Abstract
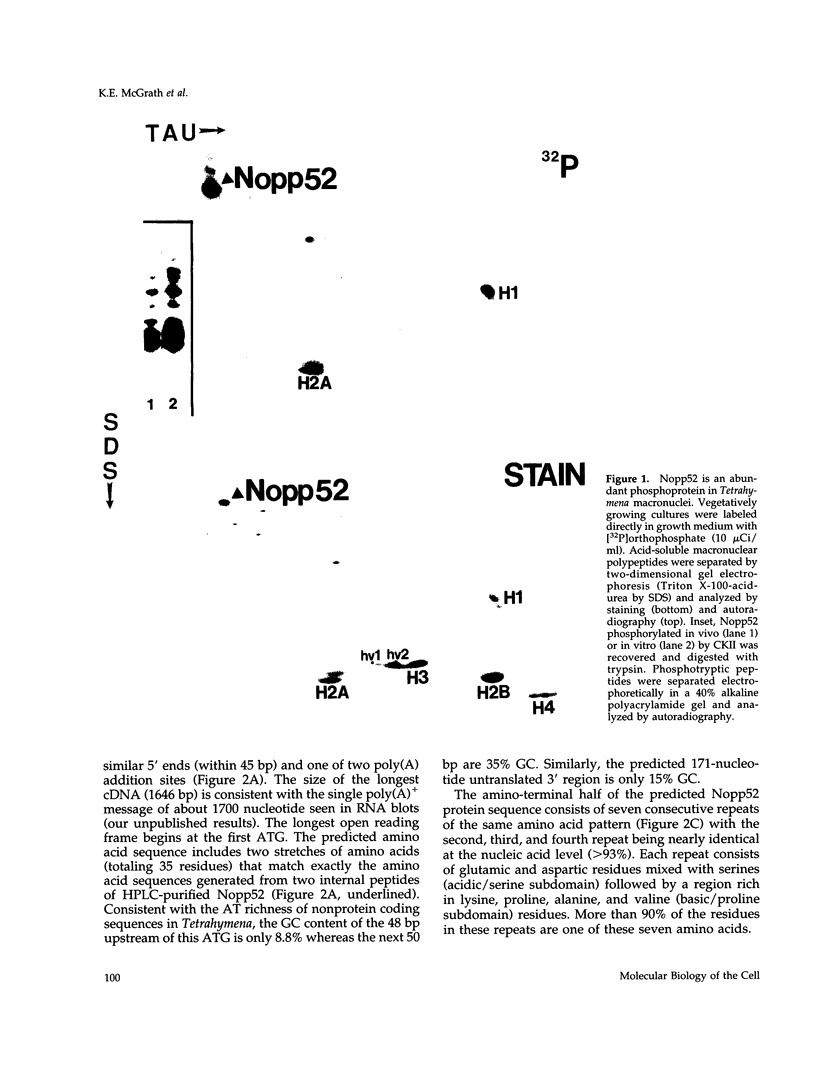
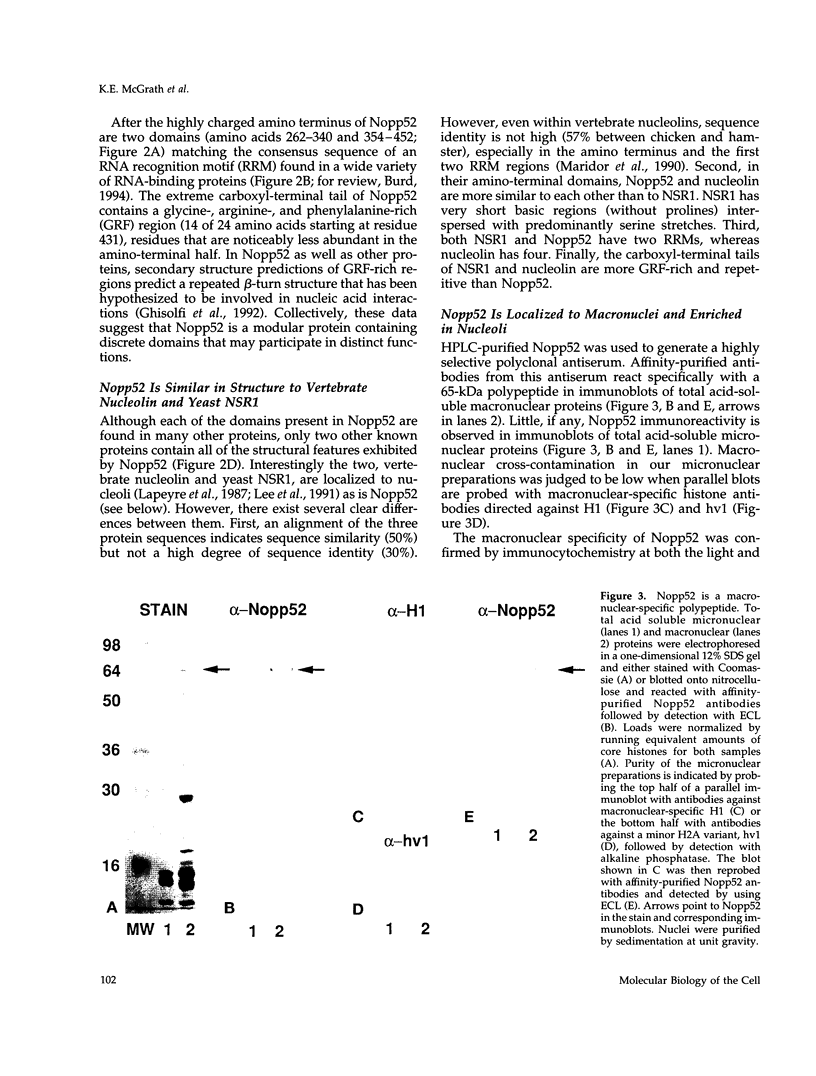
An abundant 52-kDa phosphoprotein was identified and characterized from macronuclei of the ciliated protozoan Tetrahymena thermophila. Immunoblot analyses combined with light and electron microscopic immunocytochemistry demonstrate that this polypeptide, termed Nopp52, is enriched in the nucleoli of transcriptionally active macronuclei and missing altogether from transcriptionally inert micronuclei. The cDNA sequence encoding Nopp52 predicts a polypeptide whose amino-terminal half consists of multiple acidic/serine-rich regions alternating with basic/proline-rich regions. Multiple serines located in these acidic stretches lie within casein kinase II consensus motifs, and Nopp52 is an excellent substrate for casein kinase II in vitro. The carboxyl-terminal half of Nopp52 contains two RNA recognition motifs and an extreme carboxyl-terminal domain rich in glycine, arginine, and phenylalanine, motifs common in many RNA processing proteins. A similar combination and order of motifs is found in vertebrate nucleolin and yeast NSR1, suggesting that Nopp52 is a member of a family of related nucleolar proteins. NSR1 and nucleolin have been implicated in transcriptional regulation of rDNA and rRNA processing. Consistent with a role in ribosomal gene metabolism, rDNA and Nopp52 colocalize in situ, as well as by cross-linking and immunoprecipitation experiments, demonstrating an association between Nopp52 and rDNA in vivo.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allis C. D., Dennison D. K. Identification and purification of young macronuclear anlagen from conjugating cells of Tetrahymena thermophila. Dev Biol. 1982 Oct;93(2):519–533. doi: 10.1016/0012-1606(82)90139-7. [DOI] [PubMed] [Google Scholar]
- Belenguer P., Caizergues-Ferrer M., Labbé J. C., Dorée M., Amalric F. Mitosis-specific phosphorylation of nucleolin by p34cdc2 protein kinase. Mol Cell Biol. 1990 Jul;10(7):3607–3618. doi: 10.1128/mcb.10.7.3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouche G., Caizergues-Ferrer M., Bugler B., Amalric F. Interrelations between the maturation of a 100 kDa nucleolar protein and pre rRNA synthesis in CHO cells. Nucleic Acids Res. 1984 Apr 11;12(7):3025–3035. doi: 10.1093/nar/12.7.3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd C. G., Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994 Jul 29;265(5172):615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Colavito-Shepanski M., Gorovsky M. A. The histone content of Tetrahymena ribosomal gene-containing chromatin. J Biol Chem. 1983 May 10;258(9):5944–5954. [PubMed] [Google Scholar]
- Créancier L., Prats H., Zanibellato C., Amalric F., Bugler B. Determination of the functional domains involved in nucleolar targeting of nucleolin. Mol Biol Cell. 1993 Dec;4(12):1239–1250. doi: 10.1091/mbc.4.12.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dadd C. A., Cook R. G., Allis C. D. Fractionation of small tryptic phosphopeptides by alkaline PAGE followed by amino acid sequencing. Biotechniques. 1993 Feb;14(2):266–273. [PubMed] [Google Scholar]
- Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. A simplified formaldehyde fixation and immunoprecipitation technique for studying protein-DNA interactions. Anal Biochem. 1991 Aug 15;197(1):83–90. doi: 10.1016/0003-2697(91)90359-2. [DOI] [PubMed] [Google Scholar]
- Dedon P. C., Soults J. A., Allis C. D., Gorovsky M. A. Formaldehyde cross-linking and immunoprecipitation demonstrate developmental changes in H1 association with transcriptionally active genes. Mol Cell Biol. 1991 Mar;11(3):1729–1733. doi: 10.1128/mcb.11.3.1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingwall C., Dilworth S. M., Black S. J., Kearsey S. E., Cox L. S., Laskey R. A. Nucleoplasmin cDNA sequence reveals polyglutamic acid tracts and a cluster of sequences homologous to putative nuclear localization signals. EMBO J. 1987 Jan;6(1):69–74. doi: 10.1002/j.1460-2075.1987.tb04720.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egyhazi E., Pigon A., Chang J. H., Ghaffari S. H., Dreesen T. D., Wellman S. E., Case S. T., Olson M. O. Effects of anti-C23 (nucleolin) antibody on transcription of ribosomal DNA in Chironomus salivary gland cells. Exp Cell Res. 1988 Oct;178(2):264–272. doi: 10.1016/0014-4827(88)90397-7. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nielsen H. Complete sequence of the extrachromosomal rDNA molecule from the ciliate Tetrahymena thermophila strain B1868VII. Nucleic Acids Res. 1990 Dec 11;18(23):6915–6919. doi: 10.1093/nar/18.23.6915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erard M. S., Belenguer P., Caizergues-Ferrer M., Pantaloni A., Amalric F. A major nucleolar protein, nucleolin, induces chromatin decondensation by binding to histone H1. Eur J Biochem. 1988 Aug 15;175(3):525–530. doi: 10.1111/j.1432-1033.1988.tb14224.x. [DOI] [PubMed] [Google Scholar]
- Erard M., Lakhdar-Ghazal F., Amalric F. Repeat peptide motifs which contain beta-turns and modulate DNA condensation in chromatin. Eur J Biochem. 1990 Jul 20;191(1):19–26. doi: 10.1111/j.1432-1033.1990.tb19088.x. [DOI] [PubMed] [Google Scholar]
- Ferns M. J., Hall Z. W. How many agrins does it take to make a synapse? Cell. 1992 Jul 10;70(1):1–3. doi: 10.1016/0092-8674(92)90525-h. [DOI] [PubMed] [Google Scholar]
- Frohman M. A., Dush M. K., Martin G. R. Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci U S A. 1988 Dec;85(23):8998–9002. doi: 10.1073/pnas.85.23.8998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J., Gorovsky M. A. Efficient mass transformation of Tetrahymena thermophila by electroporation of conjugants. Proc Natl Acad Sci U S A. 1992 Oct 1;89(19):9196–9200. doi: 10.1073/pnas.89.19.9196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghisolfi-Nieto L., Joseph G., Puvion-Dutilleul F., Amalric F., Bouvet P. Nucleolin is a sequence-specific RNA-binding protein: characterization of targets on pre-ribosomal RNA. J Mol Biol. 1996 Jul 5;260(1):34–53. doi: 10.1006/jmbi.1996.0380. [DOI] [PubMed] [Google Scholar]
- Ghisolfi L., Joseph G., Amalric F., Erard M. The glycine-rich domain of nucleolin has an unusual supersecondary structure responsible for its RNA-helix-destabilizing properties. J Biol Chem. 1992 Feb 15;267(5):2955–2959. [PubMed] [Google Scholar]
- Gorovsky M. A. Macro- and micronuclei of Tetrahymena pyriformis: a model system for studying the structure and function of eukaryotic nuclei. J Protozool. 1973 Feb;20(1):19–25. doi: 10.1111/j.1550-7408.1973.tb05995.x. [DOI] [PubMed] [Google Scholar]
- Gorovsky M. A., Yao M. C., Keevert J. B., Pleger G. L. Isolation of micro- and macronuclei of Tetrahymena pyriformis. Methods Cell Biol. 1975;9(0):311–327. doi: 10.1016/s0091-679x(08)60080-1. [DOI] [PubMed] [Google Scholar]
- Herrera A. H., Olson M. O. Association of protein C23 with rapidly labeled nucleolar RNA. Biochemistry. 1986 Oct 7;25(20):6258–6264. doi: 10.1021/bi00368a063. [DOI] [PubMed] [Google Scholar]
- Jordan G. At the heart of the nucleolus. Nature. 1987 Oct 8;329(6139):489–490. doi: 10.1038/329489a0. [DOI] [PubMed] [Google Scholar]
- Kondo K., Inouye M. Yeast NSR1 protein that has structural similarity to mammalian nucleolin is involved in pre-rRNA processing. J Biol Chem. 1992 Aug 15;267(23):16252–16258. [PubMed] [Google Scholar]
- Lapeyre B., Bourbon H., Amalric F. Nucleolin, the major nucleolar protein of growing eukaryotic cells: an unusual protein structure revealed by the nucleotide sequence. Proc Natl Acad Sci U S A. 1987 Mar;84(6):1472–1476. doi: 10.1073/pnas.84.6.1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Xue Z. X., Mélèse T. The NSR1 gene encodes a protein that specifically binds nuclear localization sequences and has two RNA recognition motifs. J Cell Biol. 1991 Apr;113(1):1–12. doi: 10.1083/jcb.113.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. C., Zabetakis D., Mélèse T. NSR1 is required for pre-rRNA processing and for the proper maintenance of steady-state levels of ribosomal subunits. Mol Cell Biol. 1992 Sep;12(9):3865–3871. doi: 10.1128/mcb.12.9.3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. J., Zakian V. A. Isolation and characterization of two Saccharomyces cerevisiae genes that encode proteins that bind to (TG1-3)n single strand telomeric DNA in vitro. Nucleic Acids Res. 1994 Nov 25;22(23):4906–4913. doi: 10.1093/nar/22.23.4906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madireddi M. T., Davis M. C., Allis C. D. Identification of a novel polypeptide involved in the formation of DNA-containing vesicles during macronuclear development in Tetrahymena. Dev Biol. 1994 Oct;165(2):418–431. doi: 10.1006/dbio.1994.1264. [DOI] [PubMed] [Google Scholar]
- Maridor G., Krek W., Nigg E. A. Structure and developmental expression of chicken nucleolin and NO38: coordinate expression of two abundant non-ribosomal nucleolar proteins. Biochim Biophys Acta. 1990 Jun 21;1049(2):126–133. doi: 10.1016/0167-4781(90)90032-w. [DOI] [PubMed] [Google Scholar]
- Marin O., Meggio F., Marchiori F., Borin G., Pinna L. A. Site specificity of casein kinase-2 (TS) from rat liver cytosol. A study with model peptide substrates. Eur J Biochem. 1986 Oct 15;160(2):239–244. doi: 10.1111/j.1432-1033.1986.tb09962.x. [DOI] [PubMed] [Google Scholar]
- Meggio F., Pinna L. A. Phosphorylation of phosvitin by casein kinase-2 provides the evidence that phosphoserines can replace carboxylic amino acids as specificity determinants. Biochim Biophys Acta. 1988 Sep 16;971(2):227–231. doi: 10.1016/0167-4889(88)90196-6. [DOI] [PubMed] [Google Scholar]
- Olmsted J. B. Affinity purification of antibodies from diazotized paper blots of heterogeneous protein samples. J Biol Chem. 1981 Dec 10;256(23):11955–11957. [PubMed] [Google Scholar]
- Prescott D. M. The DNA of ciliated protozoa. Microbiol Rev. 1994 Jun;58(2):233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth S. Y., Schulman I. G., Richman R., Cook R. G., Allis C. D. Characterization of phosphorylation sites in histone H1 in the amitotic macronucleus of Tetrahymena during different physiological states. J Cell Biol. 1988 Dec;107(6 Pt 2):2473–2482. doi: 10.1083/jcb.107.6.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapp M., Richter A., Weisshart K., Caizergues-Ferrer M., Amalric F., Wallace M. O., Kirstein M. N., Olson M. O. Characterization of a 48-kDa nucleic-acid-binding fragment of nucleolin. Eur J Biochem. 1989 Feb 15;179(3):541–548. doi: 10.1111/j.1432-1033.1989.tb14581.x. [DOI] [PubMed] [Google Scholar]
- Scheer U., Weisenberger D. The nucleolus. Curr Opin Cell Biol. 1994 Jun;6(3):354–359. doi: 10.1016/0955-0674(94)90026-4. [DOI] [PubMed] [Google Scholar]
- Schulman I. G., Cook R. G., Richman R., Allis C. D. Tetrahymena contain two distinct and unusual high mobility group (HMG)-like proteins. J Cell Biol. 1987 Jun;104(6):1485–1494. doi: 10.1083/jcb.104.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan X., Xue Z., Mélèse T. Yeast NPI46 encodes a novel prolyl cis-trans isomerase that is located in the nucleolus. J Cell Biol. 1994 Aug;126(4):853–862. doi: 10.1083/jcb.126.4.853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stargell L. A., Bowen J., Dadd C. A., Dedon P. C., Davis M., Cook R. G., Allis C. D., Gorovsky M. A. Temporal and spatial association of histone H2A variant hv1 with transcriptionally competent chromatin during nuclear development in Tetrahymena thermophila. Genes Dev. 1993 Dec;7(12B):2641–2651. doi: 10.1101/gad.7.12b.2641. [DOI] [PubMed] [Google Scholar]
- Stargell L. A., Karrer K. M., Gorovsky M. A. Transcriptional regulation of gene expression in Tetrahymena thermophila. Nucleic Acids Res. 1990 Nov 25;18(22):6637–6639. doi: 10.1093/nar/18.22.6637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenkert D., Allis C. D. Timing of the appearance of macronuclear-specific histone variant hv1 and gene expression in developing new macronuclei of Tetrahymena thermophila. J Cell Biol. 1984 Jun;98(6):2107–2117. doi: 10.1083/jcb.98.6.2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White E. M., Allis C. D., Goldfarb D. S., Srivastva A., Weir J. W., Gorovsky M. A. Nucleus-specific and temporally restricted localization of proteins in Tetrahymena macronuclei and micronuclei. J Cell Biol. 1989 Nov;109(5):1983–1992. doi: 10.1083/jcb.109.5.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Mélèse T. Multiple regions of NSR1 are sufficient for accumulation of a fusion protein within the nucleolus. J Cell Biol. 1993 Dec;123(5):1081–1091. doi: 10.1083/jcb.123.5.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao M. C., Yao C. H. Transformation of Tetrahymena to cycloheximide resistance with a ribosomal protein gene through sequence replacement. Proc Natl Acad Sci U S A. 1991 Nov 1;88(21):9493–9497. doi: 10.1073/pnas.88.21.9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu G. L., Hasson M., Blackburn E. H. Circular ribosomal DNA plasmids transform Tetrahymena thermophila by homologous recombination with endogenous macronuclear ribosomal DNA. Proc Natl Acad Sci U S A. 1988 Jul;85(14):5151–5155. doi: 10.1073/pnas.85.14.5151. [DOI] [PMC free article] [PubMed] [Google Scholar]