Abstract
Several serum biomarkers for recombinant human growth hormone (rhGH) have been established, however, none alone or in combination have generate a specific, sensitive, and reproducible ‘kit’ for the detection of rhGH abuse. Thus, the search for additional GH specific biomarkers continues. In this review, we focus on the use of proteomics in general and 2-dimensional electrophoresis (2-DE) in particular for the discovery of new GH induced serum biomarkers. Also, we review some of the protocols involved in 2DE. Finally, the possibility of tissues other than blood for biomarker discovery is discussed.
Keywords: proteomics, two-dimensional gel electrophoresis, growth hormone, doping, biomarker, blood, urine, skin
1. Introduction
While recombinant human GH (rhGH) has several uses for treating disorders other than GH deficiency (D), it has also been mis-used or abused with the intention of improving athletic performance. GH has been shown to have an anabolic effect in athletes with regards to protein metabolism [1], however, the actual increases in muscle mass, strength, endurance, and athletic performance have been brought into question [2, 3]. Nonetheless, rhGH remains an agent abused among individuals in sports and is on the World Anti Doping Agency (WADA) list of prohibited substances [4]. In bodybuilding, it is estimated that rhGH is abused with doses of 10–25 IU/day which is 20 times higher that therapeutic dose used for adult GHD [5]. These high doses are thought to be used three to four days a week in cycles of four to six weeks. Moreover, rhGH is believed to be used in combination with other doping agents such as anabolic steroids [6]. In endurance sports, rhGH is misused together with erythropoietin although the dosing is not known [6].
The major problem with identifying individuals who are misusing rhGH is the difficulty in detection. First, rhGH is indistinguishable from endogenous hGH which is secreted by anterior pituitary in a pulsatile pattern. Second, endogenous hGH levels are affected by many environmental factors such as exercise, sleep, stress and nutritional status [7]. Third, GH has a very short serum half life of about 15 minutes [8]. Also, rhGH injected into muscle and skin is cleared quickly to baseline values in 8–16 hours and 11–20 hours, respectively. Thus, ‘the window of opportunity’ for detection of rhGH is very short. [9]. Taken together, these factors make the detection of rhGH doping difficult. Furthermore, urine detection, often used to test drug doping among athletes, is difficult because urine GH levels are extremely low [10]. Moreover, GH in urine is poorly correlated with serum GH levels [11]. With that said, it may still be possible to use urine testing to detect GH if downstream GH-responsive biomarkers can be identified. To date, blood is the most common biological fluid used for GH doping detection methodologies.
2. Current approaches to detect GH doping
There are two approaches to detect rhGH in blood. One is based on the different isoforms of hGH. Endogenous GH has several forms including the most abundant 22 kDa isoform, a significant 20 kDa isoform generated by alternative precursor RNA splicing [12], and other minor-isoforms including a 17.08 kDa and 17.84 kDa subtype. These latter isoforms were discovered by proteomic analysis of the human pituitary gland [13]. Also, different isoforms of hGH result from differential post-translational modifications including acetylation, deamidation and phosphorylation [13]. It is well known that following rhGH injection, insulin-like growth factor 1 (IGF-1) increases which results in feedback inhibition of endogenous hGH secretion. Since rhGH comprises of only one form, i.e., the 22 kDa isoform, an imbalance in the ratio of 22 kDa isoform relative to the total GH could lead to a diagnostic test for rhGH abuse. In other words, by calculating the ratio of the 22 kDa isoform versus total GH, it is possible to distinguish whether exogenous rhGH was used [9, 14].
The limitation of this approach is that the window of opportunity to detect the rhGH/endogenous GH ratio change is short; about 24–36 hours after injection [15]. This makes it impractical to detect GH doping without daily testing. However, the GH isoform method was adopted by WADA for the 2004 Athens and 2006 Turin Olympic Games. No inappropriate results from blood samples were found possibly due to the timing of the blood tests[15].
A second approach to detect GH doping is to determine GH-dependent biomarkers that have longer half lives than GH itself. Currently there are two groups of biomarkers used for this purpose. One is IGF-1 / IGF binding proteins (IGF-1/IGFBPs) and the other includes proteins involved in bone and collagen turnover. To this end, a study entitled GH-2000 was initiated with an attempt to search for GH specific biomarkers in human serum [16, 17]. In a randomized, double blind, placebo-controlled study involving healthy volunteers of both sexes, daily rhGH administration (0.1 IU/kg /day and 0.2 IU/kg/day) for 4 weeks followed by a 8-week wash-out period resulted in detectible increases in IGF-1, acid- labile subunit (ALS) and IGFBP-3 levels [16]. Markers of bone and collagen turnover were also found to be increased. These included osteocalcin, a C-terminal propeptide of type I procollagen (P-I-P), a bone resorption marker type I collagen telopeptide (ICTP), and a soft tissue marker procollagen type III (P-III-P). Of these markers, only P-III-P and osteocalcin remained significantly elevated after the 8-week wash-out period [17]. In the GH-2000 study, females were found to be less responsive than males to GH treatment as has been reported by others [18]. This is presumably due to the antagonistic actions of estrogen on GH action [19].
As stated above, endogenous GH is regulated by many factors other than IGF-1. Thus, when attempting to detect rhGH doing, it is necessary to differentiate the effect of exogenously administered rhGH from high endogenous hGH. For example, exercise is known to induce GH. To quantify the effect of exercise on the GH/IGF-1 axis as well as the downstream markers, the effect of maximum exercise on endogenous hGH levels was examined in elite athletes [20]. hGH showed a peak concentration by the end of the exercise period and decreased to baseline levels within 30–60 min after the exercise was completed. Similarly, IGF-I, IGFBP-3, ALS and IGFBP-2 were increased following exercise. However, IGF-I, IGFBP-3, and ALS returned to baseline by 3–4 days. Also, the bone markers ICTP and P-III-P increased after exercise but decreased to baseline levels or below within 120 min whereas osteocalcin and P-I-P values were not affected by exercise as the values did not change after exercise [20]. This study demonstrated that although exercise increased the levels of several serum biomarkers, the effect was acute and diminished in a relatively short period of time [21, 22].
Another concern with the biomarkers cited above is that injury, which is quite common among athletes, can cause changes in serum bone and collagen markers. To address this, elite and amateur athletes were followed longitudinally for sport’s related injury. It was found that no change in IGF-1 occurred after injury. A rise in P-III-P serum level was found, however, this rise was minimal and did not reach the GH doping threshold as measured by the discriminate function scoring system created by the GH-2000 initiative [23].
Other factors such as age, gender, body mass index (BMI), ethnicity and type of sport activity or category were examined for their effects on the above mentioned GH responsive biomarkers. Age was the major contributor to variability in these marker values with a significant decrease occurring as age increased [24]. Gender also was a confounder that contributed significantly to the variability of IGFBP-3 and ALS, whereas BMI, ethnicity and sport category were only modest effecters [24]. Recently it was found that ethnicity did not significantly impact IGF-1 and P-III-P as markers [25]. Taken together, these results point out that reference ranges of biomarkers must be established for different factors including age and gender.
Since the discovery of these GH-responsive biomarkers in serum, more studies have confirmed that rhGH administration in either athletic adults or elite athletes increased the levels of IGF-1 [20, 26, 27], IGFBP-2 [20, 21], IGFBP-3 [20, 21], IGFBP-4 [28], IGFBP-5 [28], and ALS [20, 21], P-III-P [20, 26, 27] and ICTP [20]. However, among the handful of marker candidates, only IGF-1 and P-III-P were validated by two independent datasets derived from independent double blind, placebo controlled rhGH injection studies: namely the GH-2000 study and the Kreischa study [29]. After adjusting for assay differences, the discriminant function formula of GH-2000 was applied to Krischa data and vice versa. The GH-2000 formula was able to detect 90% of those receiving rhGH in the Kreischa study. The Kreischa formula could correctly identify 41% of individuals receiving GH in the GH-2000 study [26]. These two biomarkers, however, have relatively large variations among different individuals or even within the same person. For example, serum samples obtained over a 2- to 3-week period from 1103 elite athletes showed that the within-subject coefficient of variation (CV, a value to measure the variance relative to the mean) was nearly 21% for the IGF axis markers and 15% for the collagen markers [30]. Therefore, it is generally agreed that a single biomarker is not a robust indicator of GH doping, rather, a combination of multiple biomarkers will generate more robust and reliable results [15]. With the various confounding demographic factors as well as diversity in lifestyles, more biomarkers will help to enhance the sensitivity and specificity for rhGH detection.
Recently, hemoglobin -chain was discovered by use of a protein chip assay to be a biomarker of GH action [31]. In this regard, theoretically, all serum proteins could be profiled in individuals before and after rhGH treatment and those that are differentially regulated by exogenous rhGH administration could be identified. Thus, proteomics represents a novel and highly promising perspective to discover additional biomarkers for rhGH doping.
Previous efforts have successfully identified biomarkers for cancer and other diseases using proteomics [32–34]. There are various proteomic techniques for identifying proteins in a given sample. These methods can be classified into two major groups: two dimensional electrophoresis (2-DE) and multidimensional chromatography [35]. 2-DE is the a popular approach that is able to separate up to thousands of proteins and protein isoforms on a single gel. The chromatographic approach (for example liquid chromatography Mass Spectrometry [LCMS]) separates protein digests instead of intact proteins. This method is suitable for high-throughput screening for the establishment of biomarkers but does not necessarily identify the protein. Recently, a new technique based on antibody binding called protein arrays or protein chips has been developed [36]. Problems with this technique include difficulty in protein quantification and the ability to generate binding antibodies [37]. In this perspective, we will concentrate on 2-DE as a methodology for determining the proteome of serum/plasma as well as other tissues.
3. 2-DE as a proteomic approach to discover biomarkers of GH action
2-DE has been used to isolate proteins in a given sample for several decades. It was used to initiate resolution of proteins in human plasma as early as 1977 [38]. As the name implies, 2-DE utilizes two separations or “dimensions”. In the first dimension, proteins are separated by isoelectric focusing (IEF) which depends on their net charges. Proteins with different isoelectric points (pIs) are focused at different positions on an immobilized pH gradient (IPG) strip. IPG strips are commercially available in various lengths and pI ranges. For the second dimension, the IPG strip is placed on top of a sodium dodecyl sulfate – polyacrylamide gel (SDS-PAG) and the proteins are separated by electrophoresis (E) based on size. The protocol for SDS-PAGE is well developed with varying gel compositions to accommodate demands for separation of proteins in different molecular weight ranges [39]. Following SDS-PAGE, the gels are stained to reveal the proteins and are then imaged using software that captures the density of each spot. From these images, protein profiles and individual protein concentrations are determined and compared. In our laboratory, protein ‘spot’ detection and densitometry are performed using the Discovery Series PDQuest 2-DE analysis software package version 7.0 that accompanieds the VersaDoc 1000 Imaging System The proteins that are different between treatment groups such as rhGH-treated and non treated are then removed from the gel and identified by mass spectrometry (MS) and/or tandem MS (MS/MS).
Below we provide a more detailed description of 2-DE as a general method for proteomic studies not only for blood but also for various types of tissue samples [37]. For more comprehensive reviews on the value of proteomics, the following articles are suggested [40–43].
3.1 Sample preparation
Before 2-DE, proteins in the tissue samples must be solubilized, denatured, and reduced. In order to ensure minimum protein loss and good reproducibility of sample preparation, it is generally advised to keep procedure as simple as possible. Sample preparation usually requires lysing of the cells and/or tissues to release the cellular proteins. Body fluids such as plasma/serum, cerebral spinal fluid, or urine do not require lysing.
3.2 Cell/tissue lysis
For cultured cells, gentle lysis methods are sufficient to release cellular proteins without significant protein loss or modifications. Osmotic lysis is usually applied to blood cells and cells in culture. This involves suspension of the cells in a hypotonic solution in which the cells burst because of osmotic pressure. Subsequently different subcellular components can also be fractionated for protein analyses [44]. Another method is freeze-thaw lysis, in which cells are rapidly frozen using liquid nitrogen then thawed in one or more cycles. When harsher methods are needed (for tissues that are difficult to disrupt such as liver, kidney, and heart), a mortar and pestle can be used to grind the tissues that have been frozen in liquid nitrogen. Alternatively, various mechanical homogenization devices have been developed to efficiently lyse many types of tissues [45].
Cells and tissues are usually lysed in the presence of detergents to solubilize and denature proteins. Ionic detergents are avoided in the lysis buffer since they interfere with subsequent IEF. If ionic detergents such as SDS are used to lyse cells, then the sample must be diluted in an excess of non-ionic/ zwitterionic detergent to eliminate SDS. Alternatively, SDS can be removed by acetone precipitation. For solid tissue samples and some cells that require more vigorous lysis methods, samples can be suspended in a buffer containing detergent while being sonicated. A cautionary note; although ultrasonic waves disrupt tissues or cells, the shear forces generated by this procedure can cause heating of the sample. Therefore, samples are usually sonicated with short bursts and cooled on ice between bursts. After lysing, centrifugation is usually carried out in order to collect the protein supernatant, or differential centrifugation can be performed to recover |proteins found in various cellular organelles.
3.3 Protein solubilization/denaturation/reduction/alkylation
After proteins are collected and SDS removed (if required), the sample is then diluted in a buffer which usually contains urea, thiourea, non-ionic and/or zwitterionic detergents, a reducing agent, carrier ampholytes and protease inhibitors. Urea is typically used since it is a neutral chaotrope that denatures proteins. Heating can aid the solubilization but samples containing urea should avoid extended exposure to heat and/or temperatures above 37 ºC since at high temperatures urea in solution breaks down into ammonium cyanate which can cause protein charge modifications. Thiourea is often used in addition to urea to improve solubilization and denaturation. Non-ionic detergents such as NP-40 and Triton X-100, and/or zwitterionic detergents such as CHAPS are included to further increase solubilization and to prevent protein hydrophobic aggregation. Carrier ampholytes are added to create a pH gradient during IEF. Carrier ampholytes also serve as scavengers as they are carbamylated by any unwanted ammonium cyanate present in urea solutions. A reducing agent such as dithiothreitol (DTT), 2-mercaptoethanol, or tributylphosphine (TBP) is added to reduce disulfide bonds. Following reduction, the sulfhydryl groups on the cysteine residues should be alkylated by an agent such as iodoacetamide to prevent sulfide bond reformation. During sample preparation, proteases released from cells are always a potential problem, thus, protease inhibitor cocktails are usually added to the sample at the same time of solubilization / denaturation / reduction, and may even be added as early as cell/tissue lysis.
3.4 Contaminant removal
Often tissue or blood samples need to be further processed to remove non-protein molecules such as salt, DNA, RNA, and polysaccharides that may interfere with the separation or visualization of a 2D gel. Excess salt and other small ionic molecules can impede IEF because they make the sample more electro-conductive and should be kept at a low concentration before the first dimensional electrophoresis. Desalting can be achieved by dialysis or precipitation. DNA and RNA can increase sample viscosity and may result in background protein smearing. High molecular weight nucleic acids can also ‘clog’ gel pores and prevent focusing. Samples are often treated with DNase/RNase to hydrolyze the nucleic acid polymers. Large polysaccharides also can block gel pores and cause horizontal streaking. They can be removed by precipitation with ammonium sulphate or phenol/ammonium acetate. Also, lipids can reduce the solubility and change the pI and/or molecular weight of proteins. They can be removed by excess detergent or acetone precipitation [46]. This is especially important when analyzing adipose tissue.
3.5 Blood as a sample
As stated above, difficulties in testing for GH abuse stem mainly from rhGH being identical to endogenously produced GH as well as its short serum half life. Although there are current discriminating test for GH abuse using serum biomarkers, problems still exist related to these assays including the variability between individuals. Thus, additional GH specific biomarkers are needed. Our laboratory and others have embarked on this task.
Compared to cell culture or solid tissue samples, blood is relatively easy to manipulate and is also a rich source of proteins. It is the most common tissue collected for clinical diagnostic purposes. Centrifugation at 7,000 g for 10 minutes effectively precipitates the blood cells. Proteins in the supernatant (plasma if clotting factors are preserved, e.g. by using heparinized collecting tubes or serum if clotting factors have coagulated and precipitated together with blood cells) are readily soluble. No lysis or pre-treatment of serum/plasma is needed and the sample can be treated for denaturation and reduction directly, since normally very low concentrations of DNA, RNA, large polysaccharides or lipids are present in blood. Also, the salt concentration in the blood is well below the level that disturbs IEF.
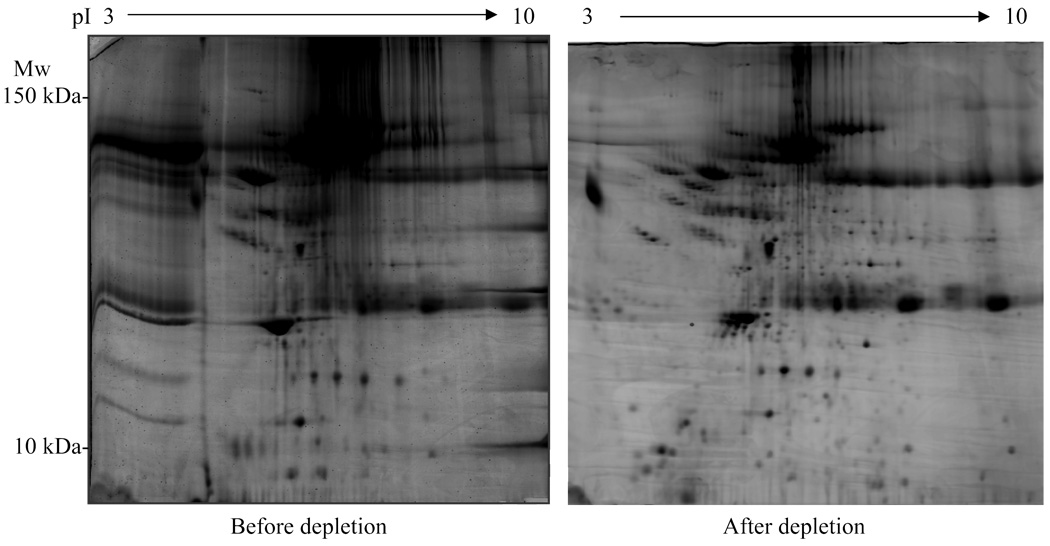
The major problem encountered when using serum/plasma is that a few abundant species of proteins obscure the presence of other less abundant proteins. For example, albumin, the most abundant plasma protein, is 10 orders of magnitude in excess of other proteins such as cytokines or hormones. Overall, 22 proteins, including albumin, transferin, haptoglobulin, immunoglobulins and lipoproteins, comprise of 99% of all proteins in plasma [47]. Out of the estimated 10,000 proteins present in human plasma, current techniques have detected 1175 distinct proteins/peptides [48]. Therefore, 1% of plasma proteome, albeit in low abundance, contains a large amount of unique proteins and therefore represents a biomarker reservoir. In a typical 2D gel of plasma proteins, albumin will appear as a huge blot masking many other proteins. Therefore it is necessary to deplete albumin and other high-abundance proteins in order to resolve the remaining proteins. Albumin can be depleted using immunoaffinity resins [49] resulting in greatly improved resolution of the 2D image [50]. Recently a new technique using a hexapeptide ligand library has been developed [51] in which serum/plasma proteins are bound by their respective hexapeptide ligands. In this scenario, the more abundant serum proteins are bound, resulting in relatively enriched population of less abundant proteins. This technique minimizes the loss of proteins, as can happen in immunodepletion, but requires a large sample volume (∼1 ml serum/plasma). This should impose no problem in the case of human samples, but for animal studies such as mice or rats, it is difficult to obtain such a large volume per animal. In figure 1, we show two plasma 2-DE protein profiles; one before albumin depletion and the other after depletion using the immunodepletion method (Fig. 1).
Fig. 1.
Human serum 2-D gels before and after albumin depletion. Gels were stained with SYPRO Orange. Abbreviations: pI, isoelectric focusing point; Mw, molecular weight.
3.6 Electrophoresis
After sample preparation, proteins are first separated by IEF. As stated above, IEF relies on movement of proteins along the pH gradient of precast gels in an electric field. Proteins migrate to a position that is equal to the pH of the respective protein. Two methods exist for creating pH gradient. One uses carrier ampholytes to generate pH gradients in polyacrylamide tube gels. A more wide-spread method is the use of immobilized pH gradients that are generated by covalently linking acidic and basic buffering groups to a polyacrylamide gel strip. This eliminates the ‘drifting’ which can occur in carrier ampholyte-based method. IPG strips are commercially available in various pH ranges with linear or non-linear pH gradients.
After IEF, proteins in the strip are subjected to the second dimension SDS-PAGE. In this procedure the proteins are coated with SDS in proportion to their mass [52] which makes them negatively charged. In an electric field, they will migrate to the cathode. Because of the ‘pores’ in the polyacrylamide, larger proteins move more slowly and smaller proteins faster; thus the separation is based on the mass of the proteins. Also, depending upon the concentration of acrylamide used for a particular gel, one can enrich the separation of proteins in a particular molecule mass range. For example, gels can be cast using 6%, 10%, or 15%, etc. polyacrylamide. Larger proteins would be separated better in lower percentages of acrylamide and vice versa for small proteins.
3.7 Staining of protein spots
Following SDS–PAGE, proteins are traditionally stained with Coomassie blue or a silver stain. Coomassie blue staining is simple and quantitative but lacks sensitivity. Silver staining is sensitive but the linearity between the staining intensity and the quantity of protein is not good. In addition, silver stain interferes with subsequent mass spectrometry (MS) analyses [53]. Fluorescent stains such as SYPRO Red, Ruby, and Orange (Molecular Probes, Inc., Eugene, OR) have been generated that possess both high sensitivity and with an excellent linear range and does not affect MS. Our laboratory routinely uses SYPRO Orange as a dye for staining 2D gels [54, 55].
3.8 MS and MS/MS
Proteins of interest (in this case of rhGH doping, those that show a different expression pattern in treated subjects compared with non treated controls) are excised from the gel and subjected to MS analysis. A common method used is ‘in-gel’ tryptic digestion of a protein followed by Matrix Assisted Laser Adsorption and Ionization – Time of Flight (MALDI-TOF). The resultant peptide fragments are ionized and then ‘fly’ through a vacuum tube, arriving at the detector where they are quantified as individual peaks ultimately generating a mass spectrum of peaks. This spectrum can then be compared to the theoretical mass finger print of proteins in a variety of data bases such as the NCBI non-redundant protein database [56] or the Swiss-Prot/TrEMBL [57]. In MS/MS analysis, two consecutive MS analyses are performed with further fragmentation of each tryptic peptide so that masses of the resulting smaller peptides and amino acid residues are determined and sequence information is obtained. Used together, MS and MS/MS generate a high degree of confidence for protein identification.
3.9 Potential biomarkers for GH using GH transgenic mice
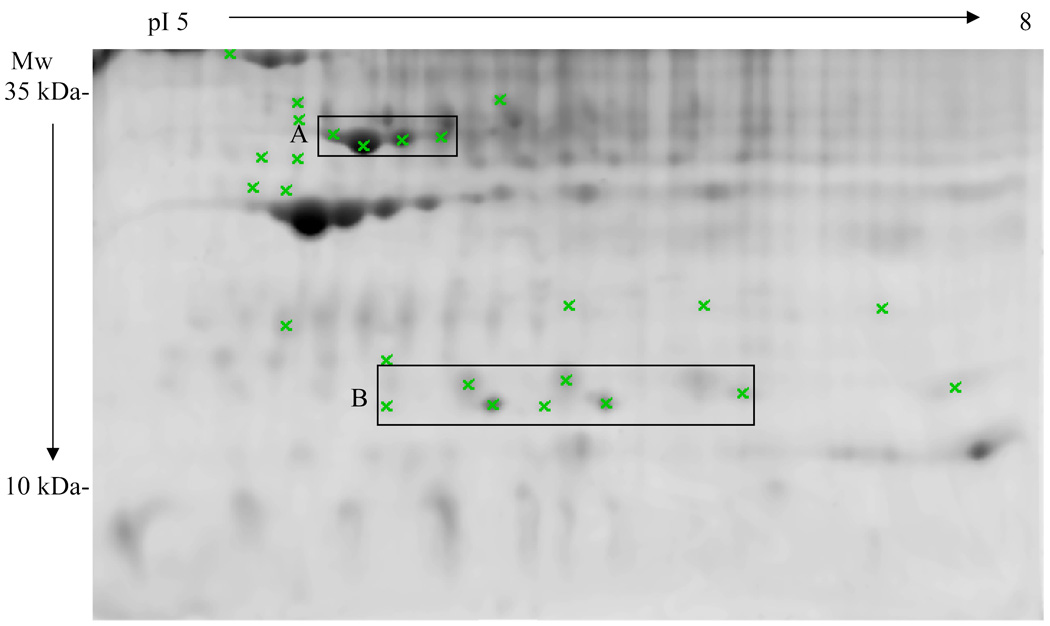
Our laboratory has created a transgenic mouse models that over expresses bovine (b) GH [58, 59]. These animals are giant with high levels of serum IGF-1 and are insulin resistant, lean, and die prematurely of liver, kidney, and heart disease. This model mimics acromegaly in humans and is useful to study the effect of GH on various tissues in the mouse including the kidney [60]. By analyzing the blood proteins of these mice by 2-DE, it is possible to determine which circulating proteins are up- or down-regulated by expression of the GH transgene. In our preliminary result, we have found several plasma proteins and protein isoforms that are differentially expressed in bGH mice comparing to their wild type littermates (Fig. 2). We are currently identifying these proteins by MS and MS/MS and the preliminary results indicate that isoforms of tranthyretin, clusterin, ApoE and ApoA1 are differentially expressed. These proteins may be potential biomarkers for initiating studies using rhGH.
Fig. 2.
2-D gel of the serum from a 4-month old male bGH transgenic mouse. The gel was stained with SYPRO Orange. Protein spots that show significant difference (p<0.05) in intensity between bGH and wild type mice are marked with Xs. Two groups of spots are enclosed by boxes labeled A and B. The four spots in box A (preliminarily identified as ApoE isoforms) are all significantly increased in bGH mice, whereas the seven spots in box B (preliminarily identified as Transthyretin isoforms) are all significantly decreased. 8 wild type mice and 10 bGH transgenic mice were included in this particular study. Abbreviations: pI, isoelectric focusing point; Mw, molecular weight.
4. Analyses of additional tissue types
In addition to blood, perhaps other tissues could be used for proteomic biomarker discover. For example, urine, and/or skin [54] holds potential for detection of rhGH abuse. By analyzing a spectrum of protein biomarkers in a sample instead of a single biomarker, the specific physiological response to rhGH doping may be resolved.
4.1 Urine biomarkers using proteomics
Urine is another noninvasive sample to detect doping and is currently widely used to detect various banned substances by the international Olympic committee and WADA. Under normal physiological conditions, urine output is approximately 1.5 L/day and contains hundreds of proteins [61]. Thus one sampling of urine of a few hundred ml can provide a sufficient amount of proteins for analysis. However, due to the low concentration of protein and high salt content, the urine needs to be concentrated and desalted in order to analyze proteins by 2-DE. Piper et al. identified 150 unique proteins using 2-DE and MALDI-TOF as well as liquid chromatography (LC)-MS/MS [62]. Several other groups also reported 200 to 400 unique proteins using various fractionation methods combined with MS/MS [63–65]. Although approximately 1400 distinct protein spots are detectable by 2-DE, identification of most of these proteins is difficult presumably due to post-translational modifications [62]. Recently, Adach et al. reported the identification of 1543 proteins from normal urine samples [66]. Interestingly nearly 50% of identified proteins were annotated as membrane proteins indicating the possibility of the presence of receptors or secretory proteins in the urine [66]. Since the effect of GH on these urinary proteins is largely unknown, quantification of these proteins in relation to GH administration may provide novel biomarkers specific for GH.
Detection of another banned substance, insulin, in urine has been reported [67]. Although the intact recombinant insulin is not detectable in urine, unique insulin degradation products are detectable using immunoaffinity chromatography (IAC) with subsequent LC-MS/MS analysis. This provides a method not only for detecting synthetic long acting insulin but also for detecting recombinant ‘native’ insulin abuse [67]. Compared to insulin, degradation products are not well characterized for GH, however, a similar methodology may be applicable. If hepatic or renal clearance produces unique GH degradation products, it may be possible to detect when rhGH is abused.
Unlike blood, urine samples are collected by the subject, thus, special care must be taken to minimize sample adulteration [68]. It is widely reported that sample substitution, dilution, and defilement occur frequently. Also, if urine samples are to be analyzed for proteins, contamination of proteases will certainly be a problem. This can be resolved by measurement of protease activity or by LC-MS/MS identification of proteases [68].
4.2 Skin biomarkers using 2-DE
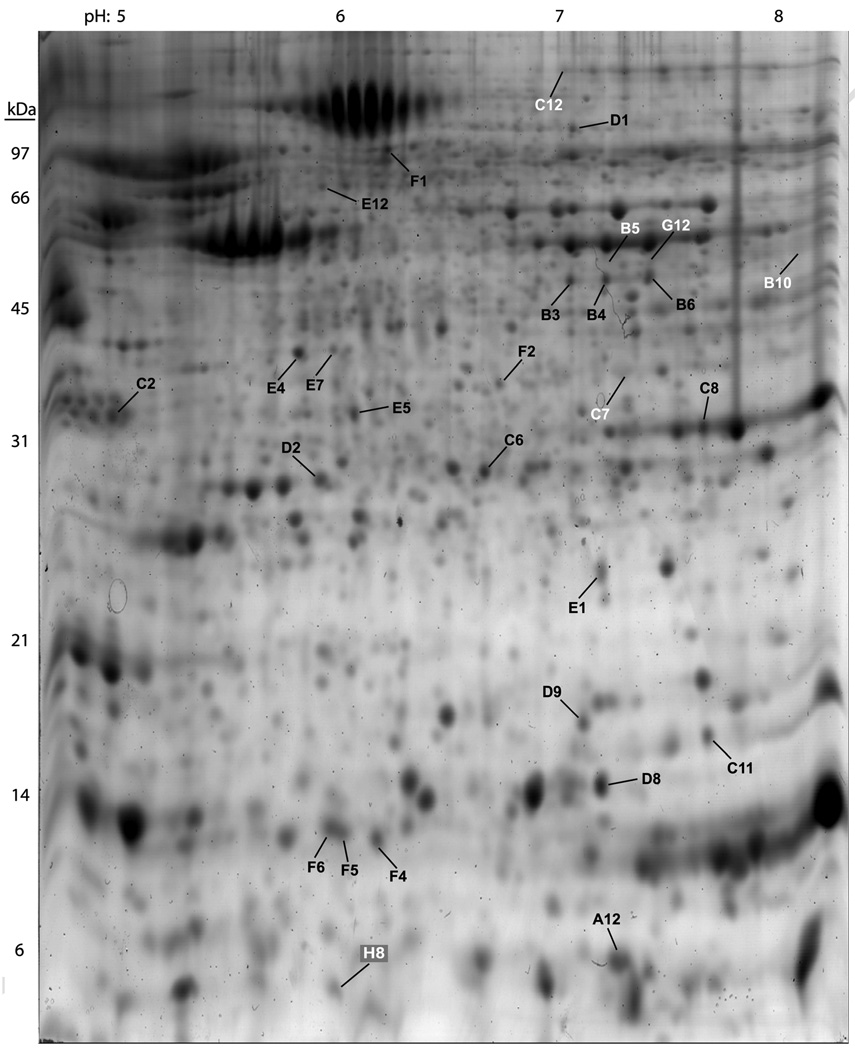
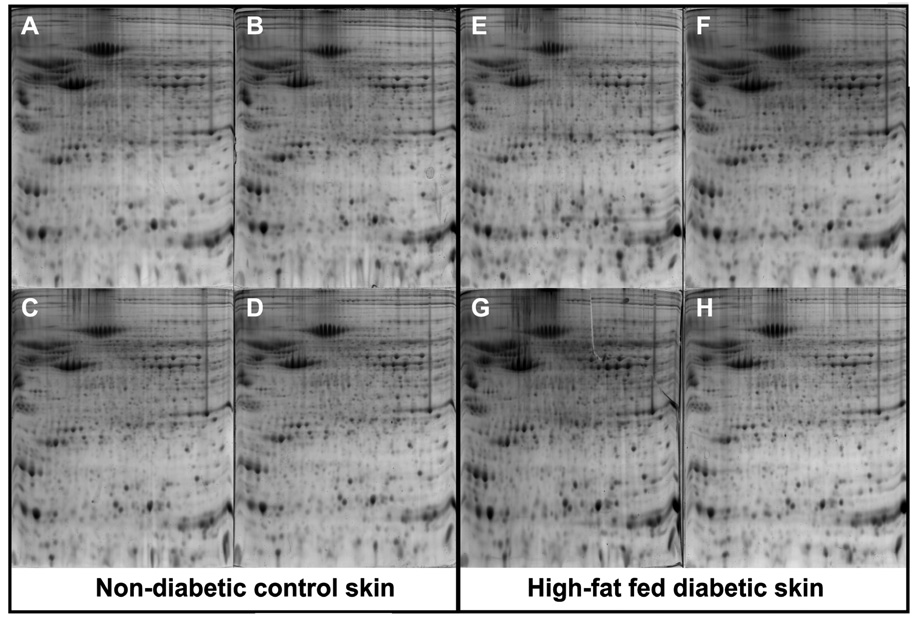
Our laboratory has developed a method for detection of skin proteins in mouse models of type 2 diabetes [69]. In this study, the comprehensive analysis of over 1,000 protein “spots” from skin of diabetic mice (Fig. 3) revealed 28 biomarkers (Table 1) that were significantly altered as compared to non-diabetic control mice. From these biomarkers it is possible to 1) select a single biomarker and develop a kit for detection or 2) to select several and consider the particular combination of up- and down- regulated proteins combined with the post-translational state of the proteins (such as the phosphorylation status of a protein) to indicate a disease state. We believe the second option holds more power as the profile of multiple biomarkers reduces the possibility of false positive. For GH doping, the same type of proteomic analysis should work since the procedure would be identical with only the protein profile changing. Excellent reproducibility was achieved using this technique (Fig. 4). However, skin biopsy analysis may be too invasive for testing athletes on multiple occasions. Therefore; serum and urine samples are still the ideal targets for similar proteomic profiling methodologies.
Fig. 3.
[69]. Two-dimensional gel of diabetic mouse skin. This SYPRO Orange stained gel contains proteins separated from the skin of diabetic mice at 19 week of age following 16 weeks on a high-fat diet. The gel was imaged using a VersaDoc 1000 Imaging System. The approximate pI and molecular weights are labeled along the top and left hand borders of the gel, respectively. Spot detection and densitometry were performed using the Discovery Series PDQuest 2-DE analysis software package version 7.0 that accompanied the VersaDoc 1000 Imaging System. Spots labeled with black letters and numbers represent proteins that were increased in the diabetic state while spots labeled with white letters and numbers represent proteins that were decreased in the diabetic state as compared to control skin samples. All labeled spots were removed from the polyacrylamide gel and analyzed by both MALDI-TOF and MS/MS mass spectrometry at the Michigan Proteome Consortium. Copyright Wiley-VCH Verlag GmbH & Co.KGaA. Reproduced with permission.
Table 1.
[69] Proteins found to be decreased or increased in the skin of high-fat fed diabetic mice following 16 weeks of feeding starting at 3 weeks of age as compared to protein level found in non-diabetic control mice fed standard rodent chow for 16 weeks starting at 3 weeks of age. Copyright Wiley-VCH Verlag GmbH & Co.KGaA. Reproduced with permission.
| Spot | Protein a | Mascot MS/MS Accession # | % Change(HF/C) b, c |
|---|---|---|---|
| B5 | creatine kinase (EC 2.7.3.2) chain M | A23590 | 936% ↓ |
| B10 | aldolase 1, A isoform | gi|42490830 | 612% ↓ |
| C7 | similar to glyceraldehyde-3-phosphate dehydrogenase | gi|6679937 | 805% ↓ |
| C12 | transferrin | gi|17046471 | 104% ↓ |
| G12 | creatine kinase (EC 2.7.3.2) chain M | A23590 | 127% ↓ |
| H8† | calpactin I light chain (Protein S100-A10) | S10AA_MOUSE | 192% ↓ |
| A12† | calpactin I light chain (Protein S100-A10) | S10AA_MOUSE | 153% ↑ |
| B3 | creatine kinase (EC 2.7.3.2) chain M | A23590 | 483% ↑ |
| B4 | creatine kinase (EC 2.7.3.2) chain M | A23590 | 413% ↑ |
| B6 | creatine kinase (EC 2.7.3.2) chain M | A23590 | 837% ↑ |
| C2 | 14-3-3 protein beta | gi|3065925 | 164% ↑ |
| C6 | peroxiredoxin 6 | gi|6671549 | 415% ↑ |
| C8 | phosphoglycerate mutase 1 | gi|10179944 | 208% ↑ |
| C11 | nucleoside-diphosphate kinase 2 | gi|6679078 | 350% ↑ |
| D1 | keratin complex 1, acidic gene 4 | gi|13386238 | 226% ↑ |
| D2 | apolipoprotein A-1 precursor | gi|109571 | 385% ↑ |
| D8 | lectin, galactose binding, soluble 7 | gi|31543120 | 120% ↑ |
| D9 | nucleoside-diphosphate kinase 1 | gi|37700232 | 63% ↑ |
| E1 | vacuolar protein sorting 29 | gi|9790285 | 64% ↑ |
| E4 | apolipoprotein E precursor | gi|114041 | 83% ↑ |
| E5 | prohibitin | gi|6679299 | 156% ↑ |
| E7 | malate dehydrogenase, cytoplasmic | gi|92087001 | 1262% ↑ |
| E12 | Keratin 1b | gi|38565071 | 808% ↑ |
| F1 | protein disulfide isomerase associated 3 | gi|112293264 | 1190% ↑ |
| F2 | proteasome (prosome, macropain) subunit, alpha type 1 | gi|33563282 | 195% ↑ |
| F4 | fatty acid-binding protein | PC4011 | 276% ↑ |
| F5 | fatty acid-binding protein, adipocyte | FABPA_MOUSE | 267% ↑ |
| F6 | fatty acid-binding protein, adipocyte | FABPA MOUSE | 403% ↑ |
A minimum of 2 significant MS/MS peptide fragments was considered sufficient to assign an ID for a spot. More detailed information can be found in the supplemental information section.
The mean decreases in spot intensities were determined using PDQuest 7.0.0 software on 24 gels from 8 mice.
Only differences that were statistically significant P<0.05 as determined by Student T-test as part of the PDQuest software are reported.
Indicates a weak identification by matching one significant fragment with at least one fragment that approached significance (P=0.014) & (P=0.089).
Fig. 4.
[69].Animal to animal reproducibility of skin samples resolved by 2-D gel electrophoresis. The 8 independent gels labeled with letters A-H represent skin proteins isolated from 8 individual animals that were sacrificed at 19 weeks of age after 16 weeks on a standard chow (A–D) or high-fat diet (E–H). Gels A, B, C and D on the left side of the figure are from 4 separate, non-diabetic control mice while gels E, F, G, and H on the right side of the figure are from 4 separate diabetic mice. Copyright Wiley-VCH Verlag GmbH & Co.KGaA. Reproduced with permission.
5. Genomics vs. Proteomics
While the concept of utilizing particular levels of multiple biomarkers as a readout for rhGH doping, we have concentrated on proteins. However, the levels of RNA is selective tissue utilizing gene chip arrays could be considered. However, we believe proteomic analysis is more appropriate.
Although genomics data are useful for many aspects of research, they are restricted to the information of the amount of RNA. While still immensely important, there are several limitations in using only genomic methods for the study of an altered physiological state (such as rhGH doping). Most importantly, proteins are the functional molecules of the cell while mRNAs serve mainly as intermediates between a given gene and its cognate protein. Quantification of mRNA in response to stimuli (such as GH doping) only approximates the level of protein. In addition, mRNA levels do not always correlate with the corresponding protein levels [70–72] and post-translational modifications of proteins can generate a variety of protein isoforms. Since genomics cannot measure the abundance or post-translational status of proteins, a different type of analysis is needed to enhance genomic information and better understand the status of a sample under a given condition (such as serum or urine of an individual that has been injecting GH).
Two excellent examples that illustrate the power of proteomics can be found in two studies about age-related changes in liver. In the first study, Keppler et al. [73] observed a four-fold increase in cathepsin B activity and a two-fold increase in cathepsin B protein level in livers of older rats compared to younger rats. Importantly, Northern blot analysis showed no change in cathepsin B mRNA levels between old and young rats. Thus, because of the sometimes poor correlation between mRNA and the cognate protein levels, this study exemplifies a major limitation of using mRNA to infer protein levels. In the second study, Helenius and colleagues [74] demonstrated that the age-related increase in nuclear factor-kappa-B (NF-κB)-binding activity in rat liver were accompanied by an increase in the abundance of p52 and p65 components. Analysis of proteins in these livers also revealed multiple post-translational modifications contributed to their regulation. The presence and abundance of various post-translational protein isoforms would not have been uncovered using RNA analysis methods. Thus, the resolution and sensitivity of proteomics-based approaches allowing for the detection of various post-translational modifications, which are often critical in elucidating the function of a protein, makes proteomics favorable to genomics for biomarker analysis at a more detailed level. Also, 2DE is the method of choice for analyses of protein isoforms, i.e. post-translationally altered proteins. Discovery of the chemical nature of the various protein isoforms is the newest challenge in the proteomic area.
6. Conclusion
To efficiently and specifically detect rhGH doping, more biomarkers are needed. Proteomic approaches provide a relatively new methodology of searching among thousands of proteins in blood, urine or skin for these new rhGH specific biomarkers. We believe that with more biomarkers, it will be possible to develop a robust, sensitive, and specific test system for rhGH using a combination of multiple markers with appropriate algorithms.
Acknowledgements
JJK is supported by funds from NIA (AG19899), NIDDK (DK075436), the State of Ohio’s Eminent Scholar Program that includes a gift from Milton and Lawrence Goll, the Diabetes Research Initiative at Ohio University, and a grant from DiAthegen LLC.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Healy ML, Gibney J, Russell-Jones DL, et al. High dose growth hormone exerts an anabolic effect at rest and during exercise in endurance-trained athletes. J. Clin. Endocrinol. Metab. 2003;88:5221–5226. doi: 10.1210/jc.2002-021872. [DOI] [PubMed] [Google Scholar]
- 2.Ehrnborg C, Ellegard L, Bosaeus I, Bengtsson BA, Rosen T. Supraphysiological growth hormone: Less fat, more extracellular fluid but uncertain effects on muscles in healthy, active young adults. Clin. Endocrinol. (Oxf) 2005;62:449–457. doi: 10.1111/j.1365-2265.2005.02240.x. [DOI] [PubMed] [Google Scholar]
- 3.Berggren A, Ehrnborg C, Rosen T, Ellegard L, Bengtsson BA, Caidahl K. Short-term administration of supraphysiological recombinant human growth hormone (GH) does not increase maximum endurance exercise capacity in healthy, active young men and women with normal GH-insulin-like growth factor I axes. J. Clin. Endocrinol. Metab. 2005;90:3268–3273. doi: 10.1210/jc.2004-1209. [DOI] [PubMed] [Google Scholar]
- 4.The world anti-doping code, THE 2008 PROHIBITED LIST, INTERNATIONAL STANDARD. 2008 http://www.wada-ama.org/rtecontent/document/2008_List_En.pdf.
- 5.Consensus guidelines for the diagnosis and treatment of adults with growth hormone deficiency: Summary statement of the growth hormone research society workshop on adult growth hormone deficiency. J. Clin. Endocrinol. Metab. 1998;83:379–381. doi: 10.1210/jcem.83.2.4611. [DOI] [PubMed] [Google Scholar]
- 6.Saugy M, Robinson N, Saudan C, Baume N, Avois L, Mangin P. Human growth hormone doping in sport. Br. J. Sports Med. 2006;40(Suppl 1):i35–i39. doi: 10.1136/bjsm.2006.027573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kopchick JJ. Discovery and development of a new class of drugs: GH antagonists. J. Endocrinol. Invest. 2003;26:16–26. [PubMed] [Google Scholar]
- 8.Holl RW, Schwarz U, Schauwecker P, Benz R, Veldhuis JD, Heinze E. Diurnal variation in the elimination rate of human growth hormone (GH): The half-life of serum GH is prolonged in the evening, and affected by the source of the hormone, as well as by body size and serum estradiol. J. Clin. Endocrinol. Metab. 1993;77:216–220. doi: 10.1210/jcem.77.1.8325945. [DOI] [PubMed] [Google Scholar]
- 9.Wu Z, Bidlingmaier M, Dall R, Strasburger CJ. Detection of doping with human growth hormone. Lancet. 1999;353:895. doi: 10.1016/S0140-6736(99)00775-8. [DOI] [PubMed] [Google Scholar]
- 10.Albini CH, Quattrin T, Vandlen RL, MacGillivray MH. Quantitation of urinary growth hormone in children with normal and abnormal growth. Pediatr. Res. 1988;23:89–92. doi: 10.1203/00006450-198801000-00020. [DOI] [PubMed] [Google Scholar]
- 11.Flanagan DE, Taylor MC, Parfitt V, Mardell R, Wood PJ, Leatherdale BA. Urinary growth hormone following exercise to assess growth hormone production in adults. Clin. Endocrinol. (Oxf) 1997;46:425–429. doi: 10.1046/j.1365-2265.1997.1410966.x. [DOI] [PubMed] [Google Scholar]
- 12.Leung KC, Howe C, Gui LY, Trout G, Veldhuis JD, Ho KK. Physiological and pharmacological regulation of 20-kDa growth hormone. Am. J. Physiol. Endocrinol. Metab. 2002;283:E836–E843. doi: 10.1152/ajpendo.00122.2002. [DOI] [PubMed] [Google Scholar]
- 13.Zhan X, Giorgianni F, Desiderio DM. Proteomics analysis of growth hormone isoforms in the human pituitary. Proteomics. 2005;5:1228–1241. doi: 10.1002/pmic.200400987. [DOI] [PubMed] [Google Scholar]
- 14.Bidlingmaier M, Wu Z, Strasburger CJ. Test method: GH, Baillieres Best Pract. Res. Clin. Endocrinol. Metab. 2000;14:99–109. doi: 10.1053/beem.2000.0057. [DOI] [PubMed] [Google Scholar]
- 15.Nelson AE, Ho KK. A robust test for growth hormone doping--present status and future prospects. Asian J. Androl. 2008;10:416–425. doi: 10.1111/j.1745-7262.2008.00395.x. [DOI] [PubMed] [Google Scholar]
- 16.Dall R, Longobardi S, Ehrnborg C, et al. The effect of four weeks of supraphysiological growth hormone administration on the insulin-like growth factor axis in women and men. GH-2000 study group. J. Clin. Endocrinol. Metab. 2000;85:4193–4200. doi: 10.1210/jcem.85.11.6964. [DOI] [PubMed] [Google Scholar]
- 17.Longobardi S, Keay N, Ehrnborg C, et al. Growth hormone (GH) effects on bone and collagen turnover in healthy adults and its potential as a marker of GH abuse in sports: A double blind, placebo-controlled study. the GH-2000 study group. J. Clin. Endocrinol. Metab. 2000;85:1505–1512. doi: 10.1210/jcem.85.4.6551. [DOI] [PubMed] [Google Scholar]
- 18.Nelson AE, Meinhardt U, Hansen JL, et al. Pharmacodynamics of growth hormone abuse biomarkers and the influence of gender and testosterone: A randomized double-blind placebo-controlled study in young recreational athletes. J. Clin. Endocrinol. Metab. 2008;93:2213–2222. doi: 10.1210/jc.2008-0402. [DOI] [PubMed] [Google Scholar]
- 19.Meinhardt UJ, Ho KK. Regulation of growth hormone action by gonadal steroids. Endocrinol. Metab. Clin. North Am. 2007;36:57–73. doi: 10.1016/j.ecl.2006.11.009. [DOI] [PubMed] [Google Scholar]
- 20.Ehrnborg C, Lange KH, Dall R, et al. The growth hormone/insulin-like growth factor-I axis hormones and bone markers in elite athletes in response to a maximum exercise test. J. Clin. Endocrinol. Metab. 2003;88:394–401. doi: 10.1210/jc.2002-020037. [DOI] [PubMed] [Google Scholar]
- 21.Wallace JD, Cuneo RC, Baxter R, et al. Responses of the growth hormone (GH) and insulin-like growth factor axis to exercise, GH administration, and GH withdrawal in trained adult males: A potential test for GH abuse in sport. J. Clin. Endocrinol. Metab. 1999;84:3591–3601. doi: 10.1210/jcem.84.10.6037. [DOI] [PubMed] [Google Scholar]
- 22.Wallace JD, Cuneo RC, Lundberg PA, et al. Responses of markers of bone and collagen turnover to exercise, growth hormone (GH) administration, and GH withdrawal in trained adult males. J. Clin. Endocrinol. Metab. 2000;85:124–133. doi: 10.1210/jcem.85.1.6262. [DOI] [PubMed] [Google Scholar]
- 23.Erotokritou-Mulligan I, Bassett EE, Bartlett C, et al. The effect of sports injury on insulin like growth factor-I and procollagen III peptide: Implications for detection of growth hormone abuse in athletes. J. Clin. Endocrinol. Metab. 2008 doi: 10.1210/jc.2007-2801. [DOI] [PubMed] [Google Scholar]
- 24.Nelson AE, Howe CJ, Nguyen TV, et al. Influence of demographic factors and sport type on growth hormone-responsive markers in elite athletes. J. Clin. Endocrinol. Metab. 2006;91:4424–4432. doi: 10.1210/jc.2006-0612. [DOI] [PubMed] [Google Scholar]
- 25.Erotokritou-Mulligan I, Bassett EE, Cowan D, et al. The influence of ethnicity on insulin like growth factor-I and procollagen III peptide in elite athletes and its effect on the ability to detect GH abuse. Clin. Endocrinol. (Oxf) 2008 doi: 10.1111/j.1365-2265.2008.03319.x. [DOI] [PubMed] [Google Scholar]
- 26.Erotokritou-Mulligan I, Bassett EE, Kniess A, Sonksen PH, Holt RI. Validation of the growth hormone (GH)-dependent marker method of detecting GH abuse in sport through the use of independent data sets. Growth Horm. IGF Res. 2007;17:416–423. doi: 10.1016/j.ghir.2007.04.013. [DOI] [PubMed] [Google Scholar]
- 27.Powrie JK, Bassett EE, Rosen T, et al. Detection of growth hormone abuse in sport. Growth Horm. IGF Res. 2007;17:220–226. doi: 10.1016/j.ghir.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 28.Ehrnborg C, Ohlsson C, Mohan S, Bengtsson BA, Rosen T. Increased serum concentration of IGFBP-4 and IGFBP-5 in healthy adults during one month's treatment with supraphysiological doses of growth hormone. Growth Horm. IGF Res. 2007;17:234–241. doi: 10.1016/j.ghir.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 29.Kniess A, Ziegler E, Kratzsch J, Thieme D, Muller RK. Potential parameters for the detection of hGH doping. Anal. Bioanal Chem. 2003;376:696–700. doi: 10.1007/s00216-003-1926-x. [DOI] [PubMed] [Google Scholar]
- 30.Nguyen TV, Nelson AE, Howe CJ, et al. Within-subject variability and analytic imprecision of insulinlike growth factor axis and collagen markers: Implications for clinical diagnosis and doping tests. Clin. Chem. 2008;54:1268–1276. doi: 10.1373/clinchem.2008.105726. [DOI] [PubMed] [Google Scholar]
- 31.Chung L, Clifford D, Buckley M, Baxter RC. Novel biomarkers of human growth hormone action from serum proteomic profiling using protein chip mass spectrometry. J. Clin. Endocrinol. Metab. 2006;91:671–677. doi: 10.1210/jc.2005-1137. [DOI] [PubMed] [Google Scholar]
- 32.Zhang R, Barker L, Pinchev D, et al. Mining biomarkers in human sera using proteomic tools. Proteomics. 2004;4:244–256. doi: 10.1002/pmic.200300495. [DOI] [PubMed] [Google Scholar]
- 33.Villanueva J, Shaffer DR, Philip J, et al. Differential exoprotease activities confer tumor-specific serum peptidome patterns. J. Clin. Invest. 2006;116:271–284. doi: 10.1172/JCI26022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang CM, Ananthaswamy HN, Barnes S, Ma Y, Kawai M, Elmets CA. Mass spectrometric proteomics profiles of in vivo tumor secretomes: Capillary ultrafiltration sampling of regressive tumor masses. Proteomics. 2006;6:6107–6116. doi: 10.1002/pmic.200600287. [DOI] [PubMed] [Google Scholar]
- 35.Righetti PG, Campostrini N, Pascali J, Hamdan M, Astner H. Eur. J. Mass. Spectrom. Vol. 10. Chichester; Eng: 2004. Quantitative proteomics: A review of different methodologies; pp. 335–348. [DOI] [PubMed] [Google Scholar]
- 36.MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760–1763. doi: 10.1126/science.289.5485.1760. [DOI] [PubMed] [Google Scholar]
- 37.Kopchick JJ, List EO, Kohn DT, Keidan GM, Qiu L, Okada S. Perspective: Proteomics--see “spots” run. Endocrinology. 2002;143:1990–1994. doi: 10.1210/endo.143.6.8882. [DOI] [PubMed] [Google Scholar]
- 38.Anderson L, Anderson NG. High resolution two-dimensional electrophoresis of human plasma proteins. Proceedings of the National Academy of Sciences. 1977;74:5421–5425. doi: 10.1073/pnas.74.12.5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallagher SR. One-dimensional SDS gel electrophoresis of proteins, Chapter 6. Curr. Protoc. Cell. Biol. 2007 doi: 10.1002/0471143030.cb0601s37. Unit 6.1. [DOI] [PubMed] [Google Scholar]
- 40.Rabilloud T. Detecting proteins separated by 2-D gel electrophoresis. Anal. Chem. 2000;72:48A–55A. doi: 10.1021/ac002709u. [DOI] [PubMed] [Google Scholar]
- 41.Banks RE, Dunn MJ, Hochstrasser DF, et al. Proteomics: New perspectives, new biomedical opportunities. Lancet. 2000;356:1749–1756. doi: 10.1016/S0140-6736(00)03214-1. [DOI] [PubMed] [Google Scholar]
- 42.Pandey AM. Mann, Proteomics to study genes and genomes. Nature. 2000;405:837–846. doi: 10.1038/35015709. [DOI] [PubMed] [Google Scholar]
- 43.Naaby-Hansen S, Waterfield MD, Cramer R. Proteomics--post-genomic cartography to understand gene function. Trends Pharmacol. Sci. 2001;22:376–384. doi: 10.1016/s0165-6147(00)01663-1. [DOI] [PubMed] [Google Scholar]
- 44.Castle JD. Purification of organelles from mammalian cells, Chapter 8. Curr. Protoc. Immunol. 2003 doi: 10.1002/0471142735.im0801bs56. Unit 8.1B. [DOI] [PubMed] [Google Scholar]
- 45.Goldberg S. Mechanical/physical methods of cell disruption and tissue homogenization. Methods Mol. Biol. 2008;424:3–22. doi: 10.1007/978-1-60327-064-9_1. [DOI] [PubMed] [Google Scholar]
- 46.Berkelman T. Removal of interfering substances in samples prepared for two-dimensional (2-d) electrophoresis. Methods Mol. Biol. 2008;424:51–62. doi: 10.1007/978-1-60327-064-9_5. [DOI] [PubMed] [Google Scholar]
- 47.Anderson NL, Anderson NG. The human plasma proteome: History, character, and diagnostic prospects. Mol Cell Proteomics. 2002;1:845–867. doi: 10.1074/mcp.r200007-mcp200. [DOI] [PubMed] [Google Scholar]
- 48.Anderson NL, Polanski M, Pieper R, et al. The human plasma proteome: A nonredundant list developed by combination of four separate sources. Mol Cell Proteomics. 2004;3:311–326. doi: 10.1074/mcp.M300127-MCP200. [DOI] [PubMed] [Google Scholar]
- 49.Steel LF, Trotter MG, Nakajima PB, Mattu TS, Gonye G, Block T. Efficient and specific removal of albumin from human serum samples. Mol Cell Proteomics. 2003;2:262–270. doi: 10.1074/mcp.M300026-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Echan LA, Tang HY, Ali-Khan N, Lee K, Speicher DW. Depletion of multiple high-abundance proteins improves protein profiling capacities of human serum and plasma. Proteomics. 2005;5:3292–3303. doi: 10.1002/pmic.200401228. [DOI] [PubMed] [Google Scholar]
- 51.Guerrier L, Righetti PG, Boschetti E. Reduction of dynamic protein concentration range of biological extracts for the discovery of low-abundance proteins by means of hexapeptide ligand library. Nat. Protoc. 2008;3:883–890. doi: 10.1038/nprot.2008.59. [DOI] [PubMed] [Google Scholar]
- 52.Shapiro AL, Vinuela E, Maizel JV., Jr Molecular weight estimation of polypeptide chains by electrophoresis in SDS-polyacrylamide gels. Biochem. Biophys. Res. Commun. 1967;28:815–820. doi: 10.1016/0006-291x(67)90391-9. [DOI] [PubMed] [Google Scholar]
- 53.Lauber WM, Carroll JA, Dufield DR, Kiesel JR, Radabaugh MR, Malone JP. Mass spectrometry compatibility of two-dimensional gel protein stains. Electrophoresis. 2001;22:906–918. doi: 10.1002/1522-2683()22:5<906::AID-ELPS906>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 54.List EO, Berryman DE, Palmer AJ, et al. Analysis of mouse skin reveals proteins that are altered in a diet-induced diabetic state: A new method for detection of type 2 diabetes. Proteomics. 2007;7:1140–1149. doi: 10.1002/pmic.200600641. [DOI] [PubMed] [Google Scholar]
- 55.Qiu L, List EO, Kopchick JJ. Differentially expressed proteins in the pancreas of diet-induced diabetic mice. Mol. Cell. Proteomics. 2005;4:1311–1318. doi: 10.1074/mcp.M500016-MCP200. [DOI] [PubMed] [Google Scholar]
- 56.NCBI non-redundant protein database. ( http://www.ncbi.nlm.nih.gov.)
- 57.Swiss-Prot/TrEMBL. ( http://www.expasy.org.)
- 58.McGrane MM, de Vente J, Yun J, et al. Tissue-specific expression and dietary regulation of a chimeric phosphoenolpyruvate carboxykinase/bovine growth hormone gene in transgenic mice. J. Biol. Chem. 1988;263:11443–11451. [PubMed] [Google Scholar]
- 59.Chen WY, Wight DC, Mehta BV, Wagner TE, Kopchick JJ. Glycine 119 of bovine growth hormone is critical for growth-promoting activity. Mol. Endocrinol. 1991;5:1845–1852. doi: 10.1210/mend-5-12-1845. [DOI] [PubMed] [Google Scholar]
- 60.Doi SQ, Jacot TA, Sellitti DF, et al. Growth hormone increases inducible nitric oxide synthase expression in mesangial cells. J. Am. Soc. Nephrol. 2000;11:1419–1425. doi: 10.1681/ASN.V1181419. [DOI] [PubMed] [Google Scholar]
- 61.Brunzel NA. 2nd. Philadelphia, PA: Saunders; 2004. Fundamentals of Urine & Body Fluid Analysis. [Google Scholar]
- 62.Pieper R, Gatlin CL, McGrath AM, et al. Characterization of the human urinary proteome: A method for high-resolution display of urinary proteins on two-dimensional electrophoresis gels with a yield of nearly 1400 distinct protein spots. Proteomics. 2004;4:1159–1174. doi: 10.1002/pmic.200300661. [DOI] [PubMed] [Google Scholar]
- 63.Sun W, Li f, Wu S, et al. Human urine proteome analysis by three separation approaches. Proteomics. 2005;5:4994–5001. doi: 10.1002/pmic.200401334. [DOI] [PubMed] [Google Scholar]
- 64.Wang L, Li F, Sun W, et al. Concanavalin A-captured glycoproteins in healthy human urine. Mol. Cell. Proteomics. 2006;5:560–562. doi: 10.1074/mcp.D500013-MCP200. [DOI] [PubMed] [Google Scholar]
- 65.Castagna A, Cecconi D, Sennels L, et al. Exploring the hidden human urinary proteome via ligand library beads. J. Proteome Res. 2005;4:1917–1930. doi: 10.1021/pr050153r. [DOI] [PubMed] [Google Scholar]
- 66.Adachi J, Kumar C, Zhang Y, Olsen JV, Mann M. The human urinary proteome contains more than 1500 proteins, including a large proportion of membrane proteins. Genome Biol. 2006;7:R80. doi: 10.1186/gb-2006-7-9-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Thomas A, Thevis M, Delahaut P, Bosseloir A, Schanzer W. Mass spectrometric identification of degradation products of insulin and its long-acting analogues in human urine for doping control purposes. Anal. Chem. 2007;79:2518–2524. doi: 10.1021/ac062037t. [DOI] [PubMed] [Google Scholar]
- 68.Thevis M, Schanzer W. Mass spectrometric identification of peptide hormones in doping-control analysis. Analyst. 2007;132:287–291. doi: 10.1039/b618748j. [DOI] [PubMed] [Google Scholar]
- 69.List EO, Berryman DE, Palmer AJ, et al. Analysis of mouse skin reveals proteins that are altered in a diet-induced diabetic state: A new method for detection of type 2 diabetes. Proteomics. 2007;7:1140–1149. doi: 10.1002/pmic.200600641. [DOI] [PubMed] [Google Scholar]
- 70.Khochbin S, Gorka C, Lawrence JJ. Multiple control level governing H10 mRNA and protein accumulation. FEBS Lett. 1991;283:65–67. doi: 10.1016/0014-5793(91)80554-g. [DOI] [PubMed] [Google Scholar]
- 71.Rousseau D, Khochbin S, Gorka C, Lawrence JJ. Induction of H1(0)-gene expression in B16 murine melanoma cells. Eur. J. Biochem. 1992;208:775–779. doi: 10.1111/j.1432-1033.1992.tb17247.x. [DOI] [PubMed] [Google Scholar]
- 72.Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- 73.Keppler D, Walter R, Perez C, Sierra F. Increased expression of mature cathepsin B in aging rat liver. Cell Tissue Res. 2000;302:181–188. doi: 10.1007/s004410000269. [DOI] [PubMed] [Google Scholar]
- 74.Helenius M, Kyrylenko S, Vehvilainen P, Salminen A. Characterization of aging-associated up-regulation of constitutive nuclear factor-kappa B binding activity. Antioxid. Redox Signal. 2001;3:147–156. doi: 10.1089/152308601750100669. [DOI] [PubMed] [Google Scholar]