Abstract
Villin-type headpiece domains are compact motifs that have been used extensively as model systems for protein folding. Although the majority of headpiece domains bind actin, there are some that lack this activity. Here, we present the first NMR solution structure and 15N-relaxation analysis of a villin-type headpiece domain natively devoid of F-actin binding activity, that of supervillin headpiece (SVHP). The structure was found to be similar to other headpiece domains that bind F-actin. Our NMR analysis demonstrates that supervillin headpiece lacks a conformationally flexible region (V-loop), present in all other villin-type headpiece domains, and which is essential to the phosphoryl regulation of dematin headpiece. In comparing the electrostatic surface potential map of SVHP, to that of other villin-type headpiece domains with significant affinity for F-actin, we identified a positive surface potential conserved among headpiece domains that bind F-actin, but absent from SVHP. A single point mutation (L38K) in SVHP, which creates a similar positive surface potential, endowed SVHP with specific affinity for F-actin that is within an order of magnitude of the tightest binding headpiece domains. We propose that this effect is likely conferred by a specific buried salt-bridge between headpiece and actin. As no high-resolution structural information exists for the villin-type headpiece: F-actin complex, our results demonstrate that through positive mutagenesis, it is possible to design binding activity into homologous proteins without structural information of the counterpart's binding surface.
Keywords: Supervillin, Electrostatic Surface Potential, Villin-Type Headpiece Domain, Protein Design, NMR Structure and Relaxation
INTRODUCTION
Supervillin (and/or its splice variant, Archvillin) is a ~205 kDa, peripheral membrane-associated protein ubiquitously expressed throughout the human body. Cellular levels of supervillin mRNA are well correlated with the level of mechanical stress experienced by the tissue: highest in skeletal muscle, bladder, heart, aorta, and uterus, and lowest in brain tissue.1 Supervillin binds extremely tightly to the plasma membrane and copurifies with detergent resistant membrane fragments that are biochemically consistent with lipid raft domains.2,3 As supervillin has also been shown to bind F-actin, it is hypothesized that supervillin may act as a direct, physical link between the membrane and the F-actin cytoskeleton.3 This hypothesis is supported by the intracellular localization of supervillin within a number of cell types.2,4-6
Cloning and subsequent sequencing of supervillin has revealed two distinct domains.1,5,7 The N-terminal two-thirds is not homologous to any other known protein, while the C-terminal third is composed of five gelsolin-like repeats and a villin-type headpiece domain. Villin-type headpiece domains are a family of small, modular, autonomously-folding domains exclusively found at the extreme C-terminus of several cytoskeletal proteins. Most, but not all, headpiece domains are thought to be F-actin binding motifs.8 The small size and high thermostability of both the full-length and the smaller C-terminal subdomain, HP36, have made these two constructs the focus of a great number of computational and experimental studies aimed at understanding the basic tenets of protein folding.
Surprisingly, domain analysis has localized the F-actin binding activity to supervillin's unique N-terminal half and demonstrated that the C-terminus, homologous to the actin binding protein, villin, is devoid of any such affinity for F-actin.4,9 To insure that remainder of supervillin did not obscure a potential F-actin binding site, the isolated headpiece domain was also tested and shown to lack F-actin affinity.8
Here, we use this well-defined system to determine why, despite 50% sequence identity to villin headpiece, supervillin is unable to bind F-actin. Through comparing the electrostatic surface potential map computed from the NMR structure of supervillin headpiece, reported here, to that of other villin-type headpiece domains with significant affinity for F-actin, we designed a single point mutation that endowed supervillin headpiece with specific affinity for F-actin.
RESULTS
Solution Structure of Human Supervillin Headpiece
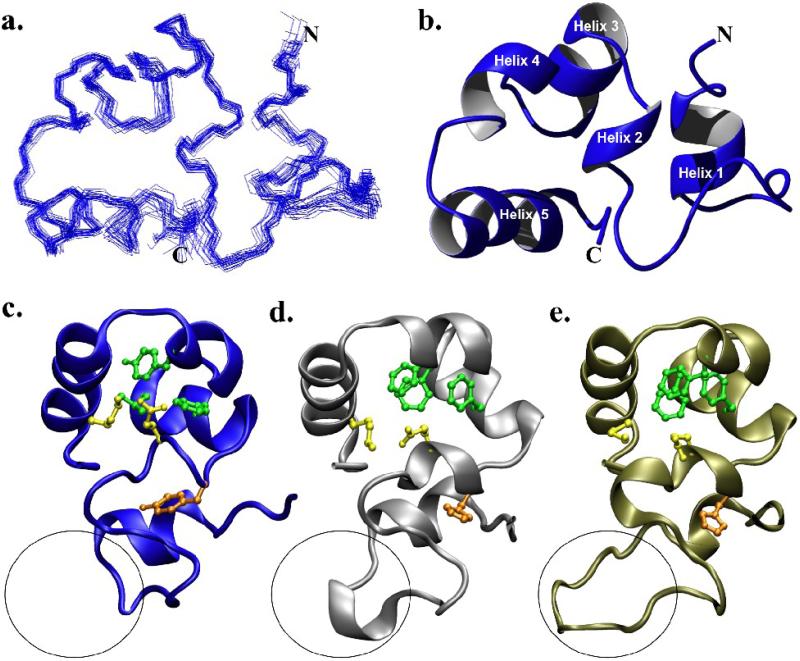
One explanation for the inability of supervillin headpiece (SVHP) to bind F-actin would be a departure from the canonical villin-type headpiece fold. To this end, we determined the NMR solution structure of supervillin headpiece (Fig. 1 A, B) and refined it to a backbone R.M.S. deviation of 0.475 Å (Table 1). Structural comparisons of SVHP to that of villin headpiece (VHP) and dematin headpiece (DHP), two proteins with high affinities for F-actin, demonstrate that SVHP possesses all of the distinguishing characteristics of the villin-type headpiece fold (Fig. 1). Briefly, the headpiece fold is formed through the association of two approximately equally sized N- and C-terminal subdomains, which are connected through a solvent-inaccessible salt-bridge and a contiguous hydrophobic core. The N-terminal subdomain is primarily composed of loops and turns while the C-terminal subdomain is a compact, three-helix bundle, enveloping a core of conserved hydrophobic residues. The C-terminal subdomains of nearly all villin-type headpiece domains contain three phenylalanine residues located at positions 47, 51, and 58 (Fig. 1) that have previously been shown to be essential to the stability of this construct.8,10 Interestingly, in supervillin headpiece, two of these three positions are occupied by other hydrophobic residues (Leu51, Tyr58; Fig. 1).
Figure 1.
The NMR solution structure of supervillin headpiece. (a) Superposition of the 22 accepted structures comprising the supervillin headpiece ensemble. (b) Ribbon diagram of the minimized average structure of supervillin headpiece aligned to (a). (c) Structural comparison of the minimized average structure of supervillin headpiece with (d) villin (1YU5) and (e) dematin headpiece (1QZP) rotated approximately 90° to (a) and (b). The characteristic, conserved residues of the headpiece fold are drawn on each ribbon model. These include His41/Tyr41 (orange), the Glu39-Lys70 salt bridge (yellow), and three conserved hydrophobic residues in the C-terminal subdomain (green). The variable length loop (V-loop) of each structure is circled.
| Supervillin Headpiece | |
|---|---|
| NMR Distance & Dihedral Constraints | |
| Total Distance Constraints | 1538 |
| Total NOE | 1502 |
| Intra-residue | 681 |
| Inter-residue | 821 |
| Sequential (|i-j|=1) | 315 |
| Medium-Range (|i-j|<4) | 271 |
| Long-Range (|i-j|≥5) | 235 |
| Hydrogen Bonds | 36 |
| Total Dihedral Angle Constraints | 124 |
| Phi | 53 |
| Psi | 53 |
| Chi1 | 28 |
| Structure Statistics | |
| Violations (r.m.s.d and s.d.) | |
| Distance Constraints (Å) | 0.0244±0.0011 |
| Dihedral Angle Constraints (°) | 0.2906±0.0483 |
| Max. Distance Constraint Violation (Å) | 0.3 |
| Max. Dihedral Angle Violation (°) | 3 |
| Deviations from Idealized Geometry | |
| Bond Lengths (Å) | 0.0029±0.00011 |
| Bond Angles (°) | 0.5052±0.0105 |
| Impropers (°) | 0.3973±0.0227 |
| Ramachandran Plot Percentages* | |
| Most Favored Regions | 1038 (81.3%) |
| Additionally Allowed Regions | 215 (16.8%) |
| Generously Allowed Regions | 23 (1.8%) |
| Disallowed Regions | 0 (0%) |
| Average Pairwise R.M.S.D. (Å)** | |
| Heavy | 0.831 |
| Backbone | 0.475 |
Calculated from the 22 accepted structures with procheck-nmr
Average pairwise R.M.S.D. of 22 accepted structures (residues 18-75)
15N-Relaxation Analysis of Human Supervillin Headpiece
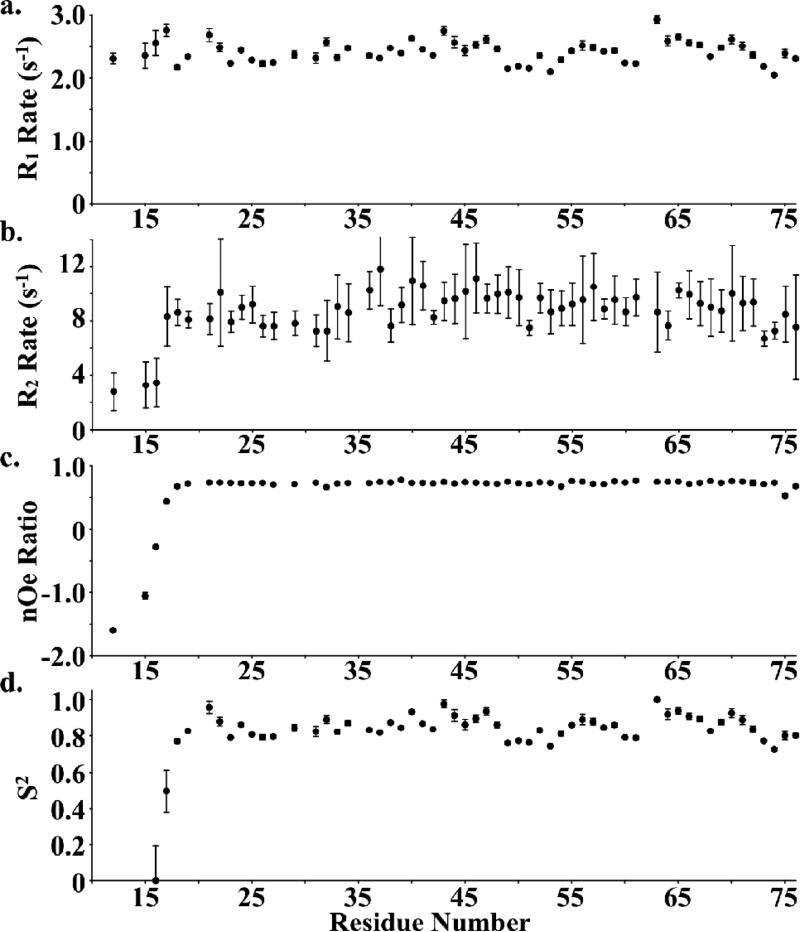
Villin-type headpiece domains possess a region, within their N-terminal subdomain, that varies in length among different headpieces and undergoes significantly more motion than the remainder of the sequence. This region, termed the variable-length loop (V-loop; 5, 8, and 10 residues long for supervillin, villin, and dematin, respectively) was originally identified by manual sequence alignment because computational sequence alignment algorithms fail to recognize this loop due to extensive gap penalties.11 No residues essential to F-actin binding have been identified in this region; however, in dematin headpiece, the V-loop changes conformation upon phosphorylation by forming a salt-bridge with the phosphorylated serine residue near the C-terminus.12 In order to determine whether the V-loop in SVHP, which is the shortest of any villin-type headpiece is also dynamic, we employed 15N-relaxation analysis to quantitate the pico- to nanosecond conformational motions of this region.
R1, R2, heteronuclear nOe ratios, and the computed S2 order parameters (without residues 10-16 as they failed to converge) for supervillin headpiece indicate no evidence of increased flexibility or mobility in any region of supervillin headpiece (Fig. 2). In contrast, these experiments demonstrated a significant, local increase in mobility in the V-loop regions of both villin and dematin headpiece.12-14 These data combined with sequence and structural alignment of SVHP with villin and dematin headpiece suggest that the latter two possess increased motion within their V-loop regions due the presence of “extra” residues beyond the 5 residues that comprise this sequence in SVHP. Further, the absence of local movement in supervillin headpiece suggests that it represents the shortest sequence compatible with the fold of the villin-type headpiece N-terminal subdomain (Fig. 1). The fact that no residues essential to F-actin binding have been identified in the V-loop and the structural similarity of SVHP to other villin-type headpiece domains, suggests that the lack of F-actin affinity should be attributed to the sequence of SVHP, not its structure.
Figure 2.
15N-relaxation analysis of supervillin headpiece. The (a) R1 relaxation rates, (b) R2 relaxation rates, (c) 15N-1H heteronuclear NOE values, and (d) S2 order parameters are plotted as a function of residue sequence number. Modelfree S2 order parameters are not displayed for residue numbers less than 16 because they failed to converge.
Computing and Modeling of Electrostatic Surface Potentials
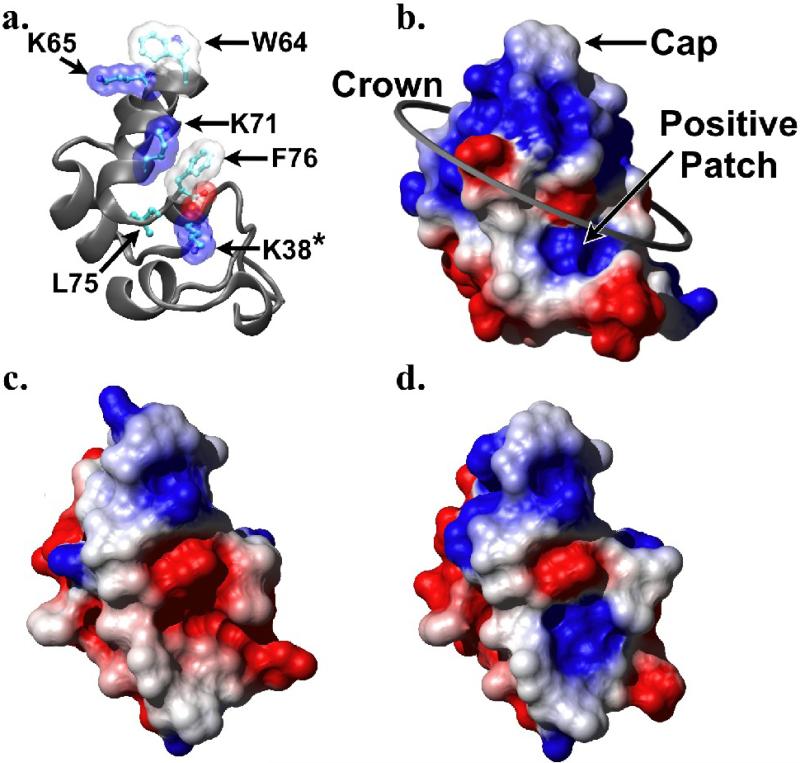
Although the binding site of villin on F-actin was reported near the completion of this work,15 this EM reconstruction is of insufficient resolution (38 Å) to provide a detailed understanding of the specific interactions which contribute to the binding affinities of these two proteins for one another. However, extensive mutagenesis performed on villin headpiece has suggested the residues required for it to bind F-actin (Fig. 3A).16-18 With the exception of two residues that form a solvent-inaccessible intramolecular salt bridge (E39, K70), all essential F-actin binding residues map to a single face of the molecule, which presumably represents the surface that contacts F-actin. Based on the electrostatic characteristics of these residues, they have previously been grouped into a tripartite schema of a hydrophobic cap (W64), a “crown” of alternating charges (K65, K70 and the C-terminal carboxylate), and a positive patch composed of R35 in dematin headpiece but K38 in villin headpiece (Fig. 3B).11 The residues contributing to each of these electrostatic surface features may be identified by comparing Figure 3A to 3B.
Figure 3.
Electrostatic surface potential maps for SVHP, SVHP L38K, and VHP. (a) Ribbon diagram of villin headpiece (1YU5) with key F-actin-binding residues depicted and labeled, drawn in the same orientation as b-d. Solvent accessible surface maps of (b) villin headpiece (1YU5), (c) the minimized average structure of supervillin headpiece, and (d) model of the L38K mutant of supervillin headpiece produced with Modeller 9v1 (Sali and Blundell, 1993) and colored by electrostatic potential from red (−0.3) though white (0.0) to blue (0.3).
Comparison of the computed electrostatic surface potential map of SVHP to that of VHP suggests that supervillin headpiece lacks only the positive patch, which is contributed by K38 in VHP (Fig. 3A,B) and that is replaced by L38 in SVHP (Fig. 3C). Although dematin headpiece also possesses a leucine at position 38, its structure revealed that it compensated for this alteration with a positive patch formed primarily by R35 and potentially also by K24.13 In silico modeling of the L38K point mutation onto the NMR solution structure of SVHP suggested that this mutation would add an appropriately positioned positive surface potential without disrupting the wild-type structure (Fig. 3D).
Engineering F-actin Binding into Supervillin Headpiece
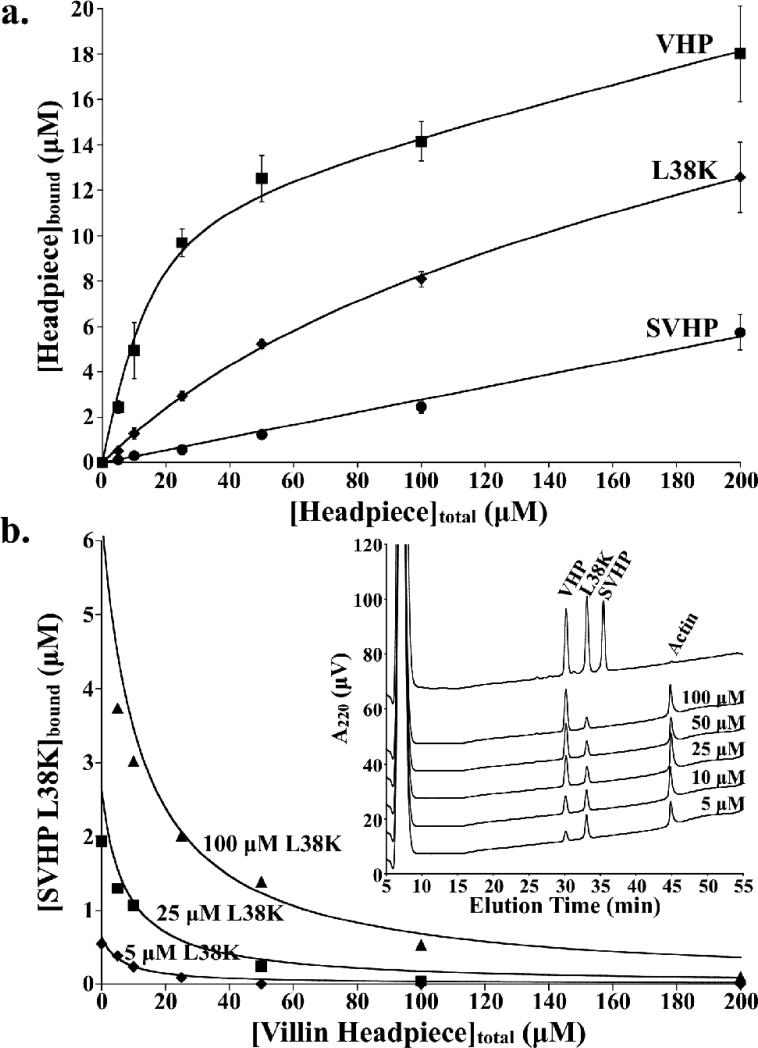
We cloned and expressed the L38K point mutant of SVHP and subsequently tested its affinity for F-actin using an F-actin sedimentation assay. In sharp contrast to wild-type supervillin headpiece, these assays demonstrated that the L38K point mutant possesses significant, specific affinity for F-actin and that its dissociation constant is decreased to just over an order of magnitude less than that determined for villin headpiece (Fig. 4A). Refined dissociation constants were 6 μM, 88 μM, and > 20,000 μM for VHP, SVHP L38K, and SVHP, respectively. Like other headpiece domains, SVHP L38K also shows a significant contribution from non-specific binding of 2.4 % which results in the upward slope of the curves past the saturation point. This non-specific binding is similar to that of villin headpiece (3.6 %) and SVHP (2.8 %).
Figure 4.
Supervillin L38K has significant and specific affinity for F-actin. (a) F-actin binding curves for villin headpiece (VHP, Kd = 6 μM), supervillin L38K (L38K, Kd = 88 μM), and wild-type supervillin headpiece (SVHP, Kd > 20,000 μM). (b) Results from F-actin competition assay performed between supervillin L38K and VHP. F-actin was incubated with a constant concentration of supervillin L38K (5 μM, diamonds; 25 μM, boxes; 100 μM, triangles) while the concentration of villin headpiece was increased from 0 to 200 μM. The amount of SVHP L38K bound to F-actin decreases with increasing concentrations of villin headpiece. The theoretical curves for pure competitive antagonism, calculated from equation 2 (See Materials and Methods) using the refined values calculated in (a), are drawn as solid lines. Inset: Representative data from F-actin competition assays performed between 100 μM SVHP L38K and increasing concentrations of VHP (5, 10, 25, 50, and 100 μM).
From the decrease in dissociation constant (~20,000 μM for SVHP to 88 μM for SVHP L38K), we can estimate that the L38K point mutant stabilizes the interaction between SVHP and F-actin by ~3 kcal/mol. This value is consistent with the free energy contribution of a buried intermolecular salt-bridges between two proteins in a complex (3-5 kcal/mol),19 and suggests that Lys38 likely forms a specific interaction with an acidic residue on the surface of actin.
The L38K Point Mutant Binds to the Same Site on F-actin as Villin Headpiece
Instead of quantitating these sedimentation assays with the traditional SDS-PAGE based methodology, we employed UV quantitation of reverse-phase HPLC separated proteins, which in addition to increasing accuracy, allowed us, for the first time, to perform F-actin competition assays on similarly sized proteins. These F-actin competition assays were performed by incubating F-actin with multiple villin-type headpiece domains. After centrifugal sedimentation of F-actin, the amount of each bound protein was individually quantitated. By monitoring the amount of SVHP L38K bound to F-actin at increasing concentrations of villin headpiece, we were able to demonstrate that it competes with villin headpiece for the same binding site on F-actin (Fig. 4B).
Preservation of the Wild-type Structure in L38K Point Mutant
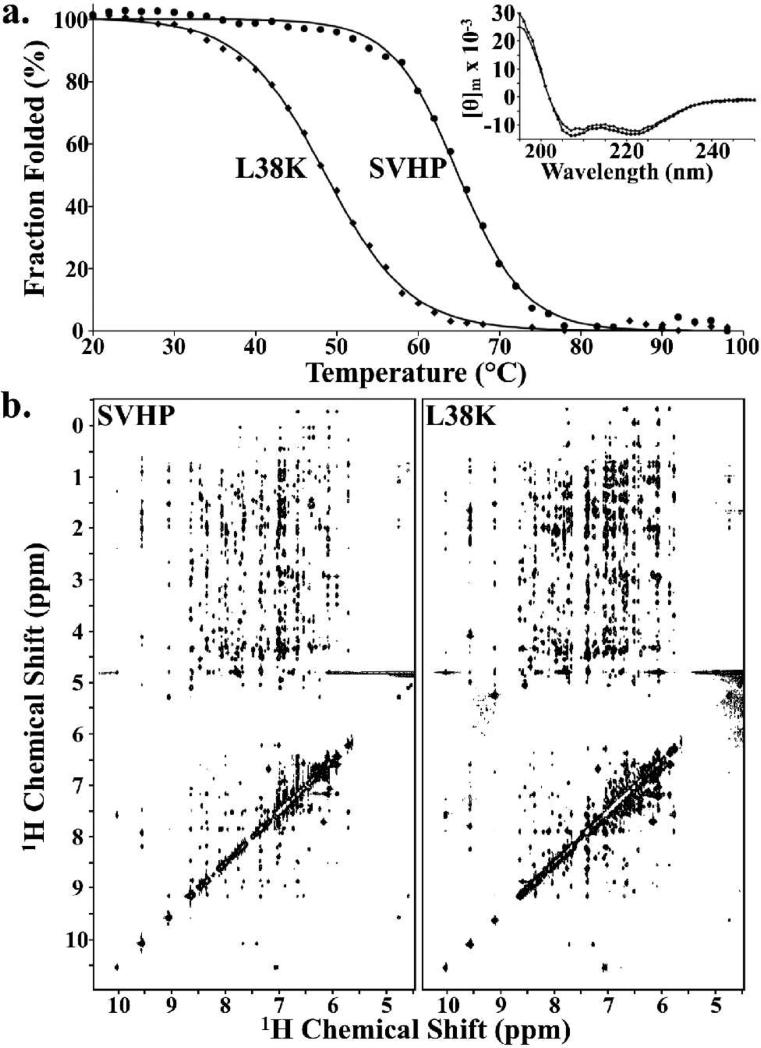
CD spectra of both wild-type and the L38K point mutant of SVHP demonstrate that these two proteins have nearly identical helical content, while thermal denaturation experiments indicate a 16 °C decrease in thermostability to 49 °C for L38K (Fig. 5A). Despite minor chemical shift differences in the immediate vicinity of the mutation, the highly similar position and pattern of nOe cross peaks in the 1H-1H NOESY NMR spectra convey that the L38K point mutation is of little or no structural consequence (Fig. 5B).
Figure 5.
The L38K point mutation retains the WT SVHP structure. (a) Thermal denaturation, monitored by CD signal at 222 nm, indicates that a lysine at position 38 decreases the thermal stability of the supervillin headpiece fold (Tm of 64.9 °C, SVHP, circles; 48.6 °C, L38K, diamonds). Inset: Plot of far-UV CD signal [Θ]m (deg cm2 dmol−1) versus wavelength for the wild-type and L38K point mutant of supervillin headpiece (circles and diamonds, respectively) demonstrate identical secondary structure. (b) The highly similar pattern and position of nOe crosspeaks in the 1H-1H NOESY spectra of wild-type supervillin headpiece and the L38K point mutant indicate that equivalent atoms experience the same chemical environment and are positioned next to the same atoms in both the wild-type and L38K point mutant of supervillin headpiece. CD and NMR spectroscopy were performed at pH 7.0 and 20 °C and NOESY spectra were acquired with a 200 ms mixing time.
Leucine 38 is Conserved Among Supervillin Sequences
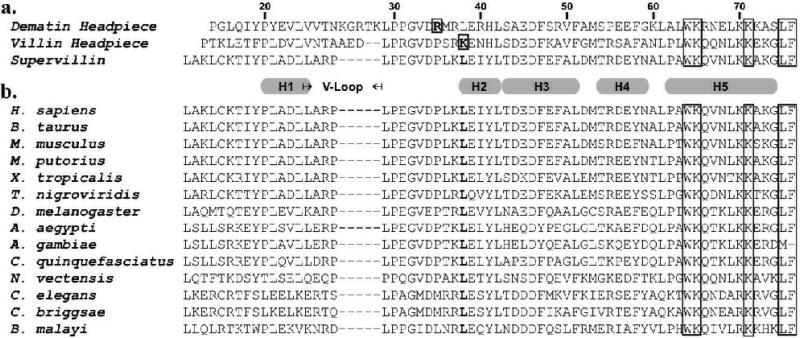
In an attempt to understand the significance of Leu38 in supervillin headpiece, we performed a multiple sequence alignment of all known supervillin headpiece domains. Interestingly, these results indicate that Leucine 38 is 100% conserved from C. elegans through H. sapiens and may represent a mechanism by which supervillin headpiece abolished affinity for F-actin, while maintaining its yet unknown functionality (Fig. 6).
Figure 6.
Sequence alignments of villin-type headpiece domains. (a) The sequence of human supervillin headpiece is aligned to that of human dematin headpiece and chicken villin headpiece. Both the numbering and position of the helices are referenced to the ensemble structure of supervillin (2K6M). Boxed letters indentify those residues believed to be involved in F-actin binding, bolded letters correspond to those residues that are positioned similarly to Lys38 (the positive patch) in villin headpiece. (b) ClustalW sequence alignment of supervillin headpiece domains from several species demonstrate 100% conservation of Leu38 (bolded, boxed). We have previously correlated the presence or absence of a positive patch with F-actin binding activity in a number of different villin-type headpiece domains.8,11
DISCUSSION
Here, we present the high-resolution NMR solution structure of supervillin headpiece, which represents the first structure of a naturally-occurring, villin-type headpiece domain natively devoid of affinity for F-actin. As might be expected from the high sequence homology to other villin-type headpiece domains, including those with high affinity for F-actin, the structure was found to be similar to those previously reported for both villin and dematin headpiece. However, as predicted by manual sequence alignment, and confirmed by both our NMR structure as well as 15N-relaxation analysis, supervillin headpiece lacks a flexible, V-loop region in its N-terminal subdomain, that in both villin and dematin is significantly more dynamic than the rest of the protein. The lack of this motile V-loop in supervillin headpiece, suggests that supervillin headpiece represents the shortest sequence compatible with the N-terminal subdomain of villin-type headpieces.
Comparison of electrostatic surface potential maps computed from our NMR structure of SVHP to that of VHP revealed the absence of a positive surface potential, contributed by K38 in VHP, a residue which has previously been shown to be important to the F-actin binding activity.18 In silico modeling of the L38K point mutation onto our NMR structure of supervillin headpiece provided evidence for the creation of an appropriately positioned positive surface potential. Comparison of the NMR spectra of SVHP and SVHP L38K indicate they adopt the same three-dimensional fold, albeit that SVHP L38K has reduced stability. The 16 °C decrease in thermostability is likely the result of mutating a gamma-branched hydrophobic leucine residue to a straight chain lysine. The introduction of the positive charge on the surface together with the changes in packing of the lysine side chain in the hydrophobic core, are the likely cause of the observed decrease in thermostability. The L38K point mutant retains a high thermostability (49 °C), which is not far below that observed of other wild-type headpiece domains (58 to 78 °C).8 Importantly, the L38K point mutant is folded at physiological temperatures and at 4 °C where we conducted our F-actin binding assays. F-actin sedimentation assays confirmed that this point mutation endowed supervillin headpiece with significant and specific affinity for F-actin. Furthermore, in F-actin competition assays, SVHP L38K acted as a competitive antagonist to VHP for F-actin indicating that these two headpiece domains share the same F-actin binding site.
The results presented here provide strong evidence that the F-actin affinity exhibited by villin-type headpiece domains is conferred by the precise placement of both hydrophobic and electrostatic potential elements about the surface of these proteins.8 In addition to confirming our hypotheses of the elements required for villin-type headpiece domains to bind F-actin, this study presents an effective strategy for designing novel proteins. In this work, we show that through positive mutagenesis, it is possible to design a novel peptide that binds specifically to a single predetermined site on a receptor, even if the binding surface of the counterpart is not known. To the best of our knowledge, this is the first reported use of identifying important protein-protein interactions by comparing electrostatic surface potentials and subsequently copying that motif onto a non-functional homolog to create a novel functional protein.
The methodology proposed here offers three advantages for designing a novel functional peptide. First, this strategy eliminates the requirement of creating a stable scaffold upon which functionality is to be engineered. Second, it utilizes a preexisting binding site on the target protein and therefore avoids developing a novel strategy of binding to a receptor in order to elicit the desired response. Third, as a detailed structure of the villin-type headpiece: F-actin complex remains to be determined, our methodology does not rely on structural knowledge of the receptor. Thus, this approach maybe useful in designing protein ligands for structurally challenging protein-protein complexes, including those involving integral membrane proteins, for which a structure may not be feasible.
Materials and Methods
Sample Preparation
The L38K point mutant was produced from wild-type supervillin headpiece (pSVHP) with QuikChange® site-directed mutagenesis (Stratagene). Prior to expression, each plasmid was sequenced at the Boston University Genetics Core Facility. Although the cloned cDNA was of bovine origin, the translated protein is 100% identical to residues 2149-2214 of human Supervillin/Archvillin. Both wild-type and the L38K point mutant of SVHP as well as villin headpiece (HP67) were expressed in and purified from E. coli BL21(DE3) cells (Novagen) as previously described.16 Briefly, after expression, cell pellets were lysed via sonication and the centrifugally cleared soluble supernatants were applied to a size exclusion chromatography column (Sephadex G-50). The fractions containing the desired protein were further purified with C18 reversed-phase high performance liquid chromatography, which resulted in each protein purified to homogeneity. Mass spectroscopy confirmed the identity of each. When 13C- and 15N-isotopically enriched samples were required, proteins were expressed in M9 minimal media containing 13C-labeled glucose and/or 15N-labeled ammonium chloride.20 Purified villin-type headpiece proteins were stored as a lyophilized powder. Actin was obtained from chicken pectoral muscle and purified from acetone powder with standard protocols.21
Multidimensional NMR Spectroscopy
All NMR spectroscopy was preformed on a Bruker DMX500 spectrometer at 20 °C. NMR spectra were taken at pH 7.0 in 5% D2O, 100 mM NaCl and 4 mM deuterated DTT with 500 μM 3-trimethylsilyl-tetradeutero sodium propionate as an internal chemical shift reference.
The following NMR spectra were acquired for 1H, 13C, and 15N chemical shift assignment and structural calculations: COSY, TOCSY, HNCA, HN(CO)CA, HNCO, HN(CA)CO, HCCH-COSY, HCCH-TOCSY, 13C-HSQC, 15N-HSQC, HMQC-J, HNHA, HNHB, NOESY, 13C-edited NOESY, and 15N-edited NOESY.22 Stereospecific assignments of leucine and valine terminal methyl groups were obtained from a 10% 13C-labeled sample using 13C-HSQC.23 When possible, stereospecific assignments for β-protons were obtained from HNHB, TOCSY, and qualitatively verified by NOESY spectra.24,25 All NMR data was processed and interpreted using NMRPipe, NMRDraw and NMRView software.26,27
Structure Calculations
1502 non-redundant NOE constraints were obtained from 2D, 1H-1H-NOESY as well as 3D, 15N- and 13C-edited NOESY spectra. Non-overlapping peaks were categorized as weak (1.8-5.2 Å), medium (1.8-3.3 Å), and strong (1.8-2.7 Å), based on the slope of their intensities calculated from a NOESY buildup series (mixing times of 50, 75, 100, 125, and 150 μs). Overlapping peaks and those only observed in 3D spectra were binned as weak. An extra 0.5 Å was added to each ambiguous restraint, which include non-stereospecifically assigned methylene, aromatic, and methyl hydrogens.
Experimentally determined ϕ angles for 23 residues were constrained according to 3J(HN,Hα) coupling constants obtained from the HNHA spectra28,29 and validated with data obtained from HMQC-J spectra.30 An additional 30 ϕ and 53 ψ angles were restrained with values calculated with TALOS using the 1Hα, 13Cα, 15N, 13CO, and 13Cβ chemical shifts.31 Finally, 10 experimentally determined χ1 angles, determined from HNHB and NOESY spectra,32 and 18 χ1 angles calculated with the PREDITOR web server33 were restrained. All angles were permitted ± 30° rotation from either their experimentally determined or computed value.
Inclusion of 18 hydrogen bond constraints, predicated on strong amide protection factors, determined from serial 15N-HSCQ spectra collected immediately following the addition of lyophilized SVHP into 100% D2O at pH 7.0 and calculated with SPHERE,34 yielded an additional 36 distance constraints. The O to N distances were restrained to values less than 3.5 Å and HN to O less than 2.5 Å.
Distance and angle constraints were inputs into the distance geometry, simulated annealing algorithm of CNS.35 After 100 computational cycles, 22 accepted structures with no NOE violations greater than 0.3 Å and no dihedral angle violations greater than 3° were obtained. These 22 structures were then used to compute the minimized average structure. The ensemble of 22 calculated structures as well as the minimized averaged structure were separately submitted to the protein data bank (PDB Identifiers 2K6M and 2K6N, respectively). The 1H, 13C, and 15N resonance assignment as well as a complete list of constraints used have been submitted to the BioMagnResBank (Accession ID: 15874).
15N Relaxation Measurements
The 15N-R1, R2, and heteronuclear NOE experiments were preformed as previously described.12 A single 1.9 mM sample of uniformly 15N-labeled SVHP in 10 mM phosphate and 1 mM DTT at pH 7.0 was used in all relaxation experiments. The relaxation delays in order of their acquisition were 100, 5, 1000, 500, 250, 50, 25, 100, 75, 125 ms for R1 experiments and 15.8, 31.7, 110.9, 63.4, 205.9, 79.2, 301.0, 158.4, 47.5, and 364.3 ms for R2 experiments. The heteronuclear nOe ratio was obtained from the average of four distinct pairs of NOE and noNOE spectra, which were interleaved and acquired within a single experiment. A relaxation delay of 5 s was executed between consecutive heteronuclear NOE experiments. The R1 and R2 rates, as well as the heteronuclear NOE ratio and their associated errors were calculated with NMRView (28). The Lipari-Szabo S2 order parameter36 was calculated for each residue using FastModelFree.37 Inclusion of a small Rex term (1.8 - 4.3 s−1) was required for residues 19, 24, 36, 39, 48, 49, 52, 58, 60, 61, 65, and 74 to convergence.
CD Spectroscopy
All CD spectra were acquired from a solution of 20 μM protein in 1 mM dithiotreitol and 10 mM phosphate at pH 7.0 on an Aviv 62DS spectrometer. Far-ultraviolet wavelength scans at 20°C were obtained from 250 to 190 nm at 1 nm increments. A 1 nm bandwidth and 15 s averaging time were used for wavelength scans in a 1 mm cell. Thermal denaturation experiments recorded the CD signal at 222 nm as the temperature was increased from 20 to 98 °C in 2 °C increments. A 1 min equilibration time, bandwidth of 3 nm, and 20 s averaging time of the sample in a 1 cm cell were employed for these scans.
F-actin Sedimentation Assays
F-actin (11.9 μM) was incubated with 0, 5, 10, 25, 50, 100, or 200 μM headpiece at 4 °C in F-Buffer (10 mM Tris-HCl at pH 8.0, 100 mM NaCl, 1 mM MgCl2, 0.1 mM CaCl2, 0.2 mM dithiotreitol, 0.1 mM adenosine triphosphate). After 1 hr, each solution was spun at 150,000g for 45 minutes and the supernatants were removed. The pellets were washed once with F-buffer and solubilized in a solution of 7% glacial acetic acid overnight at 4 °C.
Bound proteins were separated from F-actin with reverse phase high performance liquid chromatography using a linear gradient of 20% to 80% acetronitrile in 1 hr at a flow-rate of 1 ml/min through a Beckman Ultrasphere Analytical Column. Trifluoroacetate (0.1%) was used as the ion-pairing agent. The quantity of each eluting protein was determined from the UV absorption signal at 222 nm. Standard solutions of each headpiece construct were run separately to obtain a calibration curve, which related the area under the eluting peak to the concentration of protein. The equilibrium dissociation constant (Kd) and non-specific binding (NS) were obtained from fitting the average of three independent experiments to the quadratic equation (eq. 1)38 with a Bmax set to the concentration of actin, 11.9 μM, because villin-type headpiece domains are known to bind F-actin with a 1:1 stoichiometry.8,11,16,18,39
| (1) |
F-actin competition assays were preformed in an identical fashion to F-actin sedimentation assay, except, that F-actin (11.9 μM) was incubated with 0, 5, 10, 25, 50, 100, or 200 μM villin headpiece AND either 5, 25, or 100 μM supervillin L38K. Theoretical curves for competitive antagonists were calculated from equation 2,40 using the previously determined (see preceeding paragraph) F-actin dissociation constants.
| (2) |
Molecular Modeling and Electostatic Potential Calulations
We employed the program MODELLER 9v141 to model the L38K point mutation onto the coordinates of the minimized average structure of supervillin headpiece. Surface potentials were computed42 using a solvent dielectric of 80, molecular dielectric of 2, and a salt concentration 150 mM with charges specified by the “simplecharge” model using the program MOLMOL.43 Other figures were drawn with VMD,44 and all were rendered with POV-RAY version 3.6.
Data Bank Accession Codes
Protein Data Bank: Coordinates for supervillin headpiece have been deposited under accession codes 2K6M (ensemble of 22 calculated structures) and 2K6N (minimized average structure). BioMagnResBank: Assignments deposited under Accession ID 15874.
ACKNOWLEDGEMENTS
We thank Esther Bullitt for help in preparing the manuscript, Elizabeth Luna for the wild-type supervillin headpiece clone, and Jonathan Vural for assistance with NMR data collection. Financial support was provided by Boston University Graduate Student Research Fellowship to J.W.B and NIH Grant GM62886 to C.J.M.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Pope RK, Pestonjamasp KN, Smith KP, Wulfkuhle JD, Strassel CP, Lawrence JB, Luna EJ. Cloning, characterization, and chromosomal localization of human supervillin (SVIL). Genomics. 1998;52(3):342–51. doi: 10.1006/geno.1998.5466. [DOI] [PubMed] [Google Scholar]
- 2.Pestonjamasp KP, Pope RK, Wulfkuhle JD, Luna EJ. Supervillin (p205): A novel membrane-associated, F-actin-binding protein in the villin/gelsolin superfamily. J. Cell. Biol. 1997;139(5):1255–69. doi: 10.1083/jcb.139.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nebl T, Pestonmamasp KN, Leszyk JD, Crowley JL, Oh SW, Luna EJ. Proteomic analysis of a detergent-resistant membrane skeleton from neutrophil plasma membranes. J. Biol. Chem. 2002;277(45):43399–409. doi: 10.1074/jbc.M205386200. [DOI] [PubMed] [Google Scholar]
- 4.Wulfkuhle JD, Donina IE, Stark NH, Pope RK, Pestonjamasp KN, Niswonger ML, Luna EJ. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J. Cell. Sci. 1999;112(13):2125–36. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]
- 5.Oh SW, Pope RK, Smith KP, Crowley JL, Nebl T, Lawrence JB, Luna EJ. Archvillin, a muscle-specific isoform of supervillin, is an early expressed component of the costameric membrane skeleton. J. Cell. Sci. 2003;116(11):2261–75. doi: 10.1242/jcs.00422. [DOI] [PubMed] [Google Scholar]
- 6.Lee M, Joo YM, Lee YM, Kim HS, Kim J, Choi J, Ahn S, Min B, Kim C. Archvillin anchors in the Z-line of skeletal muscle via the nebulin C-terminus. Biochem. Biophys. Res. Commun. 2008;374(2):320–4. doi: 10.1016/j.bbrc.2008.07.036. [DOI] [PubMed] [Google Scholar]
- 7.Gangopadhyay SS, Takizawa N, Gallant C, Barber AL, Je H, Smith TC, Luna EJ, Morgan KG. Smooth muscle archvillin: a novel regulator of signaling and contractility in vascular smooth muscle. J. Cell. Sci. 2004;117(21):5043–57. doi: 10.1242/jcs.01378. [DOI] [PubMed] [Google Scholar]
- 8.Vardar D, Chishti AH, Frank BS, Luna EJ, Noegel AA, Oh SW, Schleicher M, McKnight CJ. Villin-type headpiece domains show a wide range of F-actin-binding affinities. Cell. Motil. Cytoskeleton. 2002;52(1):9–21. doi: 10.1002/cm.10027. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Takizawa N, Crowley JL, Oh SW, Gatto CL, Kambara T, Sato O, Li X, Ikebe M, Luna EJ. F-actin and myosin II binding domains in supervillin. J. Biol. Chem. 2003;278(46):46094–106. doi: 10.1074/jbc.M305311200. [DOI] [PubMed] [Google Scholar]
- 10.Frank BS, Vardar D, Buckley DA, McKnight CJ. The role of aromatic residues in the hydrophobic core of the villin headpiece subdomain. Protein. Sci. 2002;11(3):680–7. doi: 10.1110/ps.22202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vardar D, Buckley DA, Frank BS, McKnight CJ. NMR structure of an F-actin-binding “headpiece” motif from villin. J. Mol. Biol. 1999;294(5):1299–310. doi: 10.1006/jmbi.1999.3321. [DOI] [PubMed] [Google Scholar]
- 12.Jiang ZG, McKnight CJ. A phosphorylation-induced conformation change in dematin headpiece. Structure. 2006;14(2):379–87. doi: 10.1016/j.str.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Frank BS, Vardar D, Chishti AH, McKnight CJ. The NMR structure of dematin headpiece reveals a dynamic loop that is conformationally altered upon phosphorylation at a distal site. J. Biol. Chem. 2004;279(9):7909–16. doi: 10.1074/jbc.M310524200. [DOI] [PubMed] [Google Scholar]
- 14.Grey MJ, Tang Y, Alexov E, McKnight CJ, Raleigh DP, Palmer AG. Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: contribution of His41 to the pH-dependent stability of the N-terminal subdomain. J. Mol. Biol. 2006;355(5):1078–94. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 15.Hampton CM, Liu J, Taylor DW, DeRosier DJ, Taylor KA. The 3D Structure of Villin as an Unusual F-Actin Crosslinker. Structure. 2008;16(12):1882–91. doi: 10.1016/j.str.2008.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meng J, Vardar D, Wang Y, Guo H, Head JF, McKnight CJ. High-resolution crystal structures of villin headpiece and mutants with reduced F-actin binding activity. Biochemistry. 2005;44(36):11963–73. doi: 10.1021/bi050850x. [DOI] [PubMed] [Google Scholar]
- 17.Friederich E, Vancompernolle K, Huet C, Goethals M, Finidori J, Vandekerckhove J, Louvard D. An actin-binding site containing a conserved motif of charged amino acid residues is essential for the morphogenic effect of villin. Cell. 1992;70(1):81–92. doi: 10.1016/0092-8674(92)90535-k. [DOI] [PubMed] [Google Scholar]
- 18.Doering DS, Matsudiara P. Cysteine scanning mutagenesis at 40 of 76 positions in villin headpiece maps the F-actin binding site and structural features of the domain. Biochemistry. 1996;35(39):12677–85. doi: 10.1021/bi9615699. [DOI] [PubMed] [Google Scholar]
- 19.Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. W.H. Freeman and Company; New York: 2003. [Google Scholar]
- 20.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. J. Biomol. NMR. 2001;20(1):71–5. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 21.Pardee JD, Spudich JA. Purification of muscle actin. Meth. Enzymol. 1982;85(Pt B):164–81. doi: 10.1016/0076-6879(82)85020-9. [DOI] [PubMed] [Google Scholar]
- 22.Cavanagh J, Fairbrother WJ, Palmer AG, Rance M, Skelton NJ. Protein NMR Spectroscopy: Principles and Practice. 2nd ed Academic Press; Amsterdam: 2007. [Google Scholar]
- 23.Neri D, Szyperski T, Otting G, Senn H, Wuthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28(19):7510–6. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- 24.Archer SJ, Ikura M, Torchia DA, Bax A. An alternative 3D NMR technique for correlating backbone 15N with side chain Hβ resonances in larger proteins. J. Magn. Reson. 1991;95(3):636–41. [Google Scholar]
- 25.Clore GM, Bax A, Gronenborn AM. Stereospecific assignment of beta-methylene protons in larger proteins using 3D 15N-separated Hartmann-Hahn and 13C-separated rotating frame Overhauser spectroscopy. J. Biomol. NMR. 1991;1(1):13–22. doi: 10.1007/BF01874566. [DOI] [PubMed] [Google Scholar]
- 26.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR. 1995;6(3):277–93. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 27.Johnson BA, Blevins RA. NMRView: A computer program for the visualization and analysis of NMR data. J. Biomol. NMR. 1994;4(5):603–14. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 28.Pardi A, Billeter M, Wuthrich K. Calibration of the angular dependence of the amide proton-C alpha proton coupling constants, 3JHN alpha, in a globular protein. Use of 3JHN alpha for identification of helical secondary structure. J. Mol. Biol. 1994;180(3):741–51. doi: 10.1016/0022-2836(84)90035-4. [DOI] [PubMed] [Google Scholar]
- 29.Vuister GW, Bax A. Quantitative J correlation: a new approach for measuring homonuclear three-bond J(HNHa) coupling constants in 15n-enriched proteins. J. Am. Chem. Soc. 1993;115(17):7772–7777. [Google Scholar]
- 30.Forman-Kay JE, Gronenborn AM, Kay LE, Wingfield PT, Clore GM. Studies on the solution conformation of human thioredoxin using heteronuclear 15N-1H nuclear magnetic resonance spectroscopy. Biochemistry. 1990;29(6):1566–72. doi: 10.1021/bi00458a030. [DOI] [PubMed] [Google Scholar]
- 31.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13(3):289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 32.Arseniev A, Schultze P, Worgotter E, Braun W, Wagner G, Vasak M, Kagi JH, Wuthrich K. Three-dimensional structure of rabbit liver [Cd7]metallothionein-2a in aqueous solution determined by nuclear magnetic resonance. J. Mol. Biol. 1988;201(3):637–57. doi: 10.1016/0022-2836(88)90644-4. [DOI] [PubMed] [Google Scholar]
- 33.Berjanskii MV, Neal S, Wichart DS. PREDITOR: a web server for predicting protein torsion angle restraints. Nucleic Acids Res. 2006;34:W63–9. doi: 10.1093/nar/gkl341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bai Y, Milne JS, Mayne L, Englander SW. Primary structure effects on peptide group hydrogen exchange. Proteins. 1993;17(1):75–86. doi: 10.1002/prot.340170110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunger AT, et al. Crystallography & NMR System: A New Software Suite for Macromolecular Structure Determination. Acta Crystallogr. D. Biol. Crystallogr. 1998;54(5):905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 36.Lipari G, Szabo A. Model-free approach to the interpretation of Nuclear Magnetic Resonance relaxation in macromolecules. 1. Theory and Range of Validity. J. Am. Chem. Soc. 1982;104(17):4546–59. [Google Scholar]
- 37.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J. Biomol. NMR. 2003;26(3):203–13. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 38.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 1995;47(6):1197–203. [PubMed] [Google Scholar]
- 39.Pope B, Way M, Matsudaira P, Weeds AG. Characterization of the F-actin binding domains of villin: classification of F-actin binding proteins into two groups according to their binding site on actin. F.E.B.S. Lett. 1994;338(1):58–62. doi: 10.1016/0014-5793(94)80116-9. [DOI] [PubMed] [Google Scholar]
- 40.Buxton IL. Chapter 1. Pharmacokinetics and Pharmacodynamics: The Dynamics of Drug Absorption, Distribution, Action, and Elimination. In: Brunton LL, Lazo JS, Parker KL, editors. Goodman & Gilman's The Pharmacological Basis of Therapeutics. 11th Edition McGraw-Hill Professional; New York: 2006. pp. 1–40. [Google Scholar]
- 41.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234(3):779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 42.Nicholls A, Honig B. A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 1991;12(4):435–45. [Google Scholar]
- 43.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J. Mol. Graphics. 1996;14(1):33–8. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 44.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graphics. 1996;14(1):51–5. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]