Abstract
Objectives
The objective of this study was to compare atazanavir pharmacokinetics in genetically determined CYP3A5 expressors versus non-expressors.
Methods
HIV-negative adult volunteers were pre-screened for CYP3A5 *3, *6 and *7 polymorphisms and enrolment was balanced for CYP3A5 expressor status, gender and race (African-American versus non-African-American). Participants received atazanavir 400 mg daily for 7 days followed by atazanavir/ritonavir 300 mg/100 mg daily for 7 days with pharmacokinetic studies on days 7 and 14. Other measures collected were bilirubin, UGT1A1 *28, and ABCB1 1236C > T, 2677G > T/A and 3435C > T genotypes. Data analyses utilized least squares regression.
Results
Fifteen CYP3A5 expressors and 16 non-expressors participated. The day 7 atazanavir oral clearance (CL/F) was 1.39-fold faster (0.25 versus 0.18 L/h/kg; P = 0.045) and the Cmin was half (87 versus 171 ng/mL; P = 0.044) in CYP3A5 expressors versus non-expressors. Non-African-American CYP3A5 expressor males had 2.1-fold faster CL/F (P = 0.003) and <20% the Cmin (P = 0.0001) compared with non-African-American non-expressor males. No overall CYP3A5 expressor effects were observed during the ritonavir phase. One or two copies of wild-type ABCB1 haplotype (1236C/2677G/3435C) was predictive of slower atazanavir and ritonavir CL/F compared with zero copies (P < 0.06). Indirect bilirubin increased 1.6- to 2.8-fold more in subjects with UGT1A1 *28/*28 versus *1/*28 or *1/*1.
Conclusions
CYP3A5 expressors had faster atazanavir CL/F and lower Cmin than non-expressors. The effect was most pronounced in non-African-American men. Ritonavir lessened CYP3A5 expressor effects. The wild-type ABCB1 CGC haplotype was associated with slower CL/F and the UGT1A1 *28 genotype was associated with increased bilirubin. Thus, CYP3A5, ABCB1 and UGT1A1 polymorphisms are associated with atazanavir pharmacokinetics and pharmacodynamics in vivo.
Keywords: antiretroviral therapy, pharmacogenomics, drug metabolism, clinical pharmacology
Introduction
HIV protease inhibitors are integral components of many combination antiretroviral regimens.1 A key pharmacological feature of protease inhibitors is high pharmacokinetic variability among individuals, with AUC and trough (Cmin) values that can vary by >10-fold in controlled settings.2 Additionally, drug–drug interactions, mis-timed dosing and non-adherence to food restrictions can increase variability considerably. Pharmacokinetic variability can be clinically relevant, as maintenance of plasma concentrations above Cmin thresholds has been proposed as a strategy to maximize the probability of virological response.1
Protease inhibitors, except nelfinavir, are often dosed with ritonavir, which inhibits CYP3A in humans, resulting in higher and more sustained plasma concentrations of the protease inhibitor of interest. Atazanavir is a first-line protease inhibitor with a favourable side effect and tolerability profile.3 Although atazanavir is FDA approved as a single protease inhibitor, dosed 400 mg once daily, it is typically given with ritonavir for the reasons described above.4 Pharmacokinetic enhancement with ritonavir has some disadvantages, however, including metabolic adverse effects such as hyperlipidaemia and hyperglycaemia, as well as gastrointestinal intolerance.5 An increased understanding of the biological, genetic and environmental factors that underlie protease inhibitor pharmacokinetic variability is essential for the most informed and rational use of single and pharmacokinetically enhanced protease inhibitors in patients.
Protease inhibitors (except nelfinavir) are dedicated CYP3A substrates, which includes 3A4 and 3A5 in adults.6,7 Interindividual variability in CYP3A could be an important source of pharmacokinetic variability for protease inhibitors in vivo. CYP3A5 is polymorphically expressed in adults.8 CYP3A5 non-expressors carry two inactive alleles most commonly due to single nucleotide polymorphisms that change the mRNA splicing site (*3 or *6) or a T insertion that changes the reading frame (*7).9 The result for homozygous variant carriers is truncated and negligibly expressed CYP3A5 protein. Approximately 85% of Caucasians and 45% of African-Americans carry at least one inactive allele.9 Previous studies have shown that genetically determined CYP3A5 expression is associated with variability in the metabolism and/or oral clearance of indinavir and saquinavir as single protease inhibitors.10–14 However, studies have not investigated the influence of genetically determined CYP3A5 expression on the pharmacokinetics of atazanavir as a single protease inhibitor.
An additional source of pharmacokinetic variability for protease inhibitors may arise from P-glycoprotein, the product of the ABCB1 gene.15 The P-glycoprotein efflux transporter is expressed in the intestinal brush border, liver and renal tubule cells, thereby counteracting drug absorption and facilitating drug clearance.16 The polymorphisms most often studied in ABCB1 are 1236C > T, 2677G > T/A and 3435C > T, and these polymorphisms are often studied in haplotype form (i.e. the combination of alleles on a single chromosome). The common ABCB1 haplotype, 1236T, 2677T and 3435T (TTT), has been associated with altered protein folding (despite 1236C > T and 3435C > T being synonymous polymorphisms) resulting from slower protein synthesis compared with the wild-type haplotype (CGC). This altered protein shape may change the substrate specificity, resulting in higher or lower P-glycoprotein function depending on the substrate.17,18
A consideration aside from pharmacokinetic variability that is also important for drug response, is pharmacodynamic variability. One clear example for protease inhibitors, including atazanavir, is the inhibition of the bilirubin-conjugating enzyme, uridine-diphosphate glucuronosyl transferase isoform 1A1 (UGT1A1).19 A common polymorphism, UGT1A1 *28 (7 TA repeats in the promoter), is associated with 70% lower activity compared with the wild-type *1 allele (6 TA repeats), which can exaggerate indirect bilirubin increases associated with atazanavir therapy.19,20
The primary objective of this study was to compare the pharmacokinetics of atazanavir in genetically determined CYP3A5 expressors versus non-expressors and to evaluate the effect of pharmacokinetic enhancement with ritonavir on this relationship. Secondary objectives were to evaluate associations between ABCB1 haplotypes and atazanavir pharmacokinetics and between the UGT1A1 *28 genotype and indirect bilirubin increases caused by atazanavir.
Methods
This was a single-arm, open-labelled, two-phase study of prospective CYP3A5 genotyping for atazanavir pharmacokinetics in HIV-negative volunteers. Eligible subjects were between the ages of 18 and 55 years; with negative tests for HIV infection and pregnancy; no medical history of hepatitis B or C or autoimmune disease; no significant electrocardiography (ECG) abnormality or cardiac history; no investigational drug in the previous 30 days or chronic concomitant medication (or herbal product) contraindicated with atazanavir or ritonavir (including antacids); body mass index (BMI) 18.5–34 kg/m2; and no pre-existing haematological or metabolic abnormalities above grade I using the Division of AIDS (DAIDS) adverse event grading criteria. The study was approved by the local Institutional Review Board (IRB) and informed consent was obtained from each participant.
Following informed consent, volunteers were pre-screened for CYP3A5 expressor status by genotyping for *3, *6 and *7. Non-expressor status was assigned to homozygous variants (*3/*3, *6/*6 or *7/*7) and expressor status was assigned to those carrying at least one *1 allele.9 If a subject was a double heterozygote, they were excluded unless haplotype analysis inferred >80% probability that the subject possessed an expressor or non-expressor diplotype. In addition to expressor status, enrolment was balanced by African-American and non-African-American race (by subject self-report) and gender. Non-African-Americans could comprise all other races and ethnicities other than African-American. In addition to the CYP3A5 genotypes described above, the following genotypes were also obtained as part of the study: UGT1A1 *28 (6 > 7 TA repeats) and ABCB1 1236C > T, 2677G > T/A and 3435C > T.
The study had two sequential phases, an atazanavir alone phase followed by an atazanavir plus ritonavir phase. During the atazanavir alone phase, subjects were given a 7 day supply of atazanavir and were instructed to take 400 mg (two 200 mg capsules) by mouth each morning with breakfast. Subjects returned on day 3 of therapy for a laboratory safety assessment and a trough drug concentration. On day 7, a 24 h pharmacokinetics study was performed in the University of Colorado General Clinical Research Center Inpatient Unit following a standardized breakfast (500 kcal; 40% fat; 15% protein; 45% carbohydrates). A 5 mL sample of blood was collected at baseline and 0.5, 1, 2, 3, 5, 8, 12, 18 and 24 h after observed dosing. For the atazanavir plus ritonavir phase, starting on day 8, subjects were given a 7 day supply of atazanavir and ritonavir and were instructed to take 300 mg (two 150 mg capsules) and 100 mg (one 100 mg capsule), respectively, by mouth each morning with breakfast. Subjects returned on day 10 for a laboratory safety assessment and a trough drug concentration, and returned on day 14 for a repeat 24 h pharmacokinetics study. Study medication was counted at each study visit as a measure of adherence. Indirect bilirubin concentrations were monitored at each visit as a drug response marker; increases were expected and not considered a drug toxicity.
Plasma was harvested from blood immediately and stored at −80°C until assayed. Atazanavir and ritonavir plasma concentrations were determined with a simultaneous validated HPLC-ultraviolet (UV) procedure. Briefly, plasma (0.2 mL) was extracted with t-butylmethylether after addition of 0.4 mL of 0.0125 M NaOH and an internal standard (A86093, Abbott Laboratories). Chromatographic separation was accomplished on an YMC Octyl column (100 × 4.6 mm, 3 µm) at 35°C with a mobile phase of acetonitrile/20 mM acetate buffer. The limit of quantification was 20 ng/mL for both compounds and the within-assay and between-assay coefficients of variation for the quality controls during validation were <15%.
Genomic DNA was isolated from buccal cells in mouthwash expectorate using a commercially available kit (QIAamp® DNA Mini Kit; Qiagen, Valencia, CA, USA). Genotypes were determined by PCR-pyrosequencing analysis (PSQ 96MA genotyping system; Qiagen), as previously described.10 Genotypes were determined using PSQ™ 96MA SNP software version 2.0 (Qiagen), which provides automated genotype determinations. All samples were run in duplicate. Haplotypes were computationally assigned using HelixTree® Genetics Analysis Software (Golden Helix, Inc., Bozeman, MT, USA).
Non-compartmental methods were used to calculate pharmacokinetic parameters; the dose interval AUC was calculated with the linear–linear trapezoidal rule, and oral clearance was determined with CL/F = dose (mg/kg)/AUC.21 A value of midway between the lower limit of quantification for the assay and 0 was imputed for concentrations below the limit of quantification. Linear regression using three or more points in the log-linear elimination phase was used to determine the elimination rate constant (kel). Half-life was determined from ln 2/kel. Cmax and Cmin were taken from the raw data.
Statistics and data analyses
The primary endpoint, determined a priori, was to compare atazanavir CL/F (L/h/kg) in CYP3A5 expressors versus non-expressors at day 7. A sample size of 24 (12 expressors and 12 non-expressors) was needed to detect a 40% difference in CL/F, a value based upon experience with indinavir and CYP3A5 expressor status, and using a coefficient of variation of 30% for atazanavir clearance, a power of 80% and a two-sided alpha of 0.05.10,22 Sixteen participants per arm were sought to provide some protections if assumptions were inflated.
Secondary endpoints were to evaluate atazanavir Cmax, Cmin and half-life according to CYP3A5 expressor status at day 7; to evaluate whether ABCB1 haplotypes influenced any of the pharmacokinetic–pharmacodynamic parameters; to analyse atazanavir and ritonavir pharmacokinetic parameters at day 14; and to determine the association between the UGT1A1*28 polymorphism and the maximal indirect bilirubin changes during the atazanavir alone and atazanavir plus ritonavir phases.
Study data were inspected graphically; pharmacokinetic data were log transformed prior to statistical analysis if needed, and back transformed for presentation. ABCB1 genotypes and haplotypes were viewed graphically versus pharmacokinetic data to determine a consistent genotype or haplotype to use across the analyses. Analysis and statistical comparisons utilized Splus version 8.0 (Insightful Corp., Seattle, WA, USA). Primary comparisons of pharmacokinetic outcomes by CYP3A5 expressor status and baseline comparisons were conducted using a two-sided t-test with a significance level of 0.05. Secondary analyses, which included interactions with race and gender and additional genetic predictors, were considered hypothesis generating. Secondary outcome analysis of variance (ANOVA) models were fit using ordinary least squares regression. Other than the overall test, analyses were not adjusted for multiple comparisons (i.e. a Fisher's protected least significant difference approach). For day 7 analyses, a saturated cell means model was initiated and terms were removed in a backward stepwise approach. For consistency, day 14 analyses matched the day 7 predictors. Pharmacokinetic results are presented as geometric means with corresponding 95% confidence intervals (CIs) unless stated otherwise.
Results
Study population
A total of 15 CYP3A5 expressors and 16 non-expressors completed the day 7 primary endpoint visit. By design, 1.5 years of recruiting time was allotted to balance race and gender as much as possible, after which enrolment was opened to any race and gender. The final study population is described in Table 1. The CYP3A5 expressor and non-expressor groups were well balanced by race and gender, and did not differ by age, weight or BMI.
Table 1.
Characteristics of the study population
| Characteristic | CYP3A5 expressors (n = 15) | Non-expressors (n = 16) | P |
|---|---|---|---|
| CYP3A5 genotype (n) | *1*3 (9); *1*1 (4); *1*7 (1); *1*6 (1) | *3*3 (13); *7*7 (2); *1*3 and *1*7 (1)a | — |
| African-American | 7 (3 female) | 6 (4 female) | — |
| Non-African-American | 8 (4 female) | 10 (5 female) | — |
| Hispanic | 3 | 1 | |
| Asian | 1 | 0 | |
| Caucasian | 4 | 9 | |
| UGT1A1 *28 (n) | *1/*1 (5); *1/*28 (6); *28/*28 (4) | *1/*1 (5); *1/*28 (10); *28/*28 (1) | — |
| ABCB1 CGC copies, i.e. 1236C, 2677G and 3435C (n) | 0 copies (5); 1 copy (6); 2 copies (4) | 0 copies (6); 1 copy (7); 2 copies (3) | — |
| Age, years (median, range) | 30 (20–44) | 32 (24–51) | 0.12 |
| Weight, kg (median, range) | 73 (50–97) | 75 (55–106) | 0.78 |
| BMI (median, range) | 25.4 (19.1–31.7) | 24.6 (19.3–28.5) | 0.77 |
aComputational haplotype analysis showed >80% probability that this subject was a non-expressor (see text).
Primary endpoint
The day 7 atazanavir CL/F was 0.25 L/h/kg (0.20–0.30) in CYP3A5 expressors versus 0.18 L/h/kg (0.15–0.22) in non-expressors. This represented a 1.39-fold faster clearance in CYP3A5 expressors versus non-expressors (P = 0.045).
Additional day 7 pharmacokinetics
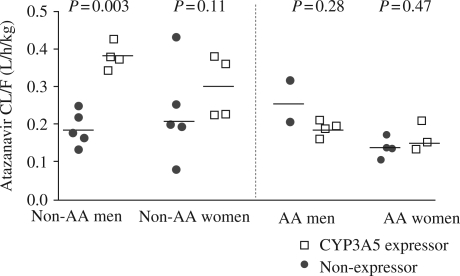
No doses were missed among the CYP3A5 expressor group whereas one individual among the non-expressor group missed one dose of medication prior to day 7, as measured by medication counts. The geometric mean Cmin was 87 ng/mL in CYP3A5 expressors versus 171 ng/mL in non-expressors (1.97-fold difference; P = 0.044). The geometric mean half-life tended to be shorter in CYP3A5 expressors versus non-expressors (5.9 h versus 7.5 h, respectively; P = 0.057). No significant differences were observed in day 7 atazanavir Cmax (P = 0.17) or AUCtau (P = 0.15) in CYP3A5 expressors versus non-expressors. The cell means model indicated that the pharmacokinetic differences were primarily in the non-African-American males, with CL/F values of 0.38 (0.27–0.53) and 0.18 (0.13–0.26) L/h/kg and Cmin values of 24 (14–41) and 131 (83–209) ng/mL in non-African-American male CYP3A5 expressors versus non-African-American male non-expressors, respectively (P < 0.003). Non-African-American women demonstrated a smaller difference with the same directionality in CL/F, 0.29 (0.21–0.40) versus 0.20 (0.15–0.27) L/h/kg in CYP3A5 expressors versus non-expressors, respectively, although this failed to reach statistical significance (P = 0.11). No significant differences in atazanavir pharmacokinetics between CYP3A5 expressors and non-expressors were observed in the African-American males (P = 0.28) or females (P = 0.47). In terms of potential gender differences, women had elevated atazanavir concentrations compared with men (1.5-fold for AUCtau; P = 0.003), but weight-adjusted oral clearance was not different (P = 0.19). The day 7 atazanavir CL/F values according to CYP3A5 expressor status, race and gender are shown in Figure 1.
Figure 1.
Atazanavir CL/F on day 7 sorted by race and gender. Solid lines show cell mean estimates. AA, African-American; non-AA, non-African-American. Atazanavir CL/F differences were most pronounced in non-African-American men.
ABCB1
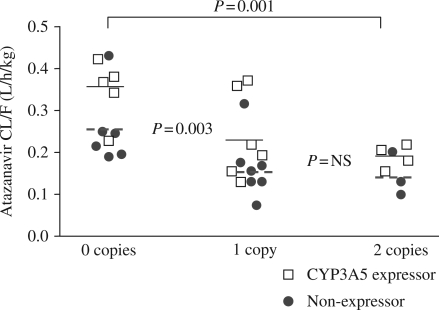
The most consistent ABCB1 findings were associated with the number of wild-type CGC haplotype copies (i.e. 1236C/2677G/3435C). The estimated overall atazanavir CL/F in subjects with zero CGC copies was 0.29 (0.23–0.36) L/h/kg. This value was 1.6- and 1.7-fold faster compared with those carrying one or two CGC copies, respectively; 0.18 (0.15–0.23) and 0.17 (0.13–0.22) L/h/kg; P < 0.007. The effects of CYP3A5 genotype, race and sex on this relationship were considered next. After controlling for the number of CGC copies, race and sex failed to reach statistical significance (P = 0.58 and P = 0.10, respectively). CYP3A5 expressors demonstrated a 1.39-fold faster CL/F compared with non-expressors in each CGC stratum; P = 0.01, Figure 2.
Figure 2.
Atazanavir CL/F on day 7 according to number of CGC copies in the ABCB1 gene (1236C, 2677G, 3435C). Lines show cell mean estimates; solid lines are for CYP3A5 expressors and dashed lines are for non-expressors (see text). Atazanavir CL/F was significantly faster in those with zero CGC copies versus those with one or two CGC copies. CYP3A5 expressors had a faster CL/F than non-expressors in each CGC category. NS, non-significant.
The overall estimated atazanavir Cmin in subjects with zero CGC copies was 66 (39–110) ng/mL. This was significantly lower than the value in those with one and two CGC copies; 159 (99–255) and 209 (110–399) ng/mL, respectively; P < 0.02. In a model that included CGC with the effects of CYP3A5 expressor status and gender (race was not significant), the estimated Cmin was ∼51% lower in CYP3A5 expressors versus non-expressors (P = 0.0062), and women had 2.33-fold higher Cmin compared with men in each CGC stratum (P = 0.0008).
Day 14 pharmacokinetics
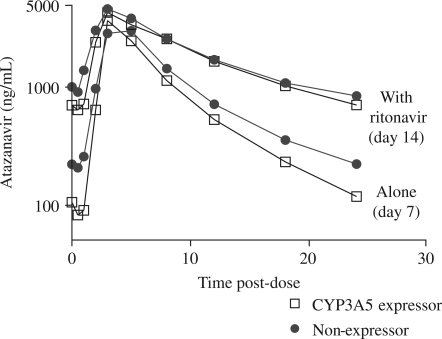
Five subjects withdrew from the study between day 7 and day 14, one expressor and four non-expressors (reasons for withdrawal are described in the Safety and tolerability section). No doses were missed among the 14 CYP3A5 expressors and half of one atazanavir dose (150 mg) was missed in one individual among the 12 non-expressors between days 8 and 14, as measured by medication counts. At the day 14 visit, ritonavir significantly reduced atazanavir CL/F by 61% and raised the Cmin by a factor of 5.7-fold (P < 0.0001). Figure 3 shows the atazanavir concentration–time curves for day 7 (atazanavir alone) and day 14 (atazanavir plus ritonavir) in CYP3A5 expressors and non-expressors.
Figure 3.
Geometric mean atazanavir plasma concentrations for genetically determined CYP3A5 expressors and non-expressors during the atazanavir alone (day 7) and atazanavir plus ritonavir (day 14) phases.
Pharmacokinetic enhancement with ritonavir weakened the association between CYP3A5 expressor status and atazanavir pharmacokinetics. The atazanavir CL/F was 0.09 (0.07–0.11) versus 0.07 (0.06–0.09) L/h/kg in CYP3A5 expressors versus non-expressors at day 14; 1.29-fold, P = 0.15. Additionally, no overall differences were found between CYP3A5 expressors and non-expressors in atazanavir Cmax, Cmin or half-life on day 14. However, the CL/F continued to be faster in the CYP3A5 expressor non-African-American males versus non-expressor non-African-American males; 0.13 (0.10–0.18) versus 0.08 (0.06–0.10) L/h/kg, 1.62-fold difference, P = 0.017, and the corresponding Cmin values were 238 (131–433) versus 570 (313–1038) ng/mL, respectively; 2.39-fold difference, P = 0.047. Atazanavir concentrations continued to be higher in women compared with men (1.4-fold for AUCtau; P = 0.009), but weight-adjusted oral clearance was not different (P = 0.32).
The ritonavir CL/F on day 14 was 0.12 versus 0.13 L/h/kg for CYP3A5 expressors and non-expressors, respectively, P = 0.75. Similar to atazanavir, ritonavir CL/F was faster in the CYP3A5 expressor non-African-American males versus non-expressor non-African-American males; 0.19 (0.12–0.30) versus 0.10 (0.06–0.15) L/h/kg, 1.90-fold difference; P = 0.036.
ABCB1
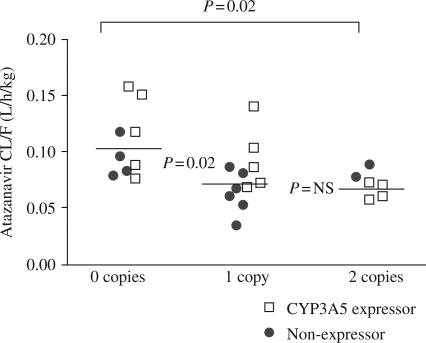
The number of copies of the ABCB1 CGC haplotype continued to be associated with atazanavir pharmacokinetics at day 14 although the effects were attenuated compared with the day 7 results. The overall atazanavir CL/F in subjects with zero CGC copies was 0.1 (0.08–1.3) L/h/kg, which was 1.4- and 1.5-fold faster compared with those carrying one or two CGC copies; 0.073 (0.060–0.88) and 0.070 (0.054–0.091), respectively, P < 0.03. After controlling for the number of CGC copies, there was no longer a CYP3A5 genotype effect on this relationship; P = 0.11 (Figure 4), and race and sex also failed to reach statistical significance (P = 0.86 and P = 0.21, respectively).
Figure 4.
Atazanavir CL/F on day 14 according to number of CGC copies in the ABCB1 gene (1236C, 2677G, 3435C). Solid lines show cell mean estimates. Atazanavir CL/F was significantly faster in those with zero CGC copies versus those with one or two CGC copies, but CYP3A5 expressor status was not predictive of CL/F in this analysis.
Atazanavir Cmin values were not significantly different according to CGC copies at day 14. However, women had 2.21-fold higher atazanavir Cmin across all numbers of CGC copy strata compared with men (P = 0.003).
Finally, overall ritonavir CL/F was ∼1.9-fold faster in subjects with zero versus two CGC haplotype copies; 0.17 (0.12–0.22) versus 0.087 (0.06–0.13), respectively; P = 0.009, and was 1.47-fold faster in subjects with zero versus one CGC copy; P = 0.059. A summary of all pharmacokinetic parameters collected in the study, stratified by genetically determined CYP3A5 expressor status, is provided in Table 2.
Table 2.
Geometric mean (95% CI) atazanavir and ritonavir pharmacokinetics sorted by CYP3A5 expressor status
| CYP3A5 expressors (n = 15) | Non-expressors (n = 16) | P | |
|---|---|---|---|
| Atazanavir alone day 7 | |||
| CL/F (L/h/kg)a | 0.25 (0.20–0.30) | 0.18 (0.15–0.22) | 0.045 |
| t1/2 (h) | 5.89 (4.95–7.01) | 7.54 (6.18–9.21) | 0.057 |
| AUCtau (h*ng/mL) | 22 151 (17 996–27 265) | 27 400 (22 407–33 504) | 0.15 |
| Cmax (ng/mL) | 3652 (2995–4453) | 4430 (3655–5368) | 0.16 |
| Cmin (ng/mL) | 87 (55–140) | 171 (109–270) | 0.044 |
| day 3 trough (ng/mL) | 108 (59–199) | 194 (92–410) | 0.21 |
| CYP3A5 expressors (n = 14) | Non-expressors (n = 12) | P | |
| Atazanavir (plus ritonavir) day 14 | |||
| CL/F (L/h/kg) | 0.09 (0.07–0.11) | 0.07 (0.06–0.09) | 0.15 |
| t1/2 (h) | 10.4 (8.5–12.8) | 13.5 (8.9–20.4) | 0.22 |
| AUCtau (h*ng/mL) | 46 022 (38 085–55 613) | 49 598 (40 426–60 850) | 0.58 |
| Cmax (ng/mL) | 4609 (4051–5244) | 4883 (4248–5613) | 0.54 |
| Cmin (ng/mL) | 605 (406–900) | 722 (470–1109) | 0.54 |
| day 10 trough (ng/mL) | 870 (537–1411) | 1159 (870–1544) | 0.30 |
| Ritonavir | |||
| CL/F (L/h/kg) | 0.12 (0.09–0.16) | 0.13 (0.09–0.17) | 0.75 |
| t1/2 (h) | 4.05 (3.48–4.71) | 4.54 (3.83–5.38) | 0.28 |
| AUCtau (h*ng/mL) | 11 706 (8657–15 832) | 11 580 (8757–15 313) | 0.95 |
| Cmax (ng/mL) | 1717 (1329–2220) | 1648 (1249–2176) | 0.82 |
| Cmin (ng/mL) | 34 (21–55) | 42 (30–58) | 0.49 |
Bold formatting indicates significant values.
aPrimary endpoint.
Pharmacodynamics: UGT1A1
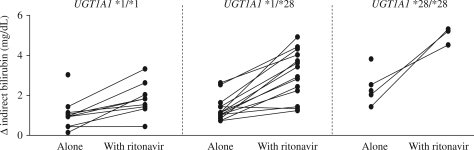
The maximum increase in indirect bilirubin was calculated for each subject in the atazanavir alone and atazanavir plus ritonavir phases. The mean increase was 1.19 (0.93–1.45) mg/dL for the atazanavir alone phase and 2.83 (2.25–3.4) mg/dL for the atazanavir plus ritonavir phase (P < 0.0001). During the atazanavir alone phase, the increase was 1.01 (0.55–1.47) mg/dL for those with UGT1A1 *1/*1, 1.23 (0.86–1.59) mg/dL for those with UGT1A1 *1/*28 and 2.38 (1.73–3.03) mg/dL for those with UGT1A1 *28/*28 (the comparisons between *28/*28 and the other two groups were significantly different; P < 0.003). CYP3A5 expressor status was not associated with bilirubin increases. Other factors that were associated with bilirubin increases at day 7 include the presence of zero versus two copies of the ABCB1 CGC haplotype, 0.95 (0.46–1.43) versus 1.79 (1.18–2.40) mg/dL, respectively (P = 0.036), and non-African-American versus African-American race, 1.04 (0.68–1.41) versus 1.75 (1.19–2.32) mg/dL, respectively (P = 0.016). For the atazanavir plus ritonavir phase, the increase was 1.79 (1.08–2.50) mg/dL for those with UGT1A1 *1/*1, 3.04 (2.46–3.61) mg/dL for those with UGT1A1 *1/*28 and 5.0 (3.76–6.24) mg/dL for those with UGT1A1 *28/*28 (the three groups were significantly different from one another; P < 0.01). Race and ABCB1 haplotype were not associated with bilirubin increases in the ritonavir phase. The indirect bilirubin changes according to UGT1A1 genotypes are summarized in Figure 5.
Figure 5.
Change in indirect bilirubin observed in the atazanavir alone and atazanavir plus ritonavir phases sorted by UGT1A1 genotypes.
Safety and tolerability
All 31 subjects completed the day 7 primary endpoint visit; 26 of the 31 completed the full 14 days of the study. Five subjects discontinued study drug between the day 10 and 14 visits. One subject experienced a grade I rash and withdrew voluntarily, one experienced a grade I elevation in aspartate aminotransferase (AST) and was removed by the investigators, two experienced grade II laboratory abnormalities (albumin and amylase) and were removed per protocol, and one was removed with a viral syndrome unrelated to study drug. No other laboratory adverse events above grade I were observed.
The most common clinical adverse events included mild stomach upset, fatigue/malaise and headache. Less common complaints (all mild) included pruritus, rash, jaundice and icteric sclera.
Discussion
In this study, subjects were pre-screened and enrolled based on genetically determined CYP3A5 expression, gender and African-American versus non-African-American race, and atazanavir pharmacokinetics were compared in CYP3A5 expressors versus non-expressors. During the atazanavir alone phase, atazanavir CL/F was 1.39-fold faster and the Cmin approximately half in CYP3A5 expressors compared with non-expressors. This CYP3A5 expressor effect was apparent in non-African-Americans, but not in African-Americans. The CYP3A5 expressor relationships did not persist overall when atazanavir was pharmacokinetically enhanced by ritonavir, but did persist in the non-African-American men. The wild-type ABCB1 haplotype (1236C/2677G/3435C) was independently associated with atazanavir pharmacokinetics on day 7 (atazanavir alone) and day 14 (atazanavir plus ritonavir); subjects with zero CGC copies had faster atazanavir CL/F and lower Cmin compared with individuals with one or two CGC copies. Finally, persons carrying two UGT1A1 *28 [(TA)7] alleles had 1.6- to 2.8-fold higher elevations in indirect bilirubin compared with those with *1/*28 or *1/*1 genotypes.
Previous studies found ∼1.4-fold faster indinavir CL/F and 1.5- to 1.7-fold faster saquinavir CL/F in CYP3A5 expressors compared with non-expressors when both were given as single protease inhibitors.10,12–14 The present study provides evidence that genetically determined CYP3A5 expression also influences atazanavir disposition when given as a single protease inhibitor. Of note, there appears to be a consistent effect size of ∼1.4- to 1.7-fold faster CL/F in CYP3A5 expressors compared with non-expressors among the studies. However, the present study could not replicate the faster CL/F observed in CYP3A5 expressor African-Americans such as was observed for indinavir and saquinavir as single protease inhibitors.10,12 In fact, this study observed the largest CYP3A5 expressor effects in the non-African-American males. CYP3A5 expression and the number of ABCB1 CGC copies remained as significant predictors of atazanavir CL/F after controlling for race and sex, suggesting that ABCB1 CGC copies may partly explain potential racial differences. However, the study was not adequately designed to address this possibility due to small subject numbers in the race strata, and other racial differences, not measured in this study, may also be important for atazanavir pharmacokinetics. Examples may include variability in the pregnane X receptor (PXR), which controls transcription of CYP3A and ABCB1, the CYP3A4 *1B genotype, and other influx or efflux transporters such as SLCO1B1, SLCO1A2, ABCC1 (MRP1), ABCC2 (MRP2) and ABCG2 (BCRP).23–27 The present study found higher atazanavir concentrations in women versus men, a finding described by others,28 but women did not have lower weight-adjusted CL/F.
The present study also adds to the evidence that pharmacokinetic enhancement with ritonavir minimizes the effects of CYP3A5 expression on protease inhibitor pharmacokinetics, although in this study the pharmacokinetic differences between expressors and non-expressors during the ritonavir phase were still apparent in non-African-American males. Several studies showed no difference in indinavir or lopinavir pharmacokinetics in CYP3A5 expressors versus non-expressors when pharmacokinetically enhanced with ritonavir.13,29,30 These findings are consistent with in vitro studies that show ritonavir inhibits both CYP3A4 and 3A5, which may largely negate the effects of CYP3A5 expression in vivo.7 It is not clear why the expressor effects were maintained in non-African-American males in the present study. It is possible these subjects exhibit biological differences other than CYP3A5 expression that persist after pharmacokinetic enhancement with ritonavir, or that large CYP3A5 expression-mediated differences can persist even in the face of ritonavir. Additional studies are needed to address these possibilities.
Multiple studies have evaluated the association between ABCB1 genotypes and haplotypes with HIV protease inhibitor pharmacokinetics and pharmacodynamics.10,13,16,19,31–39 The findings for protease inhibitors, and many other putative P-glycoprotein substrates, have been inconsistent.16,40 One study pre-screened HIV-negative subjects for the ABCB1 3435C > T polymorphism and recruited 15 T/T and 15 C/C homozygotes for a comparison of saquinavir and saquinavir/ritonavir pharmacokinetics.39 No pharmacokinetic differences were observed, including in subanalyses of 1236C > T, 2677G > T/A and 3435C > T haplotype combinations. Numerous other studies have evaluated 3435C > T and 2677G > T/A, as well as other ABCB1 genotypes, for associations with protease inhibitor concentrations (both plasma and cellular) and therapeutic effects in HIV-infected individuals. In some cases, relationships were not evident,35,38,39 in other cases carriers of C or G in 3435 or 2677 (wild-type) had faster clearances or lower drug concentrations10,37 and in other cases slower clearances or higher drug concentrations.19,33,36
The present study found slower CL/F and higher atazanavir concentrations, both for atazanavir alone and for atazanavir plus ritonavir, in subjects with one or two copies of the wild-type CGC haplotype (1236C/2677G/3435C). These findings are consistent with two previous studies that found higher atazanavir concentrations, both for atazanavir alone and for atazanavir with ritonavir, in subjects with the C/C genotype at 3435 versus those carrying the T variant at this position.19,36 Together, these findings suggest that the wild-type allele may be associated with lower P-glycoprotein activity for atazanavir in humans. This may seem counterintuitive, as the variant is often associated with lower activity. However, the T variant in 2677 encodes an Ala893Ser in exon 21, which was associated with increased activity in one in vitro study.41 Another study showed that the T variant in 2677 is associated with higher basal CYP3A4 expression and activity.42 Finally, some investigators raise the possibility that protein folding differences with the variant haplotype (1236T, 2677T, 3435T) could lead to enhanced P-glycoprotein function, depending on the substrate.17 These investigations are consistent with the findings in the present study. In summary, the pharmacology of P-glycoprotein for HIV protease inhibitors should remain a research priority, but future studies should strive to expand the number of genetic factors studied so a more complete and consistent pharmacological profile can be elucidated.
The present study adds to previous studies showing that persons carrying two UGT1A1 *28 [(TA)7] alleles had a higher risk of elevated indirect bilirubin following atazanavir therapy.19,43 Persons with *28/*28 alleles had 1.6- to 2.8-fold higher elevations in indirect bilirubin compared with *1/*28 and *1/*1 carriers for both atazanavir alone and atazanavir plus ritonavir. Additionally, indirect bilirubin increases were higher in those with two versus zero copies of ABCB1 CGC haplotypes on day 7. Pharmacokinetic enhancement with ritonavir caused ∼2-fold higher indirect bilirubin elevations versus atazanavir alone. These findings illustrate how pharmacokinetic and pharmacodynamic factors can together influence drug response.
Atazanavir is currently used with ritonavir in most HIV-infected individuals to provide higher atazanavir concentrations thereby minimizing the risks associated with low drug concentrations from pharmacokinetic variability, drug–drug interactions (e.g. tenofovir) or dosing without a meal. The currently recommended Cmin for atazanavir in individuals with wild-type HIV-1 is 150 ng/mL.1 However, this value is stringent given that the atazanavir geometric mean Cmin in HIV-infected individuals is 120 ng/mL and it is proven efficacious as a single protease inhibitor in antiretroviral-naïve individuals.22 A Cmin threshold of ≥60 ng/mL has been used as one drug exposure criterion (in addition to AUC) in paediatric dose-finding studies, a value that may better represent the lowest acceptable bound in antiretroviral-naive individuals.44 In the present study, when atazanavir was administered alone, 6 of 15 (40%) of the CYP3A5 expressor subjects had Cmin values ≤60 ng/mL versus only 1 of 16 non-expressors (6%). All four non-African-American CYP3A5 expressor males had values <60 ng/mL (range, 20–33 ng/mL). Thus, genetically determined CYP3A5 expressor status may be useful for determining which individuals would derive the greatest benefit from pharmacokinetic enhancement with ritonavir regardless of concomitant medications. Using information in addition to CYP3A5 expressor status, such as ABCB1 and other transporter data, may one day help predict individuals who do not need pharmacokinetic enhancement with ritonavir even with small drug–drug interactions such as with tenofovir. Such possibilities require validation prior to implementation, but illustrate the potential clinical utility of using genetic information to improve our understanding of protease inhibitor pharmacology in vivo.
The most important limitations in this study were the small sample sizes in the race and ethnicity subgroups including imperfect balance; the use of self-report to define race and ethnicity; and the lack of genetic information regarding other possibly important enzymes and transporters such as PXR, CYP3A4 *1B, SLCO1B1, SLCO1A2, ABCC1 (MRP1), ABCC2 (MRP2) and ABCG2 (BCRP).23–27
In conclusion, this study found that CYP3A5, ABCB1 and UGT1A1 polymorphisms were associated with atazanavir pharmacokinetics and pharmacodynamics in vivo. Further studies are warranted to increase our understanding of protease inhibitor clinical pharmacology with the goal of informing the most rational use of protease inhibitors in patients.
Funding
This research was supported by Grants R03 AI68438 (P. L. A.) and the General Clinical Research Center Grant RR000051 from the National Institutes of Health, and Bristol-Myers Squibb (P. L. A.).
Transparency declarations
This work was conceived, carried out and written by the authors without influence from the sponsors. The authors have no conflicts of interest to report.
Acknowledgements
We wish to thank Mr Dallas Hill and Drs Elizabeth Connick, Courtney V. Fletcher, Jason Hindman, Christina Lai and Kenny Utz for assistance in the conduct of the study, and the study subjects for volunteering to participate.
References
- 1.Panel on Antiretroviral Guidelines for Adults and Adolescents. Guidelines for the Use of Antiretroviral Agents in HIV-1-infected Adults and Adolescents. Department of Health and Human Services. 3 November 2008; 1–139. Available at: http://www.aidsinfo.nih.gov/ContentFiles/AdultandAdolescentGL.pdf. (30 April 2009, date last accessed)
- 2.Acosta EP, Henry K, Baken L, et al. Indinavir concentrations and antiviral effect. Pharmacotherapy. 1999;19:708–12. doi: 10.1592/phco.19.9.708.31544. [DOI] [PubMed] [Google Scholar]
- 3.Havlir DV, O'Marro SD. Atazanavir: new option for treatment of HIV infection. Clin Infect Dis. 2004;38:1599–604. doi: 10.1086/420932. [DOI] [PubMed] [Google Scholar]
- 4.Horberg M, Klein D, Hurley L, et al. Efficacy and safety of ritonavir-boosted and unboosted atazanavir among antiretroviral-naive patients. HIV Clin Trials. 2008;9:367–74. doi: 10.1310/hct0906-367. [DOI] [PubMed] [Google Scholar]
- 5.Shafran SD, Mashinter LD, Roberts SE. The effect of low-dose ritonavir monotherapy on fasting serum lipid concentrations. HIV Med. 2005;6:421–5. doi: 10.1111/j.1468-1293.2005.00328.x. [DOI] [PubMed] [Google Scholar]
- 6.Flexner C. Dual protease inhibitor therapy in HIV-infected patients: pharmacologic rationale and clinical benefits. Annu Rev Pharmacol Toxicol. 2000;40:649–74. doi: 10.1146/annurev.pharmtox.40.1.649. [DOI] [PubMed] [Google Scholar]
- 7.Koudriakova T, Iatsimirskaia E, Utkin I, et al. Metabolism of the human immunodeficiency virus protease inhibitors indinavir and ritonavir by human intestinal microsomes and expressed cytochrome P4503A4/3A5: mechanism-based inactivation of cytochrome P4503A by ritonavir. Drug Metab Dispos. 1998;26:552–61. [PubMed] [Google Scholar]
- 8.Kuehl P, Zhang J, Lin Y, et al. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 9.Lamba JK, Lin YS, Schuetz EG, et al. Genetic contribution to variable human CYP3A-mediated metabolism. Adv Drug Deliv Rev. 2002;54:1271–94. doi: 10.1016/s0169-409x(02)00066-2. [DOI] [PubMed] [Google Scholar]
- 10.Anderson PL, Lamba J, Aquilante CL, et al. Pharmacogenetic characteristics of indinavir, zidovudine, and lamivudine therapy in HIV-infected adults: a pilot study. J Acquir Immune Defic Syndr. 2006;42:441–9. doi: 10.1097/01.qai.0000225013.53568.69. [DOI] [PubMed] [Google Scholar]
- 11.Frohlich M, Hoffmann MM, Burhenne J, et al. Association of the CYP3A5 A6986G (CYP3A5*3) polymorphism with saquinavir pharmacokinetics. Br J Clin Pharmacol. 2004;58:443–4. doi: 10.1111/j.1365-2125.2004.02159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Josephson F, Allqvist A, Janabi M, et al. CYP3A5 genotype has an impact on the metabolism of the HIV protease inhibitor saquinavir. Clin Pharmacol Ther. 2007;81:708–12. doi: 10.1038/sj.clpt.6100117. [DOI] [PubMed] [Google Scholar]
- 13.Solas C, Simon N, Drogoul MP, et al. Minimal effect of MDR1 and CYP3A5 genetic polymorphisms on the pharmacokinetics of indinavir in HIV-infected patients. Br J Clin Pharmacol. 2007;64:353–62. doi: 10.1111/j.1365-2125.2007.02903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mouly SJ, Matheny C, Paine MF, et al. Variation in oral clearance of saquinavir is predicted by CYP3A5*1 genotype but not by enterocyte content of cytochrome P450 3A5. Clin Pharmacol Ther. 2005;78:605–18. doi: 10.1016/j.clpt.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 15.Choo EF, Leake B, Wandel C, et al. Pharmacological inhibition of P-glycoprotein transport enhances the distribution of HIV-1 protease inhibitors into brain and testes. Drug Metab Dispos. 2000;28:655–60. [PubMed] [Google Scholar]
- 16.Marzolini C, Paus E, Buclin T, et al. Polymorphisms in human MDR1 (P-glycoprotein): recent advances and clinical relevance. Clin Pharmacol Ther. 2004;75:13–33. doi: 10.1016/j.clpt.2003.09.012. [DOI] [PubMed] [Google Scholar]
- 17.Fung KL, Gottesman MM. A synonymous polymorphism in a common MDR1 (ABCB1) haplotype shapes protein function. Biochim Biophys Acta. 2009;1794:860–71. doi: 10.1016/j.bbapap.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimchi-Sarfaty C, Oh JM, Kim IW, et al. A ‘silent’ polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguez-Novoa S, Martin-Carbonero L, Barreiro P, et al. Genetic factors influencing atazanavir plasma concentrations and the risk of severe hyperbilirubinemia. AIDS. 2007;21:41–6. doi: 10.1097/QAD.0b013e328011d7c1. [DOI] [PubMed] [Google Scholar]
- 20.Perera MA, Innocenti F, Ratain MJ. Pharmacogenetic testing for uridine diphosphate glucuronosyltransferase 1A1 polymorphisms: are we there yet? Pharmacotherapy. 2008;28:755–68. doi: 10.1592/phco.28.6.755. [DOI] [PubMed] [Google Scholar]
- 21.Gibaldi M, Perrier D. Pharmacokinetics. New York: Marcel Dekker; 1982. [Google Scholar]
- 22.Reyataz® (atazanavir sulfate) Full Prescribing Information. Princeton, NJ: Bristol-Myers Squibb Co; April 2009. [Google Scholar]
- 23.Siccardi M, D'Avolio A, Baietto L, et al. Association of a single-nucleotide polymorphism in the pregnane X receptor (PXR 63396C→T) with reduced concentrations of unboosted atazanavir. Clin Infect Dis. 2008;47:1222–5. doi: 10.1086/592304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kwan WS, Hartkoorn RC, Salcedo-Sora E, et al. Determining the substrate specificities of SLCO1A2 and SLCO1B1 for antiretroviral drugs. Ninth International Workshop on Clinical Pharmacology of HIV Therapy; 2008; New Orleans, LA, USA. (available at: http://www.hivpresentation.com/index.cfm?vID=2E505C46-423A-F6F7-CAB7A0F1B7B0A5EC ; 19 August 2009, date last accessed) [Google Scholar]
- 25.Shallcross V, Hartkoorn RC, Egan D, et al. Influence of SLCO1B1 521T>C polymorphism on lopinavir plasma concentrations from the Liverpool TDM registry. Ninth International Workshop on Clinical Pharmacology of HIV Therapy; 2008; New Orleans, LA, USA. (available at: http://www.hivpresentation.com/index.cfm?vID=2E4FB25A-423A-F6F7-CAFE82CB53903B4E ; 19 August 2009, date last accessed) [Google Scholar]
- 26.Wandel C, Witte JS, Hall JM, et al. CYP3A activity in African American and European American men: population differences and functional effect of the CYP3A4*1B5′-promoter region polymorphism. Clin Pharmacol Ther. 2000;68:82–91. doi: 10.1067/mcp.2000.108506. [DOI] [PubMed] [Google Scholar]
- 27.Janneh O, Owen A, Chandler B, et al. Modulation of the intracellular accumulation of saquinavir in peripheral blood mononuclear cells by inhibitors of MRP1, MRP2, P-gp and BCRP. AIDS. 2005;19:2097–102. doi: 10.1097/01.aids.0000194793.36175.40. [DOI] [PubMed] [Google Scholar]
- 28.King JR, Kakuda TN, Paul S, et al. Pharmacokinetics of saquinavir with atazanavir or low-dose ritonavir administered once daily (ASPIRE I) or twice daily (ASPIRE II) in seronegative volunteers. J Clin Pharmacol. 2007;47:201–8. doi: 10.1177/0091270006296763. [DOI] [PubMed] [Google Scholar]
- 29.Wyen C, Fuhr U, Frank D, et al. Effect of an antiretroviral regimen containing ritonavir boosted lopinavir on intestinal and hepatic CYP3A, CYP2D6 and P-glycoprotein in HIV-infected patients. Clin Pharmacol Ther. 2008;84:75–82. doi: 10.1038/sj.clpt.6100452. [DOI] [PubMed] [Google Scholar]
- 30.Estrela RC, Santoro AB, Barroso PF, et al. CYP3A5 genotype has no impact on plasma trough concentrations of lopinavir and ritonavir in HIV-infected subjects. Clin Pharmacol Ther. 2008;84:205–7. doi: 10.1038/clpt.2008.12. [DOI] [PubMed] [Google Scholar]
- 31.Colombo S, Soranzo N, Rotger M, et al. Influence of ABCB1, ABCC1, ABCC2, and ABCG2 haplotypes on the cellular exposure of nelfinavir in vivo. Pharmacogenet Genomics. 2005;15:599–608. doi: 10.1097/01.fpc.0000172241.42546.d3. [DOI] [PubMed] [Google Scholar]
- 32.de la Tribonniere X, Broly F, Deuffic-Burban S, et al. ABCB1 allele polymorphism is associated with virological efficacy in naive HIV-infected patients on HAART containing nonboosted PIs but not boosted PIs. HIV Clin Trials. 2008;9:192–201. doi: 10.1310/hct0903-192. [DOI] [PubMed] [Google Scholar]
- 33.Fellay J, Marzolini C, Meaden ER, et al. Response to antiretroviral treatment in HIV-1-infected individuals with allelic variants of the multidrug resistance transporter 1: a pharmacogenetics study. Lancet. 2002;359:30–6. doi: 10.1016/S0140-6736(02)07276-8. [DOI] [PubMed] [Google Scholar]
- 34.Haas DW, Smeaton LM, Shafer RW, et al. Pharmacogenetics of long-term responses to antiretroviral regimens containing efavirenz and/or nelfinavir: an Adult Aids Clinical Trials Group Study. J Infect Dis. 2005;192:1931–42. doi: 10.1086/497610. [DOI] [PubMed] [Google Scholar]
- 35.Ma Q, Brazeau D, Zingman BS, et al. Multidrug resistance 1 polymorphisms and trough concentrations of atazanavir and lopinavir in patients with HIV. Pharmacogenomics. 2007;8:227–35. doi: 10.2217/14622416.8.3.227. [DOI] [PubMed] [Google Scholar]
- 36.Rodriguez Novoa S, Barreiro P, Rendon A, et al. Plasma levels of atazanavir and the risk of hyperbilirubinemia are predicted by the 3435C→T polymorphism at the multidrug resistance gene 1. Clin Infect Dis. 2006;42:291–5. doi: 10.1086/499056. [DOI] [PubMed] [Google Scholar]
- 37.Saitoh A, Singh KK, Powell CA, et al. An MDR1-3435 variant is associated with higher plasma nelfinavir levels and more rapid virologic response in HIV-1 infected children. AIDS. 2005;19:371–80. doi: 10.1097/01.aids.0000161766.13782.2f. [DOI] [PubMed] [Google Scholar]
- 38.Winzer R, Langmann P, Zilly M, et al. No influence of the P-glycoprotein polymorphisms MDR1 G2677T/A and C3435T on the virological and immunological response in treatment naive HIV-positive patients. Ann Clin Microbiol Antimicrob. 2005;4:3. doi: 10.1186/1476-0711-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.la Porte CJ, Li Y, Beique L, et al. The effect of ABCB1 polymorphism on the pharmacokinetics of saquinavir alone and in combination with ritonavir. Clin Pharmacol Ther. 2007;82:389–95. doi: 10.1038/sj.clpt.6100157. [DOI] [PubMed] [Google Scholar]
- 40.Chinn LW, Kroetz DL. ABCB1 pharmacogenetics: progress, pitfalls, and promise. Clin Pharmacol Ther. 2007;81:265–9. doi: 10.1038/sj.clpt.6100052. [DOI] [PubMed] [Google Scholar]
- 41.Kim RB, Leake BF, Choo EF, et al. Identification of functionally variant MDR1 alleles among European Americans and African Americans. Clin Pharmacol Ther. 2001;70:189–99. doi: 10.1067/mcp.2001.117412. [DOI] [PubMed] [Google Scholar]
- 42.Lamba J, Strom S, Venkataramanan R, et al. MDR1 genotype is associated with hepatic cytochrome P450 3A4 basal and induction phenotype. Clin Pharmacol Ther. 2006;79:325–38. doi: 10.1016/j.clpt.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 43.Rotger M, Taffe P, Bleiber G, et al. Gilbert syndrome and the development of antiretroviral therapy-associated hyperbilirubinemia. J Infect Dis. 2005;192:1381–6. doi: 10.1086/466531. [DOI] [PubMed] [Google Scholar]
- 44.Rutstein R, Samson P, Kiser J, et al. The PACTG 1020A Protocol: atazanavir with or without ritonavir in HIV-infected infants, children, and adolescents. Abstracts of the Fourteenth Conference on Retroviruses and Opportunistic Infections; 2007; Los Angeles, CA, USA. Abstract 715 (available at: http://www.retroconference.org/2007/Abstracts/28255.htm ; 19 August 2009, date last accessed) [Google Scholar]