Abstract
Native phosphodiesterase-5 (PDE5) homodimer contains distinct non-catalytic cGMP allosteric sites and catalytic sites for cGMP hydrolysis. Purified recombinant PDE5 was activated by pre-incubation with cGMP. Relatively low concentrations of cGMP produced a Native PAGE gel-shift of PDE5 from a single band position (lower band) to a band with decreased mobility (upper band); higher concentrations of cGMP produced a band of intermediate mobility (middle band) in addition to the upper band. Two point mutations (G659A and G659P) near the catalytic site that reduced affinity for cGMP substrate retained allosteric cGMP-binding affinity like that of WT PDE5 but displayed cGMP-induced gel-shift only to the middle-band position. The upper band could represent a form produced by cGMP binding to the catalytic site, while the middle band could represent a form produced by cGMP binding to the allosteric site. Millimolar cGMP was required for gel-shift of PDE5 when added to the pre-incubation before native PAGE, presumably due to removal of most of the cGMP during electrophoresis, but micromolar cGMP was sufficient for this effect if cGMP was included in the native gel buffer. cGMP-induced gel-shift was associated with stimulation of PDE5 catalytic activity, and the rates of onset and reversibility of this effect suggested that it was due to cGMP binding to the allosteric site. Incubation of PDE5 with non-hydrolyzable, catalytic site-specific, substrate analogs such as the inhibitors sildenafil and tadalafil, followed by dilution, did not produce activation of catalytic activity like that obtained with cGMP, although both inhibitors produced a similar gel-shift to the upper band as that obtained with cGMP. This implied that occupation of the catalytic site alone can produce a gel-shift to the upper band. PDE5 activation or gel-shift was reversed by lowering cGMP with dilution followed by at least one hour of incubation. Such slow reversibility could prolong effects of cGMP on PDE5 in cells after decline of this nucleotide. Reversal was also achieved by Mg++ addition to the pre-incubation mixture to promote cGMP degradation, but Mg++ addition did not reverse the gel-shift caused by sildenafil, which is not hydrolyzed by PDE5. Upon extensive dilution, the effect of tadalafil, a potent PDE5 inhibitor, to enhance catalytic-site affinity for this inhibitor was rapidly reversed. Thus, kinetic effect of binding of a high-affinity PDE5 inhibitor to the catalytic site is more readily reversible than that obtained by cGMP binding to the allosteric site. It is concluded that cGMP or PDE5 inhibitor binding to the catalytic site, or ligand binding to both the catalytic site and allosteric site simultaneously, changes PDE5 to a similar physical form; this form is distinct from that produced by cGMP binding to the allosteric site, which activates the enzyme and reverses more slowly.
Keywords: Phosphodiesterase, PDE, PDE5, PDE5 inhibitor, sildenafil, tadalafil, GAF, cyclic GMP, negative feedback, allosteric regulation
1. Introduction
Mammalian cyclic nucleotide PDEs are a superfamily (PDE1-PDE11) of metallo-phosphohydrolases that catalyze hydrolysis of cyclic nucleotides to the respective 5’-nucleotide [1–7]. They contain a conserved catalytic (C) domain that varies in specificity and affinity for cAMP and cGMP and differs in inhibitor sensitivity [3, 6, 8]. PDEs are regulated by myriad processes, including subcellular localization [9], cGMP or cAMP binding [3, 10, 11], phosphorylation [3, 8, 12–18], or Ca2+/calmodulin binding [19, 20]. Phosphodiesterase-5 (PDE5) is highly selective for cGMP breakdown, plays a prominent role in determining cGMP levels in many tissues, and is the therapeutic target for treatment of erectile dysfunction (ED) by “PDE5-selective inhibitors” such as sildenafil (Viagra™), vardenafil (Levitra ™), tadalafil (Cialis™), and udenafil (Zydena™) [21–27]. Thus far we have identified 10 mechanisms by which PDE5 is activated after cGMP elevation, and we have proposed that this brings about negative feedback control of the cGMP-signaling pathway. PDE5 is a homodimer [28] with separate cGMP sites for catalysis and allosteric regulation at the more carboxyl-terminal C domain and amino-terminal regulatory (R) domain, respectively, of each monomer. Binding of ligand to the catalytic site stimulates cGMP binding to the allosteric site [28], and conversely [29–33]. Within the R domain are two tandem GAF (cGMP-binding PDE, Anabaena adenylyl cyclase, E. coli Fh1A protein) subdomains [28, 34, 35]. Binding of cGMP to PDE5 GAF stimulates the catalytic site [30–33]. We have recently shown that this is a direct effect on the catalytic site [30, 36]. A concerted effect of catalytic-site and allosteric-site binding of ligand, along with phosphorylation [37–39], could serve for powerful negative feedback control of cGMP signaling, thereby enhancing PDE5-mediated dampening or termination of the signal. This concerted effect would also increase cGMP sequestration by the PDE5 allosteric sites [17, 40, 41], which would further reduce free cGMP and dampen cGMP-signaling. The negative feedback mechanisms should translate into positive feedback for PDE5 inhibitors that are in clinical use since these inhibitors are substrate analogs that are not metabolized in the smooth muscle cells and improve catalytic-site affinity in a time-dependent manner [42].
We have discovered two kinetic species and two physical forms of PDE5 [36, 43]. Ligand binding or phosphorylation can cause conversion of one form to another. Whether ligand binding to the allosteric site, catalytic site, or both, causes conversion of these species and forms, and whether the kinetic species represent the physical forms is unknown. The kinetic species are characterized by “high-affinity” or “low-affinity” of the catalytic site for cGMP or PDE5 inhibitors, as well as high-affinity and low-affinity of the allosteric cGMP-binding site for cGMP; the physical forms are characterized by distinct mobilities on Native PAGE. Evidence for a third physical form that is produced by ligand binding is presented herein. Establishment of the existence and mechanisms of interconversion of the kinetic species or physical forms is important in understanding regulation of cGMP action and pharmacology of inhibitor effects. Results in the present study improve understanding of molecular mechanisms that impact PDE5 inhibitor therapy and allow new directions for medications that affect PDE5 and cGMP signaling.
2. Materials and methods
2.1. Materials
Sildenafil was purified from Viagra® tablets using the method established in this laboratory [30, 44]. Purified sildenafil was radiolabeled with tritium by GE Healthcare. Tadalafil was synthesized according to Daugan [45]. After confirming the compound structure by mass spectrometry, tadalafil was submitted to GE Healthcare for radiolabeling with tritium. Vardenafil and [3H]vardenafil were provided by Bayer AG (Wuppertal, Germany). Cyclic GMP, cAMP, 3-isobutyl-1-methylxanthine (IBMX), histone type II-AS, Crotalus atrox snake venom, and bovine serum albumin (BSA) were from Sigma-Aldrich (St. Louis, MO). Pre-formed native gels and native gel sample buffer were from Bio-Rad (Hercules, CA). Sephacryl S-200 High Resolution gel filtration resin was purchased from Amersham (Piscataway, NJ). [3H]cGMP was purchased from GE Healthcare (Piscataway, NJ).
2.2. Expression and purification of PDE5A1
Human cDNA coding for full-length PDE5A1 (hPDE5A1) was ligated into the baculovirus expression vector pAcHLT-A (BD Biosciences PharMingen). The resulting plasmid pAcA-PDE5 was used to make point mutations (G659A and G659P) with the QuikChange site-directed mutagenesis kit (Stratagene). WT and mutant PDE5 proteins were obtained using the Sf-9 cell-baculovirus system (BD Biosciences PharMingen) as previously described [46, 47].
Purified His-tag recombinant bovine PDE5A1 was obtained as described [30].
Native PAGE, SDS-PAGE, enzyme phosphorylation, [3H]cGMP-binding kinetics, and catalytic properties verified structural and functional integrity of the unmutated PDE5 constructs. All recombinant PDE5A1 proteins exhibited a purity of >98% as determined by SDS/PAGE. Bovine PDE5A1 constructs were flash-frozen using liquid N2 in Tris buffer (20 mM Tris, pH 8, and 50 mM NaCl) containing a final concentration of 20% glycerol. Human PDE5A1 constructs were flash-frozen in 10 mM potassium phosphate, pH 6.8, 25 mM β-mercaptoethanol, and 1 mM EDTA (KPEM) + 150 mM NaCl and 10% sucrose. Recombinant PDE5 preparations were stored at −80 °C.
PDE activity was determined as described previously [46] with 0.4 µM [3H]cGMP as substrate.
2.3. Native polyacrylamide gel electrophoresis (Native PAGE)
20 µl samples in KPM + 100 mM NaCl were combined with 5 µl ligand and 5 µl 7 mg/ml BSA and pre-incubated as indicated. 5 µl 40 % v/v glycerol containing 0.01 % bromophenol blue (Bio-Rad, Hercules, CA) was added, and the mixture was loaded into 50-µl wells of precast Tris-HCl acrylamide (7.5 or 10 %) gels (Bio-Rad). Electrophoresis was conducted at room temperature in native-PAGE running buffer (25 mM Tris, 192 mM glycine, pH 8.3, Bio-Rad) for 40 min at 200 V, according to the manufacturer’s instructions. For studies of effects of ligands, PDE5 holoenzyme (1 µM) was incubated in 50 mM Tris-HCl, pH 7.5, and 1.75 mg/ml BSA in the absence or presence of ligands in a final volume of 10 – 30 µl for the time and temperature indicated before subjecting the proteins to electrophoresis. Proteins were visualized by either Coomassie Blue or silver staining.
2.4. [3H]cGMP Millipore membrane filtration protein binding
432 µl recombinant bovine PDE5 (225 nM) that had been diluted in 10 mM KP (pH 6.8) + 1 mg/ml BSA was added to 48 µl of a mixture containing 10 mM KP, 1 mM EDTA, 4 mg/ml histone Type IIAS (Sigma), 0.25 mM 3-isobutyl-1-methylxanthine (IBMX), and 4.9 µM [3H]cGMP. The reaction mixture was incubated for the indicated times at 30 °C and an aliquot (72 µl) was removed and pipetted into 2 ml ice-cold KP and immediately filtered under house vacuum through Millipore nitrocellulose membranes (0.45 µm pore size) that had been pre-wetted with 2 ml of ice-cold 10 mM KP. The reaction tube was then washed twice with 2-ml aliquots of ice-cold KP, and the washes were also applied to the filter. Filter membranes were removed, dried, and transferred to 6-ml scintillation vials containing 5 ml of nonaqueous scintillant for counting.
2.5. Statistical Analyses
All values are given as mean ± S.E.M. as determined by GraphPad Prism graphics software (GraphPad Software Inc., San Diego, CA). The software uses the following equation:
S.E.M. = standard deviation/n1/2, where standard deviation is determined as [Σ(yi − ymean)2/(n − 1)]1/2. All S.E.M.s fit within a 95% confidence interval, which quantifies the precision of the mean.
3. Results
3.1. Time-dependent activation of PDE5 by pre-incubation with cGMP
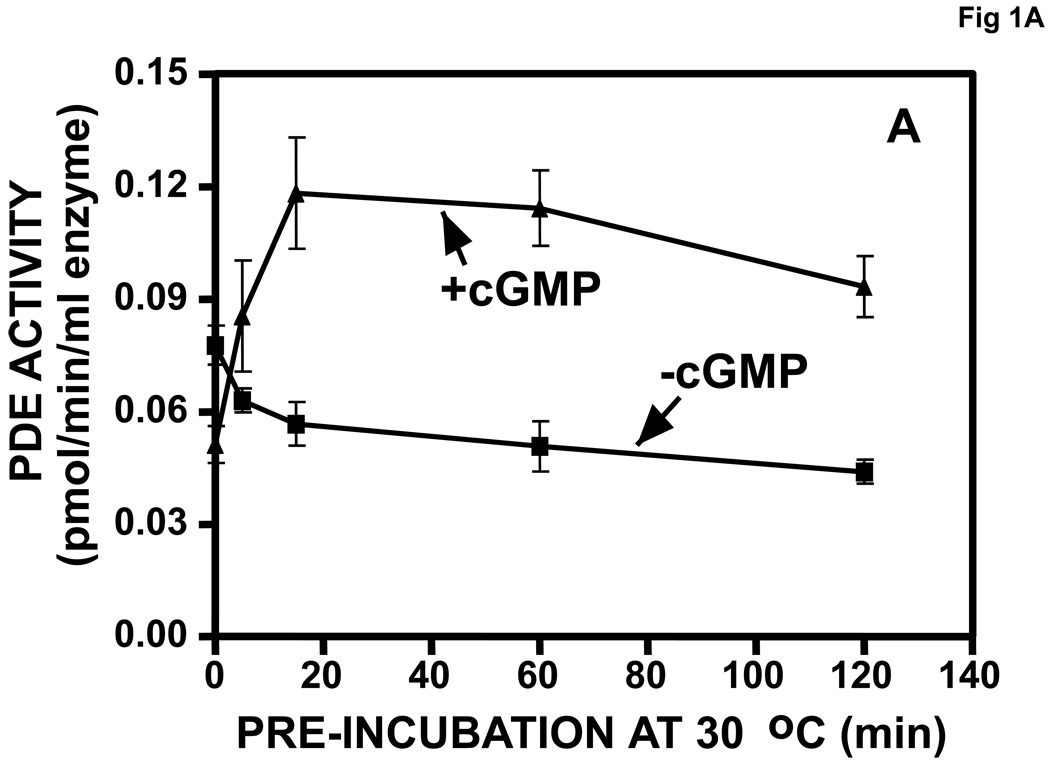
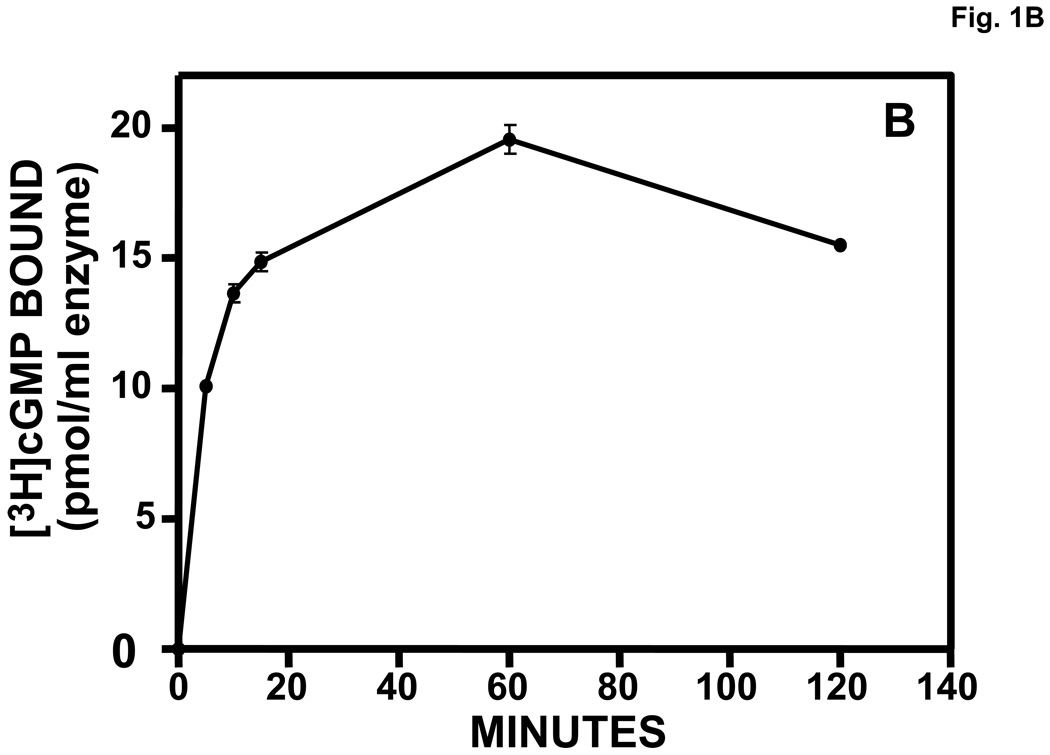
Purified, recombinant bovine PDE5 was activated in a time-dependent manner by pre-incubation with 710 µM cGMP prior to dilution and assay of PDE activity (Fig. 1A). The peak of activation was achieved by 20 min, which correlated with the relatively slow time course of binding of cGMP to the allosteric site of PDE5 (Fig. 1B). A 15-min pre-incubation with 34,000 µM cAMP did not produce significant PDE5 activation (not shown).
Fig. 1.
A. Time course of activation of PDE5 by pre-incubation with cGMP. Purified recombinant bovine PDE5 (0.061 mg/ml, 0.61 µM subunit) was pre-incubated in 1.75 mg/ml BSA and 710 µM cGMP for the indicated times at 30 °C. Each sample was diluted 300 times in KPM containing 1 mg/ml BSA and assayed immediately for cGMP-PDE activity in a 5-min incubation. n = 4. B. Time course of [3H]cGMP binding to PDE5 allosteric site. Incubation and Millipore membrane filtration were carried out as described in Materials and methods (n = 3).
3.2. Concentration-dependent cGMP activation of PDE5 by pre-incubation is associated with a Native PAGE gel-shift, while sildenafil does not cause activation but produces a similar gel shift
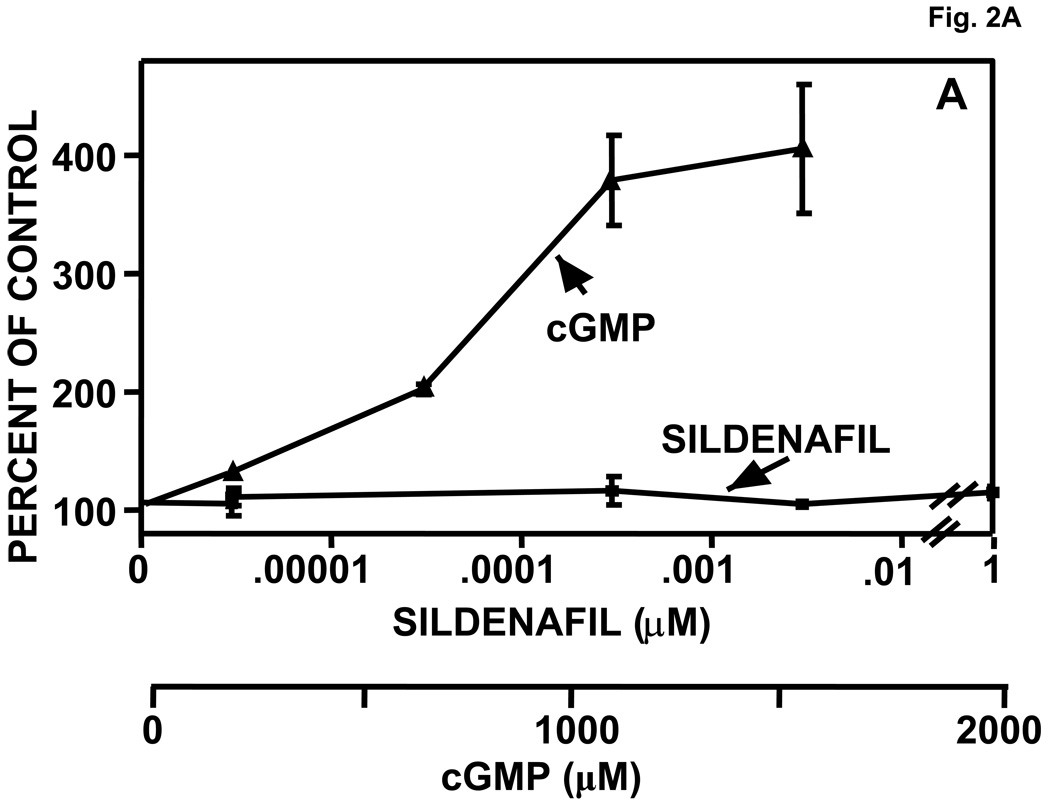
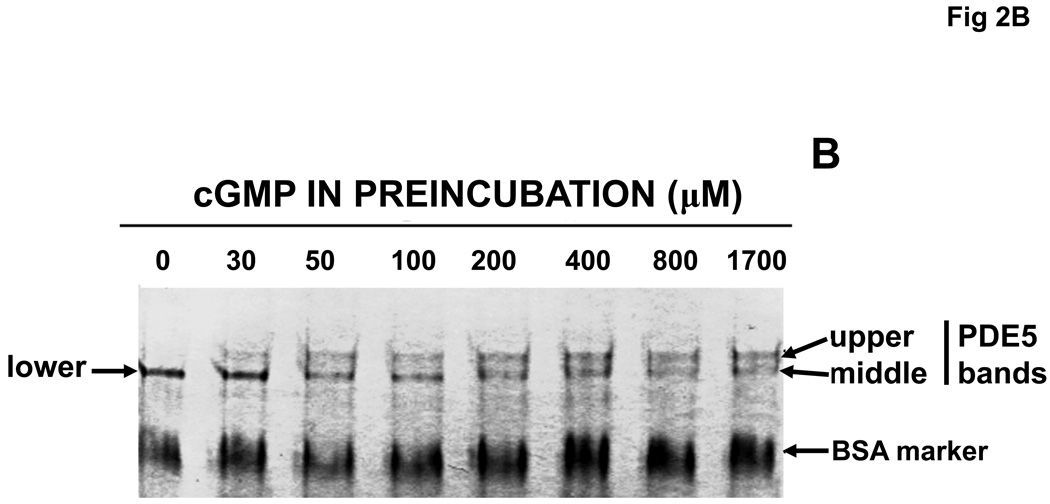
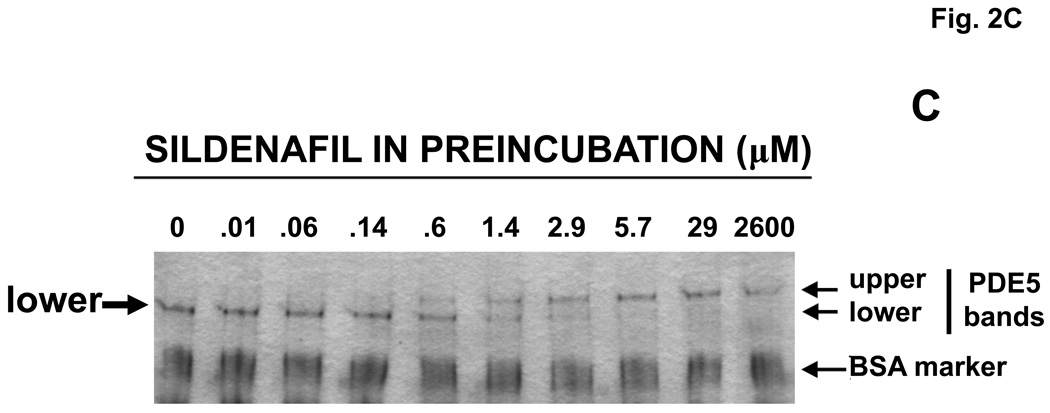
Purified, recombinant bovine PDE5 (0.61 µM) was pre-incubated with a range of concentrations of cGMP or sildenafil for 15 min at 30 °C. Aliquots of the mixtures were then either diluted extensively and assayed for cGMP PDE catalytic activity or subjected directly to Native PAGE (no SDS). In Fig. 2A it can be seen that cGMP produced a concentration–dependent activation of PDE5 catalytic activity, but sildenafil did not. PDE5 activation by cGMP was associated with shift of the PDE5 band to slower mobility on Native PAGE (Fig. 2B). In the absence of cGMP, a single band of protein was seen (labeled “lower” in the first lane), but upon pre-incubation with relatively dilute cGMP (30–50 µM) a band with significantly slower migration (labeled “upper”) was clearly evident. In addition, a “middle” band that had ~5% slower migration than that of the lower band was resolved after treatment with concentrations of cGMP higher than 200 µM, while the lower band disappeared at these concentrations. The lower band obtained with no or low cGMP had Rf relative to BSA of 0.630 ± 0.003, n = 13, while the middle band had Rf of 0.599 ± 0.002, n = 13, and the upper band had Rf of 0.538 ± 0.003, n = 18, which was 15% slower than the lower band. At >10,000 µM cGMP more than 90% of the PDE5 appeared in the upper-band position (not shown).
Fig. 2.
Activation of PDE5 by pre-incubation with cGMP, but not with sildenafil; associated Native PAGE gel-shifts with cGMP or sildenafil. Purified recombinant bovine PDE5 (0.061 mg/ml, 0.61 µM subunit) was pre-incubated in 1.75 mg/ml BSA and the indicated concentration of cGMP or sildenafil for 15 min at 30 °C. Each sample was diluted either 300 (cGMP-treated) or 10,000 (sildenafil-treated) times in KPM containing 1 mg/ml BSA and assayed immediately for cGMP-PDE activity for 5 min (cGMP-treated) or 50 min (sildenafil-treated) (Panel A, n = 4) at 30 °C or subjected to Native PAGE (Panels B and C) for 40 min at 200 V and stained with Coomassie Blue. The positions of the “lower” band in lanes containing no or low cGMP and “upper” band (plus cGMP or sildenafil) are indicated by arrows. The position of the “middle” band obtained in lanes in which the sample contained 800 or 1700 µM cGMP is indicated by arrow. Figures 2B and 2C are each representative of 3 independent experiments.
As can be seen in Fig. 2C, pre-incubation of PDE5 with sildenafil produced a concentration-dependent Native-PAGE shift from the more rapidly migrating “lower” band (lower band Rf = 0.640 ± 0.003, n = 22) to a more slowly migrating “upper” band (Rf = 0.556 ± 0.004, n = 13), which had a position similar to that of the upper band produced by cGMP (see also Fig. 2B and Fig. 4C). However, there was no discernible middle band. In head-to-head comparison (not shown, see also Fig. 4C) there was no detectable difference in mobility of the upper band produced by cGMP (Rf = 0.532 ± 0.006, n = 6) or sildenafil (Rf = 0.542 ± 0.006, n = 10). In these experiments the middle band obtained with cGMP had mobility (Rf = 0.604 ± 0.004, n = 9) that was significantly different from that of the lower band (Rf =0.630 ± 0.007, n = 22) of the untreated PDE5 or upper band (Rf = 0.538 ± 0.005, n = 20) of the PDE5 treated with sildenafil.
Fig. 4.
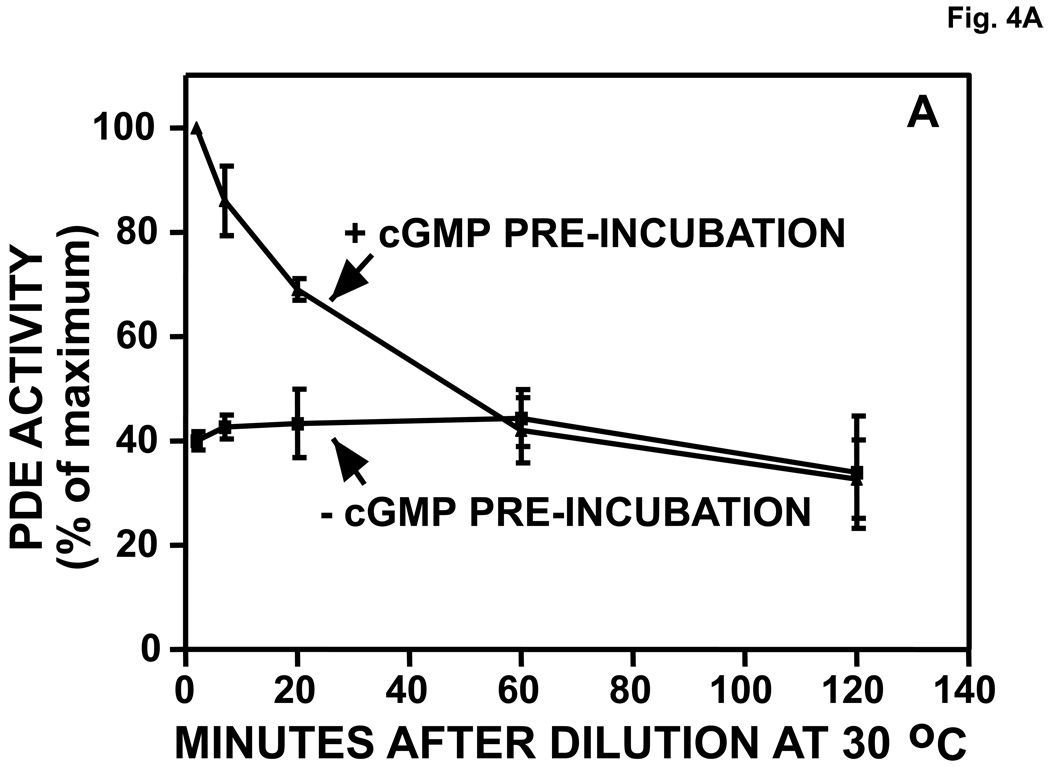
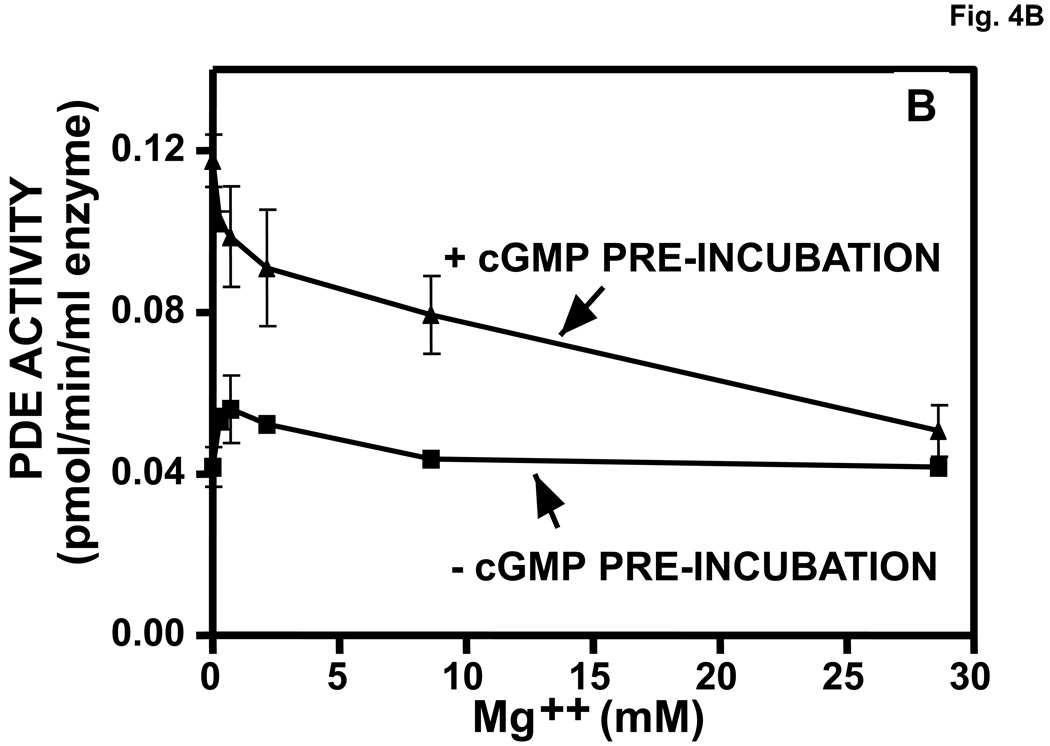
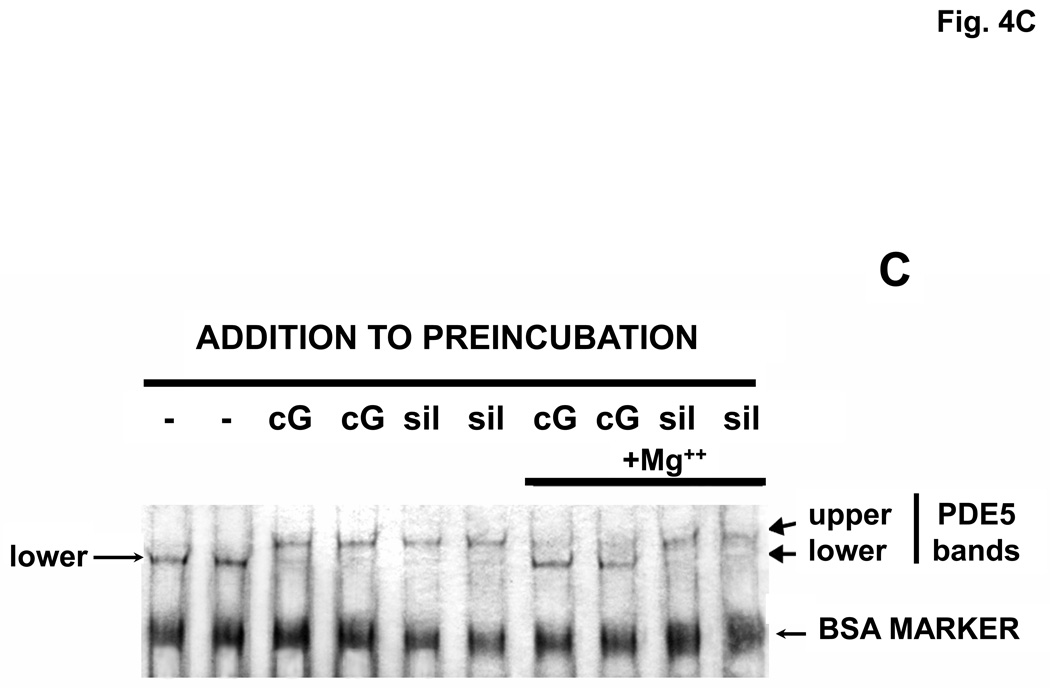
Reversal of cGMP activation and Native Page gel-shift. A. Reversal of cGMP activation induced by dilution. The experiment was performed as in Fig. 2. After pre-incubation in the presence or absence of 540 µM cGMP for 30 min, the mixture was diluted 400 times in KPM containing 1 mg/ml BSA and 20 µl aliquots were assayed (5-min assay) in a total reaction volume of 46 µl at the indicated times at 30 °C for cGMP-PDE activity (n = 3). Activity of the +cGMP sample at 0 min was taken as 100%. B. Reversal of cGMP-induced activation of PDE5 by inclusion of Mg++ in the pre-incubation mixture. Experiment performed (n = 3) as in A except that [3H]cGMP was substituted for unlabeled cGMP. After 15 min at 30 °C, 5-µl aliquots were removed and added to 95 µl KPM + 1 mg/ml BSA. 150 µl PDE assay mix (no [3H]cGMP) and 10 µl PDE assay stop mix were then added in rapid succession. The snake venom reaction and QAE columns were then carried out as in the standard PDE assay to determine [3H]cGMP breakdown. C. Reversal of cGMP-induced (cG), but not sildenafil-induced (sil), Native PAGE gel-shift by inclusion of 280 Mg+2 in the pre-incubation mixture. The experiment was performed as in Fig. 2. Sildenafil concentration in the pre-incubation was 1430 µM and cGMP was 2860 µM. Native PAGE was performed on the pre-incubation mixture (no dilution). Results were typical of those obtained in 3 independent experiments.
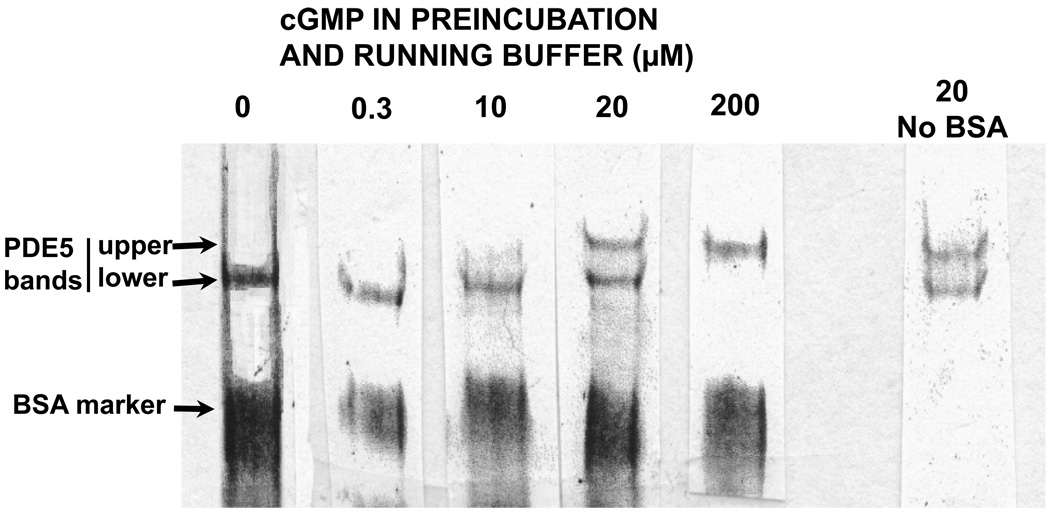
3.3. Low cGMP in the running buffer causes a gel shift of PDE5 on Native PAGE
Since millimolar concentrations of cGMP were required to produce the gelshifts seen in Fig. 2B, a physiological role for the effects based on the requirement of such high cGMP concentrations was subject to scrutiny. The allosteric binding site of PDE5 has a KD of ~0.2 µM cGMP and the catalytic site has a Km of ~2.5 µM [48]; therefore, the initial cGMP concentration in the samples was nearly 1000-times higher than should be theoretically required to bind to either site. However, it is likely that most of the cGMP in the pre-incubation was quickly removed during electrophoresis by its migration toward the anode, and that continual presence of much lower cGMP was sufficient for the effects. Therefore, a range of relatively low concentrations of cGMP were added to the pre-incubation and the electrophoresis running buffer so that cGMP was present before and during electrophoresis. It can be seen in Fig. 3 that at 20 µM cGMP (~6:1 cGMP to PDE5), approximately one-half of PDE5 was shifted to the more slowly migrating “upper band”. The protocol used for Fig. 3 required that separate gels be run for each concentration of cGMP. The lanes were then cut from the gels and placed side-by-side for photography. This procedure reduced the resolution of bands when compared with the protocol of Fig. 2B; therefore, a “middle band” could not be ascertained with suitable confidence. The combined results using the two protocols suggested that the cGMP-induced change in physical form of PDE5 is reversible. The results of Fig. 3 imply that the physical changes of PDE5 produced by cGMP as reflected by the gel-shifts occur within the physiological range of cGMP concentrations.
Fig. 3.
Low cGMP in the running buffer causes a gel-shift of PDE5 on Native PAGE. To a mixture containing 3.5 µM purified PDE5, 1 mg/ml BSA, and 60 mM NaCl was added the indicated concentration of cGMP; the respective mixtures were then incubated for 15 min at 30 °C and Native PAGE was performed as in Fig. 2 except that the running buffer contained the same concentration of cGMP as did the sample applied to the gel. A separate gel was run for each cGMP concentration indicated, and the lanes were cut out of the gels and placed side-by-side for photography. The positions of the “lower” band in lanes containing no or low cGMP and “upper” band (+ higher cGMP) are indicated by arrows.
3.4. Dilution or cGMP hydrolysis reverses cGMP activation of PDE5
If activation of PDE5 by cGMP as seen in Fig. 1A has physiological relevance it should be a reversible process. Reversibility was tested in two ways: 1) by dilution of the cGMP-PDE5 mixture after pre-incubation in order that cGMP would be lower than its affinity constant (KD) for PDE5, followed by incubation to allow for cGMP dissociation from the enzyme, and 2) by addition of Mg++ to the pre-incubation mixture [49] to foster lowering of cGMP by the hydrolytic action of the enzyme.
In Fig. 4A PDE5 (0.6 µM) was pre-incubated with ~100-fold excess of cGMP (540 µM) for 30 min and then diluted 400-fold to achieve a maximum final cGMP concentration of 1.3 µM. This dilution followed by incubation at 30 °C reversed activation of PDE5, but reversal required about 1 h. The rate of reversal was consistent with the rate of exchange-dissociation of cGMP from the allosteric cGMP-binding site of the enzyme following dilution (>90% dissociation after 30 min at 30 °C) [48]. The rate of reversal was not consistent with the rate of dissociation of cGMP from the catalytic site, which is very fast (<1 sec for >90% dissociation) (Morris et al, unpublished). The combined data suggested that activation of PDE5 by cGMP is due to binding of this ligand to the allosteric site of the enzyme. This conclusion is also in agreement with the findings of Rybalkin et al. in which antibody directed against allosteric binding site GAF A blocked cGMP activation of PDE5 [31].
In a concentration-dependent manner, Mg++ added to the pre-incubation mixture reversed cGMP-induced activation of PDE5 (Fig. 4B). Complete reversal occurred at 27 mM Mg++. Assays under these conditions indicated that >80% of the cGMP was hydrolyzed in 15 min of pre-incubation (not shown). Therefore, reversal of cGMP activation of PDE5 occurs by dilution of reactants (Fig. 4A), cGMP hydrolysis (Fig. 4B), or removal of cGMP by electrophoresis (Fig. 2B and Fig. 3).
3.5. Reversal of Native PAGE gel-shift by inclusion of Mg++ in the pre-incubation
Examination of reversal of the cGMP-induced Native PAGE gel-shift due to dilution was problematic because extensive dilution of the pre-incubation mixture compromised visualization of bands after electrophoresis. However, reversal due to incubation with Mg++ could be examined by Native PAGE since it was not necessary to dilute the pre-incubation mixture before electrophoresis. It can be seen in Fig. 4C that inclusion of Mg++ in the pre-incubation reversed most of the gel-shift produced by sufficient cGMP to shift PDE5 to the upper band. On the contrary, the gel-shift produced by non-hydrolyzable sildenafil was not reversed by Mg++. The combined results of dilution and Mg++ incubation are consistent with the interpretation that the continued presence of cGMP bound to the allosteric site is necessary for the PDE5 activation and associated change in physical form seen in Fig. 2B.
3.6. Reversal of tadalafil inhibition of PDE5
Pre-incubation of PDE5 with tadalafil increases affinity of this inhibitor as indicated by slowing of [3H]tadalafil exchange-dissociation from the enzyme [42]. Tadalafil also produces a gel-shift to the “upper-band” position that is not detectably different from that obtained by sildenafil [43]. Reversal of increased tadalafil affinity was examined by first pre-incubating PDE5 either in the absence or presence of 21 µM tadalafil followed by dilution of the reaction mixture so that the tadalafil concentration (0.07 nM) was below the KD (~2 nM) for binding to PDE5. The PDE assay was carried out immediately as described in Materials and Methods and for various times, and the IC50 for tadalafil was determined using a range of concentrations. There was no difference in the catalytic activity of the diluted enzymes (tadalafil-treated vs control) at a substrate level of 0.4 µM cGMP or affinity of the diluted enzymes for tadalafil (IC50 = 1.63 ± 0.64 nM, n = 4) compared with that of the diluted untreated enzyme (IC50 = 1.69 ± 0.46 nM, n = 3) (not shown).
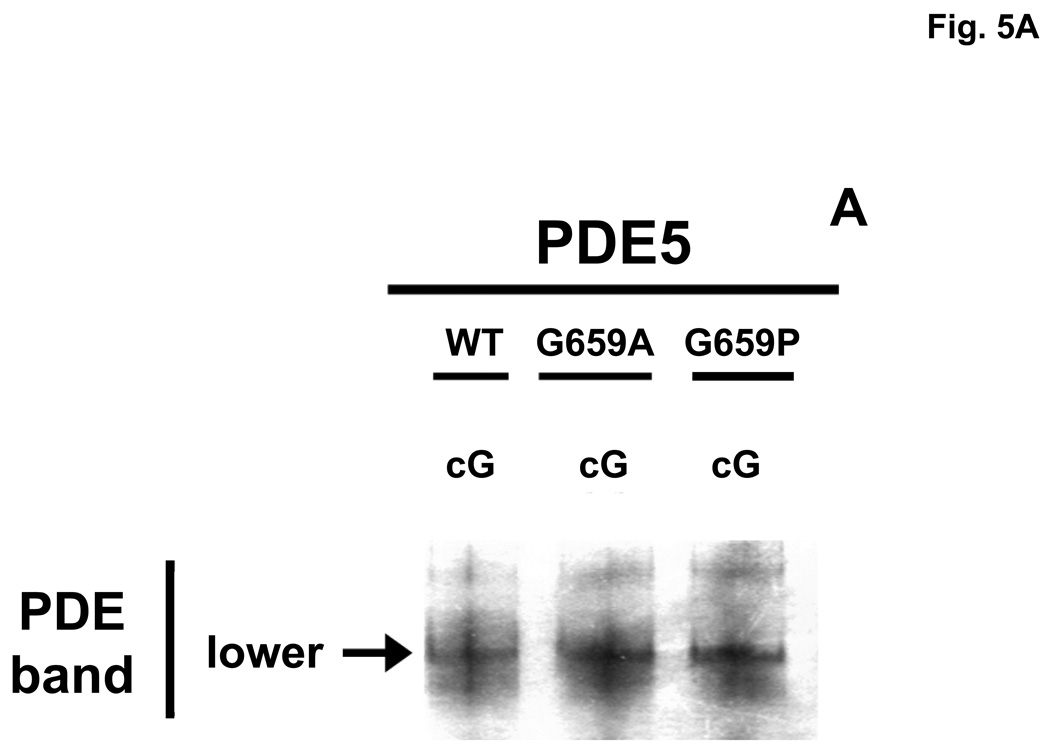
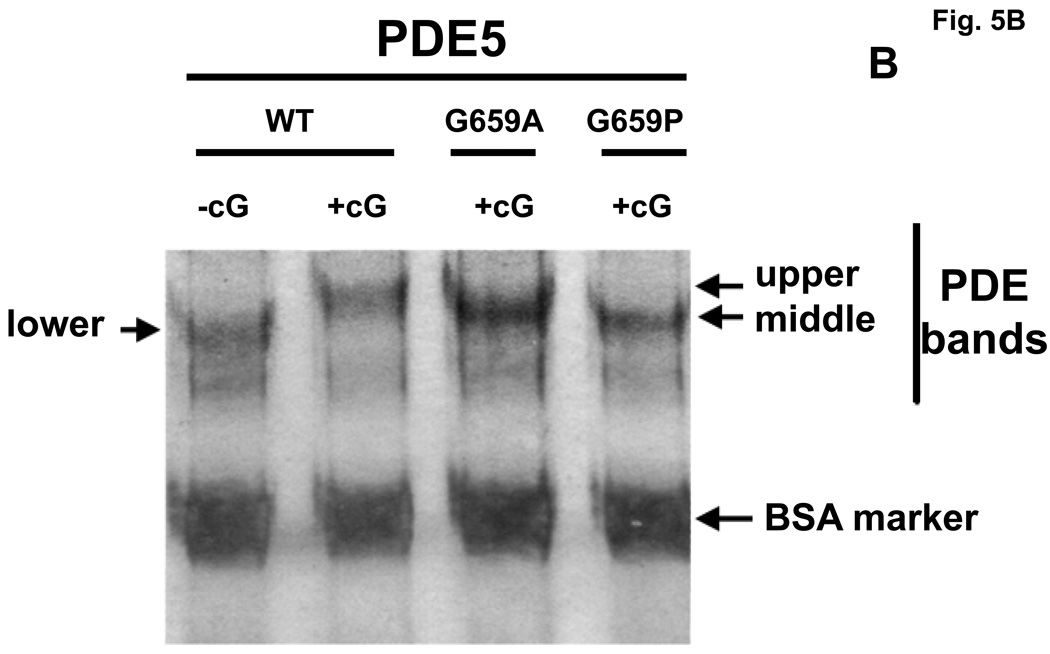
3.7. cGMP produces a Native PAGE gel-shift of PDE5 to an intermediate (middle-band) position after catalytic-site mutation
Whether or not the cGMP-induced physical changes in PDE5, as reflected by Native PAGE gel-shifts, was due to cGMP binding to the allosteric site, catalytic site, or both sites was also examined using two catalytic-domain mutants that were compromised with respect to affinity of the catalytic site for cGMP, while cGMP binding to the allosteric site was unchanged. A critical C-domain residue in human recombinant PDE5, glycine-659 [50], was mutated to alanine (G659A) or proline (G659P); these mutations produced large reductions in catalytic site affinities for cGMP (Km WT = 2.91 ± 0.8 µM, G659A = 71.2 ± 1.2 µM, G659P = 131.5 ± 7.4 µM). Native PAGE was performed as in Fig. 2B. Fig. 5A shows that in the absence of cGMP the two mutants have the same Native-PAGE mobility as does the WT enzyme. As seen in Fig. 5B, pre-incubation of WT PDE5 with 400 µM cGMP caused a shift of the PDE5 band to the upper-band position, but both mutant PDE5 bands were shifted only to the middle-band position. The Rf of the human WT PDE5 (0.604± 0.005, n = 15) relative to BSA was slightly lower than that of bovine WT PDE5 (0.630 ± 0.003, n = 13); for the human mutants the Rf was 0.571 ± 0.005, n = 22, and 0.503 ± 0.005, n = 12, for the middle and upper bands, respectively. It was concluded that binding of cGMP to the catalytic site is necessary for shift of PDE5 to the physical form reflected by the Native PAGE gel-shift to the upper-band position. This is consistent with our finding that, using the same conditions for Native PAGE as in Fig. 2B, catalytic site-specific sildenafil did not produce a perceptible gel-shift using G659P and caused only ~10% shift from the lower to the upper band using G659A (not shown). This was not consistent with lack of pronounced effect of sildenafil on IC50 of PDE5 (WT = 1.75 ± 0.29 nM; G659A = 3.45 ± 1.26 nM, n = 4), suggesting that Gly-659 is critical for cGMP, but not for sildenafil, interaction with the catalytic site. Binding of cGMP to the allosteric site causes conversion to a different physical form as reflected by shift to the middle-band position. This conclusion is consistent with reversal of the low cGMP-induced gel shift by electrophoresis seen in Fig. 2B, and with the time course of reversal of cGMP activation of PDE5 seen in Fig. 4A.
Fig. 5.
Altered cGMP-induced Native PAGE gel-shift of PDE5 after catalytic-domain mutation. Purified recombinant human PDE5 proteins (WT = 0.07 mg/ml, G659A = 0.09 mg/ml, G659P = 0.09 mg/ml) were either pre-incubated in 0.1 mg/ml BSA with no additions (Panel A) or with 400 µM cGMP (Panel B) for 15 min at 30 °C before Native PAGE and silver stain. The positions of the “lower”, “middle”, and “upper” PDE5 bands are indicated by arrows. Figure is representative of 4 independent experiments.
4. Discussion
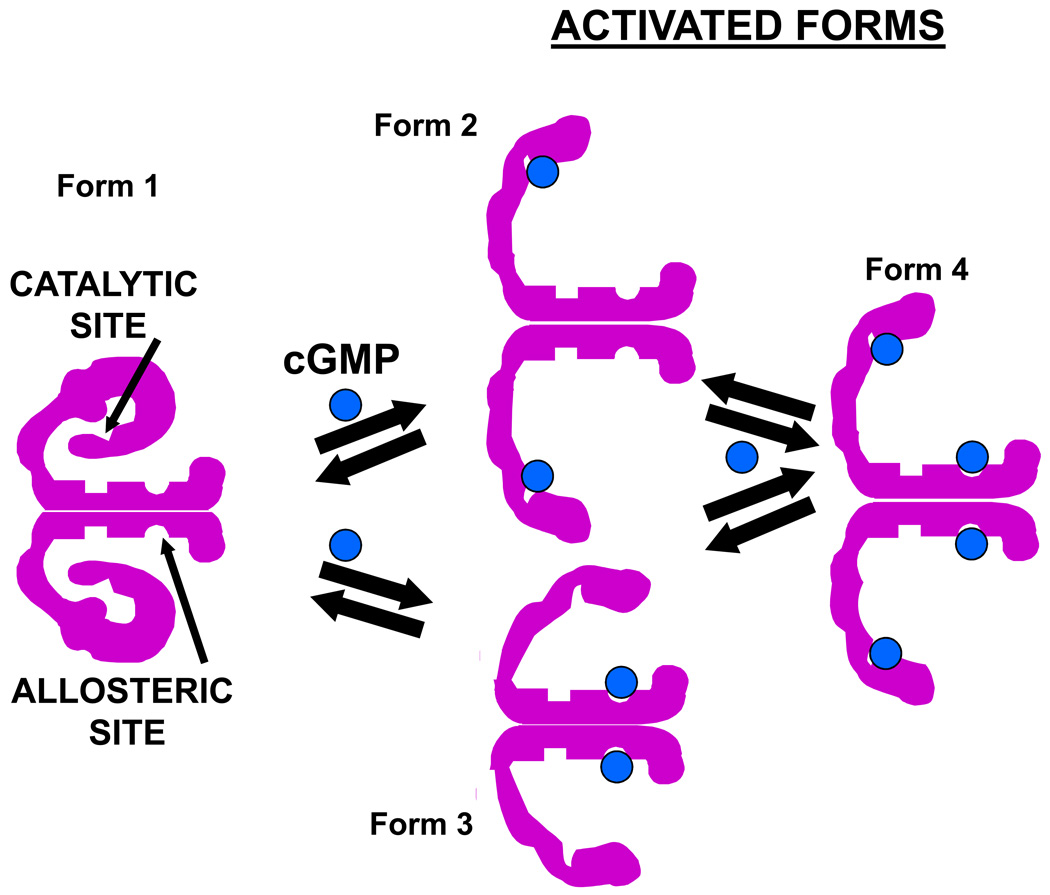
Based on the results contained herein and on our previous studies [30, 39, 42, 43] we propose the hypothesis presented in Fig. 6. It states that cGMP can convert basal PDE5 (Form 1) to Form 2 (catalytic site occupied) or Form 3 (allosteric site occupied). However, conversion to Form 2 occurs at lower cGMP concentrations than conversion to Form 3. This may seem anomalous considering the higher measured cGMP affinity for the allosteric site over the catalytic site, but it could represent the physiological situation since studies of allosteric cGMP-binding affinity of PDE5 have usually been done in the presence of a catalytic-site inhibitor such as IBMX, which is stimulatory for allosteric cGMP binding. Perhaps the allosteric site (GAF A subdomain) is unavailable for optimum cGMP binding without occupation of the catalytic site. Following conversion of PDE5 to Forms 2 and 3, each of these forms can interact with additional cGMP to produce Form 4. As compared with Form 1, Form 2 is more active catalytically, binds cGMP or PDE5 inhibitors more avidly, and is phosphorylated at increased rate by PKG. The presence of a significant level of Form 2 in cells could “prime” the enzyme for stimulation of catalytic activity by further increase in cGMP, for phosphorylation, or for inhibitor action. Like Form 2, Form 4 is more catalytically active, binds inhibitor with higher affinity, binds cGMP at the allosteric site more avidly, and is more readily phosphorylated by PKG. Based on analysis using gel-shift, Forms 2 and 4 are indistinguishable from each other in physical form. Assuming that the similar gel-shifts are not coincidental, the physical change occurring by ligand binding to the catalytic site in Form 4 may dominate over that occurring by ligand binding to the allosteric site in this form. Current PDE5 inhibitors do not bind to the allosteric site, which precludes formation of Form 3 by these agents, but these inhibitors convert the enzyme directly to Form 2, which is indistinquishable from Form 4 by gel-shift and is more active for allosteric cGMP binding and phosphorylation. It is of interest that phosphorylation also produces a PDE5 form that is indistinguishable from Form 4 based on gelshift [43]. It could not be ruled out that monomer-monomer interactions could alter the model of Fig. 6. For example, occupation of the catalytic or allosteric site of only one of the two monomers could cause conversion to Form 2 or 3, respectively.
Fig. 6.
Hypothetical model for regulation of PDE5 by cGMP binding to the allosteric site and/or catalytic site.
The hypothesis of Fig. 6 predicts several consequences. Increase in Forms 2, 3, and 4 would stimulate cGMP breakdown at the catalytic site, phosphorylation of the enzyme, and sequestration of cGMP at the allosteric site, all of which would result in increased negative feedback on the cellular cGMP pathway. PDE5 inhibitor therapy should increase formation of Forms 2 and 4, which would elevate the phosphorylation state as well as allosteric cGMP binding to PDE5. This would be expected to improve cellular catalytic activity, but occupation of the catalytic site by inhibitor following use of these medications would block this effect. Nevertheless, Forms 2 and 4 are more active for phosphorylation and allosteric cGMP binding, and both of these effects should increase further inhibitor binding to the catalytic site; thus, the inhibitor would stimulate its own binding, a positive feedback reaction that improves and prolongs inhibitor action in cells.
The present results suggest that activation of PDE5 by pre-incubation with cGMP is explained by retention of cGMP bound to the allosteric site after dilution rather than by a structural change in the enzyme that is induced by cGMP but persists after dissociation of the nucleotide. The effects of cGMP displayed in Fig. 6 are reversible at a rate that is consistent with the dissociation rate of this ligand from the allosteric site of the enzyme. The relative slowness of reversal of cGMP activation suggests that cGMP effects on PDE5 would be significantly prolonged after decline of the nucleotide in cells, which would impart resistance or “down-regulation” of the cGMP pathway. Mullershausen et al. have observed a sustained nitric oxide-induced activation of PDE5 for as long as 60 minutes in intact platelets [32].
Cyclic GMP signaling in most tissues is controlled by “graded” regulation as opposed to “switch” regulation; therefore, it is expected that factors within the pathway, e.g., PDE5, would be in a state of partial activation under most physiological conditions. This is definitely the case for PKG and PKA, which exhibit varying ratios of cyclic nucleotide-free and cyclic nucleotide-bound forms in extracts of fresh tissues [51, 52]. PDE4, a cAMP-specific PDE, exhibits two kinetic species defined as having high-affinity and low-affinity for inhibitors; the relative amounts of these species vary widely in tissues and are thought to confer different functions [53]. Depending on their physiological roles, varying ratios of the different kinetic species and physical forms of PDE5 may also occur in different tissues and could impact therapies using PDE5 inhibitors. Prevalence of the activated forms in a particular tissue might allow for specific targeting of that tissue using low doses of PDE5 inhibitors in humans, or it might cause adverse side effects due to hyper-responsiveness of an untargeted tissue to PDE5 inhibitors. The present studies lay the groundwork for investigations of the presence of different physical and kinetic forms of PDE5 in intact tissues under various physiological and pharmacological states of the organism.
Our combined results imply that the physical changes in PDE5 as reflected by ligand-induced gel-shifts cause the changes in activity of PDE5. The gel-shifts suggest that Forms 2 and 4 could have a more elongated shape than Form 1, and Form 3 could possess intermediate elongation, but changes in surface charge of the proteins, or a combination of changes in shape and charge, could also explain the differences in physical properties of these forms. Our finding of change in physical form of PDE5 by catalytic-site ligands is consistent with the observation that sildenafil binding produces a change in the x-ray crystal structure of the free catalytic domain of this enzyme [50]. The main change is movement of the “H-loop” (residues 660-683 in human PDE5A1) approximately 24Å**. However, this movement does not cause an appreciable protein elongation. Perhaps the dramatic movement of the H-loop within the C domain shifts the relative positions of the C and R domains to bring about elongation or increased electropositivity of the surface charge of the holoenzyme structure. This would explain the conversion of PDE5 to Form 2 by sildenafil binding to the catalytic site or to Form 4 by ligand binding to both the allosteric and catalytic sites. Our finding of a different physical form (Form 3) of PDE5 as reflected by an intermediate gel-shift to the “middle-band” position by cGMP binding to the allosteric site, together with the finding of reduced Native PAGE gel mobility of the cGMP-treated isolated R domain [54], predict a distinct physical change in the R domain by allosteric-site binding.
Acknowledgement
This work was generously supported by NIH grant DK40029 (J.D.C.). We thank the Vanderbilt University Diabetes Core and the Vanderbilt Ingram Cancer Center Sequencing Core for providing reagents and services. We also thank Alfreda Beasley for technical assistance, Dr. Raja Konjeti for synthesizing tadalafil, and Dr. Jennifer Busch for reading the manuscript.
Abbreviations
- PDE
cyclic nucleotide phosphodiesterase
- IBMX
3-isobutyl-1-methylxanthine
- C domain
catalytic domain
- R domain
regulatory domain
- PAGE
polyacrylamide gel electrophoresis
- KP
10 mM potassium phosphate (pH 6.8)
- GAF
cGMP-binding PDE, Anabaena adenylyl cyclase, E. coli FhlA protein.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Roya Zoraghi, Email: zoraghi@interchange.ubc.ca.
Sharron H. Francis, Email: sharron.francis@vanderbilt.edu.
References
- 1.Beavo JA, Conti M, Heaslip RJ. Mol. Pharmacol. 1994;46:399–405. [PubMed] [Google Scholar]
- 2.Conti M. Mol. Endocrinol. 2000;14:1317–1327. doi: 10.1210/mend.14.9.0534. [DOI] [PubMed] [Google Scholar]
- 3.Francis SH, Turko IV, Corbin JD. Prog. Nucleic Acid Res. Mol. Biol. 2001;65:1–52. doi: 10.1016/s0079-6603(00)65001-8. [DOI] [PubMed] [Google Scholar]
- 4.Houslay MD, Adams DR. Biochem. J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rybalkin SD, Yan C, Bornfeldt KE, Beavo JA. Circ. Res. 2003;93:280–291. doi: 10.1161/01.RES.0000087541.15600.2B. [DOI] [PubMed] [Google Scholar]
- 6.Soderling SH, Beavo JA. Curr. Opin. Cell Biol. 2000;12:174–179. doi: 10.1016/s0955-0674(99)00073-3. [DOI] [PubMed] [Google Scholar]
- 7.Omori K, Kotera J. Seikagaku. 1999;71:541–546. [PubMed] [Google Scholar]
- 8.Sette C, Conti M. J. Biol. Chem. 1996;271:16526–16534. doi: 10.1074/jbc.271.28.16526. [DOI] [PubMed] [Google Scholar]
- 9.Jin SL, Bushnik T, Lan L, Conti M, Swinnen JV. J.Biol.Chem. 1998;273:19672–19678. doi: 10.1074/jbc.273.31.19672. [DOI] [PubMed] [Google Scholar]
- 10.Beavo JA, Hardman JG, Sutherland EW. J. Biol. Chem. 1971;246:3841–3846. [PubMed] [Google Scholar]
- 11.Conti M. J. Beavo, Annu. Rev. Biochem. 2007;76:481–511. doi: 10.1146/annurev.biochem.76.060305.150444. [DOI] [PubMed] [Google Scholar]
- 12.Beltman J, Sonnenburg WK, Beavo JA. Mol. Cell. Biochem. 1993;127–128:239–253. doi: 10.1007/BF01076775. [DOI] [PubMed] [Google Scholar]
- 13.Corbin JD, Turko IV, Beasley A, Francis SH. Eur. J. Biochem. 2000;267:2760–2767. doi: 10.1046/j.1432-1327.2000.01297.x. [DOI] [PubMed] [Google Scholar]
- 14.Mullershausen F, Russwurm M, Thompson WJ, Liu L, Koesling D, Friebe A. J. Cell. Biol. 2001;155:271–278. doi: 10.1083/jcb.200107001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wyatt TA, Naftilan AJ, Francis SH, Corbin JD. Am. J. Physiol. 1998;274:H448–H455. doi: 10.1152/ajpheart.1998.274.2.H448. [DOI] [PubMed] [Google Scholar]
- 16.Degerman E, Landstr:m TR, Wijkander J, Holst LS, Ahmad, Belfrage P, Manganiello V. Methods. 1998;14:43–53. doi: 10.1006/meth.1997.0564. [DOI] [PubMed] [Google Scholar]
- 17.Kotera J, Francis SH, Grimes KA, Rouse A, Blount MA, Corbin JD. Front. Biosci. 2004;9:378–386. doi: 10.2741/1231. [DOI] [PubMed] [Google Scholar]
- 18.Liu HG, Maurice DH. J.Biol.Chem. 1999;274:10557–10565. doi: 10.1074/jbc.274.15.10557. [DOI] [PubMed] [Google Scholar]
- 19.Florio VA, Sonnenburg WK, Johnson R, Kwak KS, Jensen GS, Walsh KA, Beavo JA. Biochemistry. 1994;33:8948–8954. doi: 10.1021/bi00196a012. [DOI] [PubMed] [Google Scholar]
- 20.Sharma RK, Wang JH. Proc. Natl. Acad. Sci. USA. 1985;82:2603–2607. doi: 10.1073/pnas.82.9.2603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Conti CR, Pepine CJ, Sweeney M. Am. J. Cardiol. 1999;83:29C–34C. doi: 10.1016/s0002-9149(99)00045-4. [DOI] [PubMed] [Google Scholar]
- 22.Corbin JD, Francis SH. J. Biol. Chem. 1999;274:13729–13732. doi: 10.1074/jbc.274.20.13729. [DOI] [PubMed] [Google Scholar]
- 23.Francis SH, Corbin JD. Urol. Clin. North Am. 2005;32:419–429. doi: 10.1016/j.ucl.2005.08.001. [DOI] [PubMed] [Google Scholar]
- 24.Atz AM, Lefler AK, Fairbrother DL, Uber WE, Bradley SM. J. Thorac. Cardiovasc. Surg. 2002;124:628–629. doi: 10.1067/mtc.2002.125265. [DOI] [PubMed] [Google Scholar]
- 25.Bortolotti M, Mari C, Lopilato C, Porrazzo G, Miglioli M. Gastroenterology. 2000;118:253–257. doi: 10.1016/s0016-5085(00)70206-x. [DOI] [PubMed] [Google Scholar]
- 26.Ghofrani HA, Wiedemann R, Rose F, Schermuly RT, Olschewski H, Weissmann N, Gunther A, Walmrath D, Seeger W, Grimminger F. Lancet. 2002;360:895–900. doi: 10.1016/S0140-6736(02)11024-5. [DOI] [PubMed] [Google Scholar]
- 27.Jackson G, Chambers J. Int. J. Clin. Pract. 2002;56:397–398. [PubMed] [Google Scholar]
- 28.Thomas MK, Francis SH, Corbin JD. J. Biol. Chem. 1990;265:14964–14970. [PubMed] [Google Scholar]
- 29.Francis SH, Lincoln TM, Corbin JD. J. Biol.Chem. 1980;255:620–626. [PubMed] [Google Scholar]
- 30.Corbin JD, Blount MA, Weeks JL, 2nd, Beasley A, Kuhn KP, Ho YS, Saidi LF, Hurley JH, Kotera J, Francis SH. Mol. Pharmacol. 2003;63:1364–1372. doi: 10.1124/mol.63.6.1364. [DOI] [PubMed] [Google Scholar]
- 31.Rybalkin SD, Rybalkina IG, Shimizu-Albergine M, Tang XB, Beavo JA. Embo. J. 2003;22:469–478. doi: 10.1093/emboj/cdg051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mullershausen F, Friebe A, Feil R, Thompson WJ, Hofmann F, Koesling D. J. Cell. Biol. 2003;160:719–727. doi: 10.1083/jcb.200211041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okada D, Asakawa S. Biochemistry. 2002;41:9672–9679. doi: 10.1021/bi025727+. [DOI] [PubMed] [Google Scholar]
- 34.McAllister-Lucas LM, Sonnenburg WK, Kadlecek A, Seger D, LeTrong H, Colbran JL, Thomas MK, Walsh KA, Francis SH, Corbin JD, Beavo JA. J.Biol.Chem. 1993;268:22863–22873. [PubMed] [Google Scholar]
- 35.Aravind L, Ponting CP. Trends Biochem. Sci. 1997;22:458–459. doi: 10.1016/s0968-0004(97)01148-1. [DOI] [PubMed] [Google Scholar]
- 36.Blount MA, Beasley A, Zoraghi R, Sekhar KR, Bessay EP, Francis SH, Corbin JD. Mol. Pharmacol. 2004;66:144–152. doi: 10.1124/mol.66.1.144. [DOI] [PubMed] [Google Scholar]
- 37.Thomas MK, Francis SH, Corbin JD. J. Biol. Chem. 1990;265:14971–14978. [PubMed] [Google Scholar]
- 38.Corbin JD, Turko IV, Beasley A, Francis SH. Eur. J. Biochem. 2000;267:2760–2767. doi: 10.1046/j.1432-1327.2000.01297.x. [DOI] [PubMed] [Google Scholar]
- 39.Bessay EP, Blount MA, Zoraghi R, Beasley A, Grimes KA, Francis SH, Corbin JD. J. Pharmacol. Exp. Ther. 2008;325:62–68. doi: 10.1124/jpet.107.133405. [DOI] [PubMed] [Google Scholar]
- 40.Gopal VK, Francis SH, Corbin JD. Eur. J. Biochem. 2001;268:3304–3312. doi: 10.1046/j.1432-1327.2001.02233.x. [DOI] [PubMed] [Google Scholar]
- 41.Corbin J. Handbook of Cell. Signaling. 2004;2:465–470. [Google Scholar]
- 42.Blount MA, Zoraghi R, Bessay EP, Beasley A, Francis SH, Corbin JD. J Pharmacol. Exp. Ther. 2007;323:730–737. doi: 10.1124/jpet.107.126540. [DOI] [PubMed] [Google Scholar]
- 43.Bessay EP, Zoraghi R, Blount MA, Grimes KA, Beasley A, Francis SH, Corbin JD. Front. Biosci. 2007;12:1899–1910. doi: 10.2741/2196. [DOI] [PubMed] [Google Scholar]
- 44.Francis SH, Sekhar KR, Rouse AB, Grimes KA, Corbin JD. Int. J. Impot. Res. 2003;15:369–372. doi: 10.1038/sj.ijir.3901040. [DOI] [PubMed] [Google Scholar]
- 45.Daugan A, Grondin P, Ruault C, Le Monnier de Gouville AC, Coste H, Linget JM, Kirilovsky J, Hyafil F, Labaudiniere R. J. Med. Chem. 2003;46:4533–4542. doi: 10.1021/jm0300577. [DOI] [PubMed] [Google Scholar]
- 46.Blount MA, Zoraghi R, Ke H, Bessay EP, Corbin JD, Francis SH. Mol. Pharmacol. 2006;70:1822–1831. doi: 10.1124/mol.106.028688. [DOI] [PubMed] [Google Scholar]
- 47.Zoraghi R, Francis SH, Corbin JD. Biochemistry. 2007;46:13554–13563. doi: 10.1021/bi7010702. [DOI] [PubMed] [Google Scholar]
- 48.Thomas MK, Francis SH, Corbin JD. J. Biol.Chem. 1990;265:14964–14970. [PubMed] [Google Scholar]
- 49.Francis SH, Thomas MK, Corbin JD. In: Cyclic Nucleotide Phosphodiesterases: Structure, Regulation, and Drug Action. Beavo J, Houslay MD, editors. New York: John Wiley & Sons; 1990. pp. 117–140. [Google Scholar]
- 50.Wang H, Liu Y, Huai Q, Cai J, Zoraghi R, Francis SH, Corbin JD, Robinson H, Xin Z, Lin G, Ke H. J. Biol. Chem. 2006;281:21469–21479. doi: 10.1074/jbc.M512527200. [DOI] [PubMed] [Google Scholar]
- 51.Corbin JD, Sugden PH, Lincoln TM, Keely SL. J. Biol. Chem. 1977;252:3854–3861. [PubMed] [Google Scholar]
- 52.Jiang H, Colbran JL, Francis SH, Corbin JD. J. Biol. Chem. 1992;267:1015–1019. [PubMed] [Google Scholar]
- 53.Torphy TJ, Livi GP, Balcarek JM, White JR, Chilton FH, Undem BJ. Adv. Second Messenger Phosphoprotein Res. 1992;25:289–305. [PubMed] [Google Scholar]
- 54.Francis SH, Bessay EP, Kotera J, Grimes KA, Liu L, Thompson WJ, Corbin JD. J. Biol. Chem. 2002;277:47581–47587. doi: 10.1074/jbc.M206088200. [DOI] [PubMed] [Google Scholar]