Abstract
Compstatin is a 13-residue peptide that inhibits activation of the complement system by binding to the central component C3 and its fragments C3b and C3c. A combination of theoretical and experimental approaches has previously allowed us to develop analogs of the original compstatin peptide with up to 264-fold higher activity; one of these analogs is now in clinical trials for the treatment of age-related macular degeneration. Here we used ELISAs, surface plasmon resonance, and isothermal titration calorimetry to assess the effect of modifications at three key residues (Trp-4, Asp-6, Ala-9) on the affinity and activity of compstatin and its analogs, and we correlated our findings to the recently reported co-crystal structure of compstatin and C3c. The KD values for the panel of tested analogs ranged from 10−6 to 10−8 M. These differences in binding affinity could be attributed mainly to differences in dissociation rather than association rates, with a >4-fold range in kon values (2–10 × 105 M−1s−1) and a koff variation of >35-fold (1–37 × 10−2 s−1) being observed. The stability of the C3b-Compstatin complex seemed to be highly dependent on hydrophobic effects at position 4, and even small changes at position 6 resulted in a loss of complex formation. Induction of a β-turn shift by an A9P modification resulted in a more favorable entropy but a loss of binding specificity and stability. The results obtained by the three methods utilized here were highly correlated with regard to the activity/affinity of the analogs. Thus, our analyses have identified essential structural features of compstatin and provided important information to support the development of analogs with improved efficacy.
Keywords: Complement, Compstatin, C3, Surface Plasmon Resonance, Isothermal Titration Calorimetry, Kinetics
Introduction
The human complement system is a powerful player in the defense against pathogenic organisms and the mediation of immune responses. Complement can be activated through three different pathways: the classical, lectin, and alternative pathways. The major activation event that is shared by all three pathways is the proteolytic cleavage of the central protein of the complement system, C3, into its activation products C3a and C3b by C3 convertases. Generation of these fragments leads to the opsonization of pathogenic cells by C3b and iC3b, a process that renders them susceptible to phagocytosis or clearance, and to the activation of immune cells through an interaction with complement receptors (Markiewski and Lambris, 2007). Deposition of C3b on target cells also induces the formation of new convertase complexes and thereby initiates a self-amplification loop.
An ensemble of plasma and cell surface-bound proteins carefully regulates complement activation to prevent host cells from self-attack by the complement cascade. However, excessive activation or inappropriate regulation of complement can lead to a number of pathologic conditions, ranging from autoimmune to inflammatory diseases (Holers, 2003; Markiewski and Lambris, 2007; Ricklin and Lambris, 2007b; Sahu and Lambris, 2000). The development of therapeutic complement inhibitors is therefore highly desirable. In this context, C3 and C3b have emerged as promising targets because their central role in the cascade allows for the simultaneous inhibition of the initiation, amplification, and downstream activation of complement (Ricklin and Lambris, 2007b).
Compstatin was the first non-host-derived complement inhibitor that was shown to be capable of blocking all three activation pathways (Sahu et al., 1996). Even though the exact molecular mechanism of its inhibition is not yet fully understood, this cyclic tridecapeptide clearly binds to both C3 and C3b and prevents the cleavage of native C3 by the C3 convertases. Its high inhibitory efficacy was confirmed by a series of studies using in vitro and animal models that pointed to its potential as a therapeutic agent (Fiane et al., 1999a; Fiane et al., 1999b; Nilsson et al., 1998; Ricklin and Lambris, 2008; Schmidt et al., 2003; Soulika et al., 2000). Progressive optimization of compstatin has yielded analogs with dramatically improved activity (Ricklin and Lambris, 2008). One of these analogs is currently being tested in clinical trials for the treatment of age-related macular degeneration (AMD), the leading cause of blindness in elderly patients in industrialized nations (Coleman et al., 2008; Ricklin and Lambris, 2008). In view of its therapeutic potential in AMD and other diseases, further optimization of compstatin to achieve an even greater efficacy is of considerable importance.
Earlier structure-activity studies have identified the cyclic nature of the compstatin peptide and the presence of both a β-turn and hydrophobic cluster as key features of the molecule (Morikis et al., 1998; Morikis et al., 2002; Ricklin and Lambris, 2008). Hydrophobic residues at positions 4 and 7 were found to be of particular importance, and their modification with unnatural amino acids generated an analog with 264-fold improved activity over the original compstatin peptide (Katragadda et al., 2006).
While previous optimization steps have been based on combinatorial screening studies, solution structures, and computational models (Chiu et al., 2008; Mulakala et al., 2007; Ricklin and Lambris, 2008), the recent publication of a co-crystal structure of compstatin complexed with the complement fragment C3c (Janssen et al., 2007) represents an important milestone for initiating rational optimization. The crystal structure revealed a shallow binding site at the interface of macroglobulin (MG) domains 4 and 5 of C3c and showed that 9 of the 13 amino acids were directly involved in the binding, either through hydrogen bonds or hydrophobic effects. As compared to the structure of the compstatin peptide in solution (Morikis et al., 1998), the bound form of compstatin experienced a conformational change, with a shift in the location of the β-turn from residues 5–8 to 8–11 (Janssen et al., 2007).
In view of these findings, we have now explored the importance of the newly identified key positions by analyzing the inhibitory, kinetic, and thermodynamic properties of a panel composed of both previously described and novel compstatin analogs. In particular, we investigated the influence of hydrophobic effects at position 4, the tolerance for backbone modifications at position 6, and the effects of changing position 9 on the integrity of the β-turn. For this purpose, we used a combination of ELISAs (to assess complement inhibition), surface plasmon resonance (SPR; to measure binding affinities and kinetics), and isothermal titration calorimetry (ITC; for thermodynamics analyses). Our findings indicate that most of the applied changes had a direct effect on the stability of the compstatin-C3 complex, as evidenced by an increased dissociation rate. These findings provide key information for the further development of more effective compstatin-based therapeutics.
MATERIAL AND METHODS
Synthesis and purification of peptides and proteins
Linear peptides were synthesized using Fmoc-based solid phase synthesis as previously described (Morikis et al., 1998), with the exception of analog 4(1ForW), which was synthesized using Boc chemistry (Merrifield, 1969). All N-termini were acetylated, and the C-termini were blocked by conversion to an amide. After synthesis, the peptides were cyclized on resin using direct disulfide bond formation by oxidation with Tl(CF3CO2)3/anisole (19:1)for 3 h at room temperature in DMF, to avoid the oxidation of tryptophan. After cyclization, the resins were washed 10 times each with DMF and MeOH and 5 times with a mixture of MeOH:DCM (60:40), then dried in vacuum for 4h. Cyclic peptides were cleaved from the resin and deprotected by two different methods: for Fmoc-synthesized peptides, with a mixture of TFA:triisopropylsilane:phenol:H2O (87.5:2.5:5:5) as previously described (Sahu et al., 2000); for the Boc-based peptide, with anhydrous HF(Pennington, 1994). All peptides were purified on a C-18 Vydac C18 218TP152022 column (Western Analytical Products, Murrieta, CA) to ~95% purity, and the identity of the peptides was confirmed by laser desorption mass spectrometry (MALDI).
Fourteen compstatin analogs (Table 1 and Figure 1) were synthesized. For the purposes of this study, the parent peptide will be referred to as original compstatin, designated as 4V9H, with the sequence Ac-I[CVVQDWGHHRC]T-NH2. Modifications were made at position 4 (category I), with tyrosine (Y) and its O-methyl (MeY), O-ethyl (EtY), and O-allyl derivatives (AlY); tryptophan (W) and its 1-methyl (1MeW), 1-formyl (1ForW), and 5-methyl (5MeW) derivatives; pyrenyl-alanine (PyrA) and 2-naphthyl-alanine (2Nal). At position 6 (category II), iso-aspartic acid (IsoD) was introduced, and at position 9 (category III), alanine was changed to either proline or α-aminobutyric acid (Abu).
Table 1.
Sequence and complement inhibition activity of compstatin analogs investigated in this study.
| Peptide | Sequence1 | IC50 (μM) | RA2 |
|---|---|---|---|
| Category I | Position 4 | ||
| 4V9H5 | Ac-ICVVQDWGHHRCT | 4.5 | 12 |
| 4W | Ac-ICVWQDWGAHRCT | 1.2 | 45 |
| 4(1MeW) | Ac-ICV(1MeW)QDWGAHRCT | 0.21 | 262 |
| 4(1ForW) | Ac-ICV(1ForW)QDWGAHRCT | 0.26 | 207 |
| 4(5MeW)6 | Ac-ICV(5MeW)QDWGAHRCT | 0.87 | 62 |
| 4Y | Ac-ICVYQDWGAHRCT | 2.4 | 23 |
| 4(MeY) | Ac-ICV(MeY)QDWGAHRCT | 1.3 | 41 |
| 4(EtY) | Ac-ICV(EtY)QDWGAHRCT | 1.3 | 41 |
| 4(AlY) | Ac-ICV(AlY)QDWGAHRCT | 1.3 | 41 |
| 4(2Nal)6 | Ac-ICV(2Nal)QDWGAHRCT | 0.55 | 99 |
| 4(PyrA) | Ac-ICV(PyrA)QDWGAHRCT | 1.2 | 45 |
| Category II | Position 6 | ||
| 4(1MeW)6D3 | Ac-ICV(1MeW)QDWGAHRCT | 0.21 | 262 |
| 4(1MeW)6(isoD) | Ac-ICV(1MeW)QisoDWGAHRCT | N/A | N/A |
| Category III | Position 9 | ||
| 4W9A4 | Ac-ICVWQDWGAHRCT | 1.2 | 45 |
| 4W9Abu | Ac-ICVWQDWGAbuHRCT | 1.5 | 36 |
| 4(1MeW)9A3 | Ac-ICV(1MeW)QDWGAHRCT | 0.21 | 262 |
| 4(1MeW)9P | Ac-ICV(1MeW)QDWGPHRCT | 0.54 | 100 |
Abbreviations used: Abu, aminobutyric acid; Ac, acetyl; Al, allyl; Et, ethyl; For, formyl; Me, methyl; Nal, naphthyl-alanine; Pyr, pyrenyl
Relative activities compared to peptide H-I(CVVQDWGHHRC)T-NH2
Corresponds to peptide 4(1MeW)
Corresponds to peptide 4W
Data previously reported (Katragadda et al., 2006; Mallik et al., 2005)
Data previously reported (Katragadda et al., 2006)
Figure 1.
Structure of compstatin and its analogs. (A) The core structure of compstatin is comprised of a cyclic tridecapeptide. (The structure shown represents peptide 4W in Table 1.) In the present study, changes were made to residues 4, 6, and 9 (Table 1) by introducing several natural (P, W, Y) and the following unnatural amino acids: (b) O-methyl-tyrosine (MeY), (c) O-ethyl-tyrosine (EtY), (d) O-allyl-tyrosine (AlY), (e) 5-methyl-tryptophan (5MeW), (f) 2-naphthyl-alanine (2Nal), (g) 1-methyl-tryptophan (1MeW), (h) 1-formyl-tryptophan (1ForW), (i) 4-pyrenyl-alanine (PyrA), (j) iso-aspartic acid (IsoD), (k) α-aminobutyric acid (Abu).
C3 was purified from plasma; C3b was generated by limited trypsinization of C3 and site-specifically biotinylated at its thioester moiety as described previously (Sahu et al., 2000; Sarrias et al., 2001).
Complement inhibition assays
The ability of the compstatin analogs to inhibit the classical pathway of complement activation was assessed by ELISA as described elsewhere (Katragadda et al., 2006; Mallik et al., 2005): An antigen-antibody complex was formed on an ELISA plate, and the deposition of C3b in the presence of compstatin analogs was measured. The percentage of inhibition was plotted against the peptide concentration and fitted to the logistic dose-response function using Origin 7.0 (OriginLab).
Kinetic characterization of the compstatin analogs (SPR)
The kinetics of the interaction between C3b and each compstatin analog was analyzed by SPR on Biacore 2000 and 3000 instruments (GE Healthcare Corp., Piscataway, NJ) at 25°C. PBS-T (10 mM sodium phosphate, 150 mM NaCl, 0.005% Tween-20, pH 7.4) was used as the running buffer, and data analysis was performed using Scrubber (BioLogic Software, Campbell, Australia). Biotinylated C3b (30 μg/ml) was immobilized on a streptavidin sensor chip (GE Healthcare) at three different surface densities (approximately 3000, 4000, and 5000 resonance units [RU]). An untreated flow cell was used as a reference surface.
A kinetic ranking experiment was performed by injecting a 1-μM solution of each analog for 2 min at a flow rate of 30 μl/min and following the dissociation phase for 3 min. Signals from the reference surface and an ensemble of buffer blank injections were subtracted to correct for non-specific binding and injection artifacts (Myszka, 1999). The corrected responses of all analogs were overlaid, and differences in the binding intensities and apparent association/dissociation phases were evaluated.
For a detailed kinetic analysis, a two-fold serial dilution series consisting of 12 dilutions was prepared for each analog. The starting concentration was 64 μM for analog 4V9H, 32 μM for 4W, 4Y, and 9Abu, 8 μM for 4MeY, 4EtY, 4AlY, and 4(PyrA), and 1 μM for the 4(1MeW), 4(1ForW), 4(5MeW), 9P, and 4(2Nal) peptides. Peptide solutions were injected for 2 min at 30 μl/min, with a dissociation phase of 5 min. Peptide solutions were injected in duplicate in every experiment, and each screening assay was performed at least twice. Peptide 4(1MeW) was included in each experimental series as an internal control and reference. Biosensor data were processed as described above and globally fitted to a 1:1 Langmuir binding model. The equilibrium dissociation constant (KD) was calculated from the equation KD= koff/kon.
Thermodynamic characterization of the compstatin analogs (ITC)
ITC experiments were performed using a VP-ITC calorimeter (Microcal Inc, Northampton, MA) at 25°C as previously described (Katragadda et al., 2004; Katragadda et al., 2006). C3 concentrations of 2–5 μM in the cell and peptide concentrations of 80–200 μM in the syringe were used for these experiments. All titrations were performed in PBS (10 mM sodium phosphate, 150 mM NaCl, pH 7.4). For each experiment, 2-μl peptide injections were made into the cell containing the protein. The resulting isotherms were fit to a single set of sites model using Origin 7.0 and the Gibbs free energy was calculated as ΔG = ΔH − TΔS.
RESULTS
Kinetic ranking and profiling assays
Screening of rather small peptides (~ 1.5 kDa) on large plasma proteins such as C3b (175 kDa) by SPR puts high demands on assay design. We therefore employed several procedures in order to avoid binding artifacts: First, C3b was site-specifically immobilized to prevent surface heterogeneity (Ricklin and Lambris, 2007a). Second, a wide concentration range was covered, and both individual injections and full assays were performed at least in duplicate on C3b surfaces of different densities. Third, careful data processing that included double referencing (Myszka, 1999) was applied, and the resulting data sets were globally fitted to a Langmuir 1:1 model. Using these enhancement steps, we observed highly reproducible binding signals. A minimum surface density of 3,000 RU C3b was found to be required for achieving an acceptable signal-to-noise ratio.
A kinetic ranking study, in which a constant concentration of each compstatin analog was injected onto surface-bound C3b, allowed us to evaluate the kinetic range for the panel and further optimize the assay design. A peptide concentration of 1 μM was selected for the ranking, since it produced detectable signals for all analogs without reaching full saturation. All analogs showed rapid complex formation, reached steady-state conditions within 1 min, and the signals returned to baseline within 5 min after the injection was completed (Figure 2). The ranking study also revealed important details about the kinetic relationships of the compounds, with the dissociation phase showing a high degree of variability. In particular, the fastest dissociation rate was seen for analog 4V9H, which was included as a reference and represents an earlier step in the development of compstatin analogs; in contrast, the slowest dissociation was exhibited by the current lead compound, 4(1MeW) (Katragadda et al., 2006)(Figure 2). This interesting finding indicated that many of the previously achieved efficacy improvements are off-rate-driven and that the SPR assay is able to cover the full kinetic range of the investigated panel of compstatin analogs.
Figure 2.
Kinetic ranking of compstatin analogs. A constant concentration (1 μM) of each peptide was passed over immobilized C3b. Binding signals were overlaid to visualize relative differences in their association and dissociation phases. Both the relative signal intensities and the observed dissociation correlated well with the measured affinities (Table 2). The weakest (4V9H; red) and strongest (4(1MeW); blue) analogs are highlighted in boldface type.
We therefore extended our analysis to full kinetic profiles in order to be able to correlate the structural changes with the degree of complex formation (kon) and stability (koff). With the exception of peptide 4(1MeW)6isoD, which lacked inhibitory activity (Table 1), all the tested peptides showed dose-dependent and highly reproducible binding (Figure 3). Furthermore, data sets from C3b surfaces of increasing densities exhibited higher signal intensities but highly comparable on- and off-rates. Most importantly, the data sets for the 13 inhibitory peptides could be globally fit to simple Langmuir 1:1 kinetic models with high accuracy (Figure 3; red simulation curves). Over the entire panel of analogs, we observed a 4-fold variation in kon (2–10 × 105 M−1 s−1), while the koff values spanned a >35-fold range (1–37 × 10−2 s−1) (Table 2). In agreement with the ranking experiment, 4(1MeW) showed the highest affinity (KD = 11 nM), and 4V9H (original compstatin) was at the lower end of the binding affinities for C3b (KD = 1.6 μM; Table 2). For each category of modifications, we compared the structure-kinetic relationship data from SPR with the activity results from a complement inhibition assay (Table 1; Figure 4) and supplemented the analysis with thermodynamic parameters from ITC experiments (Table 3; Supp. Figure 1).
Figure 3.
Detailed kinetic analysis of 13 compstatin analogs. Increasing concentrations of each compstatin analog were delivered to surface-bound C3b for 2 min, and the dissociation was monitored for another 5 min. All traces have been corrected for injection artifacts and non-specific binding by subtracting responses from an untreated reference surface and several buffer blanks. Processed SPR signals (black) were globally fitted to a Langmuir 1:1 binding isotherm (red simulation curves), and the kinetic rate constants kon and koff were extracted (Table 2). A schematic representation of the assay setup for the kinetic profiling is presented at the bottom.
Table 2.
Kinetic characterization of compstatin analogs.
| Peptide1 | kon (105 M−1 s−1) | koff (10−2 s−1) | KD (nM) |
|---|---|---|---|
| 4V9H | 2.3 ± 4.6 | 37.4 ± 0.8 | 1641 ± 108 |
| 4W | 9.2 ± 3.3 | 13.4 ± 3.2 | 152 ± 29 |
| 4(1MeW) | 9.9 ± 1.8 | 1.1 ± 0.2 | 11 ± 3 |
| 4(1ForW) | 2.9 ± 0.7 | 1.2 ± 0.2 | 42 ± 7 |
| 4(5MeW) | 6.8 ± 1.5 | 8.2 ± 1.3 | 123 ± 21 |
| 4Y | 4.1± 2.2 | 15.1 ± 2.2 | 454 ± 189 |
| 4MeY | 7.0 ± 0.5 | 8.2 ± 0.8 | 118 ± 17 |
| 4EtY | 6.2 ± 1.2 | 8.6 ± 1.8 | 140 ± 1 |
| 4AlY | 5.3 ± 1.3 | 6.3 ± 1.5 | 119 ± 1 |
| 4(2Nal) | 8.1 ± 1.2 | 9.3 ± 0.2 | 109 ± 21 |
| 4(PyrA) | 7.6 ± 1.3 | 4.8 ± 1.2 | 65 ± 8 |
| 4W9Abu | 5.7 ± 0.9 | 14.1 ± 2.1 | 252 ± 25 |
| 4(1MeW)9P | 7.5 ± 0.8 | 3.0 ± 1.0 | 40 ± 9 |
Analog 6isoD did not show any inhibitory activity (see Table 1) and was therefore excluded from the panel.
Figure 4.
Complement inhibition activity of compstatin analogs. The percentage of classical pathway complement inhibition is plotted against the peptide concentration and compared to that for the lead compound 4(1MeW). (A) 1ForW at position 4, (B) tyrosine analogs at position 4 (Y, MeY, EtY, and AlY), (C) PyrA at position 4, (D) IsoD at position 6, (E) proline at position 9. (F) Schematic representation of the ELISA assay to detect classical pathway activation, where the antigen-antibody complex was formed by an albumin-coated plate recognized by a polyclonal α-ovalbumin antibody. Inhibition assays for analogs 4(5MeW), 4(2Nal), and 9Abu have been reported previously (Katragadda et al., 2006; Mallik et al., 2005).
Table 3.
Thermodynamic characterization of compstatin analogs showing changes in enthalpy (ΔH), entropy (−TΔS), and free energy (ΔG), and their differences from the values for the reference peptide 4(1MeW) (marked in boldface type).
| Peptide | KD (nM) | ΔH (kcal/mol) | ΔΔH (kcal/mol) | −TΔS (kcal/mol) | −TΔΔS (kcal/mol) | ΔG (kcal/mol) | ΔΔG (kcal/mol) |
|---|---|---|---|---|---|---|---|
| 4W1 | 140 | −18.1 | −0.5 | 8.8 | 1.9 | −9.4 | 1.3 |
| 4(1MeW) | 15 | −17.6 | 0 | 6.9 | 0 | −10.7 | 0 |
| 4(1ForW) | 82 | −14.5 | 3.1 | 4.8 | −2.1 | −9.7 | 1.0 |
| 4(5MeW)1 | 120 | −17.8 | −0.2 | 8.2 | 1.3 | −9.6 | 1.1 |
| 4Y | 556 | −13.9 | 3.7 | 5.3 | −1.6 | −8.6 | 2.1 |
| 4MeY | 115 | −18.0 | −0.4 | 8.5 | 1.6 | −9.5 | 1.2 |
| 4EtY | 134 | −16.0 | 1.6 | 6.6 | −0.3 | −9.5 | 1.2 |
| 4AlY | 135 | −13.3 | 4.2 | 3.9 | −3.0 | −9.4 | 1.3 |
| 4(2Nal)1 | 110 | −14.3 | 3.3 | 4.8 | −2.1 | −9.5 | 1.2 |
| 4PyrA2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
| 4(1MeW)9P | 43 | −15.3 | 2.3 | 5.2 | −1.7 | −10.1 | 0.6 |
ITC data are derived from a previous study (Katragadda et al., 2006) and were compared to 4(1MeW).
The solubility of analog 4PyrA in PBS was too low for performing ITC experiments.
Category I: Modifications at position 4
In our previous study (Katragadda et al., 2006), we demonstrated a good correlation between increasing hydrophobicity of position 4 and increasing inhibitory activity. However, in that study, the most active analog 4(1MeW) showed a much higher inhibitory activity and binding affinity than would have been predicted from its degree of hydrophobicity at position 4 (Katragadda et al., 2006). Thus, the increased hydrophobicity of 4(1MeW) alone is not sufficient to explain its large increase in activity and affinity. In the present study, we therefore extended our analysis of this key position in compstatin and performed substitutions using three different sets of amino acids. Derivatives of tryptophan were selected that were based on structural homology with 4(1MeW) but were variable in terms of hydrophobicity. Tyrosine derivatives, on the other hand, offered a different structural anatomy with a smaller ring size and a hydroxyl group that can be easily derivatized. Finally, PyrA and 2Nal represented bulkier, more hydrophobic residues. All peptides were tested for complement inhibition using an ELISA-based assay that detects the C3b deposition after activation by antigen-antibody complexes (Figure 4).
In case of Trp4, previous addition of a N-methyl group to its indole ring led to a unexpectedly large gain in activity (Katragadda et al., 2006). We therefore investigated the influence of a hydrophilic group at the same position by replacing the methyl group with a formyl group. In the complement inhibition assay, the presence of this higher-polarity group did not significantly influence the activity of 4(1ForW) when compared to 4(1MeW) (IC50= 226 vs. 206 nM; Table 1; Figure 1g–h), suggesting that blocking the nitrogen in the indole group of the tryptophan is important by itself. Indeed, kinetic profiling revealed that 4(1ForW) was the only analog in the study that had very similar off-rate (0.012 s−1) to that of 4(1MeW) (0.011 s−1), therefore confirming the negative effect of the indole nitrogen on complex stability (Table 2). In the SPR study, 4(1ForW) also showed a slower on-rate (2.92 × 105 M−1 s−1), resulting in a 3-fold lower binding affinity when compared to 4(1MeW). A similar difference between the KD values was also observed in the ITC experiments, in which a beneficial entropy value for 4(1ForW) could not fully compensate for the observed loss in enthalpy (Table 3). The discrepancy between the activity and affinity values may be explained by the fact that the change from methyl to formyl seems to primarily affect complex formation rather than the stability, which is less likely to influence the end-point measurement in ELISAs.
The next set of analogs tested were derivatives of the amino acid tyrosine, chosen specifically because of the progressive increase in their hydrophobic character (logP: AlY > EtY > MeY > Y: Supp. Table 1; Figure 1b–d). When compared to unmodified Y (IC50 = 2.4 μM), each of the unnatural peptides showed a two-fold increase in inhibitory activity (IC50 = 1.3 μM). Since the formation of O-alkyl ethers made the side chain more lipophilic, this result was consistent with our previous report that hydrophobicity at this position is important. However, the chain length did not seem to play an important role, since its increase from methyl to allyl did not lead to a significant difference in activity. Also, the inhibitory potencies of all the tyrosine analogs were clearly lower than that of 4(1MeW) (0.21 μM; Table 1), again demonstrating the benefits of tryptophan modifications at position 4.
The activity results for the tyrosine analogs were further confirmed and supplemented by the kinetic profiling data. For the O-alkyl derivatives, both the kon (5–7 × 105 M−1 s−1) and koff values (0.06–0.09 s−1) were similar (Table 2). However, a slight trend in the kinetic rates from MeY to AlY indicated that gains in association may be compensated by losses in complex stability, resulting in similar overall levels of inhibitory activity. However, this trend was rather small and may have been within experimental error. Affinity differences from unmodified Y were even larger and were predominantly caused by a slower off-rate (Table 2). A similar effect was observed analyzing the ITC data, where all three tyrosine derivatives showed similar changes in free energy. However, the individual enthalpic and entropic contributions to the interaction were significantly different between the tyrosine analogs: MeY showed the most favorable enthalpy change, which gradually decreased with increasing hydrophobic character of the analogs (Table 3). On the contrary, the 4(EtY) and 4(AlY) peptides had a more favorable entropy level, most likely because of their higher hydrophobicity. The less favorable enthalpy change displayed by these two analogs could possibly reflect a decrease in specific interactions with C3. These results demonstrated that the length of the alkyl chain indeed influenced the interaction of the tyrosine analogs with C3/C3b, an observation that would have been missed if only equilibrium data would have been analyzed.
Finally, 2Nal and PyrA were introduced at this position because they consist of two and four aromatic rings (Figure 1f+i), respectively, and feature higher logP values than those of tryptophan analogs (Suppl. Table 1). Despite the increased hydrophobicity, however, both analogs showed a large drop in inhibitory activity, probably because of the bulkiness of their side chains. The kinetic analysis suggested that the five- and nine-fold faster off-rates for PyrA and 2Nal, respectively, when compared to 4(1MeW) primarily accounted for this difference, whereas the on-rate was much less affected by the substitutions (Table 2).
Category II: Modifications at position 6
Iso-aspartic acid is the β-amino acid isoform of aspartic acid that allows the formation of the amidic bond via its β-carboxylic group instead of the α-carboxylic group, leading to an elongation of the peptide chain by one methylene unit (Figure 1j). As previous studies have shown that changes in the side chain at position 6 are relatively well tolerated, we investigated whether the small increase in the cyclic compstatin backbone might be beneficial for its efficacy. However, substitution of D with its homolog IsoD led to a complete loss of activity (Figure 4D; Table 1). As a consequence, no SPR or ITC studies were performed using this peptide.
Category III: Modifications at position 9
The rationale behind the modification from 9A to 9P was to induce a shift in the β-turn from residues 5–8 to residues 8–11 in order to pre-form the arrangement observed in the bound form of compstatin and thereby improve complex formation. While the substitution was tolerated, it did not yield the expected improvement and even caused a two-fold drop in activity (Table 1). Similarly, the affinity was decreased by a factor of four, with a slightly more negative effect on the off-rate (Table 2), indicating a possible perturbation in the bond formation that affected the stability of the complex. The negative impact on the enthalpy value (Table 3) further suggests that this perturbation may lead to a loss of individual contacts.
The introduction of Abu (Figure 1k) at position 9 leads to an elongation of the alanine side chain, in this case by one methylene group. This extension was very well tolerated and led to a comparatively low drop in activity when compared to the original peptide 4W (IC50 = 1.5 and 1.2 μM, respectively; (Mallik et al., 2005)). The binding affinity was affected in a similar way (KD = 250 and 150 nM, respectively), with a higher impact on kon (factor 1.6) than on koff (factor 1.05; Table 2).
Comparison of binding parameters
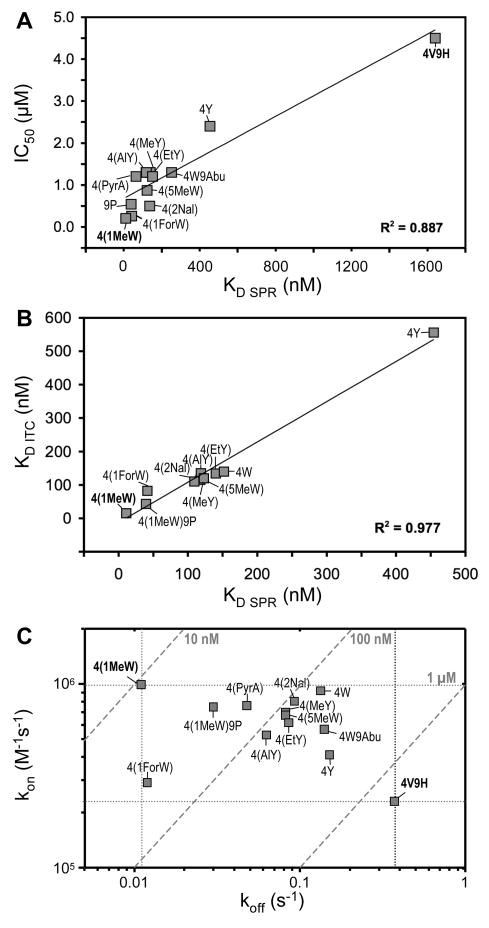
In order to validate our results and detect important connections and trends between the various binding parameters that were obtained throughout our study (by ELISA, SPR, and ITC), we performed a number of correlation analyses. In a first step, we compared the inhibitory activity (IC50) of each analog to its SPR-derived binding affinity (KD SPR; Figure 5A). As expected, an increase in affinity generally translated into enhanced activity and followed a linear trend, with a correlation constant of R2 = 0.887. Further analysis revealed that the off-rates correlate much better with the IC50 than the on-rates (Suppl. Figure 3), which we attribute to a higher dependence of ELISA on complex stability (i.e. end-point measurements). Due to technical reasons (e.g. immobilization method) and comparability with previous data (Katragadda et al., 2006), slightly different target fragments were used in SPR (i.e. C3b) and ITC (i.e. C3) experiments. We therefore performed ITC experiments with 4(1MeW) and either C3 or C3b. The comparison of the binding affinity of this compstatin analog for the two fragments revealed identical KD values (Suppl. Figure 4), confirming previous results concerning the high degree of similarity between the compstatin binding sites in C3 and C3b (Janssen et al., 2007; Sahu et al., 2000). Accordingly, the binding affinities derived from the SPR and ITC experiments over the whole panel of analogs showed a very high degree of correlation between the two methods (R2 = 0.977; Figure 5B), further indicating that the binding is highly comparable on the surface (SPR) and in solution (ITC). Finally, we analyzed the distribution of the kinetic rate constants in a kon/koff-plot (Figure 5C). As indicated by the kinetic ranking experiment, the analogs 4V9H and 4(1MeW) marked the upper and lower boundaries for both the on- and off-rates. Although 4(1ForW) had a similarly beneficial koff and 4W showed a comparable kon, the current lead compound 4(1MeW) exhibited the most favorable combination of on/off rates by far.
Figure 5.
Correlation between measured parameters. (A) Inhibitory activities and binding affinities followed a linear trend. (B) Comparison between the KD values determined by ITC and SPR showed a good correlation between solution- and surface-based binding. (C) The on/off-rate plot revealed that structural modification had distinct effects on the rate constants. Dashed diagonal lines represent binding affinity (KD). Analogs with the weakest (4V9H) and strongest 4(1MeW) binding affinity are marked in bold, and minimum/maximum rate constants are indicated with dotted lines.
DISCUSSION
The structural optimizations performed in this study are important contributions for paving the way towards more potent compstatin analogs. Previous biophysical studies (Katragadda et al., 2004; Katragadda et al., 2006; Morikis et al., 2002) and the advent of a co-crystal structure of compstatin with C3c (Janssen et al., 2007) allowed us to identify potential key areas for optimization and set the base for rational peptide design. Based on these findings, we compiled and tested a panel of compstatin analogs that covered three major aspects of the interaction: (1) the hydrophobic and hydrogen bond contact network at position 4, (2) the backbone elongation of Asp-6 that is part of the solution-based β-turn, and (3) the modulation of Ala-9 in the β-turn of bound compstatin. For this purpose, we analyzed a panel of 14 peptides bearing single mutations by various biochemical and biophysical methods in order to identify effects on their activity, affinity, kinetics, and thermodynamics. This integrated approach was chosen since simple activity or affinity measurements are often influenced by compensation effects that mask the true impact of a structural modification. We therefore focused part of our study for elucidating how structural variations can be correlated with and explained by kinetic SPR data.
The value of considering kinetic data in the development of therapeutics has long been recognized (Cooper, 2002; Huber, 2005), since it allows analyzing the effects on both complex formation (kon) and stability (koff). Examples of such structure-kinetics analyses include inhibitors of HIV protease (Markgren et al., 2002), carbonic anhydrase (Papalia et al., 2006), and estrogen receptor ligands (Rich et al., 2002). Although SPR analysis has previously been applied to the study of compstatin interactions (Sahu et al., 2000), this earlier analysis involved the immobilization of biotinylated peptides on individual flow cells, and the system was therefore not suited to the screening of a large panel of analogs. In the present study, the disadvantage of less favorable signal-to-noise ratios associated with injecting a small analyte into a large protein in the reversed assay format was compensated for by employing a series of improved assay design and data processing steps. Both the ranking and profiling assays reported here were found to be highly reproducible and capable of covering the full kinetic range of currently available compstatin analogs.
The binding affinity of an analyte-target interaction is closely related to the relative stability of the free and bound forms of the analyte and depends on the strengths of individual non-covalent contacts such as hydrogen bonds, salt bridges, van der Waals (vdW) interactions, and hydrophobic contributions (Fersht, 1999). While both complex formation and stability are influenced by all these factors, their relative contribution to the association and dissociation process may vary greatly for each interaction. Hydrogen bonds and vdW forces have been shown to be essential for the specificity of a binding event, but they provide a very small energetic contribution to the free energy of the protein-protein interactions (Freire, 2008; Hagler et al., 1979). In contrast, hydrophobic interactions have been suggested to be the major factor involved in stabilizing protein-protein complexes. (Freire, 2008; Hagler et al., 1979)
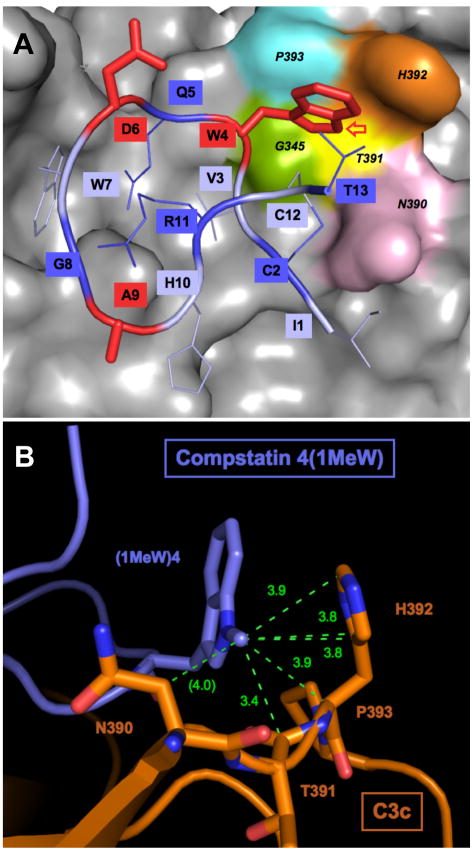
This theory fits very well with the results we observed in our analysis of the analogs bearing substitutions at position 4. The variation in association rate was rather small (three-fold), indicating a common specificity for the substrate that is primarily mediated by hydrogen bonds and vdW forces from the entire peptide, rather than only position 4. In addition, the association of peptides to a shallow binding pocket, as it is the case for the compstatin site on C3c (Janssen et al., 2007), is considered to be largely influenced by the diffusion coefficient of the peptide (Mulakala and Kaznessis, 2009). The diffusion coefficients, which depend largely on the size and shape of the entire molecules, are expected to be nearly identical for all tested analogs, which is in accord with our observation that their association constants are very similar. The off-rate, on the other hand, covered a much larger range (15-fold) when the identity of residue 4 was varied (Table 2). We observed a general trend relating increased complex stability to increased hydrophobicity of the side chain that was even more obvious within a specific scaffold as in case of tyrosine analogs (Suppl. Figure 2). The importance of the core was also shown by the 4(2Nal) and 4PyrA analogs, which were less active than would have been predicted from the increase in hydrophobicity. In case of 2Nal, a changed electrostatic distribution within the side chain may have contributed to this effect (Katragadda et al., 2006). PyrA, on the other hand, may have encountered steric limitations, even though the binding area for position 4 was expected to tolerate bulkier groups (Figure 6A). Nevertheless, this result confirms the general importance of hydrophobic interactions in position 4 for the stability of the complex. (Suppl. Figure 2)
Figure 6.
Structural base for the binding of compstatin. (A) Binding site of compstatin on C3c: The backbone of analog 4W is represented as a ribbon, with side chains as sticks in alternating colors (dark and pale blue). Positions 4, 6, and 9, which were modified in this study, are highlighted as bolded, red sticks. Contact amino acids for Trp-4 are marked with different colors on the surface of C3c (gray). (B) Structural model for the formation of hydrophobic contacts between C3c (brown) and analog 4(1MeW) (blue). The interatomic distances between the N-methyl group in compstatin analog 4(1MeW) and carbon atoms of residues 390–393 of C3c within a distance of 3.9 A are marked as dashed green lines. The binding site and the modification of Trp-4 were generated from PDB file 2QKI (Janssen et al., 2007) using PyMOL (www.pymol.org).
One notable exception to this trend was the result obtained for peptide 4(1ForW), which showed an off-rate similar to that of 4(1MeW) but had by far the most polar side chain modification at position 4 (Suppl. Figure 1). Furthermore, both indole derivatives significantly deviated from the observed trend between logP and koff (Suppl. Figure 2). This finding suggests that blockage of the indole nitrogen of tryptophan has a major effect on the high complex stability of these N-alkyl tryptophan analogs. In case of 4(1ForW), however, the positive effect on the off-rate seemed to be partially compensated by an unfavorable on-rate, most likely because of a lack of initial polar complementarity. Analysis of the crystal structure reveals that only the backbone but not the indole nitrogen forms a hydrogen bond with C3c (Janssen et al., 2007). Despite the large overall hydrophobicity of tryptophan, only two direct hydrophobic contacts within a range of 3.9 Å (Wallace et al., 1995) have been reported (Janssen et al., 2007) (Suppl. Figure 5). The indole nitrogen directly points towards a distinct cleft formed by residues 390–393 of C3c, which explains the tolerance for extending this position (Figure 6A; Suppl. Figure 6A). Strikingly, the addition of a methyl group to the indole nitrogen by molecular modeling (Suppl. Figure 6B) enables at least five additional hydrophobic contacts that surround the introduced C atom (Figure 6B). In addition, an extension into the cleft might replace solvation water from this position. Both effects may simultaneously contribute to the large effect on complex stability. Furthermore, this hypothesis is supported by the predominant effect of the 4(1MeW) mutation on the binding entropy in the ITC experiment (Table 3)(Katragadda et al., 2006), since both the formation of additional hydrophobic contacts and the replacement of water are expected to influence the entropy rather than the enthalpy (Freire, 2008). As analog 4(1ForW) extends to the same cleft in the binding site (Suppl. Figure 6C), the addition of a carbon atom at the same position is likely to be responsible for the similar effect on the off-rate. In contrast, the methyl group of analog 4(5MeW) is expected to point in a different direction (Suppl. Figure 6D). It should therefore not participate in any additional contacts but will increase the overall hydrophobicity (Suppl. Table 1). Together, these analyses provide an explanation model on why both indole-N derivatives deviate from the observed trend between hydrophobicity and complex stability (Suppl. Figure 2), and build a base for future structural optimizations at this position.
Even though the complement inhibition assay suggested a similar activity for all the O-alkyl tyrosines, their kinetic and thermodynamic profiles were clearly distinct. Formation of a methyl ether led to a significant improvement in kon and enthalpy that was gradually reversed by increasing the chain length. On the other hand, addition of longer substituents generally improved the koff and entropy values. Interestingly, MeY was more stable than was predicted by the logP trend analysis (Suppl. Figure 2), perhaps indicating that the addition of a shorter O-alkyl chain may be beneficial for tyrosine analogs.
While Trp-4 is heavily involved in contact networks at the binding interface of the compstatin-C3c co-crystal, no such contacts were observed for Asp-6 or Ala-9 (Janssen et al., 2007). Nevertheless, these two residues may be important for the correct structural orientation of the peptide, with Asp-6 taking part in the solution-based β-turn between residues 5–8 (Morikis et al., 1998), and Ala-9 found in the center of the shifted turn in the bound structure spanning positions 8–11 (Janssen et al., 2007)(Figure 6A). Although they did not directly affect individual contacts, changes in the backbone structure of these residues would be likely to influence the cyclic arrangement of the peptide and thereby its activity. Interestingly, introduction of an additional methylene moiety into the backbone was not tolerated at position 6. Because the backbones of β-amino acids such as isoD are longer, β-peptides often form different secondary structures (Steer et al., 2002). This modification may therefore disrupt the arrangement of the β-turn, which was found important for complex formation (Sahu et al., 2000). Furthermore, extension of the backbone of Asp-6 may dislocate the adjacent residues Gln-5 and Trp-7, which form a large number of individual contacts with C3c (Figure 6).
With no contacts for Ala-9 and only three total contacts for the neighboring Gly-8 and His-10 (Janssen et al., 2007), the change in 9Abu is considered less likely to disrupt essential interaction networks (Figure 6). Indeed, the complex stability, and consequently the inhibitory affinity were only slightly affected by this modification. However, the on-rate was clearly reduced when compared to the parent peptide, indicating a negative impact on the initial contact during complex formation or on the ability to perform the shift in the β-turn location. Based on these observations, we speculated that we could improve complex association by forcing the β-turn shift in solution via a proline-for-alanine substitution. Although this modification did not improve the binding, the negative impact on the affinity was comparatively small (three-fold) with a much higher impact on koff (3-fold) than on kon (factor 1.3). Furthermore, the favorable entropy change in 9P could mainly reflect contributions from conformational entropy. Pre-shaping the ligand to the conformation of the binding site has been previously described as a strategy to increase binding affinity, since it results in a minimization of the loss of conformational entropy upon binding (Leavitt and Freire, 2001). However, the 9P modification was also accompanied by an unfavorable change in enthalpy that may have been caused by a disruption of hydrogen bonds as a result of the enforced conformational change.
The inclusion of activity, affinity, kinetic, thermodynamic, and structural aspects in the characterization of compstatin analogs has proven highly effective, since it allowed for a detection of systematic trends that would have been missed if only a single method had been used. A good correlation was observed between the KD values derived from SPR and ITC; this result is in agreement with previous studies and suggests a similar mode of binding for compstatin to both surface-bound C3b and solution-based C3. As expected, the correlation with IC50 values showed a slightly lower degree of linearity, which can be explained by the differences in the setup and readout of the two assays. In fact, our ELISA-based values were generally in closer agreement with the SPR-based off-rates, with its end-point measurement, long incubation times, and wash steps making ELISA more dependent on complex stability (i.e., affected by the koff values; Suppl. Figure 3).
Despite a drug-like target affinity in the nanomolar range and even though previous optimization steps have already improved the complex stability of compstatin with C3b, the observed off-rate for the most active compound 4(1MeW) is still rather fast when compared with other therapeutic inhibitors (Markgren et al., 2002). While this profile may be suitable for acute phase situations or continuous administration, the applicability and plasma half-life of compstatin would certainly benefit from slower off-rates. The current findings may therefore well pave the way for developing compstatin analogs with improved binding properties that allow even more applications of this promising complement inhibitor.
CONCLUSION
In the present study, we have demonstrated the synergistic effect of combining different biophysical and biochemical methods to produce an enhanced structure-activity relationship analysis of compstatin analogs. The inclusion of kinetic parameters allowed us to discover effects and trends that could not be identified by activity or affinity values alone. Compensation effects between on/off rates and entropy/enthalpy values often reduced the impact of individual improvements. Most of the optimization that was achieved could be attributed to improvements in the kinetic off-rate. Increasing the overall hydrophobicity at position 4 was found to gradually improve complex stability. Even more, masking of the indole nitrogen of Trp-4 resulted in the most pronounced effect on the off-rate, most likely by formation of additional hydrophobic contacts and/or replacement of water molecules. A backbone elongation in the non-contacting residue Asp-6 was not tolerated, with a complete loss of activity in 6isoD. Induction of a β-turn at position 9 was, however, well tolerated and improved the entropy values, suggesting the usefulness of this approach for further optimization. Our work has provided new insights into the compstatin-C3 binding mechanism that will facilitate future improvements in this inhibitory peptide and the development of increasingly potent complement therapeutics.
Supplementary Material
Acknowledgments
This study was supported by the National Institutes of Health grants AI068730, GM062134, GM069736, and EB003968. We gratefully acknowledge Dr. Serapion Pyrpassopoulos for his help with the initial ITC experiments and Dr. Georgia Sfyroera for providing biotinylated C3b. We also thank Dr. Deborah McClellan for editorial assistance.
References
- Chiu TL, Mulakala C, Lambris JD, Kaznessis YN. Development of a new pharmacophore model that discriminates active compstatin analogs. Chem Biol Drug Des. 2008;72:249–56. doi: 10.1111/j.1747-0285.2008.00709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HR, Chan CC, Ferris FL, 3rd, Chew EY. Age-related macular degeneration. Lancet. 2008;372:1835–45. doi: 10.1016/S0140-6736(08)61759-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper MA. Optical biosensors in drug discovery. Nat Rev Drug Discov. 2002;1:515–28. doi: 10.1038/nrd838. [DOI] [PubMed] [Google Scholar]
- Fersht A. Structure and mechanism in protein science: a guide to enzyme catalysis and protein folding. W.H. Freeman; New York: 1999. [Google Scholar]
- Fiane AE, Mollnes TE, Videm V, Hovig T, Hogasen K, Mellbye OJ, Spruce L, Moore WT, Sahu A, Lambris JD. Compstatin, a peptide inhibitor of C3, prolongs survival of ex vivo perfused pig xenografts. Xenotransplantation. 1999a;6:52–65. doi: 10.1034/j.1399-3089.1999.00007.x. [DOI] [PubMed] [Google Scholar]
- Fiane AE, Mollnes TE, Videm V, Hovig T, Hogasen K, Mellbye OJ, Spruce L, Moore WT, Sahu A, Lambris JD. Prolongation of ex vivo-perfused pig xenograft survival by the complement inhibitor Compstatin. Transplant Proc. 1999b;31:934–5. doi: 10.1016/s0041-1345(98)01844-2. [DOI] [PubMed] [Google Scholar]
- Freire E. Do enthalpy and entropy distinguish first in class from best in class? Drug Discov Today. 2008;13:869–74. doi: 10.1016/j.drudis2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagler AT, Dauber P, Lifson S. Consistent force-field studies of inter-molecular forces in hydrogen-bonded crystals. 3. The C=O…H-O hydrogen-bond and the analysis of the energetics and packing of carboxylic-acids. J Am Chem Soc. 1979;101:5131–5141. [Google Scholar]
- Holers VM. The complement system as a therapeutic target in autoimmunity. Clin Immunol. 2003;107:140–51. doi: 10.1016/s1521-6616(03)00034-2. [DOI] [PubMed] [Google Scholar]
- Huber W. A new strategy for improved secondary screening and lead optimization using high-resolution SPR characterization of compound-target interactions. J Mol Recognit. 2005;18:273–81. doi: 10.1002/jmr.744. [DOI] [PubMed] [Google Scholar]
- Janssen BJ, Halff EF, Lambris JD, Gros P. Structure of compstatin in complex with complement component C3c reveals a new mechanism of complement inhibition. J Biol Chem. 2007;282:29241–7. doi: 10.1074/jbc.M704587200. [DOI] [PubMed] [Google Scholar]
- Katragadda M, Morikis D, Lambris JD. Thermodynamic studies on the interaction of the third complement component and its inhibitor, compstatin. J Biol Chem. 2004;279:54987–95. doi: 10.1074/jbc.M409963200. [DOI] [PubMed] [Google Scholar]
- Katragadda M, Magotti P, Sfyroera G, Lambris JD. Hydrophobic effect and hydrogen bonds account for the improved activity of a complement inhibitor, compstatin. J Med Chem. 2006;49:4616–22. doi: 10.1021/jm0603419. [DOI] [PubMed] [Google Scholar]
- Leavitt S, Freire E. Direct measurement of protein binding energetics by isothermal titration calorimetry. Curr Opin Struct Biol. 2001;11:560–6. doi: 10.1016/s0959-440x(00)00248-7. [DOI] [PubMed] [Google Scholar]
- Mallik B, Katragadda M, Spruce LA, Carafides C, Tsokos CG, Morikis D, Lambris JD. Design and NMR characterization of active analogues of compstatin containing non-natural amino acids. J Med Chem. 2005;48:274–86. doi: 10.1021/jm0495531. [DOI] [PubMed] [Google Scholar]
- Markgren PO, Schaal W, Hamalainen M, Karlen A, Hallberg A, Samuelsson B, Danielson UH. Relationships between structure and interaction kinetics for HIV-1 protease inhibitors. J Med Chem. 2002;45:5430–9. doi: 10.1021/jm0208370. [DOI] [PubMed] [Google Scholar]
- Markiewski MM, Lambris JD. The role of complement in inflammatory diseases from behind the scenes into the spotlight. Am J Pathol. 2007;171:715–27. doi: 10.2353/ajpath.2007.070166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield RB. Solid-phase peptide synthesis. Adv Enzymol Relat Areas Mol Biol. 1969;32:221–96. doi: 10.1002/9780470122778.ch6. [DOI] [PubMed] [Google Scholar]
- Morikis D, Assa-Munt N, Sahu A, Lambris JD. Solution structure of Compstatin, a potent complement inhibitor. Protein Sci. 1998;7:619–27. doi: 10.1002/pro.5560070311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morikis D, Roy M, Sahu A, Troganis A, Jennings PA, Tsokos GC, Lambris JD. The structural basis of compstatin activity examined by structure-function-based design of peptide analogs and NMR. J Biol Chem. 2002;277:14942–53. doi: 10.1074/jbc.M200021200. [DOI] [PubMed] [Google Scholar]
- Mulakala C, Kaznessis YN. Path-integral method for predicting relative binding affinities of protein-ligand complexes. J Am Chem Soc. 2009;131:4521–8. doi: 10.1021/ja807460s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulakala C, Lambris JD, Kaznessis Y. A simple, yet highly accurate, QSAR model captures the complement inhibitory activity of compstatin. Bioorg Med Chem. 2007;15:1638–44. doi: 10.1016/j.bmc.2006.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myszka DG. Improving biosensor analysis. J Mol Recognit. 1999;12:279–84. doi: 10.1002/(SICI)1099-1352(199909/10)12:5<279::AID-JMR473>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Nilsson B, Larsson R, Hong J, Elgue G, Ekdahl KN, Sahu A, Lambris JD. Compstatin inhibits complement and cellular activation in whole blood in two models of extracorporeal circulation. Blood. 1998;92:1661–7. [PubMed] [Google Scholar]
- Papalia GA, Leavitt S, Bynum MA, Katsamba PS, Wilton R, Qiu H, Steukers M, Wang S, Bindu L, Phogat S, et al. Comparative analysis of 10 small molecules binding to carbonic anhydrase II by different investigators using Biacore technology. Anal Biochem. 2006;359:94–105. doi: 10.1016/j.ab.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Pennington MW. HF cleavage and deprotection procedures for peptides synthesized using a Boc/Bzl strategy. Methods Mol Biol. 1994;35:41–62. doi: 10.1385/0-89603-273-6:41. [DOI] [PubMed] [Google Scholar]
- Rich RL, Hoth LR, Geoghegan KF, Brown TA, LeMotte PK, Simons SP, Hensley P, Myszka DG. Kinetic analysis of estrogen receptor/ligand interactions. Proc Natl Acad Sci U S A. 2002;99:8562–7. doi: 10.1073/pnas.142288199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Exploring the complement interaction network using surface plasmon resonance. Adv Exp Med Biol. 2007a;598:260–78. doi: 10.1007/978-0-387-71767-8_19. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Compstatin: a complement inhibitor on its way to clinical application. Adv Exp Med Biol. 2008;632:273–92. doi: 10.1007/978-0-387-78952-1_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricklin D, Lambris JD. Complement-targeted therapeutics. Nat Biotechnol. 2007b;25:1265–75. doi: 10.1038/nbt1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahu A, Kay BK, Lambris JD. Inhibition of human complement by a C3-binding peptide isolated from a phage-displayed random peptide library. J Immunol. 1996;157:884–91. [PubMed] [Google Scholar]
- Sahu A, Lambris JD. Complement inhibitors: a resurgent concept in anti-inflammatory therapeutics. Immunopharmacology. 2000;49:133–48. doi: 10.1016/s0162-3109(00)80299-4. [DOI] [PubMed] [Google Scholar]
- Sahu A, Soulika AM, Morikis D, Spruce L, Moore WT, Lambris JD. Binding kinetics, structure-activity relationship, and biotransformation of the complement inhibitor compstatin. J Immunol. 2000;165:2491–9. doi: 10.4049/jimmunol.165.5.2491. [DOI] [PubMed] [Google Scholar]
- Sarrias MR, Franchini S, Canziani G, Argyropoulos E, Moore WT, Sahu A, Lambris JD. Kinetic analysis of the interactions of complement receptor 2 (CR2, CD21) with its ligands C3d, iC3b, and the EBV glycoprotein gp350/220. J Immunol. 2001;167:1490–9. doi: 10.4049/jimmunol.167.3.1490. [DOI] [PubMed] [Google Scholar]
- Schmidt S, Haase G, Csomor E, Lutticken R, Peltroche-Llacsahuanga H. Inhibitor of complement, Compstatin, prevents polymer-mediated Mac-1 up-regulation of human neutrophils independent of biomaterial type tested. J Biomed Mater Res A. 2003;66:491–9. doi: 10.1002/jbm.a.10031. [DOI] [PubMed] [Google Scholar]
- Soulika AM, Khan MM, Hattori T, Bowen FW, Richardson BA, Hack CE, Sahu A, Edmunds LH, Jr, Lambris JD. Inhibition of heparin/protamine complex-induced complement activation by Compstatin in baboons. Clin Immunol. 2000;96:212–21. doi: 10.1006/clim.2000.4903. [DOI] [PubMed] [Google Scholar]
- Steer DL, Lew RA, Perlmutter P, Smith AI, Aguilar MI. Beta-amino acids: versatile peptidomimetics. Curr Med Chem. 2002;9:811–22. doi: 10.2174/0929867024606759. [DOI] [PubMed] [Google Scholar]
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–34. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.