acid and K206 donating a proton to give the enol of α-ketoadipate (IV). Tautomerization to generate the final ketone product makes use of K206 acting as a base to accept the enol proton and Y150 acting to protonate C3 (V). Release of products and binding reactants then completes the catalytic cycle; this also requires an intramolecular proton transfer from K206 to Y150, presumably mediated by solvent.
Elimination of K206 results in a low pH independent level of activity, which increases as the pH increases above 8 and Y150 becomes unprotonated and can act as a base in the hydride transfer step (II in Scheme 1), which is almost certainly rate determining for K206M. A water molecule bridging the C2 hydroxyl of HIc and Y150 may be required in this step. On the other hand the Y150F mutant enzyme exhibits behavior that is the mirror image with a low pH independent activity at high pH that increases as the pH decreases below 7. Data suggest the ability of K206 to act as an acid catalyst, donating a proton to form the enol (IV in Scheme 1). As in the case of K206M, this step may be mediated by a water molecule that bridges C2 of α-ketoadipate and K206. There are other conserved residues in the active site, but none appear to be properly positioned to carry out acid-base catalysis. However, the structure available is an apoenzyme and additional structural and mechanistic work is required to test the proposed mechanism.
Acknowledgements
We thank Rong Guan for preparing Figures 2 and 3.
This work was supported by a grant from the Oklahoma Center for the Advancement of Science and Technology to P.F.C. (HR07-016), a grant (GM 071417) from the National Institute of Health (to P. F. C. and A. H. W.), and the Grayce B. Kerr Endowment to the University of Oklahoma (to P. F. C.).
Footnotes
Abbreviations: HIcDH, homoisocitrate dehydrogenase; AAA, α-aminoadipate pathway; α-Ka, α-ketoadipate; NAD, nicotinamide adenine dinucleotide (the (+) charge is omitted for convenience); NADH, reduced nicotinamide adenine dinucleotide; NADD, reduced nicotinamide adenine dinucleotide with deuterium in the 4-R position; TDH, tartrate dehydrogenase; Hepes, N-(2-hydroxyethyl)piperazine-N'-2-ethanesulfonic acid; Ches, 2-(N-cyclohexylamino)-ethanesulfonic acid; Bis-Tris, bis(2-hydroxyethyl) imino-tris(hydroxymethyl) methane; Tris, tris(hydroxymethyl) aminomethane; Ic, isocitrate; KAc, potassium acetate; TMAC, tetramethylammonium chloride; CMP, 3-carboxypropylidenemalate; MgHIc, chelate complex of magnesium and homoisocitrate; D2O, deuterium oxide; DC1, deuterium chloride; NaOD, sodium deuterioxide; SKIE, solvent kinetic isotope effect; wt, wild type HIcDH.
The intrinsic isotope effect on the hydride transfer step is likely >3, the estimated value of D k in the 6-phosphogluconate dehydrogenase reaction (14); the estimated value of the effect in the malic enzyme reaction is about 10 (15). Given a value of 3 for D k, and the general equation for the deuterium isotope effect (D(V/K) = [Dk + (kchem/kconf)]/[1 + (kchem/kconf)]), a value of 10 for kchem/kconf would almost completely suppress the isotope effect. If the value of D k is 10, the value of kchem/kconf would have to be >100.
The Km for the first substrate bound in a rapid equilibrium ordered kinetic mechanism is zero. This results from the ability of high concentrations of B to trap A in central complexes, as long as the concentration of A is slightly greater than the concentration of enzyme.
As suggested in Footnote 2, the values are likely at least an order of magnitude lower than estimated from the observed values. This would give a value of ≥6.6 kcal/mol for K206 and ≥5.1 kcal/mol for Y150.
REFERENCES
- 1.Strassman M, Ceci LN. Enzymatic Formation of α-Ketoadipic Acid from Homoisocitric Acid. J. Biol. Chem. 1965;240:4357–4361. [PubMed] [Google Scholar]
- 2.Karsten WE, Cook PF. Pyridine Nucleotide-Dependent β-Hydroxyacid Oxidative Decarboxylases: An Overview. Pr. & Pep. Ltrs. 2000;7:281–286. [Google Scholar]
- 3.Lin Y, Alguindigue SS, Volkman J, Nicholas KM, West AH, Cook PF. Complete Kinetic Mechanism of Homoisocitrate Dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 2007;46:890–898. doi: 10.1021/bi062067q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin Y, West AH, Cook PF. Potassium Activates Homoisocitrate Dehydrogenase by Increasing the Affinity for NAD. Biochemistry. 2008;47:10809–10815. doi: 10.1021/bi801370h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin Y, Volkman J, Nicholas KM, Yamamoto T, Eguchi T, Nimmo SL, West AH, Cook PF. Chemical Mechanism of Homoisocitrate Dehydrogenase from Saccharomyces cerevisiae. Biochemistry. 2008;47:4169–4180. doi: 10.1021/bi702361j. [DOI] [PubMed] [Google Scholar]
- 6.Karsten WE, Liu DL, Rao GSJ, Harris BG, Cook PF. A Catalytic Triad is Responsible for Acid-Base Chemistry in the Ascaris suum NAD-Malic Enzyme. Biochemistry. 2005;44:3626–3635. doi: 10.1021/bi047826o. [DOI] [PubMed] [Google Scholar]
- 7.Kim TK, Lee P, Colman RF. Critical Role of Lys(212) and Tyr(140) in Porcine NADP-dependent Isocitrate Dehydrogenase. J. Biol. Chem. 2003;278:49323–49331. doi: 10.1074/jbc.M303781200. [DOI] [PubMed] [Google Scholar]
- 8.Lee P, Colman RF. Thr(373), Asp(375), and Lys(260) Are in the Coenzyme Site of Porcine NADP-dependent Isocitrate Dehydrogenase. Arch. Biochem. Biophys. 2006;450:183–190. doi: 10.1016/j.abb.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Karsten WE, Tipton PA, Cook PF. Tartrate Dehydrogenase Catalyzes a Stepwise Oxidative Decarboxylation of D-Malate with NAD+ or thioNAD+ as the Dinucleotide Substrate. Biochemistry. 2002;41:12193–12199. doi: 10.1021/bi026278g. [DOI] [PubMed] [Google Scholar]
- 10.Miyazaki J, Asada K, Fushinobu S, Kuzuyama T, Nishiyama M. Crystal Structure of Tetrameric Homoisocitrate Dehydrogenase from an Extreme Thermophile, Thermus thermophilus: Involvement of Hydrophobic Dimer-Dimer Interaction in Extremely High Thermotolerance. J. Bacteriol. 2005;187:6779–6788. doi: 10.1128/JB.187.19.6779-6788.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ceccarelli C, Grodsky NB, Ariyaratne N, Colman RF, Bahnson BJ. Crystal Structure of Porcine Mitochondrial NADP(+)-dependent Isocitrate Dehydrogenase Complexed with Mn2+ and Isocitrate - Insights Into the Enzyme Mechanism. J. Biol. Chem. 2002;277:43454–43462. doi: 10.1074/jbc.M207306200. [DOI] [PubMed] [Google Scholar]
- 12.Imada K, Inagaki K, Matsunami H, Kawaguchi H, Tanaka H, Tanaka N, Namba K. Structure of 3-Isopropylmalate Dehydrogenase in Complex with 3-Isopropylmalate at 2.0 Angstrom Resolution: the Role of Glu88 in the Unique Substrate Recognition Mechanism. Struct. Fold. Des. 1998;6:971–982. doi: 10.1016/s0969-2126(98)00099-9. [DOI] [PubMed] [Google Scholar]
- 13.Aktas DF, Cook PF. A Lysine-Tyrosine Pair Carries Out Acid-Base Chemistry in the Metal Ion-Dependent Pyridine Dinucleotide–Linked β-Hydroxyacid Oxidative Decarboxylases. Biochemistry. 2009 doi: 10.1021/bi8022976. submitted. [DOI] [PubMed] [Google Scholar]
- 14.Li L, Dworkowski FSN, Cook PF. Import of the 6-Phosphate of in the Binding of 6-Phosphogluconate in the 6-Phosphogluconate Dehydrogenase Reaction. J. Biol. Chem. 2006;281:25568–25576. doi: 10.1074/jbc.M601154200. [DOI] [PubMed] [Google Scholar]
- 15.Karsten WE, Cook PF. Stepwise versus Concerted Oxidative Decarboxylation Catalyzed by Malic Enzyme: A Reinvestigation. Biochemistry. 1994;33:2096–2103. doi: 10.1021/bi00174a016. [DOI] [PubMed] [Google Scholar]
- 16.Nelson RB, Gribble GW. On the Preparation of α-Ketoadipic Acid. Org. Prep. Proceed. Int. 1973;5:55–58. [Google Scholar]
- 17.Viola RE, Cook PF, Cleland WW. Preparation and Purification of Stereospecifically Deuterated (4-2H)-Nicotinamide Adenine Nucleotides. Anal. Biochem. 1979;96:334–340. doi: 10.1016/0003-2697(79)90590-6. [DOI] [PubMed] [Google Scholar]
- 18.Schowen KB, Schowen RL. Solvent Isotope Effects on Enzyme Systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- 19.Quinn DM, Sutton LD. In: In Enzyme Mechanism from Isotope Effects. Cook PF, editor. Boca Raton, FL: CRC press; 1991. pp. 73–126. [Google Scholar]
- 20.Bazelyansky M, Robey E, Kirsch JF. Fractional Diffusion-limited Component of Reactions Catalyzed by Acetylcholinesterase. Biochemistry. 1986;25:125–130. doi: 10.1021/bi00349a019. [DOI] [PubMed] [Google Scholar]
- 21.Cook PF, Cleland WW. Enzyme Kinetics and Mechanism. Garland Press; 2007. [Google Scholar]