Abstract
Individuals differ in their tendencies to seek positive decision outcomes or to avoid negative ones. At the neurobiological level, our model suggests that phasic changes in dopamine support learning to reinforce good decisions via striatal D1 receptors, and to avoid maladaptive choices via striatal D2 receptors. Accordingly, in a previous study individual differences in positive and negative learning were strongly modulated by two genetic polymorphisms factors related to striatal D1 and D2 function, respectively. Nevertheless, whereas the role for dopamine in positive learning is relatively well accepted, that in learning to avoid negative outcomes is more controversial. Here we further explore D2-receptor-related genetic contributions to probabilistic avoidance, in light of recent data showing that particular DRD2 polymorphisms are associated with functional modulation of receptor expression (Zhang et al 2007, PNAS). We find that a promoter polymorphism rs12364283 associated with transcription and D2 receptor density was strongly and selectively predictive of avoidance-based decisions. Two further polymorphisms (rs2283265 and rs1076560) associated with relatively reduced presynaptic relative to postsynaptic D2 receptor expression were predictive of relative impairments in negative compared to positive decisions. These previously undocumented effects of DRD2 polymorphisms were largely independent of those we reported previously for the C957T polymorphism (rs6277) associated with striatal D2 density. In contrast, effects of the commonly studied Taq1A polymorphism on reinforcement-based decisions were due to indirect association with C957T. Taken together these findings suggest multiple D2-dependent genetic mechanisms contributing to avoidance. We discuss these effects in the context of neurocomputational models of reinforcement leaning in the basal ganglia.
Keywords: genetics, basal ganglia, dopamine, reinforcement learning, computational model
Introduction
Many of the decisions we make on a day-to-day basis require balancing out factors driving the desire to achieve good results (be they financial, intellectual, scientific etc), with those associated with avoiding potentially perilous situations. Such decisions often benefit from past experience, and are therefore supported by adaptive mechanisms for learning from positive and negative outcomes. In a complex, multi-faceted world, decision outcomes are generally probabilistic rather than deterministic, and an adaptive decision making system will take this into account. Indeed, the phenomenon of relying on intuition or making “gut-level” decisions may reflect the output of an implicit system reporting its integrated value over multiple positive and negative outcomes experienced in the past, divorced from explicit memory of those outcomes (Frank, O’Reilly, & Curran, 2006; Daw, Niv, & Dayan, 2005).
When confronted with potential gains and losses, individuals differ substantially, both in their choices and in their neural responsiveness to reinforcement (e.g., Scheres & Sanfey, 2006; Frank, Woroch, & Curran, 2005; Frank, Moustafa, Haughey, Curran, & Hutchison, 2007a; Klein, Neumann, Reuter, Hennig, von Cramon, & Ullsperger, 2007). Although such choice preferences may be influenced by social and cultural factors, and are subject to changes in state (e.g., mood; Harl & Sanfey, 2007), genetic factors also contribute substantially (Frank et al., 2007a; Klein et al., 2007; Yacubian, Sommer, Schroeder, Glscher, Kalisch, Leuenberger, Braus, & Bchel, 2007; Forbes, Brown, Kimak, Ferrell, Manuck, & Hariri, 2008; Frank, Doll, Oas-Terpstra, & Moreno, in preparation). However, given the myriad of potential genes that could influence such choices, exploratory studies scanning the entire genome and identifying predictive factors x, y and z may suffer from an inability to draw substantive conclusions due to multiple comparisons, type-I errors, and the correlational nature of genetic findings. A different approach is to constrain genetic analysis to candidate genetic factors that are known to alter processing in brain regions critical for the cognitive process of interest (Green, Munafo, Deyoung, Fossella, Fan, & Gray, 2008; Tan, Chen, Goldberg, Mattay, Meyer-Lindenberg, Weinberger, & Callicott, 2007b; Tan, Callicott, & Weinberger, 2007a; Frank et al., 2007a; Frank et al., in preparation). Although not without caveats, reasonable conclusions can then be drawn if (i) focusing on functional polymorphisms that affect coding of the protein of interest; (ii) there exist theoretical or conceptual models for how that protein in the brain region(s) of interest contributes to the associated cognitive process; and (iii) a suitable task is administered which probes the specific computations of that system. An added desirable factor is that converging pharmacological data exist (ideally in combination with functional imaging or patient studies) in which manipulation of the neurotransmitter in the brain region of interest produces corresponding changes in the same cognitive process, providing a “proof of concept” that any genetic effects associated with this neurotransmitter are causal rather than simply correlational.
In the domain of reward-based decisions, considerable evidence implicates a critical role for dopaminergic processes within reinforcement learning systems of the basal ganglia (Frank, Seeberger, & O’Reilly, 2004; Pessiglione, Seymour, Flandin, Dolan, & Frith, 2006; D’Ardenne, McClure, Nystrom, & Cohen, 2008; Santesso, Dillon, Birk, Holmes, Goetz, Bogdan, & Pizzagalli, 2008; Nakamura & Hikosaka, 2006; Siessmeier, Kienast, Wrase, Larsen, Braus, Smolka, Buchholz, Schreckenberger, Rosch, Cumming, Mann, Bartenstein, & Heinz, 2006; Hariri, Brown, Williamson, Flory, de Wit, & Manuck, 2006; Santesso, Evins, Frank, Cowman, & Pizzagalli, in press; Kahnt, Park, Cohen, Beck, Heinz, & Wrase, in press; Cools, Frank, Gibbs, Miyakawa, Jagust, & D’Esposito, 2009; Moustafa, Cohen, Sherman, & Frank, 2008a). Experimental data suggest that phasic bursts and dips in midbrain dopamine (DA) firing encode “reward prediction errors” in terms of whether outcomes are better or worse than expected (Montague, Dayan, & Sejnowski, 1997; Schultz, Dayan, & Montague, 1997). These signals modify synaptic plasticity in the basal ganglia (Reynolds & Wickens, 2002) and are thought to drive adaptive learning. Although much of this work has focused on learning from positive rewards, increasing evidence also implicates a role for dopamine as supporting learning from negative prediction errors (Frank, 2005; Frank et al., 2004). Connectivity between dorsal striatum and dopaminergic midbrain areas is predictive of learning from both positive and negative outcomes (Kahnt et al., in press). Moreover, Parkinson’s patients, who while off medication have depleted DA levels, actually show enhanced ability to avoid decisions that had probabilistically been associated with negative outcomes (Frank et al., 2004), Pharmacological manipulation of DA modulates learning from both positive and negative outcomes in opposite directions, such that striatal DA elevations promote positive learning whereas striatal DA depletion promotes negative learning (Frank et al., 2004; Cools, Altamirano, & D’Esposito, 2006; Pessiglione et al., 2006; Frank & O’Reilly, 2006; Frank, Samanta, Moustafa, & Sherman, 2007b; Frank et al., in preparation; Costa, Gutierrez, de Araujo, Coelho, Kloth, Gainetdinov, Caron, Nicolelis, & Simon, 2007; Moustafa et al., 2008a; Cools et al., 2009). These data are largely consistent with theoretical models positing that phasic DA bursts facilitate positive learning in striatal “Go” neurons expressing D1 receptors, whereas DA dips promote avoidance learning in striatal “NoGo” neurons expressing D2 receptors (Frank, 2005). Direct evidence for these dual learning mechanisms in the same striatal populations posited by the models has recently been reported in rodent synaptic plasticity studies (Shen, Flajolet, Greengard, & Surmeier, 2008).
At the behavioral level in humans, we previously reported a genetic study in which we collected DNA from 69 healthy undergraduate participants and analyzed polymorphisms within three dopaminergic genes (Frank et al., 2007a). All of these genes were associated with different aspects of reinforcement-based decision making. First, a polymorphism within the DARPP-32 gene (Meyer-Lindenberg, Straub, Lipska, Verchinski, Goldberg, Callicott, Egan, Huffaker, Mattay, Kolachana, Kleinman, & Weinberger, 2007) was associated with better learning from positive outcomes. This result supports the various models implicating striatal D1 receptors in reward learning: DARPP-32 is a protein that is highly concentrated in the striatum, is activated by D1 receptor stimulation, and is required for D1-receptor mediated synaptic plasticity and reward learning in rodents (Stipanovich, Valjent, Matamales, Nishi, Ahn, Maroteaux, Bertran-Gonzalez, Brami-Cherrier, Enslen, Corbill, Filhol, Nairn, Greengard, Herv, & Girault, 2008; Calabresi, Gubellini, Centonze, Picconi, Bernardi, Chergui, Svenningsson, Fienberg, & Greengard, 2000). We also reported that a the C957T polymorphism within the DRD2 gene (Duan, Wainwright, Comeron, Saitou, Sanders, Gelernter, & Gejman, 2003), associated with increased striatal D2 receptor density (Hirvonen, Laakso, Rinne, Pohjalainen, & Hietala, 2005), was associated with enhanced learning from negative outcomes (Frank et al., 2007a; Frank et al., in preparation). Together, these findings implicate a strong genetic basis for learning from both positive and negative outcomes to maximize the probability that later decisions will yield positive outcomes and minimize the probability of negative outcomes. Moreover the results are remarkably consistent with the neural models synaptic plasticity studies noted above (Frank, 2005; Shen et al., 2008).
Theoretically, a role for D2 receptors (and therefore dopamine) in avoidance learning is controversial. As noted above, the DARPP-32 result supports the relatively well accepted notion that phasic DA bursts facilitate reward learning via activation of striatal D1 receptors (e.g., Reynolds, Hyland, & Wickens, 2001; Reynolds & Wickens, 2002), which act via DARPP-32 to initiate the cascade of intracellular events leading to synaptic weight changes and behavioral reward seeking (Stipanovich et al., 2008; Calabresi et al., 2000). In contrast, due to the low baseline firing rate and limited dynamic range of DA dips, their effectiveness as a learning signal has been questioned (Bayer & Glimcher, 2005; Daw, Kakade, & Dayan, 2002). However, we have argued that the relatively smaller changes in DA firing rates during dips compared to bursts may be compensated by the relatively enhanced sensitivity of high-affinity D2 receptors, making them particularly sensitive to the removal of DA from the synapse during DA dips (Frank, 2005; Cohen & Frank, in press). This notion is consistent with plasticity studies showing that a lack of D2 receptor stimulation promotes synaptic potentiation in striatopallidal (NoGo) cells (Shen et al., 2008). Thus the finding that DRD2 polymorphisms associated with individual differences in striatal D2 receptor density are predictive of learning from negative outcomes (including both C957T and Taq1A polymorphisms; (Frank et al., 2007a; Klein et al., 2007) is well grounded by physiologically plausible mechanisms.
Recently, Zhang, Bertolino, Fazio, Blasi, Rampino, Romano, Lee, Xiao, Papp, Wang, and Sade (2007) analyzed 23 polymorphisms within the DRD2 gene in terms of their effects on D2 receptor mRNA expression in postmortem brain tissue, in an effort to deqtermine which of them functionally controlled D2 receptor function. Interestingly, neither the Taq1A nor the C957T polymorphisms were directly responsible for changes in D2 receptor expression. Instead, a previously uncharacterized promoter SNP (rs12364283, referred to as SNP2 in Zhang et al. (2007)) within DRD2 was significantly associated with mRNA expression and transcription. Zhang et al. (2007) further showed that two other SNPs (rs2283265 and rs1076560, referred as SNPs 17/19 by Zhang et al. (2007), in complete linkage disequilibrium with each other) were predictive of relative expression of short and long isoforms of the D2 receptor. This finding is particularly informative, given that these isoforms correspond to presynaptic vs postsynaptic D2 receptors, respectively (Usiello, Baik, Roug-Pont, Picetti, Dierich, LeMeur, Piazza, & Borrelli, 2000), and thereby modulate different aspects of dopaminergic function. In particular, presynaptic receptors include autoreceptors which regulate the degree of phasic dopamine release (e.g., Wu, Reith, Walker, Kuhn, Carroll, & Garris, 2002), whereas postsynaptic receptors expressed on striatal NoGo neurons modulate changes in neural activity and plasticity resulting from changes in DA levels (Frank & O’Reilly, 2006). Thus this polymorphism has potential to explain various perplexing effects in the literature, whereby the effects of D2 dopaminergic drugs on reinforcement learning and working memory are dependent on baseline measures (Kimberg, D’Esposito, & Farah, 1998; Mattay, Callicott, Bertolino, Heaton, Frank, Coppola, Berman, Goldberg, & Weinberger, 2000; Frank & O’Reilly, 2006; Cools, Frank, Gibbs, Miyakawa, Jagust, & D’Esposito, in press; Gibbs & D’Esposito, 2005; Cohen, Krohn-Grimberghe, Elger, & Weber, 2007). These effects are potentially related to relative expression of presynaptic vs postsynaptic D2 receptors (Frank & O’Reilly, 2006; Cools et al., in press).
In light of these data, we reanalyzed data from Frank et al. (2007a), and include novel genotype data including the commonly studied Taq1A SNP, together with the 3 novel functional DRD2 SNPs identified by Zhang et al. (2007). We find that the Zhang SNPs are indeed predictive of avoidance learning, further supporting a role for D2 receptor function in accounting for individual differences in this aspect of decision making. However, our previously reported effects of C957T remained significant even when controlling for these other SNPs, suggesting that these polymorphisms contribute to independent aspects of DRD2 function. We propose differential pre-and postsynaptic D2 mechanisms which are controlled by the different polymorphisms and which can account for impairments in avoidance learning via distinct mechanisms, providing predictions for future experiments.
Materials and Methods
Sample
The sample is the same as that reported in Frank et al. (2007a). There were 69 healthy participants (30 females, 39 males), between the ages of eighteen and thirty-five (M = 21). The vast majority of participants were white, with three participants categorizing themselves as “more than one race”. We were unable to obtain genotypes for one subject for rs2283265/rs1076560, and two subjects for rs12364283.
All SNPs were in Hardy Weinberg equilibrium (p’s > 0.2). The minor allele frequencies were as follows: rs12364283: 14.3%, consistent with 15% in the population at large. rs2283265/rs1076560, 16.9%, consistent with 16.7% in the population at large. rs1800497, 15.9%, compared with 22.5% at large. For rs1677, 46.4%, compared with 46.6% at large. The gene-dose effect of rs6277 (C957T) (Frank et al., 2007a) corresponded to a breakdown of 13:38:18 (C/C:C/T:T/T), representative of the same proportions in the population.
Genotyping
Samples were genotyped using TaqMan® primer and probe pairs; the probes are conjugated to two different dyes (one for each allelic variant). Taqman assays are designed and selected using the SNPBrowser™ program (Applied Biosystems) and ordered directly from this company. The PCR reaction mixture consists of 20 ng of genomic DNA, 1x Universal PCR Master Mix, 900 nM of each primer and 200 nM of each probe in a 15 μL reaction volume. Amplification is performed using the TaqMan® Universal Thermal Cycling Protocol and fluorescence intensity will be measured using the ABI Prism 7500 Real-Time PCR System. Genotypes were acquired using the 7500 system’s allelic discrimination software (SDS version 1.2.3).
Task
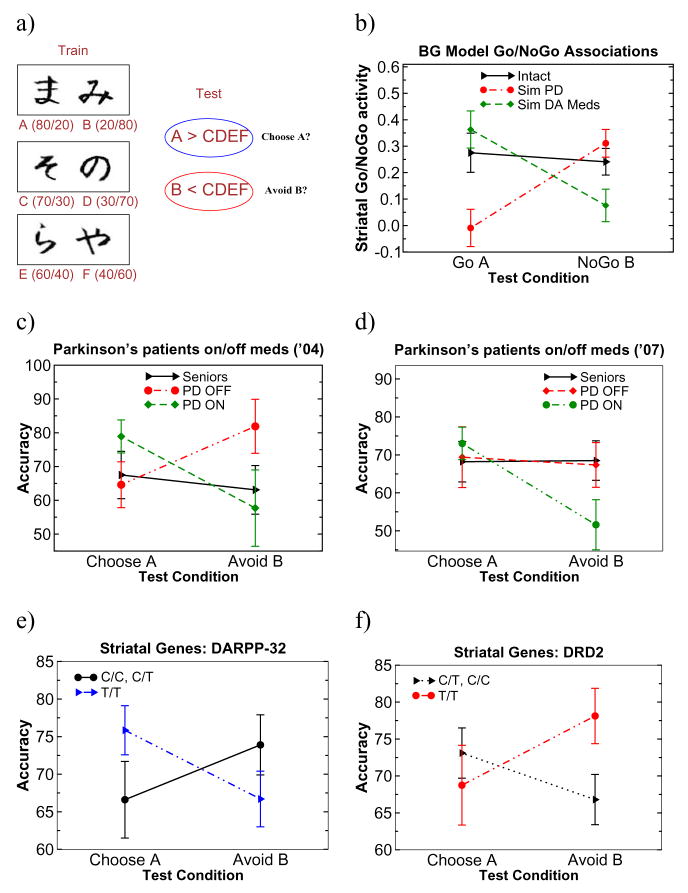
We administered the probabilistic selection reinforcement learning task (Frank et al., 2004). Three different stimulus pairs (AB, CD, EF) are presented in random order and participants have to learn to choose one of the two stimuli (Figure 1a). Feedback follows the choice to indicate whether it was correct or incorrect, but this feedback is probabilistic. In AB trials, a choice of stimulus A leads to correct (positive) feedback in 80% of AB trials, whereas a B choice leads to incorrect (negative) feedback in these trials (and vice-versa for the remaining 20% of trials). CD and EF pairs are less reliable: stimulus C is correct in 70% of CD trials, while E is correct in 60% of EF trials. Learning to choose A over B could be accomplished either by learning that A leads to positive feedback or that B leads to negative feedback (or both). To evaluate whether participants learned more about positive or negative outcomes of their decisions, we subsequently tested them with novel combinations of stimulus pairs involving either an A (AC, AD, AE, AF) or a B (BC, BD, BE, BF); no feedback was provided. If participants had learned more from positive feedback, they should reliably choose stimulus A in all novel test pairs in which it is present. On the other hand, if they learned more from negative feedback, they should more reliably avoid stimulus B. Consistent with this depiction, error-related brain activity was enhanced in participants who were particularly adept at avoiding B (Frank et al., 2005).
Figure 1.
a) Probabilistic selection reinforcement learning task. During training, participants select among each stimulus pair. Probabilities of receiving positive/negative feedback for each stimulus are indicated in parentheses. In the test phase, all combinations of stimuli are presented without feedback. “Go learning” is indexed by reliable choice of the most positive stimulus A in these novel pairs, whereas “NoGo learning” is indexed by reliable avoidance of the most negative stimulus B. b) Striatal Go and NoGo activation states when presented with input stimuli A and B respectively. Simulated Parkinson’s (Sim PD) was implemented by reducing striatal DA levels, whereas medication (Sim DA Meds) was simulated by increasing DA levels and partially shunting the effects of DA dips during negative feedback. c) Behavioral findings in PD patients on/off medication supporting model predictions (Frank et al., 2004). d) Replication in another group of patients, where here the most prominent effects were observed in the NoGo learning condition (Frank et al., 2007b). e), f) Individual differences in Go/NoGo learning in college students are predicted by genes controlling striatal D1/D2 function (Frank et al., 2007a).
Analysis
General linear models were used for all statistical analysis. The independent variables were either categorical (for most SNPs contrasting one genotype to another) or the number of T alleles in C957T to examine gene-dose effects. When considering all novel SNPs we also include secondary analysis in which the previously reported gene-dose effect of C957T is included as a regressor in the model, to determine whether effects of these polymorphisms contribute independent variance to that of C957T.
Results
There were no effects of any SNP on overall accuracy during the training phase (p’s > 0.1). The following analysis focuses on test phase reward and avoidance accuracy, as indicators of whether positive or negative aspects of stimuli drive their choices.
Taq1A, rs1800497 (Zhang et al 2007 “SNP23”)
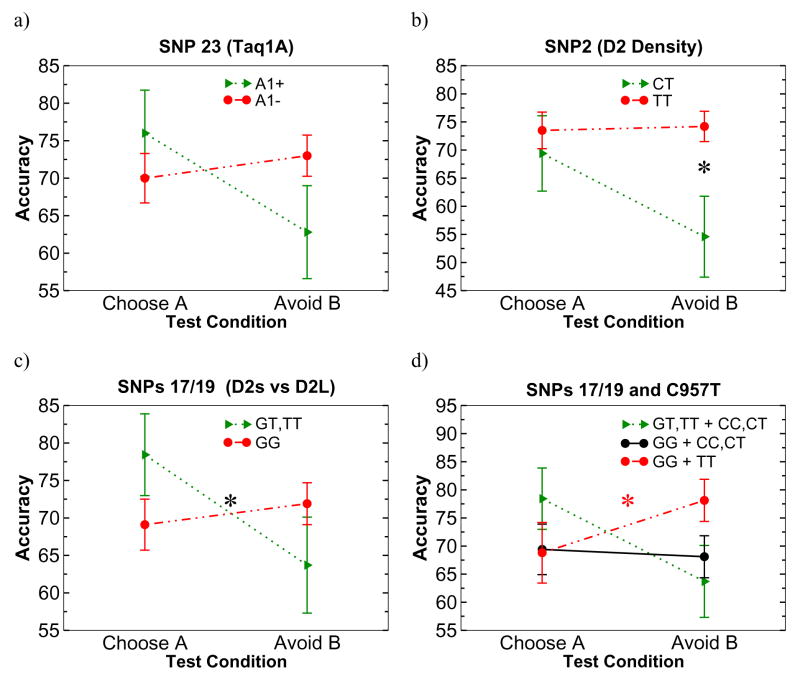
As noted above, despite being distantly downstream from the DRD2 gene, the Taq1A polymorphism is sometimes associated with reduced D2 receptor density (Pohjalainen, Rinne, Ngren, Lehikoinen, Anttila, Syvlahti, & Hietala, 1998), and in a recent study was also associated with avoidance learning in a modest sample (Klein et al., 2007). This effect was also evident in our sample (Figure 3a). While there was no overall effect of Taq1A genotype on overall performance in the test phase (F = .2), there was an interaction between Taq1A genotype and choose-A versus avoid-B condition (F[1,67] = 3.8, p =.056). Compared with A1− carriers, A1+ carriers showed numerically better positive learning (F[1,67] = 0.9, ns) but worse negative learning (F[1.67] = 3.1, p =.08). Reaction time analysis in correct trials showed a similar pattern, with A1+ carriers showing significantly slower responding in avoid-B trials (F[1,66]= 4.0, p = .05), with no difference in choose-A response times (F[1,66] = 0.9, ns).
Figure 3.
DRD2 effects on accuracy in choose-A (approach) and avoid-B (avoidance) conditions. a) Taq1A SNP, showing relatively impaired avoidance in A1+ carriers. However this effect appears to be due to indirect linkage with C957T (see text). b Promoter SNP 2 from Zhang et al, affecting D2 receptor mRNA transcription, selectively and substantially impacted avoidance learning. c) SNPs 17/19 from Zhang et al, affecting relative presynaptic autoreceptor vs postsynaptic D2 receptor expression, impacted relative reward to avoidance learning. d) Interactions between SNPs 17/19 and C957T. Carriers of the major rs2283265/rs1076560 GG genotype who were also C957T TT carriers showed relative better avoidance learning, whereas carriers of the minor rs2283265/rs1076560 allele who also carried a C957T C allele showed relatively impaired avoidance but better reward learning. The intermediate group showed no learning bias.1
However, because Taq1A is 3′ downstream from DRD2, we posited that the effects of this genotype on avoidance may be indirect, due to its linkage with other DRD2 polymorphisms, including C957T. Indeed, A1+ participants carried less C957T T alleles than did A1− participants (F = 7.4, p = .008). Moreover, when both Taq1A and C957T were included in the model, the previously described gene-dose effect of C957T on relative avoid-B than choose-A performance (Frank et al., 2007a) remained significant (F[1,66] = 5.9, p = .0178), while the effect of Taq1A vanished (F[1,66]= 1.0, ns). Similar effects were seen for avoid-B performance (F[1,66] = 3.2, p=.08 and F[1,66] = 1.0). Thus the observed effect of Taq1A on avoidance appears to be due to its indirect linkage with C957T, rather than directly controlling D2 receptor function and avoidance. This may also relate to the inconsistent association between Taq1A and D2 density, due to failed attempts to replicate this finding (e.g., Laruelle, Gelertner, & Innis, 1998). Similar to our finding, other studies have shown that while Taq1A is not directly associated with sensitivity to drugs of abuse, it is in linkage with other D2 SNPs, which themselves were associated with drug abuse (Heinz, Sander, Harms, Finckh, Kuhn, Dufeu, Dettling, Grf, Rolfs, Rommelspacher, & Schmidt, 1997; Finckh, Rommelspacher, Kuhn, Dufeu, Otto, Heinz, Dettling, Giraldo-Velasquez, Pelz, Graf, Harms, Sander, Schmidt, & Rolfs, 1997).
Promoter SNP rs12364283 (Zhang et al 2007 “SNP2”)
This SNP, 844 bp upstream of the DRD2 transcription start site, was the only SNP identified by Zhang et al. (2007) to actually influence overall D2 receptor mRNA expression in postmortem brain tissue. In terms of probabilistic learning performance, there was a main effect of this SNP (F[1,65] = 6.4, p = .01), and a trend for an interaction between positive and negative test condition (F[1,65]= 2.6, p = 0.1). Planned comparisons revealed a highly significant effect of this promoter SNP on avoid-B performance (F[1,65] = 9.5, p = .003, Figure 3b), such that C carriers performed substantially worse than T/T homozygotes, and with no significant effect on choose-A performance (F[1,65] = 0.4, ns). Similar marginal effects were seen for response times, with C carriers responding slower in avoid-B trials (F[1,64] = 3.4, p = .07), but not choose-A (F[1,64] = 1.1).
Unlike Taq1A, there was no association between rs12364283 genotype and the number of T alleles in C957T (F[1,65] = 1.8, p =.18). When both genotypes were included in the statistical model, the gene-dose effect of C957T on relative avoid-B to choose-A performance remained significant (F[1,64]=7.6, p = .008), but that of rs12364283 vanished (F[1,64] = 1.5, p > .2). However, for avoid-B performance alone, the effect of rs12364283 remained significant even when controlling for number of C957T alleles (F[1,64] = 7.7, p =.007), and the C957T gene-dose effect remained marginally significant as well (F[1,64] = 3.6, p = .06).
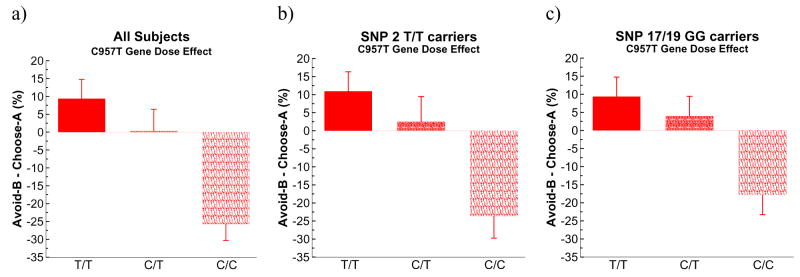
To further test whether the gene-dose effect of C957T held regardless of rs12364283, we analyzed performance only in the 51 participants carrying the major rs12364283 T/T genotype. Here, the C957T gene-dose effect continued to hold for relative avoid-B to choose-A performance (F(1,50] = 6.8, p = .01, Figure 4b) and, marginally, for avoid-B performance alone (F[1,50] = 3.5, p = .06).
Figure 4.
Gene dose effects of C957T on relative avoidance in a) all participants, b) carriers of the major T/T genotype of rs12364283, and c) carriers of the major G/G genotype of SNPS 17/19.
Similarly, we analyzed performance only in the largest C957T group (ie the C/T heterozygotes). In this population alone there was still a significant effect of rs1236283 on avoid-B performance (F[1,34] = 4.4, p = .04). Thus, these two DRD2 polymorphisms, both associated with D2 receptor density, appear to contribute independently to avoidance.
rs2283265 and rs1076560 (Zhang et al 2007 “SNPs 17/19”)
These two SNPs studied by Zhang et al. (2007) are in complete linkage disequilibrium, and accordingly there was 100% correspondence between the genotypes in our sample. In the reinforcement task, there was no overall effect of genotype on performance in the test phase (F = .01), but there was a significant interaction between rs2283265/rs1076560 genotype and choose-A versus avoid-B condition (F[1,66] = 4.3, p =.04). Compared with G/G homozygotes, T carriers showed numerically better positive learning (F[1,66] = 2.2, p =.14) but worse negative learning (F[1.66]=1.9, p =.17). G/G homozygotes have relatively more balanced expression of pre and postsynaptic D2 receptors (Zhang et al., 2007), and showed relatively similar positive and negative test performance (Figure 3c). Reaction time analysis in correct trials showed a similar pattern, with the minor T allele associated with significantly slower responding in avoid-B trials (F[1,65]= 6.4, p = .01) and no difference in choose-A (F[1,65] = 1.9, ns).
These initial findings raise the question of whether our previously reported effects of C957T on avoidance learning simply reflected a correlational effect resulting from linkage disequilibrium with these polymorphisms (Zhang et al., 2007). Although there was evidence for some degree of linkage, in that the major G/G genotype was associated with more C957T T alleles (F[1,69] = 7.5, p = .008), follow-up analysis suggests that this was not the case. First, the above linkage association was due only to the fact that all 18 C957T T/T homozygotes were also GG carriers of SNP 17/19 (but not vice-versa) – when considering the remaining 51 participants there was no association between the two SNPs (Fisher’s exact test p > 0.7). Second, when controlling for rs2283265/rs1076560 genotype in the general linear model, the C957T gene-dose effect continued to hold for relative avoid-B compared to choose-A (F[1.65] = 5.7, p = .02; Figure 4)) and marginally for avoid-B performance (F[1,65] = 3.7, p = .058). When analyzing rs2283265/rs1076560 G/G homozygotes alone, the gene-dose effect of C957T on relative avoidance learning remained significant (F[1,45] = 5.6, p=.02).1 Those G/G participants who also carried the C957T T/T genotype showed relatively better avoidance than those who did not (Figure 3d). Thus, it appears that the C957T polymorphism provides additional contributions over and beyond that indicated by SNPs 17/19, given that similar C957T gene-dose effects of the former were observed in both genotypes of the latter.
Finally, the converse analysis focused only on C/T heterozygotes for C957T, as another measure of whether the SNP17/19 effect was independent of C957T (as in the previous section). In this limited sample, the RT effects of SNPs 17/19 remained significant, with T carriers still showing slower avoid-B responding (F[1,34] = 4.7, p =.038). The effects on relative avoidance accuracy were no longer significant (p > 0.1), but the positive learning advantage of TT carriers approached significance (p=.07). Together these findings suggest independent mechanisms of the SNPs on reinforcement based mechanisms, possibly due to SNPs 17/19 effect on presynaptic autoreceptors (see discussion).
To further evaluate the nature of these two genetic effects, we tested for a gene-gene interaction. Both genotypes and their interaction were entered into the statistical general linear model. In this analysis, main effects of C957T gene-dose and rs2283265/rs1076560 genotype were both significant (F[1,64] = 8.0, p = .006, and F[1,65] = 4.1, p = .04, respectively), again suggesting independent effects. There was also a gene-gene interaction (F[1,64] = 4.2, p = .046). Post-hoc comparisons revealed that this interaction resulted from a modulation of the large avoidance deficit normally seen in C957T C/C homozygotes, such that there was a protective effect of rs2283265/rs1076560 G/G genotype (Figure 5). Those C/C participants who were also G/G for rs2283265/rs1076560 G/G performed significantly better than those carrying the minor allele (F[1,11] = 8.8, p = .01), and did not differ from C957T T/T participants (p > 0.5). Said otherwise, the greatest avoidance deficit was observed in C/C participants who also carried a rs2283265/rs1076560 T allele. Finally, there were no such interactions between any of the DRD2 SNPs and either the DARPP-32 or COMT genes on either positive or negative learning (all p’s > 0.4).
Figure 5.
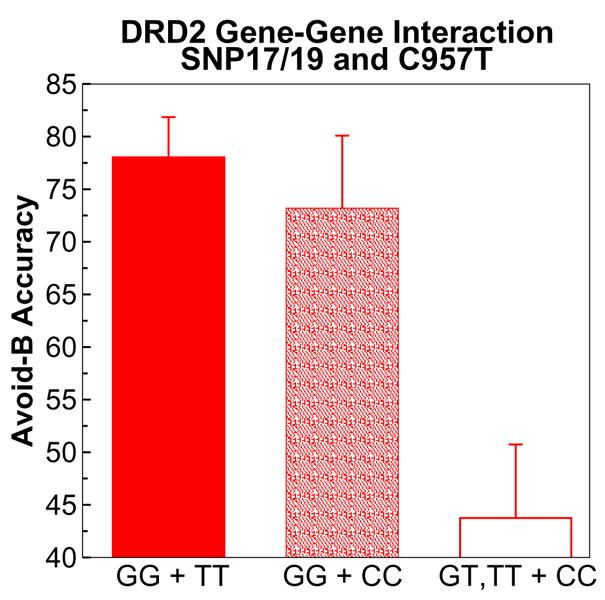
Gene-gene interactions on performance in the Avoid-B test condition. Although SNPs 17/19 and C957T were all predictive of avoidance learning, the presence of the major GG genotype in SNPs 17/19 was protective against the detrimental effects of the C957T C/C genotype.
Explained Variance due to DRD2 Genetic Factors
Given the robust genetic contributions to avoidance, it is of interest to know what proportion of the variance in overall avoidance, and in relative avoidance to reward-seeking behavior, is accounted for by these genetic variables. When all three DRD2 SNPs (2, 17/19, and C957T) are included in the general linear model, 21.2% of avoid-B variance was explained, and 16.7% of relative avoidance to reward-seeking choices. For C957T alone, the explained variance is 7.3% for avoid-B and 11.7% for relative avoidance. For SNP2 alone, the explained variance is 12.8% for avoid-B and 3.9% for relative avoidance. For SNPs 17/19 alone, the explained variance is 2.9% for avoid-B and 6.2% for relative avoidance.
Discussion
Taken together the above findings suggest multiple functional effects of DRD2 genes on brain D2 receptor function, which in turn contribute to probabilistic avoidance (Table 1). First, we find that the previously reported gene-dose effects of the C957T polymorphism (Frank et al., 2007a) continued to hold even when controlling for other DRD2 SNPs. These results imply a functional effect of C957T, consistent with observations that this SNP influences postsynaptic striatal D2 receptor density (Hirvonen et al., 2005), possibly due to alterations of mRNA stability (Duan et al., 2003).
Table 1.
Summary of genotype/phenotype effects. Single arrows indicate trends, double arrows significant effects
| Phenotype/Genotype | Choose-A | Avoid-B | Rel (Avd-B - Chs-A) | RT (Avd B) | Indep ? |
|---|---|---|---|---|---|
| Taq1A (A1+) | ↑ | ↓ | ↓↓† | ↑↑† | N |
| C957T (C carriers) | – | ↓↓ | ↓↓ | ↑↑ | Y |
| SNP2 (C carriers) | – | ↓↓ | ↓ | ↑ | Y |
| SNPs 17/19 (T carriers) | ↑ | ↓ | ↓↓ | ↑↑ | Y |
Taq1A effects disappeared when controlling for C957T, suggesting this effect was due to linkage (see text). Indep refers to whether the particular SNP showed evidence for phenotypic modulation independently of the others.
Second, we find that the Taq1A polymorphism located downstream of DRD2 also predicts relative avoidance (Klein et al., 2007), but that this effect is likely due to linkage with C957T (or other DRD2 SNPs), because the effects of this SNP vanished when controlling for C957T. Similar to our finding, other studies have shown that while Taq1A is not directly associated with sensitivity to drugs of abuse, it is in linkage with other D2 SNPs, which themselves were associated with drug sensitivity (Heinz et al., 1997; Finckh et al., 1997).
Third, we find a novel selective and substantial effect of rs12364283 on avoidance. This promoter SNP directly influences D2 receptor mRNA transcription and brain D2 receptor density (Zhang et al., 2007) and therefore provides additional supporting evidence for a role for D2 receptors in learning to avoid decisions associated with negative outcomes.
Finally, we find that the SNPs rs2283265/rs1076560, which affect splicing and relative expression of presynaptic vs postsynaptic D2 receptor mRNA (Zhang et al., 2007), are also predictive of relative avoidance to reward-seeking behavior. In this case, there was no significant effect on raw avoidance, but instead on relative avoidance to reward-seeking choices. Interestingly, those carrying the minor T allele showed numerically worse avoidance together with elevated choose-A performance. This significant interaction is intriguing, given that these participants have reduced expression of presynaptic D2 autoreceptors (Zhang et al., 2007). Because these autoreceptors are necessary for regulating phasic DA release and reuptake (Bolan, Kivell, Jaligam, Oz, Jayanthi, Han, Sen, Urizar, Gomes, Devi, Ramamoorthy, Javitch, Zapata, & Shippenberg, 2007; Meiergerd, Patterson, & Schenk, 1993; Wu et al., 2002), these participants may have enhanced phasic DA release and slower reuptake; the resulting elevated synaptic DA levels may enhance positive learning but prevent DA levels from sufficiently decreasing during negative outcomes. This explanation is consistent with the effects of pharmacologically-induced elevations in DA, which result in learning deficits from negative outcomes in this and related tasks (Frank et al., 2004; Frank et al., 2007b; Cools et al., 2006; Moustafa et al., 2008a; Cools et al., 2009).
According to the neurocomputational models, the above interpretation suggests two distinct mechanisms leading to avoidance learning deficits resulting from polymorphisms within DRD2. C957T and promoter rs12364283 may be associated with fundamental changes in postsynaptic D2 receptors expressed by striatal “NoGo” neurons (Gerfen, 1992; Frank, 2005; Shen et al., 2008), altering their modulation and plasticity in response to DA dips. In contrast, due to a shortage of presynaptic autoreceptors, SNPs rs2283265/rs1076560 may be associated with increases in presynaptic DA levels that are detrimental for learning from negative outcomes which requires DA levels to drop sufficiently. This interpretation is consistent with the gene-gene interaction, whereby those participants putatively having both reduced postsynaptic D2 density (C957T) and increased presynaptic DA levels (rs2283265/rs1076560) showed the most profound avoidance deficit. While presently speculative, this distinction predicts that if reinforcement feedback were delivered more intermittently (ie. with longer inter-trial intervals), there would be more time for DA levels to return to baseline following DA bursts, and the effects of polymorphisms affecting presynaptic receptors on avoidance would be less apparent. Conversely, if rewards were delivered more frequently, and with shorter trials, the effects may be exacerbated. Future research is necessary to test these hypotheses.
One question is why should participants with enhanced postsynaptic D2 function show the same pattern of data as Parkinson’s patients, who have depleted DA levels? Our assumption is that striatal D2 receptors, which are typically in the high-affinity state, are sensitive to, and inhibited by, even low baseline DA levels. During negative outcomes, DA dips are proposed to disinhibit striatopallidal neurons expressing D2 receptors. The greater the D2 receptor density, the more likely that these neurons are inhibited by tonic DA, and therefore the greater learning signal that would arise when DA levels drop. Furthermore, depleted DA levels as in PD would actually enhance this effect. Due to low tonic DA levels, cessation in DA firing would be associated with a greater probability that all DA would be cleared from the synapse during DA dips, thereby disinhibiting D2 receptors. Further, there is some evidence that D2 receptors are upregulated/supersensitive in PD (Seeman, 2008; Rinne, Laihinen, Rinne, Ngren, Bergman, & Ruotsalainen, 1990; Valtteri, M., Kjell, Pertti, Vesa, & O., 2000), and that the striatopallidal NoGo cells are more excitable in the DA-depleted state (Surmeier, Ding, Day, Wang, & Shen, 2007). Thus, despite the apparent paradox, both low DA levels and enhanced D2 receptor density should both be associated with enhanced avoidance, as seen empirically.
A limitation in this analysis is the sample size. Although clearly large enough to detect effects of individual SNPs on three distinct aspects of reinforcement learning (Frank et al., 2007a; Frank et al., in preparation), and here to detect a gene-gene interaction between SNPs rs2283265/rs1076560 and C957T, larger samples may be necessary to more conclusively examine interactions among the various SNPs.
In sum, we show multiple genetic mechanisms associated with probabilistic avoidance learning, all linked to factors controlling striatal D2 receptor function. These effects contrast with those of other DA-related genes, such as COMT and DARPP-32 which affect other aspects of reinforcement learning. This specificity therefore supports a role for striatal D2 receptors in NoGo learning. It remains to be seen whether these genetic mechanisms generalize to other cognitive functions thought to depend on striatal NoGo function, such as filtering out distracting information from being updated into working memory (O’Reilly & Frank, 2006; Frank & O’Reilly, 2006; Moustafa, Sherman, & Frank, 2008b; McNab & Klingberg, 2007). Moreover, these genetic effects may help explain the detrimental effects of D2 agonists on producing impulsivity and pathological gambling in a subset of Parkinson’s patients (Dodd, Klos, Bower, Geda, Josephs, & Ahlskog, 2005).
Figure 2.
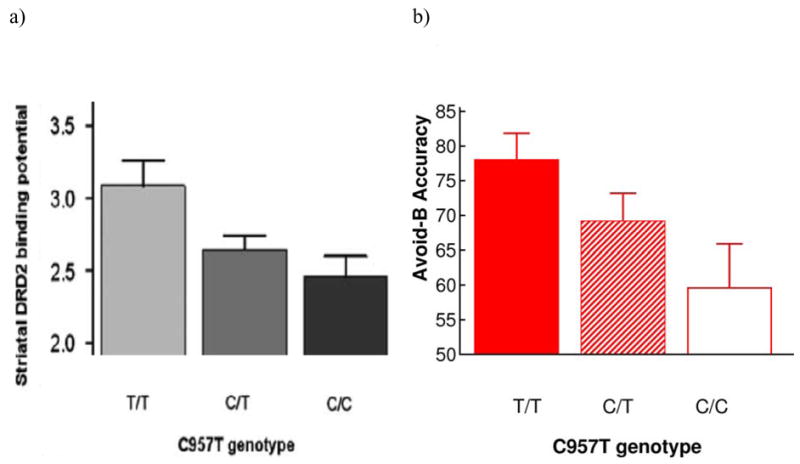
Gene dose effects. a) Monotonic effects of number of T alleles in the DRD2 gene, showing increased striatal D2 receptor density (Adapted by permission from Macmillan Publishers Ltd: Molecular Psychiatry, (Hirvonen et al., 2005), copyright 2005). b) DRD2 c957T gene dose effect on probabilistic avoidance learning (Frank et al, 2007, PNAS). Individuals with more T alleles performed better, and were relatively faster, at avoid-B test pairs. RT’s are assessed on correct trials and slowing is measured by subtracting choose-A from avoid-B RT’s. Error bars reflect standard error.
Acknowledgments
We thank Marilee Morgan and Melynda Byers for help with DNA analysis. This research was supported by NIMH grant R01 MH080066-01.
Footnotes
Such C957T gene-dose effects could not be completely examined in rs2283265/rs1076560 T-carriers, because there were no C957T T/T homozygotes in that population. Nevertheless, the same trend was observed: individuals with one copy of the C957T T allele performed relatively better than C/C homozygotes at avoid-B performance (F[1,19] = 4.5, p = .047), but not choose-A (F=0.0).
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bayer HM, Glimcher PW. Midbrain dopamine neurons encode a quantitative reward prediction error signal. Neuron. 2005;47(1):129–141. doi: 10.1016/j.neuron.2005.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolan EA, Kivell B, Jaligam V, Oz M, Jayanthi LD, Han Y, Sen N, Urizar E, Gomes I, Devi LA, Ramamoorthy S, Javitch JA, Zapata A, Shippenberg TS. D2 receptors regulate dopamine transporter function via an extracellular signal-regulated kinases 1 and 2-dependent and phosphoinositide 3 kinase-independent mechanism. Molecular pharmacology. 2007;71(5):1222–1232. doi: 10.1124/mol.106.027763. [DOI] [PubMed] [Google Scholar]
- Calabresi P, Gubellini P, Centonze D, Picconi B, Bernardi G, Chergui K, Svenningsson P, Fienberg AA, Greengard P. Dopamine and camp-regulated phosphoprotein 32 kda controls both striatal long-term depression and long-term potentiation, opposing forms of synaptic plasticity. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2000;20(22):8443–8451. doi: 10.1523/JNEUROSCI.20-22-08443.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MX, Frank MJ. Neurocomputational models of basal ganglia function in learning, memory and choice. Behavioral Brain Research. doi: 10.1016/j.bbr.2008.09.029. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen MX, Krohn-Grimberghe A, Elger CE, Weber B. Dopamine gene predicts the brain’s response to dopaminergic drug. The European journal of neuroscience. 2007;26(12):3652–3660. doi: 10.1111/j.1460-9568.2007.05947.x. [DOI] [PubMed] [Google Scholar]
- Cools R, Altamirano L, D’Esposito M. Reversal learning in parkinson’s disease depends on medication status and outcome valence. Neuropsychologia. 2006;44:1663–1673. doi: 10.1016/j.neuropsychologia.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Cools R, Frank MJ, Gibbs SE, Miyakawa A, Jagust W, D’Esposito M. Striatal dopamine predicts outcome-specific reversal learning and its sensitivity to dopaminergic drug administration. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2009;29(5):1538. doi: 10.1523/JNEUROSCI.4467-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cools R, Frank MJ, Gibbs SE, Miyakawa A, Jagust W, D’Esposito M. Striatal dopamine predicts outcome-specific reversal learning and its sensitivity to dopaminergic drug administration. Journal of Neuroscience. doi: 10.1523/JNEUROSCI.4467-08.2009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa RM, Gutierrez R, de Araujo IE, Coelho MRP, Kloth AD, Gainetdinov RR, Caron MG, Nicolelis MAL, Simon SA. Dopamine levels modulate the updating of tastant values. Genes, brain, and behavior. 2007;6(4):314–320. doi: 10.1111/j.1601-183X.2006.00257.x. [DOI] [PubMed] [Google Scholar]
- D’Ardenne K, McClure SM, Nystrom LE, Cohen JD. Bold responses reflecting dopaminergic signals in the human ventral tegmental area. Science (New York, NY) 2008;319(5867):1264–1267. doi: 10.1126/science.1150605. [DOI] [PubMed] [Google Scholar]
- Daw ND, Kakade S, Dayan P. Opponent interactions between serotonin and dopamine. Neural networks:the official journal of the International Neural Network Society. 2002;15:603–616. doi: 10.1016/s0893-6080(02)00052-7. [DOI] [PubMed] [Google Scholar]
- Daw ND, Niv Y, Dayan P. Uncertainty-based competition between prefrontal and dorsolateral striatal systems for behavioral control. Nature neuroscience. 2005;8(12):1704–1711. doi: 10.1038/nn1560. [DOI] [PubMed] [Google Scholar]
- Dodd ML, Klos KJ, Bower JH, Geda YE, Josephs KA, Ahlskog JE. Pathological gambling caused by drugs used to treat parkinson disease. Archives of neurology. 2005;62(9):1377–1381. doi: 10.1001/archneur.62.9.noc50009. [DOI] [PubMed] [Google Scholar]
- Duan J, Wainwright MS, Comeron JM, Saitou N, Sanders AR, Gelernter J, Gejman PV. Synonymous mutations in the human dopamine receptor d2 (drd2) affect mrna stability and synthesis of the receptor. Human Molecular Genetics. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- Finckh U, Rommelspacher H, Kuhn S, Dufeu P, Otto G, Heinz A, Dettling M, Giraldo-Velasquez M, Pelz J, Graf K, Harms H, Sander T, Schmidt L, Rolfs A. Influence of the dopamine D2 receptor (DRD2) genotype on neuroadaptive effects of alcohol and the clinical outcome of alcoholism. Pharmacogenetics. 1997;7(4):271. doi: 10.1097/00008571-199708000-00002. [DOI] [PubMed] [Google Scholar]
- Forbes EE, Brown SM, Kimak M, Ferrell RE, Manuck SB, Hariri AR. Genetic variation in components of dopamine neurotransmission impacts ventral striatal reactivity associated with impulsivity. Molecular psychiatry. 2008;14 doi: 10.1038/sj.mp.4002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ. Dynamic dopamine modulation in the basal ganglia: A neurocomputational account of cognitive deficits in medicated and non-medicated parkinsonism. Journal of Cognitive Neuroscience. 2005;17:51–72. doi: 10.1162/0898929052880093. [DOI] [PubMed] [Google Scholar]
- Frank MJ, Doll B, Oas-Terpstra J, Moreno F. The neurogenetics of exploration and exploitation: Striatal and prefrontal dopaminergic components in preparation. [Google Scholar]
- Frank MJ, Moustafa AA, Haughey HM, Curran T, Hutchison KE. Genetic triple dissociation reveals multiple roles for dopamine in reinforcement learning. Proceedings of the National Academy of Sciences of the United States of America. 2007a;104:16311–16316. doi: 10.1073/pnas.0706111104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank MJ, O’Reilly RC. A mechanistic account of striatal dopamine function in human cognition: Psychopharmacological studies with cabergoline and haloperidol. Behavioral Neuroscience. 2006;120:497–517. doi: 10.1037/0735-7044.120.3.497. [DOI] [PubMed] [Google Scholar]
- Frank MJ, O’Reilly RC, Curran T. When memory fails, intuition reigns: midazolam enhances implicit inference in humans. Psychological science:a journal of the American Psychological Society/APS. 2006;17:700–707. doi: 10.1111/j.1467-9280.2006.01769.x. [DOI] [PubMed] [Google Scholar]
- Frank MJ, Samanta J, Moustafa AA, Sherman SJ. Hold your horses: impulsivity, deep brain stimulation, and medication in parkinsonism. Science (New York, NY) 2007b;318:1309–1312. doi: 10.1126/science.1146157. [DOI] [PubMed] [Google Scholar]
- Frank MJ, Seeberger LC, O’Reilly RC. By carrot or by stick: Cognitive reinforcement learning in parkinsonism. Science. 2004;306:1940–3. doi: 10.1126/science.1102941. [DOI] [PubMed] [Google Scholar]
- Frank MJ, Woroch BS, Curran T. Error-related negativity predicts reinforcement learning and conflict biases. Neuron. 2005;47:495–501. doi: 10.1016/j.neuron.2005.06.020. [DOI] [PubMed] [Google Scholar]
- Gerfen CR. The neostriatal mosaic: multiple levels of compartmental organization in the basal ganglia. Annual review of neuroscience. 1992;15:285–320. doi: 10.1146/annurev.ne.15.030192.001441. [DOI] [PubMed] [Google Scholar]
- Gibbs SEB, D’Esposito M. Individual capacity differences predict working memory performance and prefrontal activity following dopamine receptor stimulation. Cognitive, affective & behavioral neuroscience. 2005;5(2):212–221. doi: 10.3758/cabn.5.2.212. [DOI] [PubMed] [Google Scholar]
- Green AE, Munafo MR, Deyoung CG, Fossella JA, Fan J, Gray JR. Using genetic data in cognitive neuroscience: from growing pains to genuine insights. Nat Rev Neurosci. 2008 doi: 10.1038/nrn2461. [DOI] [PubMed] [Google Scholar]
- Hariri AR, Brown SM, Williamson DE, Flory JD, de Wit H, Manuck SB. Preference for immediate over delayed rewards is associated with magnitude of ventral striatal activity. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2006;26(51):13213–13217. doi: 10.1523/JNEUROSCI.3446-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harl KM, Sanfey AG. Incidental sadness biases social economic decisions in the ultimatum game. Emotion (Washington, DC) 2007;7(4):876–881. doi: 10.1037/1528-3542.7.4.876. [DOI] [PubMed] [Google Scholar]
- Heinz A, Sander T, Harms H, Finckh U, Kuhn S, Dufeu P, Dettling M, Grf K, Rolfs A, Rommelspacher H, Schmidt LG. Lack of allelic association of dopamine d1 and d2 (taqia) receptor gene polymorphisms with reduced dopaminergic sensitivity to alcoholism. Alcoholism, clinical and experimental research. 1997;20(6):597–606. doi: 10.1111/j.1530-0277.1996.tb01954.x. [DOI] [PubMed] [Google Scholar]
- Hirvonen M, Laakso A, abd Nagren K, Rinne J, Pohjalainen T, Hietala J. C957t polymorphism of the dopamine d2 receptor (drd2) gene affects striatal drd2 availability in vivo (corrigendum) Molecular Psychiatry. 2005;10:889. doi: 10.1038/sj.mp.4001561. [DOI] [PubMed] [Google Scholar]
- Kahnt T, Park SQ, Cohen MX, Beck A, Heinz A, Wrase J. Dorsal striatal-midbrain connectivity in humans predicts how reinforcements are used to guide decisions. Journal of Cognitive Neuroscience. doi: 10.1162/jocn.2009.21092. in press. [DOI] [PubMed] [Google Scholar]
- Kimberg DY, D’Esposito M, Farah MJ. Effects of bromocriptine on human subjects depend on working memory capacity. Neuroreport. 1998;8:3581–3585. doi: 10.1097/00001756-199711100-00032. [DOI] [PubMed] [Google Scholar]
- Klein TA, Neumann J, Reuter M, Hennig J, von Cramon DY, Ullsperger M. Genetically determined differences in learning from errors. Science (New York, NY) 2007;318(5856):1642–1645. doi: 10.1126/science.1145044. [DOI] [PubMed] [Google Scholar]
- Laruelle M, Gelertner J, Innis RB. D2 receptors binding potential is not affected by taq1 polymorphism at the d2 receptor gene. Molecular Psychiatry. 1998;3(3):261–265. doi: 10.1038/sj.mp.4000343. [DOI] [PubMed] [Google Scholar]
- Mattay VS, Callicott JH, Bertolino A, Heaton I, Frank JA, Coppola R, Berman KF, Goldberg TE, Weinberger DR. Effects of dextroamphetamine on cognitive performance and cortical activation. NeuroImage. 2000;12:268–75. doi: 10.1006/nimg.2000.0610. [DOI] [PubMed] [Google Scholar]
- McNab F, Klingberg T. Prefrontal cortex and basal ganglia control access to working memory. Nature neuroscience. 2007;11(1):103–107. doi: 10.1038/nn2024. [DOI] [PubMed] [Google Scholar]
- Meiergerd SM, Patterson TA, Schenk JO. D2 receptors may modulate the function of the striatal transporter for dopamine: kinetic evidence from studies in vitro and in vivo. J Neurochem. 1993;61(2):764–767. doi: 10.1111/j.1471-4159.1993.tb02185.x. [DOI] [PubMed] [Google Scholar]
- Meyer-Lindenberg A, Straub RE, Lipska BK, Verchinski BA, Goldberg T, Callicott JH, Egan MF, Huffaker SS, Mattay VS, Kolachana B, Kleinman JE, Weinberger DR. Genetic evidence implicating darpp-32 in human frontostriatal structure, function, and cognition. The Journal of clinical investigation. 2007;117(3):672–682. doi: 10.1172/JCI30413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montague PR, Dayan P, Sejnowski TJ. A framework for mesencephalic dopamine systems based on predictive hebbian learning. The Journal of Neuroscience. 1997;16:1936–1947. doi: 10.1523/JNEUROSCI.16-05-01936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa AA, Cohen MX, Sherman SJ, Frank MJ. A role for dopamine in temporal decision making and reward maximization in parkinsonism. Journal of Neuroscience. 2008a;28(47):12294–12304. doi: 10.1523/JNEUROSCI.3116-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa AA, Sherman SJ, Frank MJ. A dopaminergic basis for working memory, learning, and attentional shifting in parkinson’s disease. Neuropsychologia. 2008b;46:3144–3156. doi: 10.1016/j.neuropsychologia.2008.07.011. [DOI] [PubMed] [Google Scholar]
- Nakamura K, Hikosaka O. Role of dopamine in the primate caudate nucleus in reward modulation of saccades. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2006;26(20):5360–5369. doi: 10.1523/JNEUROSCI.4853-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Reilly RC, Frank MJ. Making working memory work: A computational model of learning in the prefrontal cortex and basal ganglia. Neural Computation. 2006;18:283–328. doi: 10.1162/089976606775093909. [DOI] [PubMed] [Google Scholar]
- Pessiglione M, Seymour B, Flandin G, Dolan RJ, Frith CD. Dopamine-dependent prediction errors underpin reward-seeking behaviour in humans. Nature. 2006;442(7106):1042–1045. doi: 10.1038/nature05051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohjalainen T, Rinne JO, Ngren K, Lehikoinen P, Anttila K, Syvlahti EK, Hietala J. The a1 allele of the human d2 dopamine receptor gene predicts low d2 receptor availability in healthy volunteers. Molecular psychiatry. 1998;3(3):256–260. doi: 10.1038/sj.mp.4000350. [DOI] [PubMed] [Google Scholar]
- Reynolds JN, Wickens JR. Dopamine-dependent plasticity of corticostriatal synapses. Neural Networks. 2002;15:507–521. doi: 10.1016/s0893-6080(02)00045-x. [DOI] [PubMed] [Google Scholar]
- Reynolds JNJ, Hyland BI, Wickens JR. A cellular mechanism of reward-related learning. Nature. 2001;412:67–69. doi: 10.1038/35092560. [DOI] [PubMed] [Google Scholar]
- Rinne UK, Laihinen A, Rinne JO, Ngren K, Bergman J, Ruotsalainen U. Positron emission tomography demonstrates dopamine d2 receptor supersensitivity in the striatum of patients with early parkinson’s disease. Movement disorders:official journal of the Movement Disorder Society. 1990;5(1):55–59. doi: 10.1002/mds.870050114. [DOI] [PubMed] [Google Scholar]
- Santesso D, Evins A, Frank M, Cowman E, Pizzagalli D. Single dose of a dopamine agonist impairs reinforcement learning in humans: Converging evidence from electrophysiology and computational modeling of striatal-cortical function. Human Brain Mapping. doi: 10.1002/hbm.20642. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santesso DL, Dillon DG, Birk JL, Holmes AJ, Goetz E, Bogdan R, Pizzagalli DA. Individual differences in reinforcement learning: behavioral, electrophysiological, and neuroimaging correlates. NeuroImage. 2008;42(2):807–816. doi: 10.1016/j.neuroimage.2008.05.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheres A, Sanfey AG. Individual differences in decision making: Drive and reward responsiveness affect strategic bargaining in economic games. Behavioral and brain functions:BBF. 2006;2:35. doi: 10.1186/1744-9081-2-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz W, Dayan P, Montague PR. A neural substrate of prediction and reward. Science. 1997;275:1593. doi: 10.1126/science.275.5306.1593. [DOI] [PubMed] [Google Scholar]
- Seeman P. Dopamine d2(high) receptors on intact cells. Synapse (New York, NY) 2008;62(4):314–318. doi: 10.1002/syn.20499. [DOI] [PubMed] [Google Scholar]
- Shen W, Flajolet M, Greengard P, Surmeier DJ. Dichotomous dopaminergic control of striatal synaptic plasticity. Science (New York, NY) 2008;321(5890):848–851. doi: 10.1126/science.1160575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siessmeier T, Kienast T, Wrase J, Larsen JL, Braus DF, Smolka MN, Buchholz HG, Schreckenberger M, Rosch F, Cumming P, Mann K, Bartenstein P, Heinz A. Net influx of plasma 6-18f-fluoro-l-dopa (fdopa) to the ventral striatum correlates with prefrontal processing of affective stimuli. 2006 doi: 10.1111/j.1460-9568.2006.04903.x. [DOI] [PubMed] [Google Scholar]
- Stipanovich A, Valjent E, Matamales M, Nishi A, Ahn JH, Maroteaux M, Bertran-Gonzalez J, Brami-Cherrier K, Enslen H, Corbill AG, Filhol O, Nairn AC, Greengard P, Herv D, Girault JA. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453(7197):879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surmeier DJ, Ding J, Day M, Wang Z, Shen W. D1 and d2 dopamine-receptor modulation of striatal glutamatergic signaling in striatal medium spiny neurons. Trends Neurosci. 2007;30(5):228–235. doi: 10.1016/j.tins.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Tan H-Y, Callicott JH, Weinberger DR. Dysfunctional and compensatory prefrontal cortical systems, genes and the pathogenesis of schizophrenia. Cerebral cortex (New York, NY:1991) 2007a;17(Suppl 1):i171–i181. doi: 10.1093/cercor/bhm069. [DOI] [PubMed] [Google Scholar]
- Tan HY, Chen Q, Goldberg TE, Mattay VS, Meyer-Lindenberg A, Weinberger DR, Callicott JH. Catechol-o-methyltransferase val158met modulation of prefrontal-parietal-striatal brain systems during arithmetic and temporal transformations in working memory. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2007b;27(49):13393–13401. doi: 10.1523/JNEUROSCI.4041-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usiello A, Baik JH, Roug-Pont F, Picetti R, Dierich A, LeMeur M, Piazza PV, Borrelli E. Distinct functions of the two isoforms of dopamine d2 receptors. Nature. 2000;408:199–203. doi: 10.1038/35041572. [DOI] [PubMed] [Google Scholar]
- Valtteri KMRH, Kjell N, Pertti L, Vesa ORJ. Upregulation of putaminal dopamine d2 receptors in early parkinson’s disease: A comparative pet study with [11c]raclopride and [11c]n-methylspiperone. 2000 [PubMed] [Google Scholar]
- Wu Q, Reith MEA, Walker QD, Kuhn CM, Carroll FI, Garris PA. Concurrent autoreceptor-mediated control of dopamine release and uptake during neurotransmission: an in vivo voltammetric study. The Journal of neuroscience:the official journal of the Society for Neuroscience. 2002;22:6272–81. doi: 10.1523/JNEUROSCI.22-14-06272.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yacubian J, Sommer T, Schroeder K, Glscher J, Kalisch R, Leuenberger B, Braus DF, Bchel C. Gene-gene interaction associated with neural reward sensitivity. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(19):8125–8130. doi: 10.1073/pnas.0702029104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Bertolino A, Fazio L, Blasi G, Rampino A, Romano R, Lee MLT, Xiao T, Papp A, Wang D, Sade W. Polymorphisms in human dopamine d2 receptor gene affect gene expression, splicing, and neuronal activity during working memory. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(51):20552–20557. doi: 10.1073/pnas.0707106104. [DOI] [PMC free article] [PubMed] [Google Scholar]