Abstract
The three-dimensional organization of mitotic microtubules in a mutant strain of Saccharomyces cerevisiae has been studied by computer-assisted serial reconstruction. At the nonpermissive temperature, cdc20 cells arrested with a spindle length of approximately 2.5 microns. These spindles contained a mean of 81 microtubules (range, 56-100) compared with 23 in wild-type spindles of comparable length. This increase in spindle microtubule number resulted in a total polymer length up to four times that of wild-type spindles. The spindle pole bodies in the cdc20 cells were approximately 2.3 times the size of wild-type, thereby accommodating the abnormally large number of spindle microtubules. The cdc20 spindles contained a large number of interpolar microtubules organized in a "core bundle." A neighbor density analysis of this bundle at the spindle midzone showed a preferred spacing of approximately 35 nm center-to-center between microtubules of opposite polarity. Although this is evidence of specific interaction between antiparallel microtubules, mutant spindles were less ordered than the spindle of wild-type cells. The number of noncore microtubules was significantly higher than that reported for wild-type, and these microtubules did not display a characteristic metaphase configuration. cdc20 spindles showed significantly more cross-bridges between spindle microtubules than were seen in the wild type. The cross-bridge density was highest between antiparallel microtubules. These data suggest that spindle microtubules are stabilized in cdc20 cells and that the CDC20 gene product may be involved in cell cycle processes that promote spindle microtubule disassembly.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J Bacteriol. 1975 Oct;124(1):511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Duplication of spindle plaques and integration of the yeast cell cycle. Cold Spring Harb Symp Quant Biol. 1974;38:123–131. doi: 10.1101/sqb.1974.038.01.016. [DOI] [PubMed] [Google Scholar]
- Dawson I. A., Roth S., Artavanis-Tsakonas S. The Drosophila cell cycle gene fizzy is required for normal degradation of cyclins A and B during mitosis and has homology to the CDC20 gene of Saccharomyces cerevisiae. J Cell Biol. 1995 May;129(3):725–737. doi: 10.1083/jcb.129.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding R., McDonald K. L., McIntosh J. R. Three-dimensional reconstruction and analysis of mitotic spindles from the yeast, Schizosaccharomyces pombe. J Cell Biol. 1993 Jan;120(1):141–151. doi: 10.1083/jcb.120.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guacci V., Hogan E., Koshland D. Chromosome condensation and sister chromatid pairing in budding yeast. J Cell Biol. 1994 May;125(3):517–530. doi: 10.1083/jcb.125.3.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Mortimer R. K., Culotti J., Culotti M. Genetic Control of the Cell Division Cycle in Yeast: V. Genetic Analysis of cdc Mutants. Genetics. 1973 Jun;74(2):267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Smith D. Altered fidelity of mitotic chromosome transmission in cell cycle mutants of S. cerevisiae. Genetics. 1985 Jul;110(3):381–395. doi: 10.1093/genetics/110.3.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L. H., Weinert T. A. Checkpoints: controls that ensure the order of cell cycle events. Science. 1989 Nov 3;246(4930):629–634. doi: 10.1126/science.2683079. [DOI] [PubMed] [Google Scholar]
- Holloway S. L., Glotzer M., King R. W., Murray A. W. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor. Cell. 1993 Jul 2;73(7):1393–1402. doi: 10.1016/0092-8674(93)90364-v. [DOI] [PubMed] [Google Scholar]
- Hoyt M. A., Totis L., Roberts B. T. S. cerevisiae genes required for cell cycle arrest in response to loss of microtubule function. Cell. 1991 Aug 9;66(3):507–517. doi: 10.1016/0092-8674(81)90014-3. [DOI] [PubMed] [Google Scholar]
- Irniger S., Piatti S., Michaelis C., Nasmyth K. Genes involved in sister chromatid separation are needed for B-type cyclin proteolysis in budding yeast. Cell. 1995 Apr 21;81(2):269–278. doi: 10.1016/0092-8674(95)90337-2. [DOI] [PubMed] [Google Scholar]
- King R. W., Peters J. M., Tugendreich S., Rolfe M., Hieter P., Kirschner M. W. A 20S complex containing CDC27 and CDC16 catalyzes the mitosis-specific conjugation of ubiquitin to cyclin B. Cell. 1995 Apr 21;81(2):279–288. doi: 10.1016/0092-8674(95)90338-0. [DOI] [PubMed] [Google Scholar]
- Li R., Murray A. W. Feedback control of mitosis in budding yeast. Cell. 1991 Aug 9;66(3):519–531. doi: 10.1016/0092-8674(81)90015-5. [DOI] [PubMed] [Google Scholar]
- Mastronarde D. N., McDonald K. L., Ding R., McIntosh J. R. Interpolar spindle microtubules in PTK cells. J Cell Biol. 1993 Dec;123(6 Pt 1):1475–1489. doi: 10.1083/jcb.123.6.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. L., O'Toole E. T., Mastronarde D. N., McIntosh J. R. Kinetochore microtubules in PTK cells. J Cell Biol. 1992 Jul;118(2):369–383. doi: 10.1083/jcb.118.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison T., Evans L., Schulze E., Kirschner M. Sites of microtubule assembly and disassembly in the mitotic spindle. Cell. 1986 May 23;45(4):515–527. doi: 10.1016/0092-8674(86)90283-7. [DOI] [PubMed] [Google Scholar]
- Murray A. W. Creative blocks: cell-cycle checkpoints and feedback controls. Nature. 1992 Oct 15;359(6396):599–604. doi: 10.1038/359599a0. [DOI] [PubMed] [Google Scholar]
- Page B. D., Snyder M. Chromosome segregation in yeast. Annu Rev Microbiol. 1993;47:231–261. doi: 10.1146/annurev.mi.47.100193.001311. [DOI] [PubMed] [Google Scholar]
- Palmer R. E., Koval M., Koshland D. The dynamics of chromosome movement in the budding yeast Saccharomyces cerevisiae. J Cell Biol. 1989 Dec;109(6 Pt 2):3355–3366. doi: 10.1083/jcb.109.6.3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer R. E., Sullivan D. S., Huffaker T., Koshland D. Role of astral microtubules and actin in spindle orientation and migration in the budding yeast, Saccharomyces cerevisiae. J Cell Biol. 1992 Nov;119(3):583–593. doi: 10.1083/jcb.119.3.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts B. T., Farr K. A., Hoyt M. A. The Saccharomyces cerevisiae checkpoint gene BUB1 encodes a novel protein kinase. Mol Cell Biol. 1994 Dec;14(12):8282–8291. doi: 10.1128/mcb.14.12.8282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sethi N., Monteagudo M. C., Koshland D., Hogan E., Burke D. J. The CDC20 gene product of Saccharomyces cerevisiae, a beta-transducin homolog, is required for a subset of microtubule-dependent cellular processes. Mol Cell Biol. 1991 Nov;11(11):5592–5602. doi: 10.1128/mcb.11.11.5592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F. Getting started with yeast. Methods Enzymol. 1991;194:3–21. doi: 10.1016/0076-6879(91)94004-v. [DOI] [PubMed] [Google Scholar]
- Surana U., Amon A., Dowzer C., McGrew J., Byers B., Nasmyth K. Destruction of the CDC28/CLB mitotic kinase is not required for the metaphase to anaphase transition in budding yeast. EMBO J. 1993 May;12(5):1969–1978. doi: 10.1002/j.1460-2075.1993.tb05846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tugendreich S., Tomkiel J., Earnshaw W., Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995 Apr 21;81(2):261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Wang Y., Burke D. J. Checkpoint genes required to delay cell division in response to nocodazole respond to impaired kinetochore function in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1995 Dec;15(12):6838–6844. doi: 10.1128/mcb.15.12.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstein J., Jacobsen F. W., Hsu-Chen J., Wu T., Baum L. G. A novel mammalian protein, p55CDC, present in dividing cells is associated with protein kinase activity and has homology to the Saccharomyces cerevisiae cell division cycle proteins Cdc20 and Cdc4. Mol Cell Biol. 1994 May;14(5):3350–3363. doi: 10.1128/mcb.14.5.3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells W. A., Murray A. W. Aberrantly segregating centromeres activate the spindle assembly checkpoint in budding yeast. J Cell Biol. 1996 Apr;133(1):75–84. doi: 10.1083/jcb.133.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winey M., Goetsch L., Baum P., Byers B. MPS1 and MPS2: novel yeast genes defining distinct steps of spindle pole body duplication. J Cell Biol. 1991 Aug;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
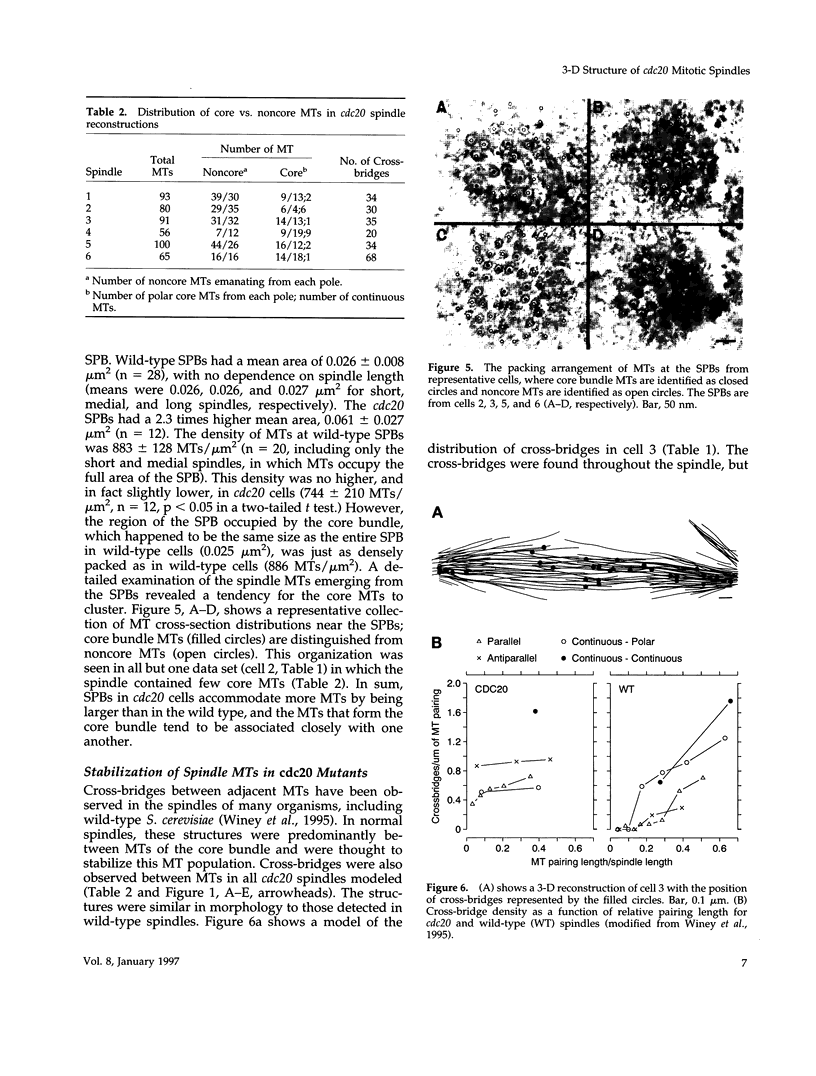
- Winey M., Mamay C. L., O'Toole E. T., Mastronarde D. N., Giddings T. H., Jr, McDonald K. L., McIntosh J. R. Three-dimensional ultrastructural analysis of the Saccharomyces cerevisiae mitotic spindle. J Cell Biol. 1995 Jun;129(6):1601–1615. doi: 10.1083/jcb.129.6.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]