Abstract
Halogenated furanones, a group of natural products initially isolated from marine red algae, are known to inhibit bacterial biofilm formation, swarming, and quorum sensing. However, their molecular targets and the precise mode of action remain elusive. Herein, we show that a naturally occurring brominated furanone covalently modifies and inactivates LuxS (S-ribosylhomocysteine lyase, EC 4.4.1.21), the enzyme which produces autoinducer-2 (AI-2).
Antibacterial activities such as inhibition of biofilm formation and swarming have been reported for halogenated furanones, a class of natural products isolated from the marine red algae Delisea pulchra.1 These compounds and their derivatives have served as lead compounds for the development of broad spectrum antibacterial agents.2 This is particularly the case against biofilm formation, a major clinical problem that currently has no effective treatment.3 Studies have shown that compounds 1–4 depicted in Figure 1 promote the clearance of Pseudomonas aeruginosa in mouse pulmonary infection models4 and furanone 1 has been further demonstrated to inhibit Bacillus anthracis growth and expression of its virulence genes.5 The effects of these compounds on several biological pathways have been investigated but the molecular targets and the precise modes of action remain elusive.6 One attractive target is bacterial quorum sensing, the process by which bacteria sense and respond to population density, and consequently behave as a coordinated community. Quorum sensing involves the synthesis, secretion and detection of signaling molecules, commonly referred to as autoinducers (AI’s).7 Typically, a particular autoinducer is recognized only by the species producing the molecule and hence, mediates intra-species communication. On the other hand, autoinducer-2 (AI-2) has been shown to mediate inter-species quorum sensing among many bacteria.8 Previous work from our laboratory and others suggested that halogenated furanones disrupt the AI-2 biosynthetic pathway.9 We present herein biochemical and chemical evidence that halogenated furanones covalently modify and inactivate the AI-2 producing enzyme LuxS.
Figure 1.
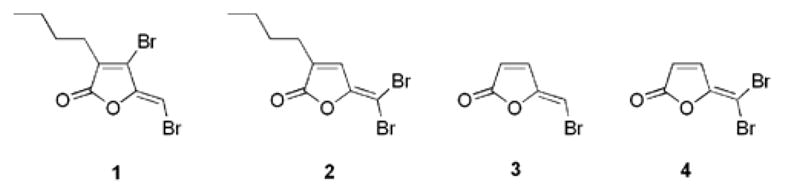
Structures of brominated furanones.
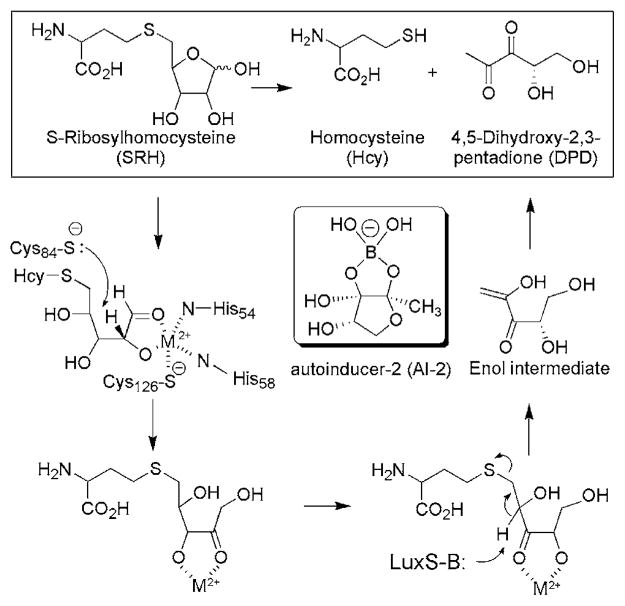
The AI-2 biosynthetic pathway is found in roughly half of all bacterial species, both Gram-positive and Gram negative.10 Biosynthesis of AI-2 begins with S-adenosyl-homocysteine (AdoHcy), the product of S-adenosyl-methionine dependent transmethylation. AdoHcy is hydrolyzed to S-ribosyl-homocysteine (SRH) and adenine by S-adenosyl-homocysteine nucleosidase (EC 3.2.2.16). LuxS (S-ribosylhomocysteine lyase, EC 4.4.1.21) then cleaves SRH to yield L-homocysteine and 4,5-dihydroxy-2,3-pentandione (DPD) which cyclizes to form various forms of AI-2,11 one of which is a borate diester shown in Scheme 1.12
Scheme 1.
Proposed mechanism of the LuxS enzyme and roles of Cys84 and Cys126.
Four brominated furanones were synthesized following literature procedures and tested for inhibition against LuxS.1a,9 Recombinant LuxS was prepared as previously described and enzyme activity was assayed after preincubation of the enzyme with furanone at a range of concentrations for 10 minutes (see SI for details).11b,13 As shown in Figure 2, furanone 1 inhibits the LuxS enzyme in a concentration-dependent manner. Comparable inhibition was observed for furanone 3 (see SI). In contrast, furanone 2 and 4 displayed much weaker inhibition (see SI). The difference in inhibitory activity may be attributed to the difference in their structures, as both furanones 1 and 3 contain a vinyl monobromide, whereas compounds 2 and 4 are substituted with a vinyl gem-dibromide (see Figure. 1).
Figure 2.

Effect of furanone 1 concentration on the initial rates of the LuxS reaction.
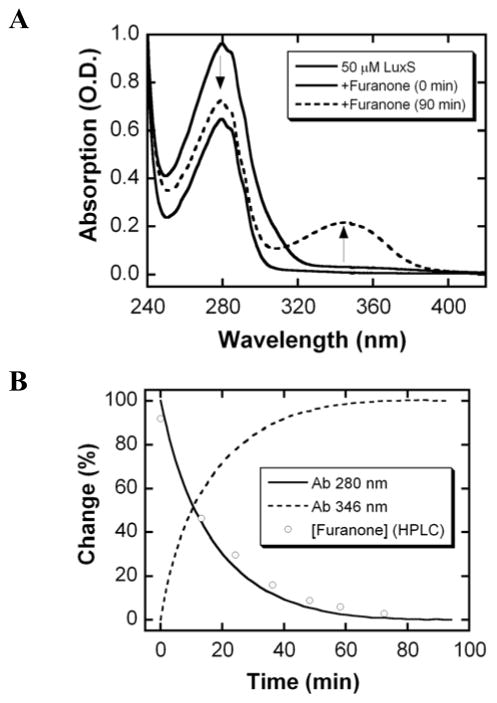
Detailed mechanistic studies were performed with furanone 1, which displayed the highest specific residual activity. It appears that relative enzyme activities after preincubation of furanone was not affected by varying SRH concentrations (data not shown), suggesting that furanone 1 covalently modifies the LuxS enzyme during preincubation. Indeed, time-dependent modification of LuxS in the presence of this furanone was observed (Figure 3). Spectroscopic analysis of the reaction of LuxS and furanone 1 reveals the formation of a new chromophore with maximal absorbance at 346 nm (Figure 3A). The formation of this new species correlates to the disappearance of the furanone, which has an absorbance maximum at 280 nm, as monitored by both HPLC/UV-vis measurements (Figure 3B). Furthermore, the absorption peak at 346 nm persisted with LuxS after the removal of excess furanone by dialysis or size-exclusion chromatography (see SI). Similar spectral changes have been observed when diethylamine is reacted with halogenated furanones to form enamines in ethereal solutions.14 Taken together, these data suggest that LuxS undergoes covalent modification by furanone 1.
Figure 3.
(A) UV/vis absorbance changes associated with the addition of furanone 1 to LuxS. (B) Time-course of absorbance changes at 346 nm (broken line) and 280 nm (solid line), and the disappearance of furanone as monitored by HPLC (circles).
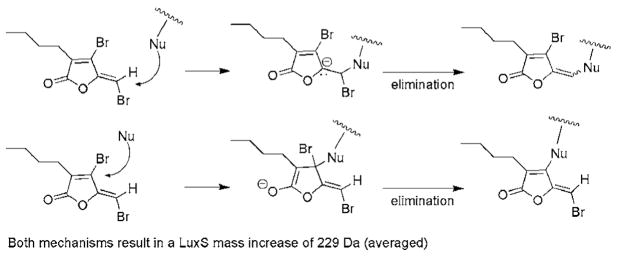
Based on these studies, an addition-elimination mechanism for the covalent modification of LuxS by furanone 1 is proposed (Scheme 2). First, a nucleophile in LuxS adds directly to the vinyl bromide to give an intermediate which subsequently collapses to eliminate the bromide. Both the exocyclic and the ring vinyl bromine may be displaced and our data cannot rule out either of these pathways. On the other hand, the aforementioned model study also indicated that the exocyclic bromide, rather than the ring vinyl bromide, was replaced with amines, suggesting that the former is more reactive. Certainly, the LuxS enzyme may alter the intrinsic chemical reactivities of the two bromides or confer steric effects. Detailed structural characterization of the complex formed between LuxS and furanone 1 may ultimately distinguish these two pathways.
Scheme 2.
Proposed mechanisms for LuxS modification by brominated furanone 1.
It is also worth noting that the exocyclic vinyl bromide appears to be essential for biological activities.1,2,15 All known naturally occurring brominated furanones display either an exocyclic vinyl monobromide or dibromide moiety, but do not require the ring vinyl bromide for activity. The exocyclic vinyl bromide groups were also determined as essential structural elements for inhibition of E. coli biofilm formation by synthetic analogs of brominated furanones.2,15b Nonetheless, since halogenated furanones may also target proteins other than LuxS, the above mentioned biological effects may or may not directly correlate to LuxS modification.
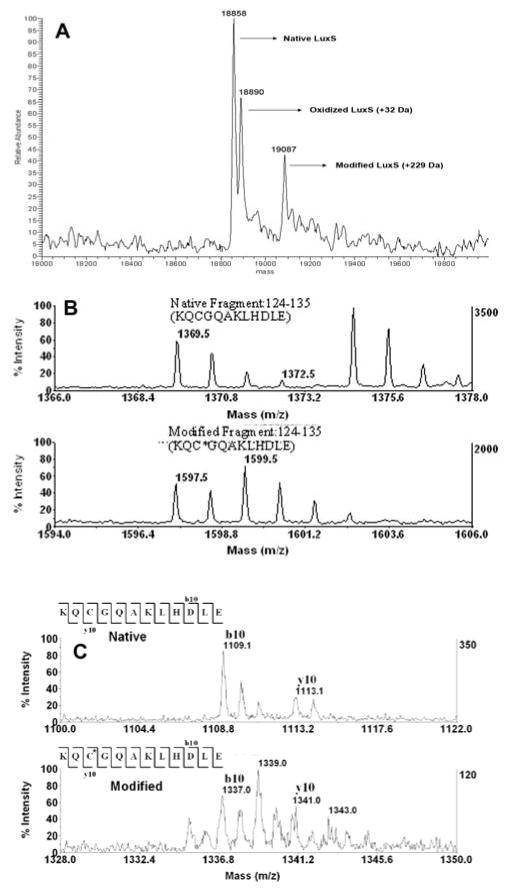
To lend additional support to the proposed mechanism and to identify the site or sites of modification in LuxS by furanone 1, the protein-inhibitor complex was analyzed by mass spectrometry. Illustrated in Figure 4A, the mass spectrum of the LuxS protein treated with furanone 1 shows peaks corresponding to the native LuxS (18,858, theoretical 18,861), its oxidized form (18,890, + 32 Da), and a new LuxS adduct (19,087, +229 Da). The 229 Da mass increase is consistent with the addition of one molecule of furanone 1 followed by the loss of one bromide (expected average mass 229 Da) and further supports the direct addition/elimination mechanism outlined in Scheme 2.
Figure 4.
A) Mass spectrum of LuxS modified by furanone 1. B) Mass spectra of native peptide fragment 124–135 (top) and modified fragment 124–135 (bottom) after Glu-C digestion C) MS/MS of native residue fragment 124–135 (top) and modified (bottom). C* indicates the site of modification by furanone 1.
The modified LuxS was subsequently digested by Glu-C endoprotease and trypsin, and the peptide fragments were analyzed by MALDI-MS. The modification site was localized to Cys126 in the 124–135 peptide fragment. Conclusively, the mass shifts of 228 and 230 Da (1597.5 and 1599.5 vs 1369.5 for the unmodified peptide) and the distinct isotopic signature shown in Figures 4B and 4C are consistent with the addition of one molecule of furanone 1 minus one bromide. As indicated by the crystal structure of LuxS, Cys126 coordinates an active site divalent metal ion, which is thought to catalyze the aldose-ketose isomerization steps depicted in Scheme 1.13,16 Alkylation of Cys126 may perturb proper metal binding and/or block substrate binding, resulting in the reduction of enzymatic activity. This reminisces the selective alkylation of a cysteine ligand to the catalytic zinc ion in methionine synthase (MetE) and other metalloenzymes.17
In addition, we find that furanone 1 modifies LuxS in a highly sequence selective manner. As shown in Figure 3A, only one adduct appeared after treatment of LuxS with furanone. In contrast, when LuxS was treated with chloroacetone, addition of four ketone moieties was observed (see SI), likely resulting from the alkylation of all four cysteine residues in LuxS. In addition, Cys84, a catalytic acid/base in the active site of LuxS, has a pKa value around 6 and thus exists predominantly in the thiolate form;18 and is also more solvent accessible than Cys126 (see SI). Yet, no modification of Cys84 was detected.
In summary, the biochemical and chemical studies reported herein identify a molecular target for halogenated furanones and suggest an inhibition mechanism. Since the LuxS enzyme and the AI-2 pathway are widespread in bacteria and absent in humans, our results open new avenues for the development of antibacterial agents.
Supplementary Material
Acknowledgments
The authors are indebted to Bonnie Bassler, Ken Cornell, Michael Riscoe, Mark Hilgers and Martha Ludwig for providing bacterial strains. This work was supported by the NIAID and the NIH (1R01AI058146 to Z.S.Z, R01EB003872 to T.K.W). L.M.C. was supported by a grant from the College of Sciences and Honors College at Washington State University. This is publication number 950 from the Barnett Institute.
Footnotes
Supplementary Data
Experimental procedures for the LuxS inactivation assays and mass spectral data.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.(a) Beechan CM, Sims JJ. Tetrahedron Lett. 1979;19:1649. [Google Scholar]; (b) Hentzer M, Riedel K, Rasmussen TB, Heydorn A, Andersen JB, Parsek MR, Rice SA, Eberl L, Molin S, Hoiby N, Kjelleberg S, Givskov M. Microbiology. 2002;148:87. doi: 10.1099/00221287-148-1-87. [DOI] [PubMed] [Google Scholar]; (c) Ren D, Sims JJ, Wood TK. Environ Microbiol. 2001;3:731. doi: 10.1046/j.1462-2920.2001.00249.x. [DOI] [PubMed] [Google Scholar]; (d) Ren D, Sims JJ, Wood TK. Lett Appl Microbiol. 2002;34:293. doi: 10.1046/j.1472-765x.2002.01087.x. [DOI] [PubMed] [Google Scholar]
- 2.(a) de Nys R, Givskov M, Kumar N, Kjelleberg S, Steinberg PD. Prog Mol Subcell Biol. 2006;42:55. doi: 10.1007/3-540-30016-3_2. [DOI] [PubMed] [Google Scholar]; (b) Ni N, Li M, Wang J, Wang B. Med Res Rev. 2009;29:65. doi: 10.1002/med.20145. [DOI] [PubMed] [Google Scholar]
- 3.(a) Davies D. Nat Rev Drug Discov. 2003;2:114. doi: 10.1038/nrd1008. [DOI] [PubMed] [Google Scholar]; (b) Hentzer M, Eberl L, Nielsen J, Givskov M. BioDrugs. 2003;17:241. doi: 10.2165/00063030-200317040-00003. [DOI] [PubMed] [Google Scholar]
- 4.Hentzer M, Wu H, Andersen JB, Riedel K, Rasmussen TB, Bagge N, Kumar N, Schembri MA, Song Z, Kristoffersen P, Manefield M, Costerton JW, Molin S, Eberl L, Steinberg P, Kjelleberg S, Hoiby N, Givskov M. EMBO J. 2003;22:3803. doi: 10.1093/emboj/cdg366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones MB, Jani R, Ren D, Wood TK, Blaser MJ. J Infect Dis. 2005;191:1881. doi: 10.1086/429696. [DOI] [PubMed] [Google Scholar]
- 6.(a) Manefield M, Rasmussen TB, Henzter M, Andersen JB, Steinberg P, Kjelleberg S, Givskov M. Microbiology. 2002;148:1119. doi: 10.1099/00221287-148-4-1119. [DOI] [PubMed] [Google Scholar]; (b) Manefield M, Welch M, Givskov M, Salmond GP, Kjelleberg S. FEMS Microbiol Lett. 2001;205:131. doi: 10.1111/j.1574-6968.2001.tb10936.x. [DOI] [PubMed] [Google Scholar]; (c) Defoirdt T, Miyamoto CM, Wood TK, Meighen EA, Sorgeloos P, Verstraete W, Bossier P. Environ Microbiol. 2007;9:2486. doi: 10.1111/j.1462-2920.2007.01367.x. [DOI] [PubMed] [Google Scholar]
- 7.(a) Camilli A, Bassler BL. Science. 2006;311:1113. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Jayaraman A, Wood TK. Annu Rev Biomed Eng. 2008;10:145. doi: 10.1146/annurev.bioeng.10.061807.160536. [DOI] [PubMed] [Google Scholar]
- 8.Waters CM, Bassler BL. Annu Rev Cell Dev Biol. 2005;21:319. doi: 10.1146/annurev.cellbio.21.012704.131001. [DOI] [PubMed] [Google Scholar]
- 9.Ren D, Bedzyk LA, Ye RW, Thomas SM, Wood TK. Biotechnol Bioeng. 2004;88:630. doi: 10.1002/bit.20259. [DOI] [PubMed] [Google Scholar]
- 10.(a) Xavier KB, Bassler BL. Curr Opin Microbiol. 2003;6:191. doi: 10.1016/s1369-5274(03)00028-6. [DOI] [PubMed] [Google Scholar]; (b) Winzer K, Hardie KR, Williams P. Adv Appl Microbiol. 2003;53:291. doi: 10.1016/s0065-2164(03)53009-x. [DOI] [PubMed] [Google Scholar]; (c) Sun J, Daniel R, Wagner-Dobler I, Zeng AP. BMC Evol Biol. 2004;4:36. doi: 10.1186/1471-2148-4-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.(a) Alfaro JF, Zhang T, Wynn DP, Karschner EL, Zhou ZS. Org Lett. 2004;6:3043. doi: 10.1021/ol049182i. [DOI] [PubMed] [Google Scholar]; (b) Zhao G, Wan W, Mansouri S, Alfaro JF, Bassler BL, Cornell KA, Zhou ZS. Bioorg Med Chem Lett. 2003;13:3897. doi: 10.1016/j.bmcl.2003.09.015. [DOI] [PubMed] [Google Scholar]; (c) Gopishetty B, Zhu J, Rajan R, Sobczak AJ, Wnuk SF, Bell CE, Pei D. J Am Chem Soc. 2009;131:1243. doi: 10.1021/ja808206w. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Wnuk SF, Lalama J, Garmendia CA, Robert J, Zhu J, Pei D. Bioorg Med Chem. 2008;16:5090. doi: 10.1016/j.bmc.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Wnuk SF, Lalama J, Robert J, Garmendia CA. Nucleosides Nucleotides Nucleic Acids. 2007;26:1051. doi: 10.1080/15257770701513190. [DOI] [PubMed] [Google Scholar]
- 12.Chen XSS, Potier N, Van Dorsselaer A, Pelczer I, Bassler BL, Hughson FM. Nature. 2002;415:545. doi: 10.1038/415545a. [DOI] [PubMed] [Google Scholar]
- 13.Hilgers MT, Ludwig ML. Proc Natl Acad Sci USA. 2001;98:11169. doi: 10.1073/pnas.191223098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kazlauskas R, Murphy PT, Quinn RJ, Wells RJ. Tetrahedron Lett. 1977;18:37. [Google Scholar]
- 15.(a) Lonn-Stensrud J, Petersen FC, Benneche T, Aamdal Scheie A. Oral Microbiology Immunology. 2007;22:340. doi: 10.1111/j.1399-302X.2007.00367.x. [DOI] [PubMed] [Google Scholar]; (b) Han Y, Hou S, Simon KA, Ren D, Luk Y. Bioorg Med Chem Lett. 2008;18:1006. doi: 10.1016/j.bmcl.2007.12.032. [DOI] [PubMed] [Google Scholar]
- 16.Ruzheinikov SN, Das SK, Sedelnikova SE, Hartley A, Foster SJ, Horsburgh MJ, Cox AG, McCleod CW, Mekhalfia A, Blackburn GM, Rice DW, Baker PJ. J Mol Biol. 2001;313:111. doi: 10.1006/jmbi.2001.5027. [DOI] [PubMed] [Google Scholar]
- 17.(a) Gonzalez JC, Banerjee RV, Huang S, Sumner JS, Matthews RG. Biochemistry. 1992;31:6045. doi: 10.1021/bi00141a013. [DOI] [PubMed] [Google Scholar]; (b) Matthews RG, Goulding CW. Curr Opin Chem Biol. 1997;1:332. doi: 10.1016/s1367-5931(97)80070-1. [DOI] [PubMed] [Google Scholar]; (c) Zhou ZS, Peariso K, Penner-Hahn JE, Matthews RG. Biochemistry. 1999;38:15915. doi: 10.1021/bi992062b. [DOI] [PubMed] [Google Scholar]
- 18.Zhu J, Knottenbelt S, Kirk ML, Pei D. Biochemistry. 2006;45:12195. doi: 10.1021/bi061434v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.