Abstract
Objectives
To characterize isolates of Klebsiella pneumoniae producing KPC carbapenemase (KPC-Kp) associated with an outbreak in a long-term acute care hospital (LTACH) in South Florida.
Methods
During 21 March to 20 April 2008, 241 K. pneumoniae isolates detected at Integrated Regional Laboratories (Ft. Lauderdale, FL) for which the ertapenem MICs were ≥4 mg/L were studied. PCR, cloning and sequence analysis were used to detect blaKPC and to characterize the β-lactamase and outer membrane proteins (Omps). The expression level of KPC enzymes was studied by immunoblotting. Genetic relatedness of isolates was investigated with rep-PCR and PFGE. Clinical records of patients were investigated.
Results
Seven KPC-Kp strains were isolated from different patients located at a single LTACH, with a further three isolates being recovered from patients at different hospitals. All KPC-Kp isolates in patients from the LTACH and from one hospital patient were genetically related and shared PFGE patterns that clustered with known sequence type (ST) 258 strains. These strains were highly resistant to carbapenems (MICs ≥ 32 mg/L) due to an increased level of KPC expression and loss of Omps. Rectal colonization was documented in all LTACH patients with KPC-Kp isolates. Treatment failures were common (crude mortality rate of 69%). Active surveillance and enhanced infection control practices terminated the KPC-Kp outbreak.
Conclusions
The detection of KPC-Kp in an LTACH represents a serious infection control and therapeutic challenge in a new clinical setting. The speed at which the epidemic of KPC-Kp is spreading in our healthcare system mandates urgent action.
Keywords: LTCF, porins, carbapenemases, Enterobacteriaceae, outbreak
Introduction
The global spread of Gram-negative pathogens producing KPC carbapenemases represents a potential clinical threat with devastating effects on patient outcomes.1 Since the first detection of a blaKPC-positive Klebsiella pneumoniae (KPC-Kp) isolate in 1996 in North Carolina, KPC β-lactamases have been identified in many Enterobacteriaceae and Pseudomonas spp.2 However, K. pneumoniae still remains the most common species possessing these clinically important enzymes.3 To date, KPC-Kp isolates have been responsible for many hospital outbreaks on the east coast of the USA, in Israel and in Greece.3–6 Sporadic KPC-Kp isolates have also been detected in Canada, South America, China, western Europe and India.6–10
Long-term care facilities (LTCFs) are nursing homes that take care of stable patients for long periods of time, sometimes serving as the patients' lifetime residences. In contrast, long-term acute care hospitals (LTACHs) administer care to patients with more complex medical problems, multiple co-morbidities (including respiratory failure that requires weaning off a mechanical ventilator) and acute medical needs.11 Initially, KPC-Kp strains appeared to be limited to causing hospital-acquired infections. Now, KPC-Kp isolates are being described in LTCFs.12 Despite excessive antibiotic use in LTACHs, KPC-Kp isolates have yet to be reported from these facilities. As LTACHs represent an increasingly important component of the healthcare network in the USA,11,13,14 the occurrence of KPC-Kp isolates in this setting is very worrisome.
According to the CLSI interpretative criteria, the majority of KPC-Kp isolates do not manifest full resistance to carbapenems (e.g. imipenem and meropenem MICs of 2–8 mg/L).3,15 However, Endimiani et al. recently reported that carbapenem MICs were elevated (i.e. imipenem MICs of >64 mg/L) for approximately 20% of KPC-Kp strains detected in the USA.4 We hypothesized that the higher MIC values for this subpopulation may be due to either: (i) increased levels of expression of the KPC β-lactamase; or (ii) reduced penetration of the antibiotics into the bacterial periplasmic space. With regards to the latter possibility, we suspected that the three main outer membrane protein (Omp) channels of K. pneumoniae (i.e. OmpK-35, -36 and -37) play an important role in β-lactam antibiotic susceptibility, including that of carbapenems.16,17 To our knowledge, the impact on susceptibility to carbapenems of KPC expression levels combined with changes in the background of OmpKs has not been extensively explored among KPC-Kp isolates.2,18
In the present work, we communicate the first description of genetically related KPC-Kp isolates recovered from patients residing in an LTACH. We further demonstrate their genetic similarity to the dominant strain observed throughout the USA.4,6 In addition, molecular characterization of strains demonstrates that the high-level resistance to carbapenems (i.e. MICs ≥ 32 mg/L) is due to the combination of increased expression levels of KPC β-lactamase and loss of the three OmpKs. The convergence of these clinical, epidemiological and genetic determinants emphasizes the importance of the ‘reservoirs of resistance’ in the spread of KPC-Kp.
Methods
This investigation was initiated because a patient died from pneumonia, empyema and respiratory failure on 2 March 2008. After residing in the LTACH for 15 days, the patient had been transferred to a Regional Medical Center (RMC) 1 day prior to death. A carbapenem-resistant K. pneumoniae isolate from urine was sent to the CDC, where it was identified by PCR as blaKPC positive (data not shown). The RMC notified the LTACH about the KPC-Kp isolate from the transferred patient. In response, an investigation of other K. pneumoniae isolates from the LTACH and other facilities in South Florida served by the Integrated Regional Laboratories (IRL, Fort Lauderdale, FL, USA) was initiated.
Clinical isolates
All K. pneumoniae isolates were collected at the IRL. This laboratory serves a population of 2.5 million people located in four counties in South Florida, and analyses clinical samples collected in 13 hospitals, 8 rehabilitation hospitals and 14 LTCFs. Identification (ID) and antimicrobial susceptibility testing (AST) of the clinical isolates were routinely performed using the Vitek-2 System (bioMérieux, Durham, NC, USA).
During a 1 month period (i.e. 21 March 2008–20 April 2008) all K. pneumoniae isolates showing reduced susceptibility to ertapenem (i.e. MICs ≥ 4 mg/L) were screened for the production of carbapenemases using the modified Hodge test.15 Positive isolates were further investigated with molecular methods to confirm the presence of blaKPC (see below). For blaKPC-positive isolates, MICs of colistin, tigecycline, imipenem, meropenem, ertapenem and doripenem were determined by the Etest method (AB Biodisk, Solna, Sweden) using cation-adjusted Mueller–Hinton agar (BBL, Becton Dickinson, Sparks, MD, USA). In addition, imipenem MICs were also determined using agar dilution (ranges, 0.06–2048 mg/L), with a Steers replicator.4
All AST data for samples collected from January to 20 March 2008 (before the beginning of the study period) at the IRL were also retrospectively analysed.
Molecular methods
PCR and DNA sequence analysis for blaKPC, blaTEM, blaSHV, blaCTX-M, blaCMY-2-like, blaP99, blaMIR-3/4, blaACT-1-like, blaVIM and blaIMP genes were performed using primers and conditions previously reported.4 In cases where multiple alleles were suspected (i.e. double or multiple ‘peaks’ in the DNA sequencing traces at a single position), cloning experiments with pCR-XL-TOPO vector (TOPO XL PCR Cloning Kit; Invitrogen Corporation, Carlsbad, CA, USA) and direct sequencing of the plasmid preparations was done.4 PCR for ompK-35, ompK-36 and ompK-37 genes was also performed.19 DNA sequence analysis was performed using Lasergene 7.2 (DNASTAR, Madison, WI, USA). The final amino acid sequences were determined using the ExPASy Proteomics Server (http://ca.expasy.org), and compared with those previously described (i.e. GenBank accession numbers: OmpK-35, AJ303057; OmpK-36, AJ344089; and OmpK-37, AJ011502; www.lahey.org/Studies for β-lactamases).
Analytical isoelectric focusing (aIEF)
β-Lactamase preparations for aIEF were obtained as previously described and loaded onto 5% polyacrylamide gels containing ampholines (pH range, 3.5–9.5; GE Healthcare Bio-Sciences AB, Uppsala, Sweden).20 Gels were electrophoresed at 4°C with 8 W for 150 min using a Multiphor II apparatus (Amersham Biosciences, Piscataway, NJ, USA). Detection of β-lactamases was obtained by the addition of 1 mM nitrocefin (Becton Dickinson Biosciences, Cockeysville, MD, USA) onto the gel. The following purified β-lactamases were used as controls: TEM-1 (pI 5.4); SHV-1 (pI 7.6); KPC-2 (pI 6.7); and CMY-2 (pI 9.0).
Genotyping by rep-PCR
KPC-Kp isolates were examined using the automated rep-PCR-based strain typing system (DiversiLab™, bioMérieux, Athens, GA, USA) to establish their genetic similarity. Extraction of DNA was performed using the UltraClean™ Microbial DNA Isolation Kit (Mo Bio Laboratories, Inc., Carlsbad, CA, USA). Rep-PCR was performed using the DiversiLab Klebsiella kit. DNA fragment separation and detection were done using the Agilent® 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA) and results were analysed and interpreted with the DiversiLab web-based software using the Kullback–Leibler method, as previously validated by Endimiani et al.4 Isolates were defined as ‘genetically related’ when ≥95% similarity was identified. Notably, isolates collected during the present study were compared with 39 KPC-Kp strains previously characterized and collected in the eastern USA.4 A comparison with the first KPC-Kp isolate (i.e. Kp1534) detected in 1996 in North Carolina was also done.2
PFGE
Three representative KPC-Kp isolates from the Florida LTACH were sent to the CDC for PFGE typing using XbaI as previously described for Escherichia coli (http://www.cdc.gov/pulsenet/protocols.htm).21 PFGE patterns were subsequently compared with the CDC's KPC-Kp database consisting of >400 isolates from each of the major geographic regions of the continental USA and including strains previously characterized with multilocus sequence typing (MLST).6
Immunoblotting
In order to measure the expression of KPC β-lactamase in mid-log phase, we raised polyclonal antibodies to this protein. The blaKPC-2 gene in E. coli DH10B was cloned and expressed using the pET24a(+) vector, then isolated and purified as previously described.22 The polyclonal anti-KPC-2 rabbit antibodies were raised by SIGMA-Genosys (The Woodlands, TX, USA), and isolated from serum using protein G column purification (SIGMA-Genosys).22 Isolates expressing blaKPC were grown in lysogeny broth to an OD600 of 0.8. Preparation of samples, immunoblotting and recognition of β-lactamase expression were performed as previously described.23
Clinical data
Clinical records of patients developing infection or colonization due to KPC-Kp or ertapenem-resistant strains were examined retrospectively as a public health activity. The following data were recorded: age; sex; antimicrobial agents administered during the infectious episode; patient location (e.g. hospital ward) 72 h prior to first positive KPC or ertapenem-resistant K. pneumoniae isolate; and cause of death. Co-morbidity score was determined according to the Charlson weighted index of co-morbidity.24 Predisposing conditions of infection (when present for at least 72 h before KPC-Kp isolation) were also investigated, including mechanical ventilation, intravascular and bladder catheters, and thoracic, abdominal and other drainages.25 In addition, prior use of antibiotics, history of surgery, and use of corticosteroids or antineoplastic drugs were taken into account when administered for at least 2 weeks before KPC-Kp isolation. Antibiotic treatment was defined as ‘empirical’ when given before microbiological results were available. Treatment outcome was classified as complete response, partial response, relapse, treatment failure or not assessable as formerly reported.25
Intervention
To identify the extent of intestinal colonization among patients, active surveillance rectal cultures were collected from all LTACH patients and Intensive Care Unit (ICU) patients at three RMCs which serve as referral facilities for the LTACH. Samples were collected for a 3 day period from the LTACH (i.e. 9, 10 and 14 April) and a 5 day period at the RMCs (i.e. 28 and 30 April, and 5, 13 and 15 May). Screening was performed using chromogenic CHROMagar KPC plates.26 Infection control measures, including instituting contact precautions, isolating patients infected and colonized with KPC-Kp, and using dedicated staff and equipment as much as possible, were implemented as indicated in recent CDC guidelines for control of infection with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities.27
Results
Clinical isolates and susceptibility testing
During the study period (21 March 2008–20 April 2008), 241 non-duplicate K. pneumoniae isolates were sent to IRL from all institutions served. Among them, 10 isolates with reduced susceptibility to ertapenem (i.e. MIC ≥ 4 mg/L) were identified. Each of these isolates was from a different patient and tested positive by the modified Hodge test for carbapenemase production. PCR analysis showed that all 10 isolates harboured the blaKPC gene. Thus, the overall prevalence of blaKPC-positive strains among the total number of K. pneumoniae isolates in this time period was 4.1% (10/241).
The facilities where the 10 KPC-Kp strains were isolated, along with the results of AST, are shown in Table 1. Seven of the KPC-Kp isolates (FL-1–FL-6 and FL-9) were detected in patients residing in a single 100 bed LTACH. The remaining isolates (FL-7, FL-10 and FL-11) were collected from patients located in three different hospitals (Table 1). One isolate (FL-7) was recovered from a urine sample of a patient previously admitted to a hospital in Cleveland, OH, USA.
Table 1.
Characteristics of the KPC-producing K. pneumoniae (KPC-Kp) isolates recognized at the Integrated Regional Laboratories (IRL)
| Isolate: | FL-1 | FL-2 | FL-3 | FL-4 | FL-5 | FL-6 | FL-9 | FL-7 | FL-10 | FL-11 |
|---|---|---|---|---|---|---|---|---|---|---|
| Institution: | LTACH | LTACH | LTACH | LTACH | LTACH | LTACH | LTACH | Hosp #1 | Hosp #2 | Hosp #3 |
| Antibiotic, MIC (mg/L)a | ||||||||||
| ciprofloxacin | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R | ≥4 R |
| gentamicin | 4 S | 4 S | 4 S | 4 S | 4 S | 4 S | ≤1 S | 4 S | ≥16 R | ≥16 R |
| amikacin | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | 32 I | ≥64 R | 16 S | ≤2 S |
| tobramycin | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R | ≥16 R |
| trimethoprim/sulfamethoxazole | ≥320 R | ≥320 R | ≥320 R | ≥320 R | ≥320 R | ≥320 R | ≥320 R | ≥320 R | ≥80 R | ≥320 R |
| colistinb,c | 1.0 | 1.0 | 0.5 | 0.5 | 1.0 | 0.75 | 0.38 | 0.75 | 0.38 | 0.25 |
| tigecyclineb,c,d | 1.5 S | 2.0 S | 2.0 S | 1.5 S | 1.5 S | 1.5 S | 1.5 S | 2.0 S | 1.5 S | 0.25 S |
| piperacillin/tazobactam | ≥128 R | ≥128 R | ≥128 R | ≥128 R | ≥128 R | ≥128 R | ≥128 R | ≥128 R | 64 I | 64 I |
| ceftazidime | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R |
| cefepime | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≤1 S | 2 S |
| aztreonam | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | ≥64 R | 16 I | ≥64 R |
| meropenemb | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | 24 R | ≥32 R |
| imipenemb | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | 12 R | 12 R |
| doripenemb,d | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R |
| ertapenemb | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R | ≥32 R |
| bla genese | KPC-3 | KPC-3 | KPC-3 | KPC-3 | KPC-3 | KPC-3 | KPC-3 | KPC-3 | KPC-2 | KPC-3 |
| SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-11 | SHV-7 | |
| TEM-1 | TEM-1 | TEM-1 | TEM-1 | TEM-1 | TEM-1 | TEM-1 | TEM-1 | SHV-33 | TEM-1 | |
| TEM-1 | ||||||||||
| aIEF | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 6.7, 7.6 | 5.4, 5.8, 6.7, 7.6 | 5.4, 6.0, 6.7, 7.6 | 5.4, 6.7, 7.6 |
aAntimicrobial susceptibility tests were performed with the VITEK 2 System using AST-GN-13 cards (bioMérieux, Durham, NC, USA). Results were interpreted according to CLSI criteria:15 S, susceptible; I, intermediate; and R, resistant.
bMICs were determined by the Etest method (AB Biodisk, Solna, Sweden) on Mueller–Hinton agar.
cInterpretative criteria have not yet been released by the CLSI.15
dSusceptibility tests were interpreted according to FDA criteria (i.e. tigecycline: S ≤ 2 mg/L, R > 4 mg/L; doripenem: S ≤ 0.5 mg/L).
eSHV-11 and SHV-33 are broad-spectrum β-lactamases, whereas SHV-7 is an extended-spectrum β-lactamase (ESBL).
All KPC-Kp isolates detected at the LTACH were highly resistant to carbapenems (i.e. MICs ≥ 32 mg/L), whereas FL-10 and FL-11 showed lower MIC values for imipenem and meropenem using the Etest method. The seven strains from the LTACH were susceptible to gentamicin. However, only colistin and tigecycline were active in vitro against all tested KPC-Kp isolates (Table 1).
Retrospective analysis
As a result of the retrospective analysis of the AST data (January–20 March 2008), six additional cases of infection due to ertapenem-resistant K. pneumoniae were identified. In particular, isolates were recognized in five additional LTACH patients (one in January, three in February and one in March), and from one RMC patient (February, patient #6) 1 day after transfer from the LTACH. The above six K. pneumoniae isolates were not available for molecular characterization.
Characterization of β-lactamases
PCR amplification, cloning and sequencing revealed that blaKPC-3, blaTEM-1 and blaSHV-11 genes were present in the seven KPC-Kp isolates detected in the LTACH and in FL-7. The remaining two KPC-Kp strains possessed blaKPC-2 or blaKPC-3, blaTEM-1 and different blaSHV genes. The aIEF results were consistent with the production of the above bla genes (Table 1).
rep-PCR
As shown in Figure S1 [available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)], all KPC-Kp isolates detected at the LTACH, and FL-7, were genetically related (similarity >95%). Notably, these eight isolates were also genetically related to most of the eastern USA KPC-Kp isolates previously analysed.4 The remaining two isolates (i.e. FL-10 and FL-11) and the index strain Kp1534 were not related (similarity < 90%) to these strains (Figure S1).
PFGE
The three KPC-Kp isolates sent to the CDC (i.e. FL-1, FL-2 and FL-3) shared PFGE patterns that were indistinguishable from one another, confirming that they are genetically related. In addition, when compared with the rest of the CDC's KPC-Kp PFGE database, these isolates shared PFGE patterns that clustered with isolates previously recognized by MLST as sequence type (ST) 258 (data not shown), the dominant strain type of KPC-Kp observed throughout the USA.6 ST258 isolates have also been identified in Israel and Europe.9 Notably, the index KPC-Kp isolate (Kp1534) was previously identified as ST37.6
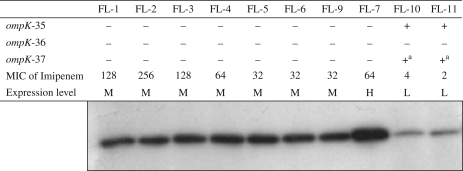
Expression levels of KPC enzymes and background of OmpKs
Figure 1 shows the results of the immunoblotting and analyses of OmpKs along with the MIC results for imipenem by agar dilution. Isolates collected in the LTACH and FL-7 showed high MICs of imipenem (i.e. MICs ≥ 32 mg/L), medium to high levels of KPC enzyme expression and genes unable to encode functional OmpK proteins. In contrast, for FL-10 and FL-11 the imipenem MICs were within the susceptible range (i.e. MICs ≤ 4 mg/L), the isolates expressed low levels of KPC enzyme and functional ompK-35 genes were found (Figure 1).
Figure 1.
Western blotting analysis of KPC level of expression (L, low; M, medium; H, high) and interpretation of DNA sequence analysis of ompK genes. MICs of imipenem (mg/L) were determined using the agar dilution method. +, complete gene when compared with the deposited GenBank sequence (see the Methods section); –, gene sequence disrupted (i.e. insertions, deletions and point mutations). aOnly two point mutations were detected (i.e. I70M and I128M).
Notably, the anti-KPC antibodies did not show cross-reactivity with TEM-1, CTX-M-9, OXA-1, IMP-2, VIM-2, CMY-2 and ADC-7 β-lactamases; only a very weak cross-reactivity with SHV-1 was observed (data not shown).
Clinical data
Table 2 summarizes the clinical characteristics of LTACH patients from whom KPC-Kp isolates were obtained from 21 March 2008 to 20 April 2008. Data for those patients from whom ertapenem-resistant K. pneumoniae isolates were collected before 20 March, 2008 are summarized in Table S1 [available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)].
Table 2.
Clinical characteristics and outcomes of patients with KPC-Kp infection or colonization from the South Florida LTACH
| Isolate | Age/sex | Underlying conditionsa | Previous use of antibiotics (last 2 weeks)b | Charlson weighted index | Risk factorsc | Site of infection/colonizationd | Rectal colonization | Empirical treatment [agent (daily dose)]b | Antimicrobial therapy administered after ID/AST |
Treatment outcome | Patient outcome and comment | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| agentb | timing from infection onset | duration (days) | daily dose | |||||||||||
| FL-1 | 85/M | RF, CRF, DW | FEP, IMP | 9 | Bc, CVC, Trac and MV, haemodialysis, TPN | UTI, BSI | not tested | FEP (500 mg), IMP (250 mg every 12 h) | FEP TZP GEN COL TMP |
first day 6 days after 6 days after 16 days after 9 days after |
11 1 10 5 12 |
500 mg every 12 h 2250 mg every 8 h 100 mg every 48 h 100 mg every 36 h 80 mg every 12 h |
failure | death 17 days after onset of infection; multiorgan failure, sepsis, and pneumonia |
| FL-2 | 76/M | RF, DM2, HTN, HF, PVD, CRF | none | 10 | Bc, CVC, Trac and MV, haemodialysis, thoracocentesis, G-tube, ICU stay | UTI | present | none | GEN COL |
4 days after 10 days after |
6 9 |
130 mg every 48 h 150 mg every 24 h |
relapse | death 32 days after onset of infection; cardiopulmonary arrest and sepsis due to Enterococcus spp. |
| FL-3 | 83/M | RF, DM2, HF, PVD, CAD, AAV | IMP, TZP, TOB | 6 | Bc, CVC, Trac and MV | BSI | not tested | none | CIP TOB FEP LEV GEN TIG COL |
2 days after 2 days after 4 days after 4 days after 8 days after 8 days after 9 days after |
6 2 4 1 1 3 2 |
400 mg every 12 h 130 mg every 24 h 1000 mg every 12 h 250 mg every 24 h 340 mg every 24 h 50 mg every 12 h 115 mg every 12 h |
failure | death 11 days after onset of infection; multiorgan failure, sepsis and pneumonia |
| FL-4 | 41/M | RF, ALS, DW | TOB, FEP | 0 | Bc, CVC, Trac and MV | urine (colonized) | present | TZP (3375 mg every 6 h), CIP (400 mg every 12 h) | GEN FEP |
6 days after 6 days after |
1 9 |
390 mg every 24 h 1000 mg every 12 h |
not assessable | death 30 days after onset of infection; sepsis due to Enterobacter spp. |
| FL-5 | 78/M | RF, DM2, HTN, CVA, colon CA | TOB, CIP, TZP | 5 | Bc, CVC, Trac and MV | sputum (colonized), BSI | present | none | IPM COL |
6 days after 8 days after |
2 9 |
500 mg every 12 h 115 mg every 12 h |
relapse | alive and in LTACH at the end of study period |
| FL-6 | 82/F | RF, CAD, HF, PVD, CRF, CLD | none | 4 | Bc, CVC, Trac and MV, haemodialysis, ICU stay | urine (colonized) | present | CIP (500 mg every 12 h) | LEV CEF CIP |
4 days after 5 days after 2 days before |
1 2 10 |
500 mg every 24 h 500 mg every 12 h 500 mg every 12 h |
not assessable | alive and in LTACH at the end of study period; persistent urinary tract colonization |
| FL-9 | 65/F | RF, DM2, HTN, HF, CRF, DW | IMP | 3 | Bc, CVC, MV, ICU stay | urine (colonized) | present | AMP (2000 mg every 6 h), LEV (500 mg every 12 h) | AMP LEV |
1 day after 1 day after |
8 8 |
2000 mg every 6 h 250 mg every 24 h |
not assessable | alive and in LTACH at the end of study period; persistent urinary tract colonization |
aAAV, artificial aortic valve; ALS, amyotrophic lateral sclerosis; CA, carcinoma; CAD, coronary artery disease; CLD, chronic lung disease; CRF, chronic renal failure; CVA, cerebrovascular accident; DM2, type II diabetes mellitus; DW, decubitus wounds; HF, heart failure; HTN, hypertension; PVD, peripheral vascular diseases; RF, respiratory failure.
bAMP, ampicillin; CEF, cefalexin; CFZ, cefazolin; CIP, ciprofloxacin; COL, colistin; FEP, cefepime; GEN, gentamicin; IPM, imipenem; LEV, levofloxacin; TMP, trimethoprim; TIG, tigecycline; TOB, tobramycin; TZP, piperacillin/tazobactam. Notably, only antimicrobial treatments for Gram-negative bacteria were taken into consideration.
cBc, bladder catheter; CVC, central vascular catheter; ICU, intensive care unit; MV, mechanical ventilation; TPN, total parenteral nutrition; Trac, tracheostomy.
dUTI, urinary tract infection; BSI, bloodstream infection.
In general, the LTACH patients (n = 13) were elderly (mean age = 71 years), possessed multiple underlying illnesses (average Charlson weighted index = 7.7), had many invasive devices and were extensively treated with numerous courses of antibiotics (including carbapenems in 31% of cases). Rectal colonization was present in each of 6 patients tested (three of whom were also colonized in the urinary tract) (Table 2). None of the other LTACH (n = 68) or ICU patients at an RMC (n = 43) in whom active surveillance was performed was colonized with KPC-Kp. Treatment failures were common, and nine patients died (crude mortality rate = 69%). One patient colonized with a KPC-Kp strain was eventually discharged to the community, one patient died after the study period ended and two patients identified as colonized were still in the LTACH 1 year later. All these patients had been hospitalized in Florida hospitals during the previous year (data not shown). Clinical information regarding the non-LTACH patients (i.e. FL-7, FL-10 and FL-11) was not available.
After enhanced infection-control practices were implemented, the outbreak was brought under control, with only one instance of transmission of KPC-Kp infection after early April 2008. The active surveillance programme was terminated on 15 May 2008.
Discussion
K. pneumoniae is an important pathogen responsible for 6% of infections in LTCFs.28 Here, we describe several cases of infection and/or colonization due to genetically related KPC-Kp strains occurring in a single LTACH. Our study emphasizes the following four important points.
First, the clinical isolates collected at the LTACH are genetically related to ST258, the dominant strain of KPC-Kp seen throughout the USA.4,6 This supports the earlier hypothesis by Endimiani et al. that the acquisition of a blaKPC gene, in addition to other potential resistant determinants, may confer on this isolate of K. pneumoniae a marked selective advantage that is leading to its successful dissemination.
Secondly, we noted that the seven KPC-Kp strains found in the LTACH and FL-7 showed high-level resistance to carbapenems, whereas the remaining two KPC-Kp strains (FL-10 and FL-11) had imipenem MICs in the susceptible range by agar dilution.15 We show that the higher MIC levels observed in the first group are a result of the combination of an increased level of KPC expression and loss of the three OmpK proteins. Possessing at least one porin (e.g. OmpK-35) and having low-level KPC expression does not raise the carbapenem MICs within the resistant range. The loss of porins is very common in K. pneumoniae.16,17,29,30 However, this phenomenon combined with the presence of a KPC β-lactamase has been observed infrequently.2,18
Thirdly, the AST results support the notion that the therapeutic options against serious infections due to KPC-Kp strains are limited to tigecycline and colistin.4 However, tigecycline may not reach desired serum levels to treat systemic infections, leaving colistin as the ‘last choice’.31,32 Unfortunately, colistin-resistant KPC-Kp isolates have also been reported.33 Therefore, novel antibiotics or combinations with potent activity against infections due to KPC-Kp isolates need to be considered.34,35
Finally, we conclude this analysis by noting that the spread of KPC-Kp strains outside of the hospital setting (e.g. LTCFs, including LTACHs) is particularly alarming. As illustrated in this report, these patients moved frequently among hospitals, LTCFs and the community. This, like other studies, shows these patients to be elderly, with serious co-morbidities, and frequently requiring intravascular and bladder catheters.11,36 The occurrence of multidrug-resistant organisms (MDROs) in this population has been reported previously,37 mostly resulting from the transfer of patients colonized in the hospital setting to LTCFs and potentially to the community. The experiences with community-associated methicillin-resistant Staphylococcus aureus and CTX-M-producing E. coli isolates serve as harsh reminders that the community could potentially represent a critical reservoir for MDROs.38,39 On the other hand, the frail nature of patients in LTACHs, the excessive use of antibiotics and the difficulty in diagnosing infection versus colonization fosters an environment where isolates containing resistance mechanisms are selected and transmitted to other components of the healthcare system (the primary reservoir of resistance).11 Understanding the transmission dynamics of KPC-Kp among these different settings has therefore become critical.
We realize that the presence of KPC-Kp isolates in LTACHs poses a series of new and frightening challenges. The clinical outcome associated with KPC-Kp infections in hospitalized patients is poor.1,40–42 Although the high mortality reported here suggests this may be the case in other settings, the clinical impact of KPC-Kp on infected or colonized patients remains to be determined. Furthermore, the isolation of KPC-Kp strains results in therapeutic and infection control responses that require considerable effort, expertise and resources often not available outside of hospitals. For instance, colistin has emerged as the last option for infections caused by KPC-Kp.3 However, this antibiotic is perceived as a potentially nephrotoxic medication and patients usually need to be closely monitored while they receive this cationic polypeptide.32 Moreover, it is a common practice in hospitals to isolate patients harbouring KPC-Kp, a measure that may be difficult to implement in LTCFs.
A special challenge illustrated here is the control of the inter-institutional transfer of KPC-Kp. In this report we demonstrate that one patient (i.e. the patient harbouring isolate FL-7) was previously admitted to a hospital in Cleveland, OH, an endemic area of KPC-Kp transmission, and was subsequently transferred to one of the hospitals associated with the South Florida outbreak. Similar occurrences have been reported elsewhere.9,43 Furthermore, all patients tested at the LTACH were colonized in their gastrointestinal and/or urinary tract. However, we do not have a clear explanation why active surveillance did not detect any colonized patient apart from those detected by clinical samples. It should be noted that colonization might promote the spread of KPC-Kp isolates in the community.1 In one recent report, a patient residing in the community probably acquired the KPC-Kp from his wife, who was a documented carrier of this organism as a result of her hospitalization in a tertiary-care hospital in Israel.44
As demonstrated previously, performing surveillance in patients admitted from geographic areas with high-level rates of KPC-Kp infection/colonization (e.g. eastern USA) or before discharging them into the community might be a necessary infection control practice.27,45 In this case, implementation of CDC guidelines (i.e. active surveillance with rectal swabs along with enhanced infection-control practices) helped end the KPC-Kp outbreak at the LTACH in early April 2008.27 Whether attempts should be made to decolonize patients with such MDROs still remains an open question, and procedures and guidelines may need to be established. The speed and fury by which the epidemic of KPC-Kp is spreading in North America mandates urgent action.
Funding
This work was supported in part by the Veterans Affairs Merit Review Program (R. A. B.), the National Institutes of Health (grant RO1-AI063517 and grant RO3-AI081036 to R. A. B.) and Geriatric Research Education and Clinical Center VISN 10 (R. A. B.).
Transparency declarations
R. A. B. has received research funding and speaking invites from various pharmaceutical companies. None of these poses a conflict of interest with the present work. Other authors: none to declare.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the CDC.
Supplementary data
Supplementary Material
Acknowledgements
We thank Drs Arjun Srinivasan, Richard Hopkins, Robert Freedman, Ronald Baker, John Livengood, Roger Sanderson, Robin Kay, Mayda Cruz Solex, Linda McQuade and Kristine M. Hujer for their help in the preparation of this work. We also thank Dr Jean B. Patel for critical review of the manuscript prior to submission.
References
- 1.Schwaber MJ, Carmeli Y. Carbapenem-resistant Enterobacteriaceae: a potential threat. JAMA. 2008;300:2911–3. doi: 10.1001/jama.2008.896. [DOI] [PubMed] [Google Scholar]
- 2.Yigit H, Queenan AM, Anderson GJ, et al. Novel carbapenem-hydrolyzing β-lactamase, KPC-1, from a carbapenem-resistant strain of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:1151–61. doi: 10.1128/AAC.45.4.1151-1161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nordmann P, Cuzon G, Naas T. The real threat of Klebsiella pneumoniae carbapenemase-producing bacteria. Lancet Infect Dis. 2009;9:228–36. doi: 10.1016/S1473-3099(09)70054-4. [DOI] [PubMed] [Google Scholar]
- 4.Endimiani A, Hujer AM, Perez F, et al. Characterization of blaKPC-containing Klebsiella pneumoniae isolates detected in different institutions in the Eastern USA. J Antimicrob Chemother. 2009;63:427–37. doi: 10.1093/jac/dkn547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maltezou HC, Giakkoupi P, Maragos A, et al. Outbreak of infections due to KPC-2-producing Klebsiella pneumoniae in a hospital in Crete (Greece) J Infect. 2009;58:213–9. doi: 10.1016/j.jinf.2009.01.010. [DOI] [PubMed] [Google Scholar]
- 6.Kitchel B, Rasheed JK, Patel JB, et al. Molecular epidemiology of KPC-producing Klebsiella pneumoniae in the United States: clonal expansion of MLST sequence type 258. Antimicrob Agents Chemother. 2009;53:3365–70. doi: 10.1128/AAC.00126-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goldfarb D, Harvey SB, Jessamine K, et al. Detection of plasmid-mediated KPC-producing Klebsiella pneumoniae in Ottawa, Canada: evidence of intrahospital transmission. J Clin Microbiol. 2009;47:1920–22. doi: 10.1128/JCM.00098-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pavez M, Mamizuka EM, Lincopan N. Early dissemination of KPC-2-producing Klebsiella pneumoniae strains in Brazil. Antimicrob Agents Chemother. 2009;53:2702. doi: 10.1128/AAC.00089-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Samuelsen O, Naseer U, Tofteland S, et al. Emergence of clonally related Klebsiella pneumoniae isolates of sequence type 258 producing plasmid-mediated KPC carbapenemase in Norway and Sweden. J Antimicrob Chemother. 2009;63:654–8. doi: 10.1093/jac/dkp018. [DOI] [PubMed] [Google Scholar]
- 10.Roche C, Cotter M, O'Connell N, et al. First identification of class A carbapenemase-producing Klebsiella pneumoniae in the Republic of Ireland. Euro Surveill. 2009;14 pii=19163. [PubMed] [Google Scholar]
- 11.Munoz-Price LS. Long-term acute care hospitals. Clin Infect Dis. 2009;49:438–43. doi: 10.1086/600391. [DOI] [PubMed] [Google Scholar]
- 12.McGuinn M, Hershow RC, Janda WM. Escherichia coli and Klebsiella pneumoniae carbapenemase in long-term care facility, Illinois, USA. Emerg Infect Dis. 2009;15:988. doi: 10.3201/eid1506.081735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eskildsen MA. Long-term acute care: a review of the literature. J Am Geriatr Soc. 2007;55:775–9. doi: 10.1111/j.1532-5415.2007.01162.x. [DOI] [PubMed] [Google Scholar]
- 14.Gould CV, Rothenberg R, Steinberg JP. Antibiotic resistance in long-term acute care hospitals: the perfect storm. Infect Control Hosp Epidemiol. 2006;27:920–5. doi: 10.1086/507280. [DOI] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100-S19. Wayne, PA, USA: CLSI; 2009. [Google Scholar]
- 16.Martinez-Martinez L. Extended-spectrum β-lactamases and the permeability barrier. Clin Microbiol Infect. 2008;14(Suppl 1):82–9. doi: 10.1111/j.1469-0691.2007.01860.x. [DOI] [PubMed] [Google Scholar]
- 17.Doumith M, Ellington MJ, Livermore DM, et al. Molecular mechanisms disrupting porin expression in ertapenem-resistant Klebsiella and Enterobacter spp. clinical isolates from the UK. J Antimicrob Chemother. 2009;63:659–67. doi: 10.1093/jac/dkp029. [DOI] [PubMed] [Google Scholar]
- 18.Woodford N, Tierno PM, Jr, Young K, et al. Outbreak of Klebsiella pneumoniae producing a new carbapenem-hydrolyzing class A β-lactamase, KPC-3, in a New York Medical Center. Antimicrob Agents Chemother. 2004;48:4793–9. doi: 10.1128/AAC.48.12.4793-4799.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urban C, Bradford PA, Tuckman M, et al. Carbapenem-resistant Escherichia coli harboring Klebsiella pneumoniae carbapenemase β-lactamases associated with long-term care facilities. Clin Infect Dis. 2008;46:e127–30. doi: 10.1086/588048. [DOI] [PubMed] [Google Scholar]
- 20.Paterson DL, Rice LB, Bonomo RA. Rapid method of extraction and analysis of extended-spectrum β-lactamases from clinical strains of Klebsiella pneumoniae. Clin Microbiol Infect. 2001;7:709–11. [PubMed] [Google Scholar]
- 21.Ribot EM, Fair MA, Gautom R, et al. Standardization of pulsed-field gel electrophoresis protocols for the subtyping of Escherichia coli O157:H7, Salmonella, and Shigella for PulseNet. Foodborne Pathog Dis. 2006;3:59–67. doi: 10.1089/fpd.2006.3.59. [DOI] [PubMed] [Google Scholar]
- 22.Hujer AM, Bethel CR, Bonomo RA. Antibody mapping of the linear epitopes of CMY-2 and SHV-1 β-lactamases. Antimicrob Agents Chemother. 2004;48:3980–8. doi: 10.1128/AAC.48.10.3980-3988.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hujer AM, Page MG, Helfand MS, et al. Development of a sensitive and specific enzyme-linked immunosorbent assay for detecting and quantifying CMY-2 and SHV β-lactamases. J Clin Microbiol. 2002;40:1947–57. doi: 10.1128/JCM.40.6.1947-1957.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Charlson ME, Pompei P, Ales KL, et al. A new method of classifying prognostic comorbidity in longitudinal studies: development and validation. J Chronic Dis. 1987;40:373–83. doi: 10.1016/0021-9681(87)90171-8. [DOI] [PubMed] [Google Scholar]
- 25.Endimiani A, Luzzaro F, Perilli M, et al. Bacteremia due to Klebsiella pneumoniae isolates producing the TEM-52 extended-spectrum β-lactamase: treatment outcome of patients receiving imipenem or ciprofloxacin. Clin Infect Dis. 2004;38:243–51. doi: 10.1086/380645. [DOI] [PubMed] [Google Scholar]
- 26.Samra Z, Bahar J, Madar-Shapiro L, et al. Evaluation of CHROMagar KPC for rapid detection of carbapenem-resistant Enterobacteriaceae. J Clin Microbiol. 2008;46:3110–1. doi: 10.1128/JCM.00249-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Centers for Disease Control and Prevention. Guidance for control of infections with carbapenem-resistant or carbapenemase-producing Enterobacteriaceae in acute care facilities. MMWR Morb Mortal Wkly Rep. 2009;58:256–60. [PubMed] [Google Scholar]
- 28.Viray M, Linkin D, Maslow JN, et al. Longitudinal trends in antimicrobial susceptibilities across long-term-care facilities: emergence of fluoroquinolone resistance. Infect Control Hosp Epidemiol. 2005;26:56–62. doi: 10.1086/502487. [DOI] [PubMed] [Google Scholar]
- 29.Leavitt A, Chmelnitsky I, Colodner R, et al. Ertapenem resistance among extended-spectrum-β-lactamase-producing Klebsiella pneumoniae isolates. J Clin Microbiol. 2009;47:969–74. doi: 10.1128/JCM.00651-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gulmez D, Woodford N, Palepou MF, et al. Carbapenem-resistant Escherichia coli and Klebsiella pneumoniae isolates from Turkey with OXA-48-like carbapenemases and outer membrane protein loss. Int J Antimicrob Agents. 2008;31:523–6. doi: 10.1016/j.ijantimicag.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 31.Peterson LR. A review of tigecycline—the first glycylcycline. Int J Antimicrob Agents. 2008;32(Suppl 4):S215–22. doi: 10.1016/S0924-8579(09)70005-6. [DOI] [PubMed] [Google Scholar]
- 32.Li J, Nation RL, Turnidge JD, et al. Colistin: the re-emerging antibiotic for multidrug-resistant Gram-negative bacterial infections. Lancet Infect Dis. 2006;6:589–601. doi: 10.1016/S1473-3099(06)70580-1. [DOI] [PubMed] [Google Scholar]
- 33.Bratu S, Tolaney P, Karumudi U, et al. Carbapenemase-producing Klebsiella pneumoniae in Brooklyn, NY: molecular epidemiology and in vitro activity of polymyxin B and other agents. J Antimicrob Chemother. 2005;56:128–32. doi: 10.1093/jac/dki175. [DOI] [PubMed] [Google Scholar]
- 34.Livermore DM, Mushtaq S, Warner M, et al. NXL104 combinations versus Enterobacteriaceae with CTX-M extended-spectrum β-lactamases and carbapenemases. J Antimicrob Chemother. 2008;62:1053–6. doi: 10.1093/jac/dkn320. [DOI] [PubMed] [Google Scholar]
- 35.Endimiani A, Choudhary Y, Bonomo RA. In vitro activity of NXL104 in combination with β-lactams against Klebsiella pneumoniae isolates producing KPC carbapenemases. Antimicrob Agents Chemother. 2009;53:3599–601. doi: 10.1128/AAC.00641-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pop-Vicas A, Mitchell SL, Kandel R, et al. Multidrug-resistant gram-negative bacteria in a long-term care facility: prevalence and risk factors. J Am Geriatr Soc. 2008;56:1276–80. doi: 10.1111/j.1532-5415.2008.01787.x. [DOI] [PubMed] [Google Scholar]
- 37.O'Fallon E, Pop-Vicas A, D'Agata E. The emerging threat of multidrug-resistant gram-negative organisms in long-term care facilities. J Gerontol A Biol Sci Med Sci. 2009;64:138–41. doi: 10.1093/gerona/gln020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Helfand MS, Bonomo RA. Extended-spectrum β-lactamases in multidrug-resistant Escherichia coli: changing the therapy for hospital-acquired and community-acquired infections. Clin Infect Dis. 2006;43:1415–6. doi: 10.1086/508891. [DOI] [PubMed] [Google Scholar]
- 39.Seybold U, Kourbatova EV, Johnson JG, et al. Emergence of community-associated methicillin-resistant Staphylococcus aureus USA300 genotype as a major cause of health care-associated blood stream infections. Clin Infect Dis. 2006;42:647–56. doi: 10.1086/499815. [DOI] [PubMed] [Google Scholar]
- 40.Mathers AJ, Cox HL, Bonatti H, et al. Fatal cross infection by carbapenem-resistant Klebsiella in two liver transplant recipients. Transpl Infect Dis. 2009;11:257–65. doi: 10.1111/j.1399-3062.2009.00374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisenberg SA, Morgan DJ, Espinal-Witter R, et al. Clinical outcomes of patients with Klebsiella pneumoniae carbapenemase-producing K. pneumoniae after treatment with imipenem or meropenem. Diagn Microbiol Infect Dis. 2009;64:233–5. doi: 10.1016/j.diagmicrobio.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Patel G, Huprikar S, Factor SH, et al. Outcomes of carbapenem-resistant Klebsiella pneumoniae infection and the impact of antimicrobial and adjunctive therapies. Infect Control Hosp Epidemiol. 2008;29:1099–106. doi: 10.1086/592412. [DOI] [PubMed] [Google Scholar]
- 43.Woodford N, Zhang J, Warner M, et al. Arrival of Klebsiella pneumoniae producing KPC carbapenemase in the United Kingdom. J Antimicrob Chemother. 2008;62:1261–4. doi: 10.1093/jac/dkn396. [DOI] [PubMed] [Google Scholar]
- 44.Gottesman T, Agmon O, Shwartz O, et al. Household transmission of carbapenemase-producing Klebsiella pneumoniae. Emerg Infect Dis. 2008;14:859–60. doi: 10.3201/eid1405.071340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kochar S, Sheard T, Sharma R, et al. Success of an infection control program to reduce the spread of carbapenem-resistant Klebsiella pneumoniae. Infect Control Hosp Epidemiol. 2009;30:447–52. doi: 10.1086/596734. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.