Abstract
The human genome, human endogenous retroviruses (HERV), of which HERV-K113 and HERV-K115 are the only known full-length proviruses that are insertionally polymorphic. Although a handful of previously published papers have documented their prevalence in the global population; to date, there has been no report on their prevalence in the United States population. Here, we studied the geographic distribution of K113 and K115 among 156 HIV-1+ subjects from the United States, including African Americans, Hispanics, and Caucasians. In the individuals studied, we found higher insertion frequencies of K113 (21%) and K115 (35%) in African Americans compared with Caucasians (K113 9% and K115 6%) within the United States. We also report the presence of three single nucleotide polymorphism sites in the K113 5′ long terminal repeats (LTRs) and four in the K115 5′ LTR that together constituted four haplotypes for K113 and five haplotypes for K115. HERV insertion times can be estimated from the sequence differences between the 5′ and 3′ LTR of each insertion, but this dating method cannot be used with HERV-K115. We developed a method to estimate insertion times by applying coalescent inference to 5′ LTR sequences within our study population and validated this approach using an independent estimate derived from the genetic distance between K113 5′ and 3′ LTR sequences. Using our method, we estimated the insertion dates of K113 and K115 to be a minimum of 800,000 and 1.1 million years ago, respectively. Both these insertion dates predate the emergence of anatomically modern Homo sapiens.
Keywords: insertion frequencies, haplotype, HERV-K113 and HERV-K115, hervotype, human evolution, Homo erectus
Introduction
Human endogenous retroviruses (HERV) are relics of ancient exogenous retroviruses that integrated in the germ line millions of years ago and are now vertically inherited (Gifford and Tristem 2003; Bannert and Kurth 2004). In humans, there are three classes of endogenous retroviruses consisting of more than 31 families and subfamilies, and together, they comprise 8.29% of the human genome (Lander et al. 2001; Bannert and Kurth 2006).
Most endogenous retroviruses (ERV) are millions of years old, and insertions are shared by multiple animal species (Mariani-Costantini et al. 1989; Lander et al. 2001). ERV-K, a transcriptionally active family of endogenous retroviruses, are at least 28 million years old and can be found in the genomes of humans, apes, and Old World monkeys (Costas 2001; Reus et al. 2001). However, a few ERV-K members are unique to the human genome, indicating that HERV-K members retained the potential to infect until recently in evolutionary history (Mariani-Costantini et al. 1989; Steinhuber et al. 1995; Simpson et al. 1996; Barbulescu et al. 1999). Previous reports have shown that HERV-K family members have continued to evolve postintegration due to insertion, transposition, and/or solo-long terminal repeat (LTR) formation (Costas 2001; Turner et al. 2001; Hughes and Coffin 2004; Macfarlane and Simmonds 2004; Belshaw et al. 2005; Mayer et al. 2005). In 2001, Turner et al. discovered two HERV-K family members with complete 5′ and 3′ LTR, HERV-K113 and HERV-K115, which are insertionally polymorphic in humans (fig. 1). These youngest members of the HERV-K family, K113 and K115, are unique to humans, and the fact that they are not yet fixed in the human population indicates a very recent insertion. The age of an HERV insertion can be estimated by comparison of the 5′ and 3′ LTR sequences of a specific retroviral insertion locus, with the assumption that the LTR were identical in sequence at the time of insertion (Dangel et al. 1995). Using this dating methodology, Turner et al. (2001) sequencing data that revealed no sequence differences between the 5′ and 3′ LTR led them to estimate an insertion time for K113 of 100,000–200,000 years ago. The age of K115 estimated by using this LTR sequence comparison method is highly inaccurate because the K115 3′ LTR appears to have undergone gene conversion with another HERV-K locus, and therefore, the sequence differences between LTR's have not accrued in a clocklike manner (Turner et al. 2001).
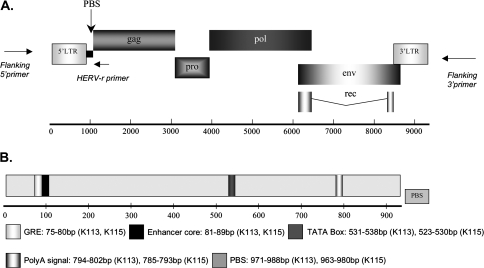
FIG. 1.—
Genome structures of K113 and K115 are shown. (A) The proviruses are flanked by a short duplicated host DNA also known as preintegration site. LTRs line the viral genomes both at 5′ and 3′ ends. The primer positions of the flanking primers (specific to human DNA sequences flanking the proviral insertions) and HERV-K–specific primer (annealing to HERV-K insertions between the 5′ LTR and gag gene) are also included. (B) LTR of K113 and K115 indicating loci for various regulatory regions. An additional 17 HERV-K sequences (supplementary table 1, Supplementary Material online) were used to validate the coordinates of the known regulatory regions within the 5′ LTR.
Various recent reports have shown that some HERV-Ks may play a role in disease pathogenesis (Lower 1999), especially in some cancers (Contreras-Galindo et al. 2008; Golan et al. 2008; Wang-Johanning et al. 2008), autoimmune diseases such as rheumatoid arthritis (Sicat et al. 2005; Ehlhardt et al. 2006), type 1 diabetes (Marguerat et al. 2004), schizophrenia (Otowa et al. 2006), and possibly also in HIV-1 infection (Contreras-Galindo et al. 2007; Garrison et al. 2007). Like other HERV-K family members, K113 and K115 have also been predicted to play a role in diseases. Burmeister et al. (2004) suggested a pathological role for K113 in non-European breast cancer patients and observed that the prevalence of K115 is 2-fold lower in age-matched healthy controls compared with breast cancer patients. Moyes et al. (2005) suggested that the prevalence of K113 is increased in people with Sjorgen's syndrome.
HERV insertions could affect the human genome by expression of retroviral genes as shown by studies investigating HERV–disease associations and by gene reshuffling or possibly by reinfection. Also, HERV LTRs that include regulatory elements (fig. 1B) with the potential to regulate the genes in their vicinity are suspected of playing a role in speciation (Lebedev et al. 2000). All reports pertaining to K113 and K115 have only investigated their presence or absence in the human genome with an assumption that these recent insertions have remained monomorphic postinsertion (the only possible exception being the formation of a solo-LTR due to the total loss of the coding regions of the insertion). However, like any sequence in the genome, there is a potential for these loci to accumulate mutations postinsertion. Thus, testing for the presence or absence of the insertion would not reveal the entire complexity of an individual's HERV genotype. Exploring the variations within these HERV insertions may shed more light on their associations with diseases and any role in human evolution. To explore the possibility of single nucleotide polymorphisms (SNPs) within the K113 and K115 genomes, we sequenced the 5′ LTR of both K113 and K115. Polymorphisms in the known regulatory regions of the LTR would be of great interest in disease studies. Also, the traditional method of comparing the sequences of the 5′ and 3′ LTR to date an insertion is not applicable for solo-LTRs or to estimate the insertion dates of HERV insertions such as K115 that show evidence of gene conversion in one LTR sequence. Sequence polymorphisms within the insertions could be used to develop a method to estimate the age of HERV loci that are not amenable to current estimation methods.
Materials and Methods
Peripheral blood samples were obtained from a total of 156 HIV-1 positive individuals from two cohorts. A total of 96 subjects were perinatally HIV-1–infected participants born to HIV-1+ mothers, seen at the pediatric clinic at Jacobi Medical Center, Bronx, NY. A total of 60 samples of HIV-1–infected adults were obtained from the SCOPE cohort in San Francisco, CA. The study was approved by the Institutional Review Boards at Jacobi Medical Center and the University of California at San Francisco (UCSF). Primary data analysis that included K113 and K115 “hervotyping” (determination of specific HERV sequence of a particular endogenous retrovirus) and haplotype construction for this study was carried out without prior knowledge of the ethnicity of the participants. All participants self-identified their ethnicities.
Each individual's DNA was genotyped for null, heterozygous, or homozygous insertion of K113 and K115 insertions by using two different polymerase chain reaction (PCR) amplifications for each insertion using two different sets of primers (Turner et al. 2001; Burmeister et al. 2004). The first PCR amplification using primers derived from human DNA sequences flanking the proviral insertion demonstrated heterozygosity of the insertion. The second PCR using 5′ primer annealing to the host flanking DNA upstream of the proviral insertion and a 3′ primer downstream of the 5′ LTR between the 5′ LTR and gag gene was used to amplify the 5′ LTR of the proviral insertion. This PCR was repeated with a set of confirmation primers on insertion positive individuals to produce an amplicon for sequencing of the 5′ region of the HERV insertion. A fourth and final PCR using a 5′ primer upstream of 3′ LTR (in the env region of the insertion) along with the 3′ primer annealing to the flanking host DNA was used to screen for solo-LTR formation in individuals positive for insertions. PCR amplification primers for each HERV-K insertions are listed in supplementary table 2 (Supplementary Material online). In order to refer to the individuals who possessed at least one retroviral insertion positive allele as a group, we define the term “insertion frequency” as the percentage of individuals carrying at least one retroviral insertion at the genomic insertion site, which is equal to the number of heterozygotes plus the number of homozygotes divided by the size of the total population studied. Subsequently, we will use the terms “insertion frequency of K113” or “insertion frequency of K115” to refer to this population of individuals.
The K113 integration site was detected by primer pairs K113-f/K113-r, which amplified a band of 303 bp in K113 null and heterozygous individuals; no bands were seen in individuals homozygous for the viral insertion, and heterozygotes cannot be discriminated from K113 null individuals (thus the need for amplification with HERV-specific primers). The K113 5′ LTR was amplified using primer pairs K113-f/HERV-r, which amplified a band of 1,146 bp in individuals heterozygous and homozygous for a K113 insertion; no bands were seen in null individuals. The K113 3′ LTR was detected using K113-3′f/K113-r primer pairs and amplified a band sized 1,200 bp in individuals heterozygous and homozygous for a K113 insertion but not in K113 null individuals. The K115 integration site was detected by primer pairs K115-f/K115-r, which amplified a band of 557 bp in K-115 null and heterozygous individuals. The K115 5′ LTR was amplified using primer pairs K115-f/HERV-r and amplified a band sized 1,269 bp in individuals heterozygous for a K115 insertion; no bands were seen in null individuals. The K115 3′ LTR was detected using K115-3′f/K115-r primer pairs, which amplified a band of 1,200 bp in heterozygous individuals but not in K115 null individuals.
PCR was performed in volume of 50 μl with 25 ng of genomic DNA in 1.0 μM of each primer, 200 μM of each deoxynucleoside triphosphate, 25 mM of MgCl2, and 0.5 units of AmpliTaq gold DNA polymerase (Applied Biosystems, Foster City, CA). PCR was conducted with 10 min of initial denaturing at 95 °C, 30 cycles of 10 s at 94 °C, 30 s at annealing temperature, and 59 s of elongation at 72 °C followed by a 10 min final extension at 72 °C. Amplified PCR product was stored at 4 °C. The amplified PCR products were analyzed by electrophoresis on 1–2% agarose gel. The 100-bp ladder (Invitrogen, Carlsbad, CA) was used to verify the product size.
Sequencing was done at MCLAB, South San Francisco, using ABI 3730XL sequencer. Sequences were aligned, and SNPs were identified using Sequencher version 5.0. Computerized base calls were verified by visual inspection of chromatograms. Three methods were used to rule out the PCR error: Only those SNPs verifiable by bidirectional sequencing were used. Although some SNPs at early sites at the 5′ LTR were not always verifiable by bidirectional sequencing of the original amplicon, these SNPs were verified by performing PCR on the genomic DNA with confirmation primers (supplementary table 2, Supplementary Material online) and sequencing the verification amplicons. PCRs and sequencing were performed at least twice on each individual positive for the insertions.
The Fisher's exact test was implemented using the GraphPad Prism v5.0b statistical software package. In all cases, the two-tailed (nondirectional) test statistic was reported when P was below 0.05.
Initial multiple sequence alignments were generated using Multalin (Corpet 1988), with default gap parameters and the “DNA 5.0” weight matrix. Subsequent manual aligning was performed using the Se–Al sequence alignment editor v2.0 (Rambaut 1996). All phylogenetic analyses were performed using the HyPhy software package (http://www.hyphy.org; Pond et al. 2005). Phylogenetic trees were built using a heuristic maximum likelihood (ML) search procedure under the general reversible substitution model with site-to-site rate variation corrected for with the beta–gamma distribution (Pond and Frost 2005), performing randomized sequential addition with nearest neighbor interchange branch swapping after every 10 sequences were added.
Blast (Altschul et al. 1990) and Primer-Blast (Rozen and Skaletsky 2000) were used to confirm that K113 and K115 insertion sites (flanking regions targeted by PCR primers) were unique in the human genome, neutralizing the possibility that retroviral replication and transposition contributed to the observed variation in proviral sequences (supplementary fig. 1, Supplementary Material online). Sequences of the most recent common ancestors (MRCAs) of K113 and K115 insertions were inferred using ML reconstruction (as described above). K115 (GenBank accession number AY037929, haplotype CAGA) was chosen as an outgroup taxon for K113 haplotypes, and K113 (GenBank accession number AY037928, haplotype GCC) served as the outgroup for K115 haplotypes in the trees built for MRCA inference (supplementary fig. 2, Supplementary Material online). Coalescence (insertion) times were approximated using the formula: , where T = is the time (My) passed since the insertion event, n is the number of observed taxa, D is the divergence associated with a descendant taxon (cumulative branch lengths between observed haplotype and inferred MRCA), and R represents one of two previously reported divergence (evolutionary) rates (substitutions/base/My; Kimura 1968). We used two different values for the R parameter (2.2 × 10−9 and 1.3 × 10−9 mutations/site/year) to generate upper and lower bound estimates of insertion times (Kumar and Subramanian 2002; Lebedev et al. 2000).
Results
Previous reports indicate that the rate of both K113 and K115 proviral insertion is highest in Africans (table 1). All current published reports of K113 and K115 have included participants from outside the United States. Due to the lack of information on the prevalence of K113 and K115 insertions in North American populations, we examined genomic DNA from African Americans, Caucasians, and Hispanics from two groups of HIV-1–positive individuals in New York and San Francisco for K113 and K115 insertions. Based on previously reported observations, we hypothesized that there would be a higher insertion frequency of K113 and K115 in the African American population compared with Caucasians and Hispanics. One hundred and fifty-six randomly selected and unrelated individuals comprising three major ethnic groups (African American, Hispanic, and Caucasian) were hervotyped for K113 and K115 insertions using a PCR-based assay (Turner et al. 2001). Two sets of primers for each of these two insertions were used to determine their HERV insertion genotypes (null/heterozygous/homozygous) in each individual. Of the 156 individuals, none were homozygous for K115 insertion, but six individuals were homozygous for K113 insertion. Those homozygous for a K113 insertion included individuals of all three major ethnicities: one Hispanic, four African Americans, and one Caucasian. Interestingly, a Hispanic, four African Americans, and a Caucasian were also positive for at least one copy of both the proviruses.
Table 1.
Summary of Previously Reported Frequencies of K113 and K115 in Global Populations
| K113 |
K115 |
||||
| Geographic Region | Study Size (n) | Insertion Frequency (%) | Study Size (n) | Insertion Frequency (%) | Reference |
| Africa | |||||
| Unspecifieda | 25 | 20 | 25 | 20 | Macfarlane and Simmonds (2004) |
| Malawi | 60 | 27 | 54 | 30 | Moyes et al. (2005) |
| Cote d'Ivoire | 64 | 19 | 60 | 43 | Moyes et al. (2005) |
| Kenya | 50 | 20 | 50 | 28 | Moyes et al. (2005) |
| Kenya | 46 | 36 | 50 | 22 | Herrera et al. (2006) |
| Rwanda | 49 | 22 | 49 | 22 | Herrera et al. (2006) |
| Cameroon | 16 | 9 | 16 | 21 | Herrera et al. (2006) |
| Meanb | 22 | 27 | |||
| Middle East | |||||
| Yemen | 50 | 8 | 56 | 7 | Moyes et al. (2005) |
| Egypt | 43 | 5 | 45 | 13 | Herrera et al. (2006) |
| Oman | 43 | 3 | 57 | 14 | Herrera et al. (2006) |
| Meanb | 5 | 11 | |||
| Europe | |||||
| Unspecifieda | 22 | 0 | 22 | 0 | Macfarlane and Simmonds (2004) |
| United Kingdom | 96 | 4 | 96 | 1 | Moyes et al. (2005) |
| Galicia | 48 | 2 | 50 | 7 | Herrera et al. (2006) |
| Basque | 50 | 2 | 49 | 5 | Herrera et al. (2006) |
| Meanb | |||||
| Asia | |||||
| Unspecifieda | 28 | 10 | 28 | 0 | Macfarlane and Simmonds (2004) |
| China | 44 | 12 | 42 | 3 | Herrera et al. (2006) |
| Taiwan | 47 | 16 | 49 | 4 | Herrera et al. (2006) |
| Japan | — | — | 359 | 9 | Otowa et al. (2006) |
| Meanb | 13 | 4 | |||
| Oceania | |||||
| Papua New Guinea | 26 | 0.09 | 26 | 0.2 | Macfarlane and Simmonds (2004) |
| Papua New Guinea | 54 | 0 | 52 | 0 | Moyes et al. (2005) |
| Meanb | 0.04 | 0.10 | |||
The specific countries were not specified.
Mean frequencies of K113 and K115 in each geographical area calculated from specified previous reports. Based on the previously published data, the average insertion frequency of K113 is 22% in Africans, 5% in Middle Easterners, 2% in Europeans, 13% in Asians, and <0.1% in Papua New Guineans. Similarly, average insertion frequency of K115 is 27% in Africans, 11% in Middle Easterners, 3% in Europeans, 4% in Asians, and 0.1% in Papua New Guineans.
Our analysis of the PCR results revealed significant differences in the insertion frequencies of K115 between the ethnic groups involved in this study. We found that the insertion frequency of K115 was higher in African Americans than in Caucasians (P < 0.0013, Fisher's exact test). Additionally, we found a statistically significant difference in the insertion frequency of K115 in Hispanics and Caucasians (P < 0.0015, Fisher's exact test). In African Americans, the insertion frequency of K115 varied between geographic areas sampled within the United States, but this variation was not statistically significant. Variation in the insertion frequency of K113 between ethnic groups did not reach statistical significance, and the percentages did not vary between cohorts from the two geographic locations studied (table 2).
Table 2.
Insertion Frequencies of K113 and K115 in Three Major Ethnicities in the United States
| K113 |
K115 |
||||||
| Ethnicity | M/F/na | n | +Insertion | % | Homozygous (n) | +Insertion | % |
| Jacobi Cohort, New York | |||||||
| African Americans | 34/22/0 | 56 | 11 | 20 | 3 | 22 | 39 |
| Hispanics | 17/18/1 | 36 | 6 | 17 | 1 | 14 | 39 |
| Multiracial | — | 4 | 0 | 0 | 0 | 0 | 0 |
| Total | 96 | 17 | 18 | 4 | 36 | 38 | |
| Scope Cohort, San Francisco | |||||||
| African Americans | 16/3/0 | 19 | 5 | 26 | 1 | 4 | 21 |
| Caucasians | 25/7/0 | 32 | 3 | 9 | 1 | 2 | 6 |
| Multiracial | — | 9 | 2 | 22 | 0 | 2 | 22 |
| Total | 60 | 10 | 17 | 2 | 8 | 13 | |
| Combined (New York and San Francisco) | |||||||
| African Americans | 50/25/0 | 75 | 16 | 21 | 4 | 26 | 35 |
| Hispanics | 17/18/1 | 36 | 6 | 17 | 1 | 14 | 39 |
| Caucasians | 25/7/0 | 32 | 3 | 9 | 1 | 2 | 6 |
| Multiracial | — | 13 | 2 | 15 | 0 | 2 | 15 |
| Total | 156 | 27 | 17 | 6 | 44 | 28 | |
NOTE.—M/F/na male/female/not available. When both parents of a participant reported different ethnicities for themselves, the participants were considered multiracial. Frequencies of K113 and K115 in three major ethnicities from two geographical regions are shown. Both the insertions were more common in African Americans in both geographical regions. The frequency of K115 insertion was higher in African Americans from New York than those from San Francisco. There were a total of five individuals, mostly African Americans, homozygous for K113 insertions. None were homozygous for K115 insertions.
We used PCR to amplify the 5′ LTR of HERV-K113 and K115 and sequenced the amplified DNA. Three sites in the K113 5′ LTR, loci 174, 581, and 629 (named for their position in the sequence amplicon as aligned to the GenBank reference sequence), had SNPs, whereas the 5′ LTR of K115 had SNP in four loci: 268, 385, 410, and 687 (table 3). Alleles at all three SNP sites in K113 5′ LTR consisted of at least two alternate nucleotide bases with one being more common than the other (174: A > G, 581: T > C, and 629: C > T) in all the ethnicities examined in this study. However, occasionally, African Americans also had a G instead of a T or a C at locus 581 of the K113 5′ LTR. All four loci for the K115 5′ LTR consisted of two different nucleotide bases (268: C > T, 385: A > C, 410: G > A, and 687: A > C). In African Americans, variation in the sequence of the K115 5′ LTR consisted of almost equal frequencies of the two bases at positions 268, 385, and 687, whereas in Hispanics, the base frequencies at all these positions were biased toward the most common bases. The base frequencies at these sites are summarized in table 3.
Table 3.
SNP in Various Positions of 5′ LTR of K113 and K115
| African Americans |
Hispanics |
Caucasians |
|||||
| Position | Base | n | f | n | f | n | f |
| A. Frequencies of bases at various SNP positions in 5′ LTR of K113 | |||||||
| 174 | A | 8 | 0.89 | 3 | 0.60 | 2 | 1 |
| G | 1 | 0.11 | 2 | 0.40 | 0 | 0 | |
| 581 | T | 7 | 0.78 | 3 | 0.60 | 2 | 1 |
| C | 1 | 0.11 | 2 | 0.40 | 0 | 0 | |
| G | 1 | 0.11 | 0 | 0 | 0 | 0 | |
| 629 | C | 7 | 0.78 | 3 | 0.60 | 2 | 1 |
| T | 2 | 0.22 | 2 | 0.40 | 0 | 0 | |
| Total | 9 | 5 | 2 | ||||
| B. Frequencies of bases at various SNP positions in 5′ LTR of K115 | |||||||
| 268 | C | 9 | 0.56 | 8 | 0.89 | 1 | 1 |
| T | 7 | 0.44 | 1 | 0.11 | 0 | 0 | |
| 385 | A | 9 | 0.56 | 8 | 0.89 | 1 | 1 |
| C | 7 | 0.44 | 1 | 0.11 | 0 | 0 | |
| 410 | G | 14 | 0.88 | 8 | 0.89 | 1 | 1 |
| A | 2 | 0.13 | 0 | 0 | 0 | 0 | |
| 687 | A | 8 | 0.50 | 8 | 0.89 | 1 | 1 |
| C | 8 | 0.50 | 1 | 0.11 | 0 | 0 | |
| Total | 16 | 9 | 1 | ||||
NOTE.—(A) Three sites with SNP (174, 581, and 629) were observed in the 5′ LTR of K113. None of the Caucasians had any SNP in any of these three sites indicating all of them had the same allele of K113. Both African Americans and Hispanics had at least two SNPs in each locus. There was an additional base at position 581 in African Americans. Base frequencies at each base were biased toward one common base. (B) Four SNP sites (286, 385, 410, and 687) were identified in the 5′ LTR of K115. All the Caucasians had the same allele of K115. African Americans had two bases at each of the SNP sites with a similar base frequency except at position 410 in which base frequency was biased. Hispanics did show diversity at all three sites but did not have an SNP at position 410. All SNPs were numbered according to their position in GenBank sequences AY037928 for K113 and AY037929 for K115.
Once we identified the SNP sites in the 5′ LTR of both these proviruses, we constructed haplotypes for various alleles of K113 and K115. We identified common haplotypes as well as ethnicity-specific private alleles for both K113 and K115 (table 4). Despite only three SNP sites in the K113 5′ LTR, we could identify four haplotypes for K113 (fig. 2). African Americans had both the common haplotypes A–T–C (f = 0.57) and A–T–T (f = 0.14) in addition to a rare private allele exclusively found within the African American population A–G–C (f = 0.14) that was not present in Hispanics and Caucasians. Hispanics had three haplotypes including the two common haplotypes A–T–C (f = 0.25) and A–T–T (f = 0.25) and a Hispanic-specific private allele G–C–C (f = 0.25). Caucasians contained the A–T–C common haplotype exclusively. We sequenced three individual homozygous for the K113 insertion. An African American and a Hispanic were heterozygous at each of the three polymorphic sites (A/G–C/T–C/T). One Caucasian individual homozygous for the K113 insertion had only A–T–C at the polymorphic sites, consistent with the common Caucasian haplotype. Sequence alignments of all the K113 haplotypes are shown in supplementary figure 3 (Supplementary Material online).
Table 4.
Haplotypes and Haplotype Frequencies of K113 and K115 Based on Variations in the 5′ LTR
| Polymorphic Loci on LTR |
|||||||
| n | 174 | 581 | 629 | f | |||
| A. Haplotypes of K113 in various ethnic groups in the United States | |||||||
| African Americans | 7 | ||||||
| 4 | A | T | C | 0.57 | |||
| 1 | A/G | C/T | C/T | 0.14 | |||
| 1 | A | T | T | 0.14 | |||
| 1 | A | G | C | 0.14 | |||
| Hispanics | 4 | ||||||
| 1 | A | T | C | 0.25 | |||
| 1 | A/G | C/T | C/T | 0.25 | |||
| 1 | A | T | T | 0.25 | |||
| 1 | G | C | C | 0.25 | |||
| Caucasians | 2 | ||||||
| 2 | A | T | C | 1 | |||
| n | 268 | 385 | 410 | 687 | f | ||
| B. Haplotypes of K115 in various ethnic groups in the United States | |||||||
| African Americans | 16 | ||||||
| 6 | C | A | G | A | 0.38 | ||
| 6 | T | C | G | C | 0.38 | ||
| 2 | C | A | A | A | 0.13 | ||
| 1 | C | C | G | G | 0.06 | ||
| 1 | T | A | G | C | 0.06 | ||
| Hispanics | 9 | ||||||
| C | A | G | A | 0.89 | |||
| T | C | G | C | 0.11 | |||
| Caucasians | 1 | ||||||
| 1 | C | A | G | A | 1 | ||
NOTE.—Four haplotypes were seen for K113 of which two were common in African Americans and Hispanics. Both African Americans and Hispanics also had one unique private allele for K113. Caucasians had only one haplotype. Five different haplotypes were also observed in K115. The most diverse ethnic group with all five haplotypes was African American. African Americans also had two private alleles. Hispanics had two different haplotypes of K115, whereas the Caucasians had only a single haplotype.
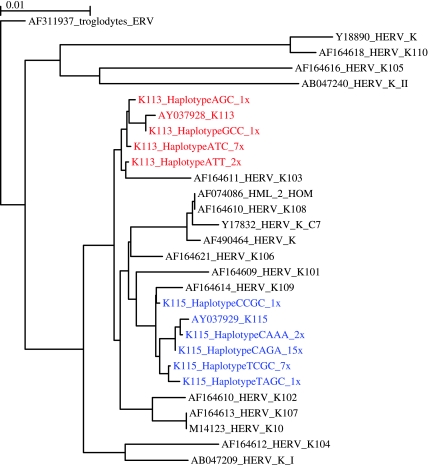
FIG. 2.—
ML phylogeny of HERV-K 5′ LTR sequences including K113 and K115 haplotypes. Taxon names of all reference sequences include GenBank accession numbers. K113 and K115 sequences are highlighted in red and blue, respectively, and the number of times each haplotype was observed in this study is listed in taxon labels. Scale bar represents 1% genetic distance.
The K115 5′ LTR was also very diverse and consisted of a total of five different haplotypes (fig. 2). African Americans had all five haplotypes with two common haplotypes C–A–G–A (f = 0.38) and T–C–G–C (f = 0.38) being the most prevalent followed by three private alleles, C–A–A–A (f = 0.13), C–C–G–G (f = 0.7), and T–A–G–C (f = 0.7). Hispanics only had the two common haplotypes with C–A–G–A being more prevalent (f = 0.89) than T–C–G–C (f = 0.11). Caucasians had a very low insertion frequency and had only one common haplotype C–A–G–A. The haplotypes of K113 and K115 along with their frequencies are summarized in table 4, and the sequence alignments of all the K115 haplotypes are shown in supplementary figure 3 (Supplementary Material online).
Substitutions that generate differences between LTR of the same provirus are expected to result from mutation postinsertion. Therefore, the inter-LTR genetic divergence can provide an estimate of insertion time, given that mutations accrue after insertion in a clocklike manner. HERV insertion times have previously been estimated by measuring the genetic distance between 5′ and 3′ LTR sequences (Dangel et al. 1995; Johnson and Coffin 1999; Turner et al. 2001). We compared the 5′ and 3′ LTR of K113 (AY037928) and found three single-base differences between the two LTR sequences. Using 0.13% My as the rate of divergence (Lebedev et al. 2000), we calculated K113 inserted in the human genome 1.19 Ma. This approach cannot be applied to K115, due to evidence that gene conversion has played a considerable role in the evolutionary history of its 3′ LTR (Turner et al. 2001). In this study, we modified the method to exploit genetic variation between the 5′ LTR sequences of multiple proviral haplotypes. Flanking genomic sequence for the K113 and K115 insertions in each individual was compared with eliminate the possibility of characterizing polymorphisms in HERV-K HML-2 insertions occurring at other loci (supplementary fig. 1, Supplementary Material online). Additionally, the clustering of all haplotypes of K113 and K115 LTRs indicates that gene conversion has not played a role in generating any of the observed sequence variants of the 5′ LTRs (fig. 2).
We used measurements of genetic divergence from inferred ancestral 5′ LTR sequences (supplementary fig. 2, Supplementary Material online) to estimate K113 and K115 insertion times. Upper and lower bounds for insertion times were obtained by applying two different previously reported evolutionary rates to translate genetic distance into coalescence time. An upper bound was generated using an inferred evolutionary rate specific to the HERV LTR of 0.13%/My (1.3 × 10−9 mutations/site/year; Lebedev et al. 2000). A lower bound was generated using the inferred mammalian genome mutation rate of 2.2 × 10−9 mutations/site/year, which is reported to be relatively invariant within and between primate genomes (Bulmer et al. 1991; Kumar and Subramanian 2002). Based on these rates, K113 was integrated into the human genome between 800,000 and 1.3 Ma (mean divergence in K113 cluster = 0.17%). K115 was integrated between 1.1 and 1.9 Ma (mean divergence in K115 cluster = 0.24%). Estimating the insertion date for HERV-K113 using the traditional method of comparing the two LTRs (1.19 My) falls within the range of our cross-sectional sequence comparison insertion date estimate. This finding further validates our method and increases confidence in the insertion date estimate for HERV-K115.
Discussion
Recent studies have measured the insertion frequencies of HERV-K113 and K115 in diverse population groups on various continents, with Africa hosting the highest prevalence of these insertions and Papua New Guinea with the lowest (table 1). We observed that within the United States, the insertion frequency of HERV-K115 varied among ethnicities and insertion frequencies was higher in African Americans and Hispanics than in Caucasians. The insertion frequency of K115 also varied between the two geographical locations within the United States, although the difference was not significant and may be due to sampling bias.
HERV-K113 and K115 along with other full-length proviruses have been utilized as markers to investigate human evolution. In addition to examining solo-LTR formation of other HERV-K HML-2 members, Herrera et al. (2006) and Macfarlane and Simmonds (2004) have also used insertion frequencies of K113 and K115 to examine the potential use of HERV-K as novel genetic markers for the study of human evolution. Results from both these studies were consistent with previously used genetic markers to concur with the “out of Africa” theory of human evolution. We identified multiple SNP loci in both the proviral insertions. This is, to the best of our knowledge, the first evidence that K113 and K115 are not only insertionally polymorphic at the population level but also genetically polymorphic at the sequence level. In this study, we demonstrated that both K113 and K115 have multiple haplotypes, and haplotype diversity was greatest within the African American study participants. Our polymorphism data also confirm the occurrence of the insertion in Africa and its spread with human migrations.
Although none of the SNPs were in previously characterized transcription factor or transcriptional enhancer binding sites within the LTR (fig. 1C), our identification of sequence polymorphism is relevant for future study of the association between HERV and disease. Although previously published reports have investigated the role of K113 and K115 insertion in different diseases such as cancers and autoimmune diseases (Burmeister et al. 2004; Sicat et al. 2005; Ehlhardt et al. 2006; Contreras-Galindo et al. 2008; Golan et al. 2008; Wang-Johanning et al. 2008), no study prior to ours has been conducted to investigate polymorphisms within the LTR of K113 or K115. Studies to detect disease associations with HERV-K113 and 115 should include a component of sequence analysis, to discover any role for sequence variants in disease susceptibility or progression.
Various previously-published studies have reported that K113 is less than 450,000 years old (Turner et al. 2001; Moyes et al. 2005). The reported age of K115 was based on an estimation because the method of dating endogenous retroviruses by inter-LTR comparison (Dangel et al. 1995) is not applicable to K115 due to evidence of gene conversion (Turner et al. 2001). Our upper- and lower-bound insertion time estimates suggest that both K113 and K115 may have integrated into the human genome significantly earlier than previously reported. Our upper-bound estimates of integration time are based on an inferred HERV-K LTR mutation rate calibrated using the divergence time between humans and Old World monkeys (Lebedev et al. 2000). We chose this rate because it has been used in multiple studies of endogenous retroviral insertion times (Reus et al. 2001; Lavie et al. 2004), and it was derived specifically for the HERV-K LTR loci. However, recent research has suggested that particular interspecies comparisons may result in an underestimate of the mutation rate (Ho and Endicott 2008). We obtained a lower bound estimate of insertion times based on an inferred universal mammalian genome mutation rate derived using inter- and intragenomic comparison (Kumar and Subramanian 2002). This mutation rate relies on a large number of external calibration points and has also been used to date HERV-K insertion times (Costas 2001). We cannot exclude the possibility that these mutation rates used to generate our insertion times underestimate the true mutation rate. A significantly higher mutation rate would shorten our predicted insertion times and alter our conclusions. Alternatively, the insertion (coalescence) times reported here may still represent underestimates because our sampling may not have adequately captured the entire range of K113 and K115 proviral sequence variants in the global population.
It is curious that K113 and K115 remain insertionally polymorphic despite their persistence in the human population for a minimum of 0.8 and 1.1 Ma, respectively. A variety of factors are known to influence the fixation time and persistence of endogenous retroviruses, including local recombination rate and gene density (Katzourakis et al. 2007; Belshaw et al. 2007). The presence of an HERV-K insertion in the human genome may be of selective consequence to the host individual, and therefore, neutral evolutionary theory may not provide the best framework to predict HERV-K fixation times. A formal evaluation of the factors involved in the fixation of K113 and K115 are beyond the scope of this study, but this observation warrants further investigation.
Our minimum estimates of the age of K113 (800,000 years) and K115 (1.1 My) put the insertion dates for both at a time well before the emergence of anatomically modern humans. Current fossil evidence dates the emergence of anatomically modern humans in Ethiopia to 154–195,000 years ago (White et al. 2003; McDougall et al. 2005). Our estimates date the insertions of K113 and K115 to the time of Homo erectus (Clark et al. 1994). It will also be of interest to explore the Neanderthal genome for the existence of these insertions or the preintegration site, since our insertion time estimates also predate the currently published divergence time of Homo sapiens neanderthalensis and Homo sapiens sapiens (Noonan et al. 2006). Furthermore, our method of dating could be used to infer the age of other HERV insertions in the human genome with LTR sequences that have been modified by gene conversion and even that of solo-LTRs to analyze their role in evolution and speciation.
Conclusion
K113 and K115 proviruses are not monomorphic and their ages indicate that the insertions occurred prior to the emergence of anatomically modern H. sapiens. Simply screening for the presence or absence of a provirus may not provide sufficient information to evaluate their impact on an individual's phenotype. Considering hervotypes, that is, the genetic variation between haplotypes, of K113 and K115 will be essential in evaluating their relationship to human disease and their true contribution to evolutionary history.
Supplementary Material
Supplementary figures 1–3 and tables 1 and 2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
This study was conducted in part from funds from the UCSF AIDS Research Institute, a sponsored research agreement from Pfizer, Inc., and National Institutes of Health grants 1K01DA024654 (S.K.P.) and AI76059 (D.F.N.). Author's contributions: A.R.J., D.F.N., and K.E.G. designed the study; A.R.J. performed the experiments; E.C.S., D.J.W., and A.R.J. performed confirmatory experiments; A.R.J., K.E.G., and S.K.P. analyzed the data; V.A.Y., E.R.C., J.N.M., M.G.R., and S.G.D. provided the reagents; D.F.N. and K.E.G. supervised the project; and A.R.J., S.K.P., D.F.N., and K.E.G. wrote the article with input from all the coauthors. This study was conducted in part from funds from the University of California San Francisco AIDS Research Institute, a sponsored research agreement from Pfizer, Inc., and National Institutes of Health grants 1K01DA024654 (S.K.P.) and AI76059 (D.F.N.).
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Bannert N, Kurth R. Retroelements and the human genome: new perspectives on an old relation. Proc Natl Acad Sci USA. 2004;101(Suppl 2):14572–14579. doi: 10.1073/pnas.0404838101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannert N, Kurth R. The evolutionary dynamics of human endogenous retroviral families. Annu Rev Genomics Hum Genet. 2006;7:149–173. doi: 10.1146/annurev.genom.7.080505.115700. [DOI] [PubMed] [Google Scholar]
- Barbulescu M, Turner G, Seaman MI, Deinard AS, Kidd KK, Lenz J. Many human endogenous retrovirus K (HERV-K) proviruses are unique to humans. Curr Biol. 1999;9:861–868. doi: 10.1016/s0960-9822(99)80390-x. [DOI] [PubMed] [Google Scholar]
- Belshaw R, Dawson AL, Woolven-Allen J, Redding J, Burt A, Tristem M. Genomewide screening reveals high levels of insertional polymorphism in the human endogenous retrovirus family HERV-K(HML2): implications for present-day activity. J Virol. 2005;79:12507–12514. doi: 10.1128/JVI.79.19.12507-12514.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Watson J, Katzourakis A, Howe A, Woolven-Allen J, Burt A, Tristem M. Rate of recombinational deletion among human endogenous retroviruses. J Virol. 2007;81:9437–9442. doi: 10.1128/JVI.02216-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulmer M, Wolfe KH, Sharp PM. Synonymous nucleotide substitution rates in mammalian genes: implications for the molecular clock and the relationship of mammalian orders. Proc Natl Acad Sci USA. 1991;88:5974–5978. doi: 10.1073/pnas.88.14.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister T, Ebert AD, Pritze W, Loddenkemper C, Schwartz S, Thiel E. Insertional polymorphisms of endogenous HERV-K113 and HERV-K115 retroviruses in breast cancer patients and age-matched controls. AIDS Res Hum Retroviruses. 2004;20:1223–1229. doi: 10.1089/aid.2004.20.1223. [DOI] [PubMed] [Google Scholar]
- Clark JD, de Heinzelin J, Schick KD, et al. (11 co-authors) African Homo erectus: old radiometric ages and young Oldowan assemblages in the Middle Awash Valley, Ethiopia. Science. 1994;264:1907–1910. doi: 10.1126/science.8009220. [DOI] [PubMed] [Google Scholar]
- Contreras-Galindo R, Kaplan MH, Leissner P, et al. (11 co-authors) Human endogenous retrovirus-K (HML-2) elements in the plasma of people with lymphoma and breast cancer. J Virol. 2008;82:9329–9336. doi: 10.1128/JVI.00646-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Galindo R, Lopez P, Velez R, Yamamura Y. HIV-1 infection increases the expression of human endogenous retroviruses type K (HERV-K) in vitro. AIDS Res Hum Retroviruses. 2007;23:116–122. doi: 10.1089/aid.2006.0117. [DOI] [PubMed] [Google Scholar]
- Corpet F. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 1988;16:10881–10890. doi: 10.1093/nar/16.22.10881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costas J. Evolutionary dynamics of the human endogenous retrovirus family HERV-K inferred from full-length proviral genomes. J Mol Evol. 2001;53:237–243. doi: 10.1007/s002390010213. [DOI] [PubMed] [Google Scholar]
- Dangel AW, Baker BJ, Mendoza AR, Yu CY. Complement component C4 gene intron 9 as a phylogenetic marker for primates: long terminal repeats of the endogenous retrovirus ERV-K (C4) are a molecular clock of evolution. Immunogenetics. 1995;42:41–52. doi: 10.1007/BF00164986. [DOI] [PubMed] [Google Scholar]
- Ehlhardt S, Seifert M, Schneider J, Ojak A, Zang KD, Mehraein Y. Human endogenous retrovirus HERV-K(HML-2) Rec expression and transcriptional activities in normal and rheumatoid arthritis synovia. J Rheumatol. 2006;33:16–23. [PubMed] [Google Scholar]
- Garrison KE, Jones RB, Meiklejohn DA, et al. (14 co-authors) T cell responses to human endogenous retroviruses in HIV-1 infection. PLoS Pathog. 2007;3:e165. doi: 10.1371/journal.ppat.0030165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford R, Tristem M. The evolution, distribution and diversity of endogenous retroviruses. Virus Genes. 2003;26:291–315. doi: 10.1023/a:1024455415443. [DOI] [PubMed] [Google Scholar]
- Golan M, Hizi A, Resau JH, Yaal-Hahoshen N, Reichman H, Keydar I, Tsarfaty I. Human endogenous retrovirus (HERV-K) reverse transcriptase as a breast cancer prognostic marker. Neoplasia. 2008;10:521–533. doi: 10.1593/neo.07986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrera RJ, Lowery RK, Alfonso A, McDonald JF, Luis JR. Ancient retroviral insertions among human populations. J Hum Genet. 2006;51:353–362. doi: 10.1007/s10038-006-0370-0. [DOI] [PubMed] [Google Scholar]
- Ho SY, Endicott P. The crucial role of calibration in molecular date estimates for the peopling of the Americas. Am J Hum Genet. 2008;83:142–146. doi: 10.1016/j.ajhg.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Coffin JM. Human endogenous retrovirus K solo-LTR formation and insertional polymorphisms: implications for human and viral evolution. Proc Natl Acad Sci USA. 2004;101:1668–1672. doi: 10.1073/pnas.0307885100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Coffin JM. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci USA. 1999;96:10254–10260. doi: 10.1073/pnas.96.18.10254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katzourakis A, Pereira V, Tristem M. Effects of recombination rate on human endogenous retrovirus fixation and persistence. J Virol. 2007;81:10712–10717. doi: 10.1128/JVI.00410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci USA. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, et al. (255 co-authors) Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lavie L, Medstrand P, Schempp W, Meese E, Mayer J. Human endogenous retrovirus family HERV-K(HML-5): status, evolution, and reconstruction of an ancient betaretrovirus in the human genome. J Virol. 2004;78:8788–8798. doi: 10.1128/JVI.78.16.8788-8798.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebedev YB, Belonovitch OS, Zybrova NV, Khil PP, Kurdyukov SG, Vinogradova TV, Hunsmann G, Sverdlov ED. Differences in HERV-K LTR insertions in orthologous loci of humans and great apes. Gene. 2000;247:265–277. doi: 10.1016/s0378-1119(00)00062-7. [DOI] [PubMed] [Google Scholar]
- Lower R. The pathogenic potential of endogenous retroviruses: facts and fantasies. Trends Microbiol. 1999;7:350–356. doi: 10.1016/s0966-842x(99)01565-6. [DOI] [PubMed] [Google Scholar]
- Macfarlane C, Simmonds P. Allelic variation of HERV-K (HML-2) endogenous retroviral elements in human populations. J Mol Evol. 2004;59:642–656. doi: 10.1007/s00239-004-2656-1. [DOI] [PubMed] [Google Scholar]
- Marguerat S, Wang WY, Todd JA, Conrad B. Association of human endogenous retrovirus K-18 polymorphisms with type 1 diabetes. Diabetes. 2004;53:852–854. doi: 10.2337/diabetes.53.3.852. [DOI] [PubMed] [Google Scholar]
- Mariani-Costantini R, Horn TM, Callahan R. Ancestry of a human endogenous retrovirus family. J Virol. 1989;63:4982–4985. doi: 10.1128/jvi.63.11.4982-4985.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer J, Stuhr T, Reus K, Maldener E, Kitova M, Asmus F, Meese E. Haplotype analysis of the human endogenous retrovirus locus HERV-K (HML-2.HOM) and its evolutionary implications. J Mol Evol. 2005;61:706–715. doi: 10.1007/s00239-005-0066-7. [DOI] [PubMed] [Google Scholar]
- McDougall I, Brown FH, Fleagle JG. Stratigraphic placement and age of modern humans from Kibish, Ethiopia. Nature. 2005;433:733–736. doi: 10.1038/nature03258. [DOI] [PubMed] [Google Scholar]
- Moyes DL, Martin A, Sawcer S, Temperton N, Worthington J, Griffiths DJ, Venables PJ. The distribution of the endogenous retroviruses HERV-K113 and HERV-K115 in health and disease. Genomics. 2005;86:337–341. doi: 10.1016/j.ygeno.2005.06.004. [DOI] [PubMed] [Google Scholar]
- Noonan JP, Coop G, Kudaravalli S, et al. (11 co-authors) Sequencing and analysis of Neanderthal genomic DNA. Science. 2006;314:1113–1118. doi: 10.1126/science.1131412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otowa T, Tochigi M, Rogers M, Umekage T, Kato N, Sasaki T. Insertional polymorphism of endogenous retrovirus HERV-K115 in schizophrenia. Neurosci Lett. 2006;408:226–229. doi: 10.1016/j.neulet.2006.09.004. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD. A simple hierarchical approach to modeling distributions of substitution rates. Mol Biol Evol. 2005;22:223–234. doi: 10.1093/molbev/msi009. [DOI] [PubMed] [Google Scholar]
- Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- Reus K, Mayer J, Sauter M, Zischler H, Muller-Lantzsch N, Meese E. HERV-K(OLD): ancestor sequences of the human endogenous retrovirus family HERV-K(HML-2) J Virol. 2001;75:8917–8926. doi: 10.1128/JVI.75.19.8917-8926.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambaut A. Se-Al: sequence alignment editor v2.0 [Internet] 1996 [cited 2005 March 1]; Available from: http://evolve.zoo.ox.ac.uk/ [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Sicat J, Sutkowski N, Huber BT. Expression of human endogenous retrovirus HERV-K18 superantigen is elevated in juvenile rheumatoid arthritis. J Rheumatol. 2005;32:1821–1831. [PubMed] [Google Scholar]
- Simpson GR, Patience C, Lower R, Tonjes RR, Moore HD, Weiss RA, Boyd MT. Endogenous D-type (HERV-K) related sequences are packaged into retroviral particles in the placenta and possess open reading frames for reverse transcriptase. Virology. 1996;222:451–456. doi: 10.1006/viro.1996.0443. [DOI] [PubMed] [Google Scholar]
- Steinhuber S, Brack M, Hunsmann G, Schwelberger H, Dierich MP, Vogetseder W. Distribution of human endogenous retrovirus HERV-K genomes in humans and different primates. Hum Genet. 1995;96:188–192. doi: 10.1007/BF00207377. [DOI] [PubMed] [Google Scholar]
- Turner G, Barbulescu M, Su M, Jensen-Seaman MI, Kidd KK, Lenz J. Insertional polymorphisms of full-length endogenous retroviruses in humans. Curr Biol. 2001;11:1531–1535. doi: 10.1016/s0960-9822(01)00455-9. [DOI] [PubMed] [Google Scholar]
- Wang-Johanning F, Radvanyi L, Rycaj K, Plummer JB, Yan P, Sastry KJ, Piyathilake CJ, Hunt KK, Johanning GL. Human endogenous retrovirus K triggers an antigen-specific immune response in breast cancer patients. Cancer Res. 2008;68:5869–5877. doi: 10.1158/0008-5472.CAN-07-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White TD, Asfaw B, DeGusta D, Gilbert H, Richards GD, Suwa G, Howell FC. Pleistocene Homo sapiens from Middle Awash, Ethiopia. Nature. 2003;423:742–747. doi: 10.1038/nature01669. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.