Abstract
We describe the testing and release of AutoDock4 and the accompanying graphical user interface AutoDockTools. AutoDock4 incorporates limited flexibility in the receptor. Several tests are reported here, including a redocking experiment with 188 diverse ligand-protein complexes and a cross-docking experiment using flexible sidechains in 87 HIV protease complexes. We also report its utility in analysis of covalently-bound ligands, using both a grid-based docking method and a modification of the flexible sidechain technique.
Keywords: AutoDock, computational docking, protein flexibility, covalent ligands, computer-aided drug design
INTRODUCTION
Automated docking is widely used for prediction of biomolecular complexes in structure/function analysis and in molecular design. Dozens of effective methods are available, incorporating different trade-offs in molecular representation, energy evaluation, and conformational sampling to provide predictions with a reasonable computational effort 1–8. AutoDock combines an empirical free energy force field with a Lamarckian Genetic Algorithm, providing fast prediction of bound conformations with predicted free energies of association9.
In our hands, AutoDock3 has proven to be effective in roughly half of complexes that we have studied. The remaining half show significant motion of the receptor upon binding, and thus have required a more sophisticated model of motion in the receptor, typically performed outside of AutoDock3. The new version of AutoDock described here--AutoDock4--incorporates explicit conformational modeling of specified sidechains in the receptor to address this problem. This capability also provides an effective method for analysis of covalently-attached ligands.
METHODS
Overview of AutoDock4
Since its release in 199010, AutoDock has proven to be an effective tool capable of quickly and accurately predicting bound conformations and binding energies of ligands with macromolecular targets9,11–14. In order to allow searching of the large conformational space available to a ligand around a protein, AutoDock uses a grid-based method to allow rapid evaluation of the binding energy of trial conformations. In this method, the target protein is embedded in a grid. Then, a probe atom is sequentially placed at each grid point, the interaction energy between the probe and the target is computed, and the value is stored in the grid. This grid of energies may then be used as a lookup table during the docking simulation.
The primary method for conformational searching is a Lamarckian genetic algorithm, described fully in Morris et al.9. A population of trial conformations is created, and then in successive generations these individuals mutate, exchange conformational parameters, and compete in a manner analogous to biological evolution, ultimately selecting individuals with lowest binding energy. The “Lamarckian” aspect is an added feature that allows individual conformations to search their local conformational space, finding local minima, and then pass this information to later generations. A simulated annealing search method and a traditional genetic algorithm search are also available in AutoDock4.
AutoDock4 uses a semiempirical free energy force field to predict binding free energies of small molecules to macromolecular targets. Development and testing of the force field has been described elsewhere11. The force field is based on a comprehensive thermodynamic model that allows incorporation of intramolecular energies into the predicted free energy of binding. This is performed by evaluating energies for both the bound and unbound states. It also incorporates a new charge-based desolvation method that uses a typical set of atom types and charges. The method has been calibrated on a set of 188 diverse protein-ligand complexes of known structure and binding energy, showing a standard error of about 2–3 kcal/mol in prediction of binding free energy in cross-validation studies.
Receptor Flexibility
AutoDock4 allows fully flexible modeling of specific portions of the protein, in a similar manner as the ligand. The user selects specific sidechains and these are separated from the protein. During the simulation, these are treated explicitly, allowing rotation around torsional degrees of freedom using the same methods used to explore the conformational space of the flexible ligand. The remaining portion of the protein is represented using the affinity grids described above.
Methods for Covalent Docking
We have developed and tested two methods for docking of covalently-attached complexes: a grid-based approach and a modification of the flexible sidechain method. The grid-based approach calculates a special map for the site of attachment of the covalent ligand. A Gaussian function is constructed with zero energy at the site of attachment and steep energetic penalties at surrounding areas. The docking analysis is then performed by assigning a special atom type in the ligand for the atom that forms the covalent linkage. The docking simulation places this within the Gaussian well. One caveat is that this does not constrain the geometry of the covalent attachment to reasonable bond angles. To overcome this limitation, we tested the method using two Gaussian grids to define the bond that is formed during covalent linkage. Note, however, that the conformational freedom allowed with a single Gaussian grid may be an advantage if the method is used, for instance, to target ligands to metal coordination sites.
We also tested use of the flexible sidechain method for docking of covalent ligands. In this case, a coordinate file is created with the ligand attached to the proper sidechain in the protein, by overlapping ideal coordinates of the ligand onto the proper bond in the protein. This sidechain-ligand structure is then treated as flexible during the docking simulation, searching torsional degrees of freedom to optimize the interaction with the rest of the protein.
Overview of AutoDockTools
With the release of AutoDock3, it became apparent that the tasks of coordinate preparation, experiment design, and analysis required an effective graphical user interface to make AutoDock a widely accessible tool. AutoDockTools was created to fill this need. AutoDockTools facilitates formatting input molecule files, with a set of methods that guide the user through protonation, calculating charges, and specifying rotatable bonds in the ligand and the protein (described below). To simplify the design and preparation of docking experiments, it allows the user to identify the active site and determine visually the volume of space searched in the docking simulation. Other methods assist the user in specifying search parameters and launching docking calculations. Finally, AutoDockTools includes a variety of novel methods for clustering, displaying, and analyzing the results of docking experiments.
AutoDockTools is implemented in the object-oriented programming language Python and is build from reusable software components15,16. The easy-to-use graphical user interface has a gentle learning curve and an effective self-taught tutorial is available online. Reusable software components are used to represent the flexible ligand, the sets of parameters and the docking calculation, enabling a range of uses from a single use to thousands of docking experiments involving many different sets of molecules, facilitating automated high-throughput applications. For example, converting the NCI diversity database of small molecules into AutoDock-formatted ligand files was possible with a short Python script of less than 20 lines by leveraging the existing software components underlying AutoDockTools.
AutoDockTools exists in the context of a rich set of tools for molecular modeling, the Python Molecular Viewer (PMV)16,17. PMV is a freely distributed Python-based molecular viewer. It is built with a component-based architecture with the following software components: ViewerFramework, a generic OpenGL-based 3-dimensional viewing component; and MolKit, a hierarchical data representation of molecules. AutoDockTools consists of a set of commands dynamically extending PMV with commands specific to the preparation, launching and analysis of AutoDock calculations. Hence, all PMV commands (such as reading/writing files, calculating and displaying secondary structure, adding or deleting hydrogens, calculating charges and molecular surfaces, and many others) are also naturally available in AutoDockTools. PMV also provides access to the Python-interpreter so that commands or scripts can be called interactively. PMV commands log themselves, producing a session file that can be rerun. In summary, AutoDockTools is an example of a specialization of the generic molecular viewer PMV for the specific application of AutoDock.
Specification of Ligand and Receptor Flexibility in AutoDockTools
AutoDockTools provides an interactive method for defining the torsional tree for a given ligand and receptor. Each step in this process has been automated to allow automatic assignment, for use in batch processes such as virtual screening. Ligand flexibility is assigned in several steps. First, a root atom is chosen, which will act as the center of rotation during coordinate transformation in the docking simulation. To find the optimal atom, we evaluate the number of atoms in each branch, and choose the root atom that minimizes the size of the largest branch. In some cases, the user may wish to limit the flexibility of the ligand. AutoDockTools provides two choices to do this assignment automatically. One choice selects the set of torsional degrees of freedom that will move the largest number of atoms (torsions near the root), the other adds torsions progressively from the leaves, moving the fewest number of atoms and leaving the core of the molecule rigid.
Validation Data Sets
Two sets of complexes were used for the validation of AutoDock4, both of which have been described previously11. A set of 188 diverse protein-ligand complexes were taken from the Ligand-Protein Database (http://lpdb.scripps.edu) and a set of 87 HIV protease complexes where taken from the PDBBind database (http://www.pdbbind.org). All coordinates were checked manually for the proper biological unit and inconsistency in naming schemes. Hydrogen atoms and charges were added in AutoDockTools, using Babel for hydrogens and the Gasteiger PEOE18 method for charges. Several misassigned charges were modified manually, as described in the previous report.
The 87 HIV protease complexes were aligned to allow easy comparison of docked conformations during cross dockings. An analysis of steric clashes was performed by swapping ligands within the set of 87 aligned complexes. ARG8 showed the largest number of bad contacts, with 877 cases where a ligand atom contacted a sidechain atom with less than 2Å distance. Other contacts occurred with ILE50 (140 cases), PRO81 (95 cases) and PHE82 (88 cases, in two V82F mutants).
Docking Experiments
Docking experiments were performed with AutoDock4 and compared with docking experiments with AutoDock3. For each complex, 50 docking experiments were performed using the Lamarckian genetic algorithm with the default parameters from AutoDock3. A maximum of 25 million energy evaluations was applied for each experiment. The results were clustered using a tolerance of 2.0 Å.
In the HIV cross dockings, ligand flexibility was limited to 10 torsional degrees of freedom, picking torsions that allowed the fewest number of atoms to move (freezing the core of the molecule). Flexible docking was performed allowing three torsions to rotate in residue ARG8, in both the A and B chains. The structural water (water 301) was included in complexes that included this water in the crystallographic structure, and hydrogen atoms were added in geometry that allowed hydrogen bonding to the flaps.
We have also changed the default model for the unbound system in the current version of AutoDock. Our previous method calculated internal energies for an extended form of the molecule, mimicking a conformation that might be expected when fully solvated11. Results from beta testers, however, showed that this protocol has severe limitations when used for virtual screening. In cases where the ligand is sterically crowded, the artificial force field used to drive the ligand into an extended conformation tends to lead to conformations with sub-optimal energy. When the difference is calculated between this unbound conformation and the bound conformation, it leads to artificially favorable predictions of the free energy of binding. In response to this problem, we have returned to the default model of assuming that the unbound conformation of the ligand is the same as the bound conformation. Other options in AutoDock allow the user to use an energy-minimized conformation of the ligand as the unbound model.
Implementation and Availability
AutoDock 4.2 is currently being distributed free of charge as open source under a GPL license at the WWW site: http://autodock.scripps.edu. ADT is being distributed free of charge as part of the MGLTools package, at the WWW site: http://mgltools.scripps.edu/downloads.
RESULTS AND DISCUSSION
Redocking Analysis
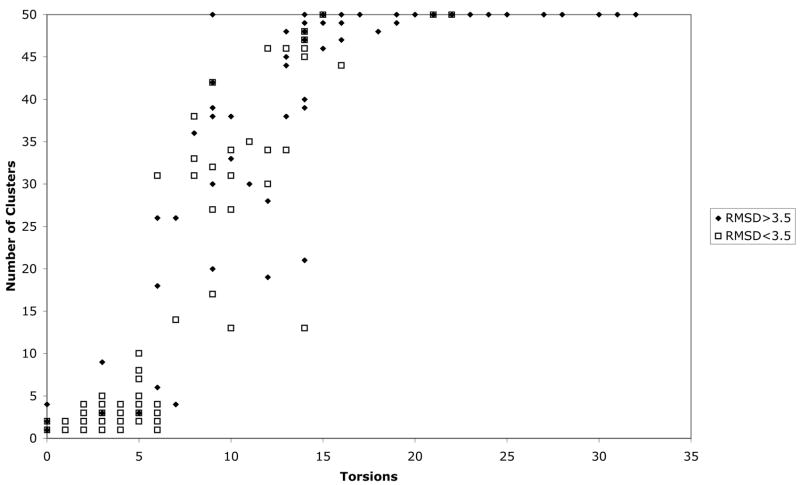
Our first test of AutoDock4 is a redocking experiment using a set of 188 diverse protein-ligand complexes. The results, presented in Figure 1, are similar to the redocking study performed during the energy function calibration11. AutoDock4 successfully redocks most complexes with about 10 or fewer torsional degrees of freedom, but fails for most complexes with higher conformational flexibility. In 100 of 188 complexes, the docked conformation with lowest energy was within 3.5Å RMSD of the crystallographic conformation. This is slightly better than a similar redocking experiment with AutoDock3. AutoDock3 successfully docked complexes with about 10 or fewer torsional degrees of freedom, and in 97 of 188 complexes, the lowest energy conformation showed RMSD less than 3.5 Å (data not shown).
Figure 1. Redocking results.
Results of redocking of 188 diverse ligand-protein complexes. Open squares represent complexes where the docked conformation with best predicted energy was less that 3.5Å RMSD relative to the experimentally observed conformation. Dots are complexes where the best docked conformation was greater than 3.5Å RMSD from the experimental structure.
Cross Docking Analysis with Protein Flexibility
To validate the use of flexible sidechains in docking, we have used a set of 87 retroviral proteases with inhibitors. These proteins have a tunnel-shaped active site that wraps around a peptidomimetic inhibitor. An arginine-aspartate salt bridge forms at each end of the tunnel, bridging the two subunits. In complexes with large inhibitors, these arginines move to make space, whereas they adopt a more closed position in complexes with small inhibitors. In previous work, we have demonstrated that steric clash with these arginines prevents the docking of large inhibitors to proteins in the more closed conformation.
Using a distributed computing environment, we performed a large cross docking experiment, taking inhibitors from 87 crystallographic structures and docking them to the protein conformations from these structures. We performed two parallel experiments, one with a rigid protein target, and one modeling the two ARG8 amino acids as flexible. Results are shown in Figure 2. Parameters were chosen that correspond to a typical computational effort for an individual docking experiment, with 25,000,000 evaluations and 50 docked conformations. The torsional degrees of freedom in the ligand were limited to 10, with an added 6 degrees of freedom for the two arginine sidechains in the flexible sidechain tests.
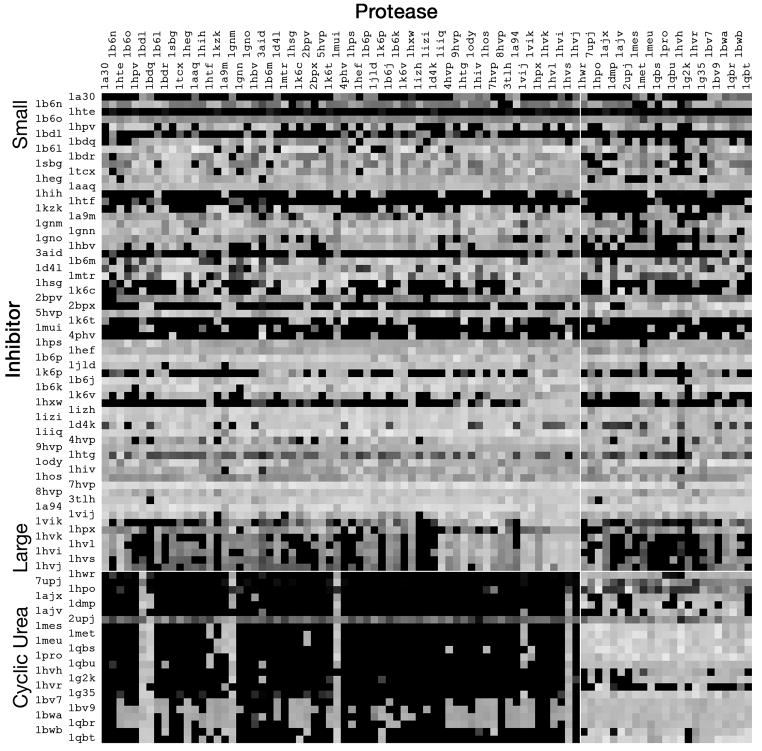
Figure 2. Cross docking results.
(A) Difference in energy between rigid docking and flexible docking. White points are docking experiments where the flexible docking showed 20 kcal/mol more favorable predicted free energy of docking and black points are docking experiments where the rigid docking showed similar or worse free energy of docking. Inhibitors are ordered from small to large, with the cyclic urea inhibitors separated at the bottom. The protease structures are ordered similarly. (B) Cross docking with rigid protease structures. Each point is colored by the RMSD of ligand atoms from the crystallographic structure, with RMSD=0 Å in white and RMSD>5 Å in black. (C) Cross docking with ARG8 treated as flexible in the protease. Each point is colored with the same scale as in (B).
The complexes in Figure 2 are arranged to highlight the structural features of the complexes. The complexes are arranged from top to bottom based on the size of the inhibitor, with small inhibitors at the top and large inhibitors at the bottom. The cyclic urea inhibitors and similar inhibitors that are designed to displace the structural water 301 are separated at the bottom, also ordered from small to large. The protease structures are arranged in identical order from left to right, so the diagonal corresponds to redocking experiments. Note that the large block at lower left corresponds to cyclic urea inhibitors docked to protease structures that include water 301, so we don’t expect that they will dock correctly. The large block at upper right includes peptidomimetic inhibitors docked to protease structure that do not include water 301, so we might expect some loss of specificity in docking, although this is not observed.
As seen in Figure 2A, flexible docking improves docking in the complexes that are most expected to benefit. The white band below center corresponds to large inhibitors binding to protease structures that were solved with small inhibitors. These complexes show the most favorable change in docking energy with flexible docking, as unfavorable contacts with the ARG8 residues are relaxed.
Looking to the RMSD results in Figure 2B, we see that rigid docking fails with several types of complexes. The large block of cyclic urea inhibitors at lower left show poor RMSD values, as expected. Roughly 2/3 of the small inhibitors were docked successfully, and the mid-size ones were very successful. The horizontal block just below center (inhibitors 1hvk through1hvj) are cases where large inhibitors are docked to active sites with ARG8 in a non-permissive position, so these are the complexes where we expect improvement with a flexible protease model.
These results underscore the fact that in this experiment, the larger inhibitors are actually an easier problem, for two reasons. First, the space searched in each experiment includes the active site and a small region outside the protein. In some cases, small ligands find incorrect conformations entirely outside the active site, but the larger ligands are forced to be at least partially inside the active site, due to their size. Second, several torsional degrees of freedom are frozen in the proper orientation in the large inhibitors, and this tends to favor the crystallographic conformation.
The results for the cyclic urea inhibitors follow a similar trend. The block at lower left are results for docking cyclic urea inhibitors into protease structures that include the structural water, so it is not surprising that the experiments do not find the proper answer. The block at lower right shows docking of cyclic urea inhibitors with protease structures without the structural water. It shows a similar trend of large inhibitors docking more consistently than the smaller inhibitors. The block at upper right shows docking of other inhibitors to proteases without the structural water. These experiments perform similarly to the experiments with the structural water.
When we add flexibility to the two ARG8 at the ends of the active site tunnel, we achieve the desired result with the large inhibitors that contact this residue. AutoDock4 successfully docks the large inhibitors in cases where the docking failed with rigid receptors (Figure 2C). Unfortunately, the experiments with flexible sidechains showed significantly worse results for the small inhibitors, failing in over half of the cases. This underscores another limitation of use of flexible sidechains. They increase the size of the conformational space to be searched—in these cases, increasing it from a problem that is accessible to AutoDock to a problem with too many degrees of freedom.
Covalent Docking
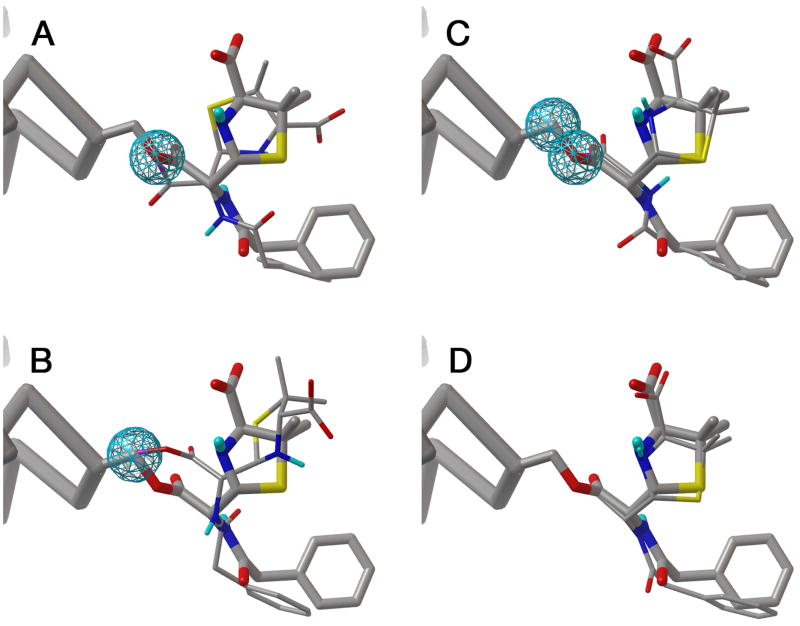
We tested two methods for docking covalently-attached ligands: a grid-based approach and an approach using flexible sidechains. The results are shown in Figure 3 for docking of penicillin in a covalent complex with D-alanyl-D-alanine carboxypeptidase. In the first two experiments, we created a Gaussian map centered on the CB or OG atoms of SER62 (Figure 3A and 3B). We then performed docking experiments with the appropriate atoms in the ligand targeted to that map. Poor results were obtained: a wide range of conformation was found and the conformation of best energy showed only slight resemblance to the crystallographic conformation. Better results were obtained when two Gaussian maps were used, corresponding to CB and OG of the serine (Figure 3C). This forced the docked ligand to adopt the appropriate geometry of bonding with the serine, and resulted in docked conformations that were much more similar to the crystallographic conformation.
Figure 3. Results of covalent docking.
(A) Using a Gaussian map centered on serine OG. The crystallographic structure is shown in large bonds and the best docked conformation is shown in thinner bonds. The blue sphere surrounds the region of most favorable energy in the Gaussian map. (B) Using a Gaussian map centered on serine CB. (C) Using two Gaussian maps. (D) Using a flexible sidechain to model the covalent ligand.
Use of a flexible sidechain to model the covalent ligand gave excellent results. All 10 docking runs gave similar conformations, all of which were very similar to the crystallographic conformation (Figure 3D).
CONCLUSIONS
Dependence on grid-based energy evaluation is a major limitation of AutoDock4. It is required to allow rapid evaluation of binding energies during the docking simulation, but it places a severe restriction on the representation of the target macromolecule: all of the atoms included in the grid must be treated as rigid. The off-grid modeling of specific sidechains is a method for incorporating limited flexibility within this paradigm, and the results presented here show that it will be effective in some cases. However, adding flexibility presents several problems: 1) the calculation of the receptor energy is more computationally-intensive since flexible regions must be evaluated by a full pair wise energy evaluation, and 2) the conformational space is larger and hence there is more potential for false positives. Future solutions to these problems will require advances in energetic functions, simplifying and/or speeding up pair wise approaches, and use of hierarchal approaches19,20 that allow different levels of sophistication in a docking simulation.
Acknowledgments
This work was supported under grant RO1 GM069832 from the National Institutes of Health. We thank the FightAIDS@Home volunteers and World Community Grid for computer resources. This is manuscript 19885 from the Scripps Research Institute.
References
- 1.Leach AR, Shoichet BK, Peishoff CE. J Med Chem. 2006;49:5851–5855. doi: 10.1021/jm060999m. [DOI] [PubMed] [Google Scholar]
- 2.Coupez B, Lewis RA. Curr Med Chem. 2006;13:2995–3003. doi: 10.2174/092986706778521797. [DOI] [PubMed] [Google Scholar]
- 3.Sousa SF, Fernandes PA, Ramos MJ. Proteins. 2006;65:15–26. doi: 10.1002/prot.21082. [DOI] [PubMed] [Google Scholar]
- 4.Mohan V, Gibbs AC, Cummings MD, Jaeger EP, DesJarlais RL. Curr Pharm Des. 2005;11:323–333. doi: 10.2174/1381612053382106. [DOI] [PubMed] [Google Scholar]
- 5.Kitchen DB, Decornez H, Furr JR, Bajorath J. Nature Rev Drug Discov. 2004;3:935–949. doi: 10.1038/nrd1549. [DOI] [PubMed] [Google Scholar]
- 6.Brooijmans N, Kuntz ID. Annu Rev Biophys Biomol Struct. 2003;32:335–373. doi: 10.1146/annurev.biophys.32.110601.142532. [DOI] [PubMed] [Google Scholar]
- 7.Taylor RD, Jewsbury PJ, Essex JW. J Comp Aided Mol Design. 2002;16:151–166. doi: 10.1023/a:1020155510718. [DOI] [PubMed] [Google Scholar]
- 8.Halperin I, Ma B, Wolfson H, Nussinov R. Proteins: Struct Funct Genet. 2002;47:409–443. doi: 10.1002/prot.10115. [DOI] [PubMed] [Google Scholar]
- 9.Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. J Comp Chem. 1998;19:1639–1662. [Google Scholar]
- 10.Goodsell DS, Olson AJ. Proteins: Struct Funct Genet. 1990;8:195–202. doi: 10.1002/prot.340080302. [DOI] [PubMed] [Google Scholar]
- 11.Huey R, Morris GM, Olson AJ, Goodsell DS. J Comput Chem. 2006;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 12.Osterberg F, Morris GM, Sanner MF, Olson AJ, Goodsell DS. Proteins: Struct Funct Genet. 2001 in press. [Google Scholar]
- 13.Morris GM, Goodsell DS, Huey R, Olson AJ. J Comput-Aided Molec Design. 1996;10:293–304. doi: 10.1007/BF00124499. [DOI] [PubMed] [Google Scholar]
- 14.Goodsell DS, Morris GM, Olson AJ. J Molec Recognition. 1996;9:1–5. doi: 10.1002/(sici)1099-1352(199601)9:1<1::aid-jmr241>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- 15.Lutz M, Asher D. Learning Python; O’Reilly & Associates Sebastopol CA. 1999 [Google Scholar]
- 16.Sanner MF. J Mol Graphics Mod. 1999 February;17:57–61. [PubMed] [Google Scholar]
- 17.Sanner MF. Structure. 2005;13:447–462. doi: 10.1016/j.str.2005.01.010. [DOI] [PubMed] [Google Scholar]
- 18.Gasteiger J, Marsili M. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 19.Zhao Y, Stoffler D, Sanner MF. Bioinformatics. 2006;22:2768–2774. doi: 10.1093/bioinformatics/btl481. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y, Sanner MF. J Comput Aided Mol Des. 2007;22:673–679. doi: 10.1007/s10822-007-9148-5. [DOI] [PMC free article] [PubMed] [Google Scholar]