Abstract
Centrosomes are microtubule-organizing centers that must be precisely duplicated prior to mitosis. Centrosomes regulate mitotic spindle assembly, and the presence of excess centrosomes leads to the production of aberrant mitotic spindles that generate chromosome segregation errors. Many human tumors possess excess centrosomes that lead to the production of abnormal spindles in situ. In some tumors these extra centrosomes appear before aneuploidy, suggesting that defects in centrosome duplication might promote genomic instability and tumorigenesis. The Mps1 protein kinase is required for centrosome duplication, and preventing the proteasome-dependent degradation of Mps1 at centrosomes increases its local concentration and causes the production of excess centrosomes during a prolonged S-phase. Here we show that Mps1 degradation is misregulated in two tumor-derived cell lines, and that the failure to appropriately degrade Mps1 correlates with the ability of these cells to produce extra centrosomes during a prolonged S-phase. In the 21NT breast-tumor derived cell line a mutant Mps1 protein that is normally constitutively degraded can accumulate at centrosomes and perturb centrosome duplication, suggesting that these cells have a defect in the mechanisms that target Mps1 to the proteasome. In contrast, the U2OS osteosarcoma cell line expresses a non-degradable form of Mps1, which we show causes the dose-dependent over duplication of centrioles even at very low levels of expression. Our data demonstrate that defects in Mps1 degradation can occur through multiple mechanisms, and suggest that Mps1 may provide a link between the control of centrosome duplication and genomic instability.
Introduction
Centrosomes are microtubule-organizing centers that consist of a pair of centrioles surrounded by a pericentriolar matrix responsible for microtubule nucleation. Through cytokinesis each daughter cell inherits one copy of the genome and a single centrosome, and faithful maintenance of genome integrity requires that both be precisely replicated prior to the next mitosis (Fukasawa 2007). During mitosis the mitotic spindle segregates one copy each of the genome and centrosome into daughter cells. Because centrosomes become mitotic spindle poles during mitosis, the presence of extra centrosomes leads to the formation of aberrant mitotic spindles that generate errors in chromosome segregation (Azimzadeh and Bornens 2007; Fisk et al. 2002; Fukasawa 2007). Accordingly, proper regulation of centrosome duplication controls not just centrosome number but also the integrity of the genome.
Centrosome duplication is coordinated with DNA replication, and like DNA replication is a semi-conservative process. Within the centrosome it is the centrioles that are replicated through a tightly controlled process (Azimzadeh and Bornens 2007; Fisk et al. 2002; Fukasawa 2007). The assembly of a single daughter centriole (called a procentriole) at a single site adjacent to each mature centriole ensures that the single parent centrosome is faithfully replicated to produce two daughter centrosomes, each with two centrioles (Nigg 2007). This so called templated replication mechanism is temporally controlled by Cdk2 such that it occurs only once during a limited window in the cell cycle. Although recent evidence demonstrates that centrioles can form de-novo in human cells (La Terra et al. 2005) as in algae (Marshall et al. 2001), in the presence of existing centrioles the templated replication mechanism predominates (La Terra et al. 2005; Marshall et al. 2001; Tsou and Stearns 2006). Templated replication is critical for genomic integrity, because the presence of extraneous centrioles leads to the formation of aberrant mitotic spindles (Lingle and Salisbury 1999) that can missegregate chromosomes and cause aneuploidy (Lingle et al. 2002).
Most human tumors are aneuploid, and many human tumors possess extra centrosomes that lead to the production of multipolar mitotic spindles in situ (Lingle and Salisbury 2000). Extra centrosomes are thought to arise by one of two mechanisms; an aborted mitosis and/or a cytokinesis failure that produces polyploid cells that have inherited extra centrosomes, or a defect in centrosome duplication (Doxsey 2002; Nigg 2002). While both mechanisms will ultimately lead to the production of aberrant spindles that generate aneuploid cells with extra centrosomes, the presence of extra centrosomes in cells that are not aneuploid can only be explained by defects in centrosome duplication. At least in some breast (Lingle et al. 2002) and prostate (Pihan et al. 2001; Pihan et al. 2003) tumors extra centrosomes appear prior to aneuploidy, suggesting that in such tumors the extra centrosomes arose via defects in centrosome duplication. In addition, centrosome defects precede tumor formation in a mouse model of hormone-induced breast tumorigenesis (Milliken et al. 2008), and their presence strongly correlates with aneuploidy in higher grade tumors (Lingle et al. 2002; Lingle et al. 1998). Together these observations suggest that errors in centrosome duplication might promote the genetic instability that is thought to be important in tumorigenesis (Ellsworth et al. 2004a; Ellsworth et al. 2004b; Lengauer et al. 1998; Tsikitis and Chung 2006). In fact, many tumor-derived cell lines are capable of centrosome re-duplication, a phenomenon wherein cells produce extra centrioles during a single, prolonged S-phase (Fisk et al. 2002; Lingle and Salisbury 2000; Nigg 2002; Salisbury et al. 1999). While this might reflect the execution of extra rounds of the canonical templated duplication pathway in mouse cells (Fisk and Winey 2001), in human cells centrosome re-duplication appears instead to reflect an aberration of this pathway wherein existing parental centrioles generate multiple procentrioles (Duensing et al. 2007; Kleylein-Sohn et al. 2007). This can occur “in parallel” by the simultaneous formation of multiple procentrioles (Duensing et al. 2007; Kleylein-Sohn et al. 2007), but in principle could also occur “in series” by the successive formation and release of procentrioles.
The Mps1 protein kinase was initially described in the budding yeast by virtue of its requirement in the duplication of the fungal centrosome equivalent (Schutz and Winey 1998; Winey et al. 1991), and was subsequently shown to be required for the spindle assembly checkpoint (Hardwick et al. 1996; Weiss and Winey 1996). Mps1 kinases have since been found in virtually all eukaryotes and their function in the spindle checkpoint is clearly conserved (Abrieu et al. 2001; Fisk et al. 2003; Liu et al. 2003; Stucke et al. 2002). However, the function of Mps1 in centrosome duplication is controversial (Stucke et al. 2002) and has received less attention (Fisk et al. 2003; Fisk and Winey 2001; Kanai et al. 2007; Kasbek et al. 2007; Stucke et al. 2002). Our data demonstrates that Mps1 is required for centrosome duplication in human cells (Fisk et al. 2003; Kasbek et al. 2007), and that this function of Mps1 is regulated by Cdk2 (Kasbek et al. 2007); Cyclin A-associated Cdk2 (Cdk2/A) phosphorylates Mps1 at T468, and this phosphorylation suppresses the proteasome-mediated degradation of Mps1 to allow accumulation of a centrosomal pool of Mps1 (Kasbek et al. 2007). This Cdk2-regulated Mps1 degradation pathway appears to be specific to centrosomes, and when Mps1 cannot be phosphorylated at T468 it is lost from centrosomes but not from other locations (Kasbek et al. 2007). We also demonstrated that the level of this pool is critical for the proper regulation of centrosome duplication; a version of Mps1 that cannot be phosphorylated at T468 cannot substitute for endogenous Mps1 in centrosome duplication, while mimicking constitutive phosphorylation at T468 or removing the Mps1 degradation signal causes centrosome re-duplication (Kasbek et al. 2007).
Our recent data demonstrates that suppressing the degradation of Mps1 is sufficient to cause centrosome re-duplication, and we observed three potential mechanisms by which this degradation can be suppressed; inhibiting the degradation machinery (e.g. proteasome inhibition), up-regulation of Cdk2 activity (e.g. overexpressing cyclin A1 or A2, which suppresses the degradation of Mps1 at centrosomes), or Mps1 mutations that mimic Cdk2 phosphorylation or remove the Mps1 degradation signal (Mps1 T468D/T468E or Mps1Δ12/13, respectively) (Kasbek et al. 2007). Because many signal transduction events impact the regulation of cyclin dependent kinases, our data suggests that Mps1 degradation might be misregulated in a variety of tumors. This further suggests that Mps1 might contribute to the extra centrosomes found in some tumors. To test these suggestions we sought to examine the regulation of Mps1 in human tumor-derived cells. For this analysis we chose the U2OS osteosarcoma cell line that is well known to undergo centrosome re-duplication (Fisk et al. 2003; Kleylein-Sohn et al. 2007; Stucke et al. 2002), and two mammary carcinoma cell lines that misregulate Mps1 or Cdk2; MCF7 that has been shown to overexpress the Mps1 message by 8.4-fold compared to other mammary epithelial cell lines (Yuan et al. 2006), and 21NT that overexpresses cyclin A1 (Coletta et al. 2004; Ford et al. 2000). We have found that the U2OS and 21NT tumor-derived cell lines fail to appropriately degrade Mps1 in the absence of Cdk2. This misregulation of Mps1 degradation occurs through a distinct mechanism in each cell line and correlates with the misregulation of centrosome duplication. Accordingly, Mps1 may provide a link between the control of centrosome duplication and genomic instability.
Results
Centrosome re-duplication in tumor-derived cells
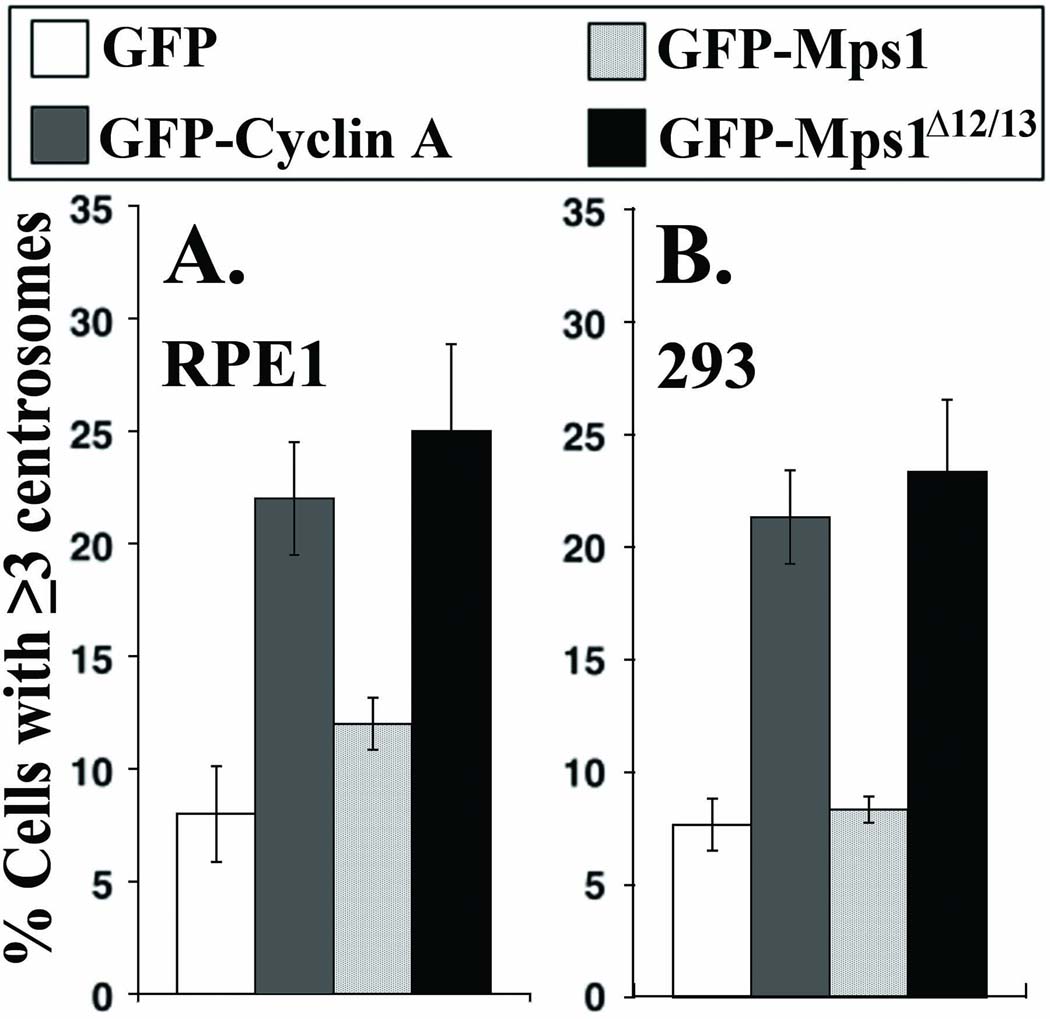
To test the suggestion that Mps1 might contribute to the centrosome defects observed in human tumors we have examined the regulation of Mps1 and centrosome duplication in the following human cell lines; RPE1 telomerase-immortalized retinal pigment epithelia cells, 293 virally transformed human embryonic kidney cells, HeLa cervical carcinoma cells, U2OS osteosarcoma cells, and two mammary carcinoma cell lines, MCF7 and 21NT. Although HeLa cells do not normally undergo centrosome re-duplication (Fisk et al. 2003; Kasbek et al. 2007), preventing the degradation of Mps1 is sufficient to cause centrosome re-duplication in HeLa cells (Kasbek et al. 2007). Similarly, neither RPE1 (Figure 1 A) nor 293 (Figure 1 B) cells undergo centrosome re-duplication, and overexpression of GFP-Mps1 is not sufficient to cause centrosome re-duplication in these cells. However, as in HeLa cells, overexpression of either cyclin A or the non-degradable Mps1Δ12/13 (see below) causes centrosome re-duplication in both RPE1 and 293 cells. This suggests that the regulation of centrosome duplication by Mps1 degradation is a consistent feature of human cells.
Figure 1. Centrosome re-duplication in non tumor-derived RPE1 and 293 cells.
(A and B) Preventing Mps1 degradation causes centrosome re-duplication in RPE1 and 293 cells. RPE1 or 293 cells transfected with the indicated expression constructs were arrested in S-phase with a 24 hr HU treatment. Centrosome number was determined after an additional 48 hr of S-phase arrest. Bar graphs showing the percentage of (A) RPE1 cells or (B) 293 cells expressing GFP (white), GFP-Cyclin A (dark gray), GFP-Mps1 (light gray), or GFP-Mps1Δ12/13 (black) that have three or more centrosomes.
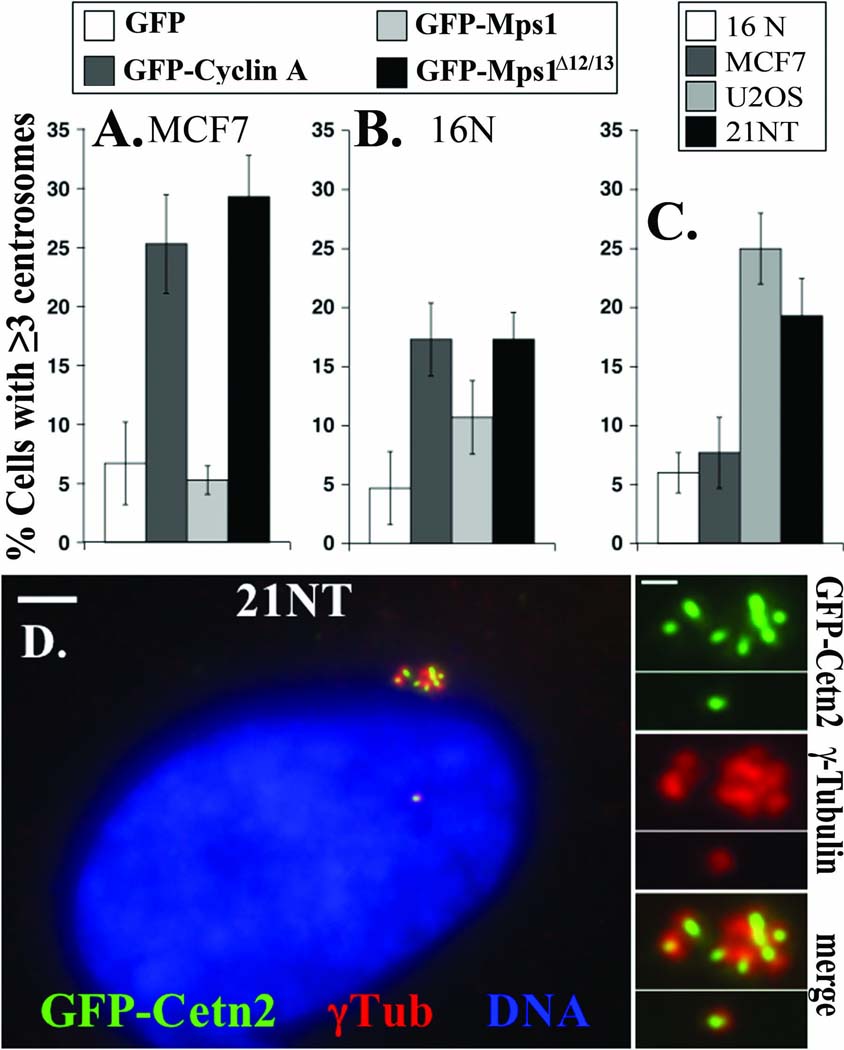
In contrast, the U2OS osteosarcoma cell line is well known to undergo centrosome re-duplication (Fisk et al. 2003; Guarguaglini et al. 2005; Kleylein-Sohn et al. 2007; Stucke et al. 2002). Centrosome re-duplication in U2OS cells appears to be Mps1-dependent, because it can be prevented by overexpression of a catalytically inactive version of Mps1 (Fisk et al. 2003; Kasbek et al. 2007). Moreover, overexpression of wild type Mps1 accelerates the onset of centrosome re-duplication in U2OS cells (Fisk et al. 2003; Kanai et al. 2007; Kasbek et al. 2007). Given the Mps1-dependence of centrosome re-duplication in U2OS cells, we examined centrosome re-duplication in MCF7 and 21NT cells that overexpress the Mps1 message or cyclin A1, respectively. As a control we also tested the 16N cell line that is isogenic with 21NT; while 21NT was derived from a primary tumor site (Band and Sager 1991; Band et al. 1990), 16N is a papilomavirus-immortalized derivative of the normal mammary epithelium of the same breast cancer patient (Band and Sager 1991). Neither MCF7 cells (Figure 2 A) nor 16N (Figure 2 B) cells undergo centrosome re-duplication, and overexpression of GFP-Mps1 is not sufficient to cause centrosome re-duplication in either cell line. However, as observed for HeLa (Kasbek et al. 2007), 293, and RPE1 cells, preventing the degradation of Mps1 by overexpressing cyclin A or Mps1Δ12/13 is sufficient to cause centrosome re-duplication in both MCF7 (Figure 2 A) and 16N (Figure 2 B) cells. In contrast, like U2OS cells, 21NT cells are capable of centrosome re-duplication on their own (Figure 2 C, D).
Figure 2. Centrosome re-duplication in tumor-derived cells.
(A and B) Centrosome re-duplication in (A) MCF7 and (B) 16N cells requires overexpression of Cyclin A or Mps1Δ12/13. MCF7 or 16N cells transfected with the indicated expression constructs were analyzed as described in Figure 1. (C and D) Like U2OS cells, 21NT cells naturally undergo centrosome re-duplication. (C) Untransfected 16N, MCF7, U2OS, or 21NT cells were arrested in S-phase with a 24 hr HU treatment, and centrosome number was determined as described in Figure 1. 16N (white), MCF7 (dark gray), U2OS (light gray), 21NT (black). (D) 21NT cells were transfected with GFP-Centrin 2 (Cetn2), then arrested in S-phase and analyzed as in (C). Shown is a representative 21NT cell with extra centrosomes. Green, GFP-Cetn 2; red, γ-Tubulin; Blue, DNA; bar=5μm. Insets show 4-fold magnified images of centrosomes, bar=2µm.
Misregulation of Mps1 degradation in human tumor-derived cell lines
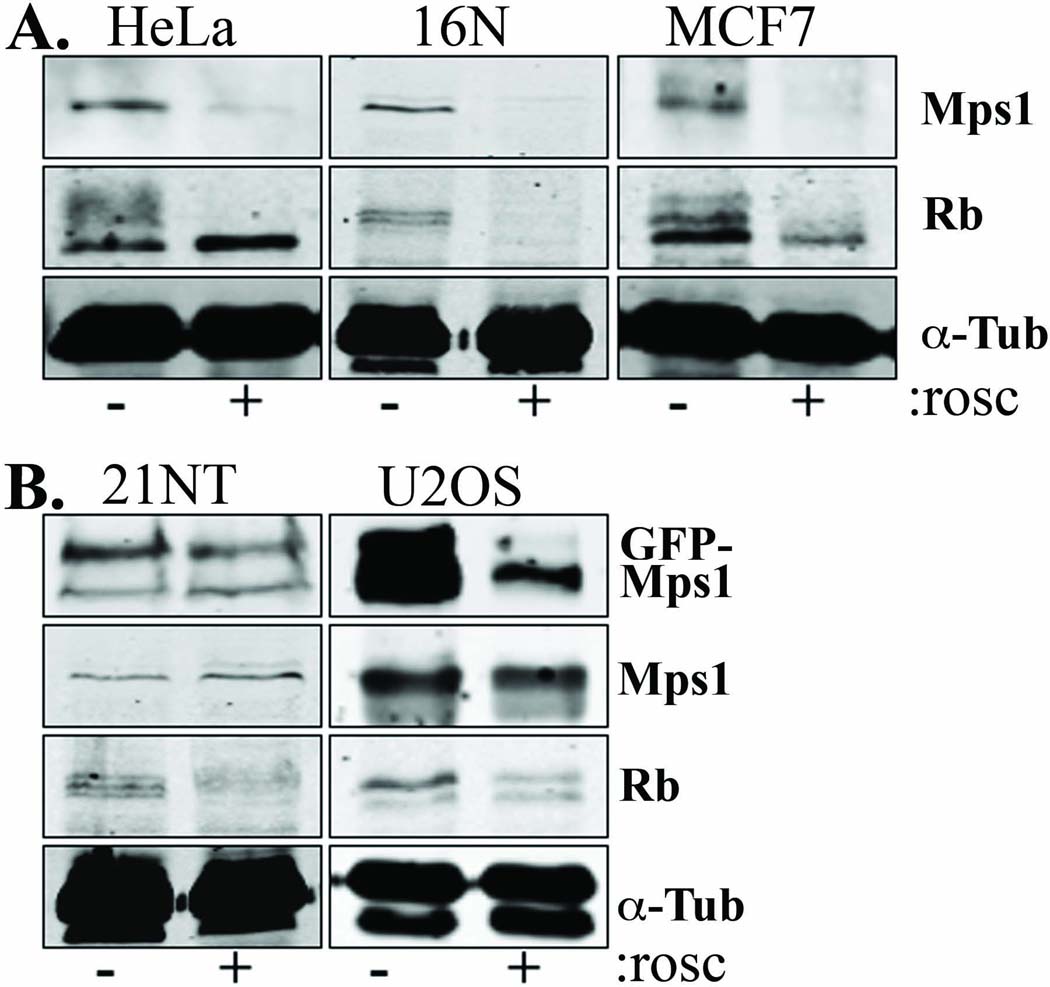
Consistent with our previous observations that Mps1 is degraded by the proteasome in the absence of Cdk2 activity (Fisk and Winey 2001; Kasbek et al. 2007), Mps1 is degraded in response to the Cdk2 inhibitor roscovitine in HeLa, 16N, and MCF7 cells (Figure 3 A). However, we found that Mps1 is not degraded in the presence of roscovitine in U2OS or 21NT cells (Figure 3 B). To rule out the possibility that U2OS and 21NT cells were refractory to roscovitine, we examined the hyperphosphorylation of the Cdk2 substrate Rb. By inhibiting Cdk2 activity roscovitine reverses Rb hyperphosphorylation, and has been shown to cause either the loss of slower mobility forms of Rb, or a reduction in total Rb on immunoblots, depending on the time, concentration, and cell line used (Whittaker et al. 2004). Rb hyperphosphorylation is lost in response to roscovitine in HeLa and MCF7 cells as judged by the loss of slower mobility forms, and in 16N cells as judged by the loss of the Rb protein (Figure 3 A). Rb hyperphosphorylation is similarly lost in response to roscovitine in U2OS cells as judged by both reduction of slower mobility forms and reduction of total Rb protein, and in 21NT cells as judged by the loss of the Rb protein (Figure 3 B). Therefore, the failure to degrade Mps1 in response to roscovitine is not due to the failure of roscovitine to inhibit Cdk2 in U2OS and 21NT cells.
Figure 3. Misregulation of Mps1 degradation in tumor-derived cells.
(A) Mps1 is degraded appropriately in the presence of roscovitine in HeLa, 16N, and MCF7 cells. Cells were arrested in S-phase with a 24 hr HU treatment, treated with 50 µM roscovitine for an additional 24 hr in the presence of HU, then analyzed by immunoblot with antibodies against Mps1, Rb, or α-Tubulin (loading control) as indicated. (B) Mps1 is not degraded in the presence of roscovitine in 21NT or U2OS cells. 21NT cells also fail to degrade GFP-Mps1, while GFP-Mps1 is appropriately degraded in U2OS cells. 21NT or U2OS cells were transfected with GFP-Mps1, then arrested in S-phase, treated with roscovitine, and analyzed as in (A) with antibodies against GFP, Mps1, Rb, or α-Tubulin (loading control) as indicated.
21NT cells cannot target centrosomal Mps1 for proteasome-mediated degradation
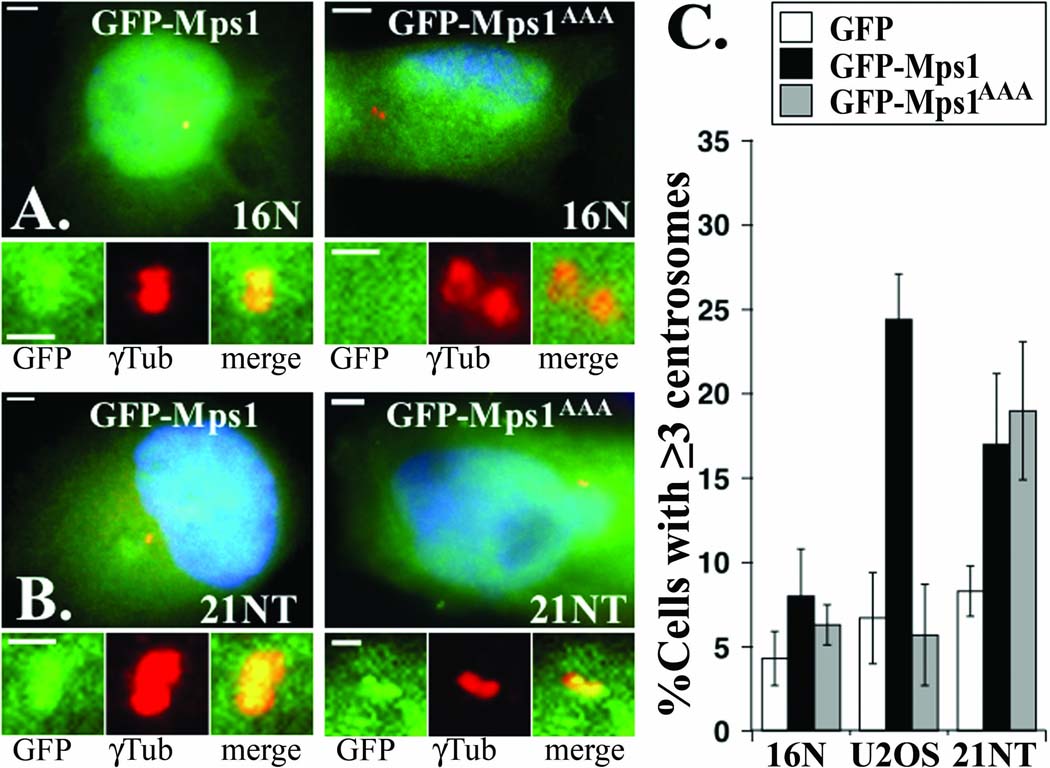
In order to assess the mechanisms responsible for the misregulation of Mps1, we examined the regulation of transgenic Mps1. The 21NT cell line also fails to degrade GFP-Mps1 in response to roscovitine (Figure 3 B), suggesting that these cells have a defect either in the upstream regulation of Mps1 (e.g. in the Cdk2 pathway) or in the downstream machinery that targets Mps1 for proteasome-dependent degradation. In order to distinguish between these possibilities we examined the centrosomal accumulation of GFP-Mps1AAA, an Mps1 mutant that is constitutively degraded at centrosomes in the presence of Cdk2 activity because it contains the T468A mutation (Kasbek et al. 2007). Because GFP-Mps1AAA cannot be phosphorylated at T468 it should not be responsive to Cdk2 inhibition and its whole cell levels would be uninformative. However, its ability to accumulate at centrosomes should be informative, because even in the presence of elevated Cdk2 activity GFP-Mps1AAA should only accumulate at centrosomes if the machinery that targets Mps1 for degradation is defective. As expected, GFP-Mps1AAA did not localize to centrosomes in 16N cells that regulate Mps1 appropriately (Figure 4 A). In contrast, the localization of GFP-Mps1AAA was indistinguishable from that of GFP-Mps1 in 21NT cells (Figure 4 B). This ability of GFP-Mps1AAA to accumulate at centrosomes in 21NT cells suggests that these cells cannot properly degrade Mps1 at centrosomes.
Figure 4. The Mps1 degradation machinery is defective in 21NT cells.
(A and B) GFP-Mps1AAA accumulates at centrosomes in 21NT cells. (A) 16N or (B) 21NT cells were transfected with GFP-Mps1 or GFP-Mps1AAA as indicated, arrested in S-phase with a 24 hr HU treatment, and analyzed by fluorescence microscopy as described in Materials and Methods. The GFP-Mps1 signal satisfied the previously established objective criteria for centrosome localization in 60% of 16N cells, as well as in 60% of 21NT cells (compared to 40% of HeLa cells (Kasbek et al. 2007)). The GFP-Mps1AAA signal did not correlate with centrosomes in any 16N cell analyzed, but correlated with centrosomes in 66% of 21NT cells. Green, GFP-Mps1 or GFP-Mps1AAA; red, γ-Tubulin; Blue, DNA; bar=5µm. Insets show 4-fold magnified images of centrosomes, bar=2µm. (C) GFP-Mps1AAA accelerates centrosome re-duplication in 21NT cells. 16N, U2OS, and 21NT cells transfected with the indicated expression constructs were arrested in S-phase with a 24 hr HU treatment. Centrosome number was determined after an additional 24 hr of S-phase arrest. Bar graph shows the percentage of cells expressing GFP (white), GFP-Mps1 (black), or GFP-Mps1AAA (gray) that have three or more centrosomes.
To further test this suggestion, we examined the function of GFP-Mps1AAA at centrosomes in 21NT cells. In U2OS cells overexpression of Mps1 accelerates the onset of centrosome re-duplication; after only 24 hours of S-phase arrest roughly five-fold more U2OS cells expressing GFP-Mps1 have excess centrosomes compared to cells expressing GFP alone (Fisk et al. 2003; Kanai et al. 2007; Kasbek et al. 2007). However, GFP-Mps1AAA cannot accelerate the onset of centrosome re-duplication in U2OS cells, because it cannot accumulate at centrosomes to levels sufficient to affect centrosome duplication (Kasbek et al. 2007). As shown in Figure 4 C, GFP-Mps1 similarly accelerates the onset of centrosome re-duplication in 21NT cells. However, GFP-Mps1AAA is as effective as GFP-Mps1 at accelerating centrosome re-duplication in 21NT cells (Figure 4 C). Together, these data suggest that 21NT cells are deficient in some factor that targets Mps1 for degradation at centrosomes, such as an E3 ubiquitin ligase. While we look forward to identifying the molecular nature of the defect in 21NT cells, the molecules that target Mps1 for proteasome-mediated degradation at centrosomes are currently unknown.
U2OS Cells express a non-degradable Mps1 allele
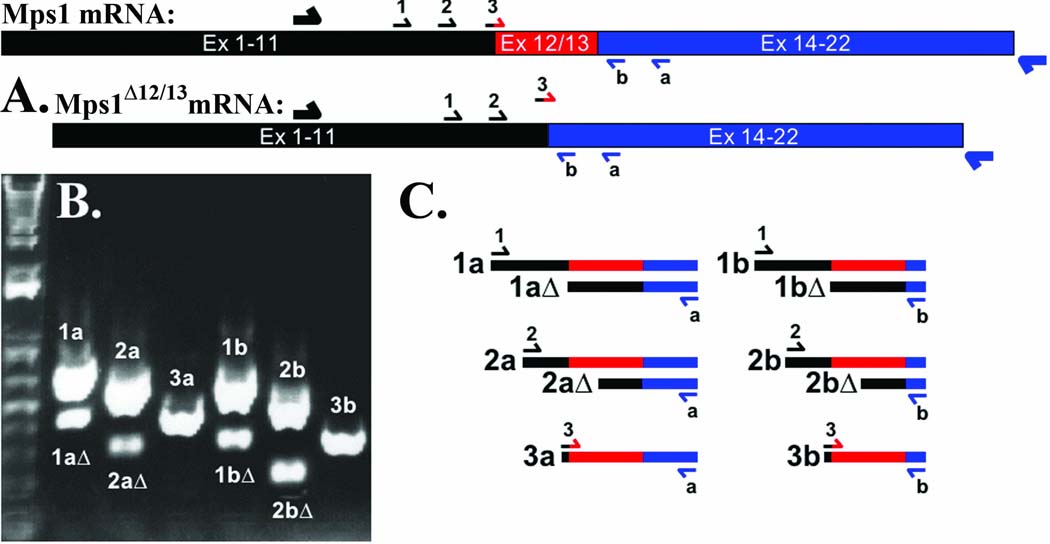
Despite failing to appropriately degrade endogenous Mps1, U2OS cells retain the ability to degrade transgenic wild type GFP-Mps1 in response to roscovitine (Figure 3 B). This suggests that the failure to degrade Mps1 in U2OS cells is not the consequence of a defect in the regulation or execution of Mps1 degradation, but is intrinsic to Mps1 itself. Because this suggested that U2OS cells might harbor an Mps1 mutation, we sequenced the Mps1 coding region in PCR products derived from U2OS genomic DNA and cDNA preparations. While we found no Mps1 coding mutations in U2OS cells, we were unable to sequence across the exon 11–12 junction in Mps1 RT-PCR products. When we examined the region surrounding the exon 11–12 junction in mRNA from U2OS using the primers shown in Figure 5 A, we identified the presence of two RT-PCR products (Figure 5 B). The smaller of these products disappeared when a primer spanning both exons 11 and 12 was used, and sequence analysis demonstrated that this smaller product represented a message lacking exons 12 and 13 (Figure 5 C shows a schematic of the resulting PCR products). This message, which we have designated as Mps1Δ12/13, encodes for an internally truncated protein lacking amino acids 420–507 (Kasbek et al. 2007). Identified in U2OS cells as described herein, Mps1Δ12/13 was pivotal in our recent study demonstrating that preventing Mps1 degradation is sufficient to cause centrosome re-duplication (Kasbek et al. 2007); the region missing in Mps1Δ12/13 contains both the Mps1 degradation signal and the T468 phosphorylation site, and is required for the degradation of Mps1 at centrosomes (Kasbek et al. 2007). Accordingly, GFP-Mps1Δ12/13 is sufficient to cause centrosome re-duplication in a variety of cell types ((Kasbek et al. 2007) and Figure 1 and Figure 2 above).
Figure 5. Identification of Mps1Δ12/13 from U2OS cells.
(A) Schematic diagram showing the Mps1 and Mps1Δ12/13 mRNAs (black, exons 1–11; red, exons 12 and 13; blue, exons 14–22), the Mps1 primer used for reverse transcription (large half arrows), and the primers used for PCR (small half arrows). Primers 1, 2, a, and b have binding sites in Mps1 and Mps1Δ12/13, but primer 3 (black/red) only binds to the Mps1 cDNA because it contains sequence from both exons 11 and 12. (B) Two Mps1 messages exist in U2OS cells. mRNA was isolated from U2OS cells, processed, reverse transcribed, and analyzed by PCR as described in Materials and Methods with the primers indicated in (A). (C) Schematic diagram of the RT-PCR products in (B), colors as in (A).
Mps1Δ12/13 causes dose-dependent centrosome re-duplication
While the expression of the Mps1Δ12/13 protein could potentially explain both centrosome re-duplication and the misregulation of Mps1 degradation in U2OS cells, Figure 5 B shows that the Mps1Δ12/13 message is expressed at a significantly lower level than that of the wild type message. Although this RT-PCR analysis was not quantitative, the major Mps1 species is the ~97 kDa size expected for full length Mps1 and we were unable to detect the ~87 kDa Mps1Δ12/13 protein in U2OS cells unless we forced its overexpression by transient transfection. Thus, we did not report U2OS cells as the source of Mps1Δ12/13 in our previous study detailing its affects on centrosome duplication (Kasbek et al. 2007) because it did not appear to be expressed in U2OS cells.
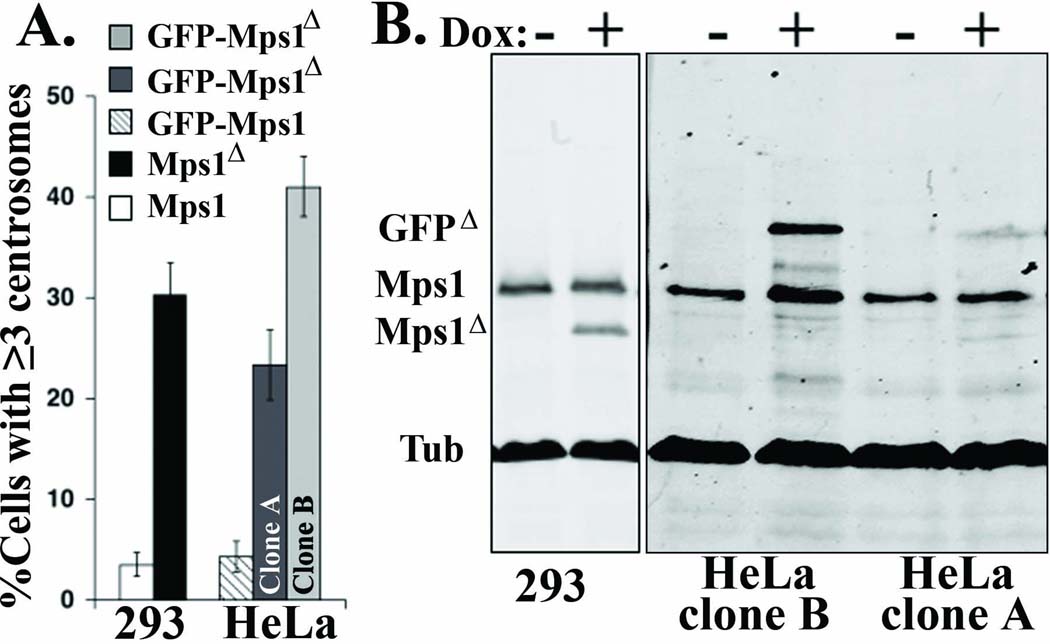
However, observations we have since generated using stable cell lines suggest that even very low levels of Mps1Δ12/13 are sufficient to cause centrosome re-duplication. In order to obtain stable cell lines that inducibly express wild type or non-degradable Mps1, we transfected 293 T-REx and HeLa T-REx cells that harbor the native Tetracycline (Tet) Repressor with constructs that express Mps1 or Mps1Δ12/13, and GFP-Mps1 or GFP-Mps1Δ12/13, respectively, under the control of the Tet operator. We then isolated stable clones and examined their ability to undergo centrosome re-duplication. As with transient expression experiments (Kasbek et al. 2007), Tet-inducible expression of Mps1 in 293 clones, or of GFP-Mps1 in HeLa clones, did not cause centrosome re-duplication (Figure 6 A, white and striped bars, respectively), despite a roughly 5-fold increase in Mps1 levels under inducing conditions (not shown). In contrast, Tet-inducible expression of Mps1Δ12/13 in 293 clones (Figure 6 A, black bar, Mps1Δ), and of GFP-Mps1Δ12/13 in HeLa clones (Figure 6 A, light and dark gray bars, GFP-Mps1Δ) caused centrosome re-duplication equal to or greater than that caused by transient expression of GFP-Mps1Δ12/13 from a viral promoter (see e.g. Figure 2), even though the levels of Mps1Δ12/13 or GFP-Mps1Δ12/13 were between two and five fold lower than that of endogenous Mps1 (Figure 6 B).
Figure 6. Mps1Δ12/13 causes dose-dependent centrosome re-duplication.
(A) Centrosome re-duplication in 293- and HeLa-derived clones. Stable clones generated as described in Materials and Methods were arrested in S-phase with a 24 hr HU treatment in the presence of Dox. Centrosome number was determined after an additional 48 hr of S-phase arrest. Bar graph showing the percentage of cells from 293 T-REx- or HeLa T-REx-derived clones with three or more centrosomes. White bar, 293 Mps1 (Mps1); black bar, 293 Mps1Δ12/13 (Mps1Δ); striped bar, HeLa GFP-Mps1; dark gray bar, HeLa GFP-Mps1Δ12/13 clone A (GFP-Mps1Δ-A); dark gray bars, HeLa GFP-Mps1Δ12/13 clone B (GFP-Mps1Δ-B). (B) Expression levels of Mps1Δ12/13 and GFP-Mps1Δ12/13 are lower than that of endogenous Mps1. The cell lines from (A) were arrested in S-phase with a 24 hr HU treatment in the presence or absence of 1 µg/ml Dox, and analyzed by simultaneous immunoblot with the mouse antibodies N1 (anti-Mps1) and DM1A (anti-α-Tubulin) as described in Materials and Methods. The ratio of Mps1Δ12/13 (Mps1Δ) to endogenous Mps1 in the 293 T-REx-derived Mps1Δ12/13 clone (293) is 0.5, and the ratio of GFP-Mps1Δ12/13 (GFPΔ) to endogenous Mps1 in HeLa T-REx-derived GFP-Mps1Δ12/13 expressing clones A (HeLa clone A) and B (HeLa clone B) is 0.2 and 0.4, respectively.
While all 293 T-REx derived clones expressed Mps1Δ12/13 at a level roughly two-fold lower than that of endogenous Mps1 in S-phase arrested cells, expression of GFP-Mps1Δ12/13 was variable among HeLa T-REx derived clones. Figure 6 B shows quantitative immunoblot analysis (Kasbek et al. 2007; Yang et al. 2008) of two representative HeLa T-REx clones. The ratio of GFP-Mps1Δ12/13 to endogenous Mps1 is 0.2 in clone A and 0.4 in clone B (e.g. the level of GFP-Mps1Δ12/13 is 5-fold and 2.5-fold lower than that of endogenous Mps1, respectively), indicating that the relative overexpression of GFP-Mps1Δ12/13 in clone B is roughly 2-fold higher than that in clone A. As shown in Figure 6 A clone B (light gray bar, Clone B) also exhibits a roughly two-fold higher level of centrosome re-duplication than does clone A (dark gray bar, Clone A), demonstrating that GFP-Mps1Δ12/13 causes centrosome re-duplication in a dose dependent manner.
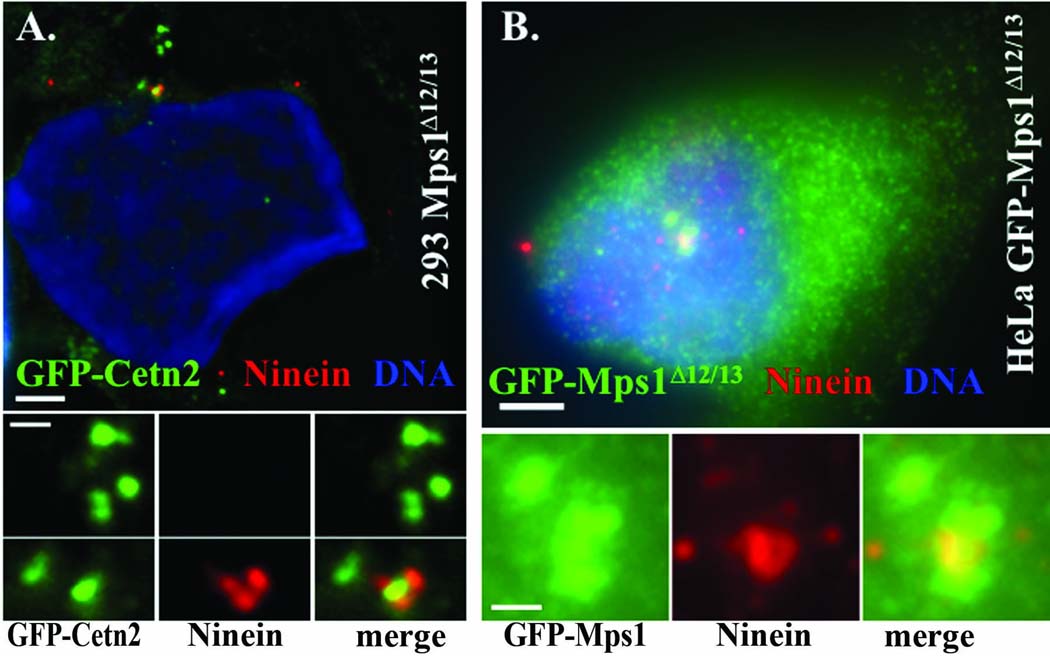
In order to determine the basic mechanism of Mps1Δ12/13-dependent centrosome re-duplication, we analyzed these stable clones with an antibody against Ninein, a marker of mature centrioles (Ou et al. 2002; Piel et al. 2000). While Ninein is not absolutely restricted to the maternal centriole, Ninein should be associated with more than one centrosome if centrosome re-duplication had occurred via an extra round of the canonical templated replication cycle or via mitotic failure (Guarguaglini et al. 2005). We would also expect an even number of centrioles that would be found in a characteristic two-by-two configuration if extra centrosomes were produced via the canonical duplication pathway. We found that Ninein was largely restricted to a single centriole in 293 T-REx clones expressing Mps1Δ12/13 (Figure 7 A) and HeLa T-REx- clones expressing GFP-Mps1Δ12/13 (Figure 7 B), and that centrioles were often found in apparent isolation (e.g. Figure 7 A). We obtained similar data using both HeLa T-REx clones described above, and clone A that has the lowest level of GFP-Mps1Δ12/13 is shown in Figure 7B. Together, this data demonstrates both that very low levels of Mps1Δ12/13 can cause centrosome re-duplication, and that Mps1Δ12/13 causes the production of multiple daughter centrioles from one or more parental centriole.
Figure 7. Mps1Δ12/13 causes centriole overproduction.
(A and B) The mature centriole marker Ninein is predominantly associated with a single centriole in cells expressing Mps1Δ12/13, indicating that extra centrioles arose by formation of more than one procentriole by parental centriole(s). (A) 293 Mps1Δ12/13 cells were transfected with GFP-Centrin 2 (Cetn2), arrested in S-phase with a 24 hr HU treatment in the presence of Dox, and centrosomes were analyzed with an antibody against Ninein after an additional 48 hr of S-phase arrest in the presence of Dox. (B) HeLa GFP-Mps1-A cells were arrested in S-phase with a 24 hr HU treatment and analyzed as in (A). Shown are representative (A) 293 Mps1Δ12/13 or (B) HeLa GFP-Mps1Δ12/13 cells that have extra centrioles. Green, GFP-Cetn 2 or GFP-Mps1Δ12/13; red, Ninein; Blue, DNA. Bar=5 µm. Insets show 4-fold magnified images of centrosomes, bar=2µm.
Mps1Δ12/13 influences wild type Mps1 and migrates anomalously on immunoblots
As discussed above, we have been unable to document the presence of the predicted ~87 kDa Mps1Δ12/13 protein in U2OS cell lysates. In an attempt to verify that Mps1Δ12/13 can be detected in U2OS lysates, we transiently expressed the untagged protein in U2OS cells from the pT-REx-Mps1Δ12/13 plasmid. While the predicted ~87 kDa band is detected, we also observed a greatly increased Mps1 signal at ~97 kDa, the predicted molecular weight of wild type Mps1 (Figure 8). This increased signal might be due to an increase in the stability of the endogenous Mps1 protein caused by the presence of Mps1Δ12/13, or to an anomalous migration of the Mps1Δ12/13 protein itself. As shown above in Figure 6 B, the level of the ~97 kDa endogenous Mps1 band was indeed increased in the presence of Dox in HeLa GFP-Mps1Δ12/13 cells (this is most obvious in cell line B that expresses a higher level of GFP-Mps1Δ12/13). This suggests that the presence of Mps1Δ12/13 can influence the levels of endogenous Mps1.
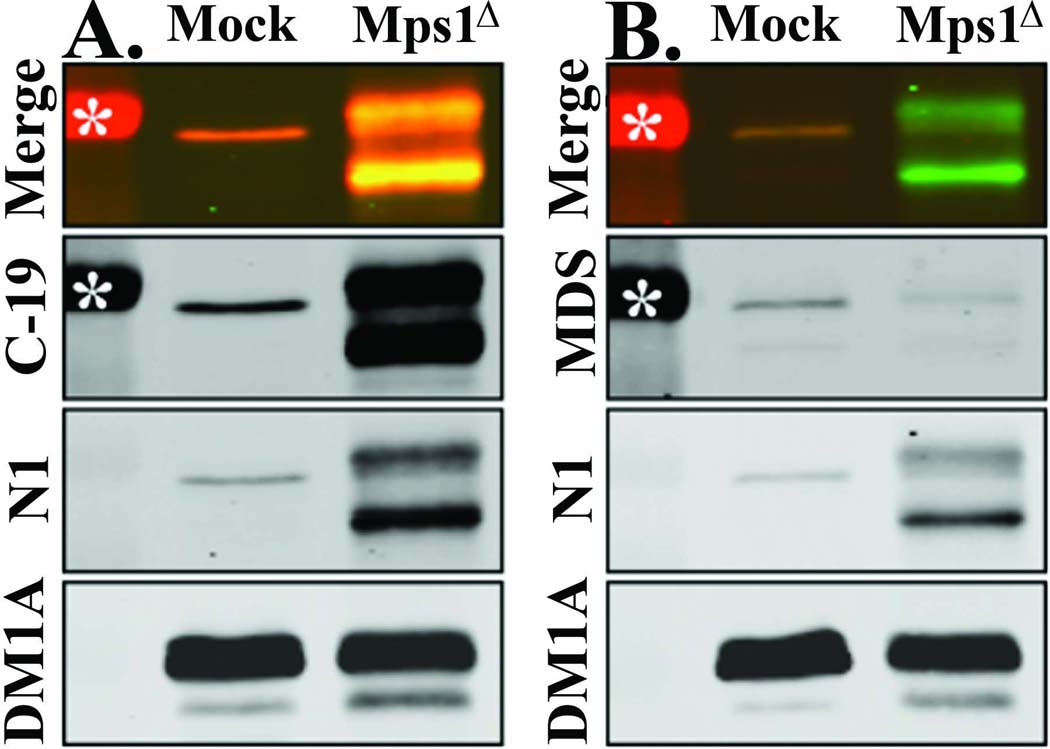
Figure 8. Mps1Δ12/13 runs anomalously in immunoblots.
(A) Expression of Mps1Δ12/13 leads to an increased Mps1 signal at ~97 kDa. (B) The increased Mps1 signal is likely to be a modified form of Mps1Δ12/13 because it is not recognized by an antibody directed against the region missing in Mps1Δ12/13. Untransfected (Mock) or Mps1Δ12/13-transfected (Mps1Δ) U2OS cells were arrested in S-phase with a 24 hr HU treatment, and identical immunoblots were analyzed by quantitative dual-color immunoblot with (A) N1 mouse anti-Mps1, DM1A mouse anti-α-Tubulin, and C-19 rabbit anti-Mps1, or (B) N1, DM1A, and MDS rabbit anti-Mps1 Degradation Signal as described in Methods and Materials. Asterisks represent the 95 kDa molecular weight marker.
In order to determine whether Mps1Δ12/13 itself could account for the increased ~97 kDa signal observed in U2OS cells transfected with pT-REx-Mps1Δ12/13, we analyzed lysates by simultaneous dual-color immunoblot with N1 mouse anti-Mps1 that recognizes the Mps1 N-terminus (Stucke et al. 2002), and one of two rabbit anti-Mps1 antibodies; C-19 (Figure 8 A) that recognizes the Mps1 C-terminus, and our own MDS antibody (Figure 8 B) that recognizes Mps1 amino acids 420–507, the region missing in Mps1Δ12/13 (referred to as MDS because amino acids 420–507 contain the Mps1 Degradation Signal (Kasbek et al. 2007)). As shown in Figure 8 A, both N1 (green) and C-19 (red) recognize both the ~87 kDa Mps1Δ12/13 protein, and the increased ~97 kDa signal. In contrast MDS (red, Figure 8 B) does not recognize any additional Mps1 species upon transfection with Mps1Δ12/13, as expected for an antibody directed against the region missing in Mps1Δ12/13. Because C-19 and N1 recognize the additional species at ~97 kDa while MDS does not, we conclude that it must represent a modified form of Mps1Δ12/13 with slower mobility. This slower mobility form of Mps1Δ12/13 appears distinct from endogenous Mps1 when stained with the N1 antibody, as evidenced by the presence of a doublet near 97 kDa (Figure 8 A and B; most obvious in Figure 8 B, “Merged”). However, the affinity of N1 for the slower mobility form of Mps1Δ12/13 is much lower than its affinity for the ~87 kDa Mps1Δ12/13 protein. The ability of N1 to bind to bacterially expressed Mps1 is blocked by phosphorylation (Fisk et al. 2003), providing precedence for the reduced affinity of N1 for modified forms of Mps1. However, the identity of the blocking phosphorylation site and its relevance in vivo are not known. In contrast, C-19 recognizes both forms of Mps1Δ12/13 with equal affinity, but cannot distinguish the slower mobility form from endogenous Mps1. We note that we still have not documented the presence of the Mps1Δ12/13 protein in U2OS cells. However, because the two most common Mps1 antibodies might fail to distinguish the Mps1Δ12/13 protein from the wild type Mps1 protein depending on its modification state, our data suggest that we cannot rule out its presence by the absence of the predicted ~87 kDa band.
Together, our observations with stable Mps1Δ12/13-expressing cell lines demonstrate that very low levels of Mps1Δ12/13 cause centrosome re-duplication and affect the behavior of the endogenous wild type Mps1 protein. Moreover, the Mps1Δ12/13 protein can display an anomalous mobility that is similar to the wild type protein. Accordingly, it remains possible that Mps1Δ12/13 could account for both centrosome re-duplication and misregulation of Mps1 degradation in U2OS cells. Regardless, the observation that GFP-Mps1 is appropriately degraded in U2OS cells, combined with the identification of the non-degradable Mps1Δ12/13 in these cells, suggests that the misregulation of Mps1 degradation in U2OS cells is intrinsic to Mps1 itself.
Discussion
The presence of extra centrosomes and/or centrioles leads to the production of aberrant mitotic spindles that can generate aneuploid daughter cells by causing errors in chromosome segregation (Azimzadeh and Bornens 2007; Fisk et al. 2002; Fukasawa 2007). Extra centrosomes are observed in a variety of human tumors where it is assumed they contribute to aneuploidy (Lingle and Salisbury 2000). However, the precise mechanisms responsible for generating extra centrosomes are not known. In both mouse and human cells Mps1 is degraded by the proteasome in the absence of Cdk2 activity (Fisk and Winey 2001; Kasbek et al. 2007). Our recent data suggests that by attenuating Mps1 degradation Cdk2 controls the accumulation of a centrosomal Mps1 pool, and that preventing the degradation of Mps1 at centrosomes increases this pool and causes production of extra centrosomes in vitro (Kasbek et al. 2007). This further suggests the possibility that defects in Mps1 degradation represent one possible mechanism for generating extra centrosomes in human tumors. Here we have taken the first step toward testing this possibility by demonstrating that the 21NT mammary carcinoma and U2OS osteosarcoma cell lines display defects in the regulation of Mps1 degradation.
The failure to degrade Mps1 in 21NT and U2OS cells is not due to a failure to inhibit Cdk2, nor is it likely to be a consequence of immortalization or adaptation to culture; Mps1 is degraded appropriately in a number of tumor- and non tumor-derived cell lines, including the 16N cell line that is isogenic to 21NT. Moreover, in the cell lines from this study the regulation of centrosome duplication correlates with the regulation of Mps1 degradation. U2OS cells are well known to undergo centrosome re-duplication, and of the cells used in this study only 21NT cells share this capability; HeLa, 293, RPE1, 16N, and MCF7 cells that properly regulate Mps1 degradation are not capable of centrosome re-duplication. Because preventing Mps1 degradation is sufficient to cause centrosome re-duplication (Kasbek et al. 2007), our observations with U2OS and 21NT suggest that the failure to appropriately degrade Mps1 in these cells might account for their ability to undergo centrosome re-duplication. However, significant work remains to determine whether defects in the regulation of Mps1 contribute to extra centrosomes in situ.
MCF7 cells have been shown to overexpress the Mps1 message (Yuan et al. 2006) yet regulate both centrosomes and Mps1 appropriately, and whole-cell Mps1 levels are indistinguishable between 16N that regulate Mps1 appropriately and 21NT cells that fail to degrade Mps1 in the absence of Cdk2. These observations underscore the point that centrosome re-duplication is unrelated to whole-cell Mps1 levels. Rather, we suggest that the critical factor for the regulation of centrosome duplication is the level of Mps1 at centrosomes, which depends not on whole-cell Mps1 levels but on the regulation of Mps1 degradation at centrosomes (Kasbek et al. 2007). Our recent study suggesting that the centrosomal pool accounts for perhaps 10% of total cellular Mps1 used siRNA to deplete Cdk2 activity, which caused the loss of Mps1 specifically from centrosomes with little effect on whole-cell Mps1 levels (Kasbek et al. 2007). Here we have probed the regulation of Mps1 degradation using roscovitine, which greatly reduces the whole-cell levels of Mps1 (Fisk and Winey 2001; Kasbek et al. 2007). The difference between the two methods may be explained by the presence of the roscovitine-inhibited kinase that presumably mimics a dominant negative Cdk2 allele, but we cannot rule out the existence of off-target affects of roscovitine. Regardless, roscovitine has clearly proven a useful tool for probing the regulation of Mps1 degradation.
In principle there are three obvious mechanisms that might result in the misregulation of Mps1 degradation; 1) up-regulation of Cdk2 activity; 2) defects in the molecules that target Mps1 for proteasome-mediated degradation; and 3) Mps1 mutations that render it non-degradable. In our previous study we demonstrated that each of these mechanisms can prevent the degradation of Mps1 at centrosomes in HeLa cells (Kasbek et al. 2007); overexpression of cyclin A led to a 2.4-fold increase in the level of Mps1 at centrosomes, inhibition of the proteasome with MG115 led to a similar increase in centrosomal Mps1, and Mps1 mutations that removed the degradation signal (Mps1Δ12/13) or mimicked Cdk2 phosphorylation (Mps1T468D and Mps1T468E) prevented the removal of Mps1 from centrosomes in the absence of Cdk2 activity. While we have shown that the first of these mechanisms can prevent the degradation of Mps1 at centrosomes, up-regulation of Cdk2 activity should be invisible to the experiments described here unless it also rendered Cdk2 insensitive to roscovitine. We chose to examine 21NT cells because they overexpress cyclin A1 (Coletta et al. 2004; Ford et al. 2000), which prevents Mps1 degradation and causes centrosome re-duplication in HeLa cells (Kasbek et al. 2007). However, while we cannot rule out a role for cyclin A1 in preventing Mps1 degradation in 21NT cells, our data indicate that increased Cdk2 activity is not responsible. Instead we found that an Mps1 mutant that is normally degraded even in the presence of Cdk2 activity accumulates at centrosomes and accelerates centrosome re-duplication in 21NT cells. It is unlikely that the proteasome itself is defective in 21NT cells, because Rb is degraded in response to roscovitine. Rather, it seems likely that 21NT cells are deficient in a factor that serves to target centrosomal Mps1 to the proteasome. While the molecules that target Mps1 for degradation at centrosomes are currently unknown, 16N and 21NT may be helpful in their identification. For example, the use of single nucleotide polymorphism or methylation arrays to identify genomic regions that differ between 16N and 21NT might lead to the identification of interesting candidate regulatory proteins.
In contrast, U2OS cells retain the ability to degrade transgenic wild type Mps1. This demonstrates that both the upstream regulation (e.g. Cdk2) and downstream machinery of Mps1 degradation remain intact, and suggests that the defect in U2OS cells is intrinsic to Mps1. Consistent with this suggestion, the Mps1Δ12/13 message that encodes the non-degradable Mps1Δ12/13 protein was discovered in U2OS cells. While Mps1Δ12/13 represents a small fraction of total Mps1 message and U2OS cells lack a detectable signal at the predicted ~87 kDa size of the Mps1Δ12/13 protein, data provided here suggest that we cannot rule out a role for Mps1Δ12/13 in both centrosome re-duplication and misregulation of Mps1 degradation in U2OS cells. Mps1Δ12/13 causes dose-dependent centrosome re-duplication by perturbing the canonical templated centrosome replication pathway, the mechanism established for centrosome re-duplication in U2OS cells (Guarguaglini et al. 2005). Moreover, it does this at very modest expression levels that can also influence the wild type Mps1 protein, and the presence of a slower mobility form of Mps1Δ12/13 that we observed in this study might go undetected by the two most common Mps1 antibodies. A final resolution as to the presence of Mps1Δ12/13 in U2OS cells will likely require mass spectrometry that is beyond the scope of this report.
While the data presented here demonstrate that misregulation of Mps1 degradation correlates with centrosome defects in tumor-derived cells, there remains a large gap to be bridged in order to determine whether defects in the regulation of Mps1 contribute to extra centrosomes in situ, and we look forward to examining Mps1 regulation in tumor tissue. Such an analysis presents many challenges, the greatest of which will be the need to examine Mps1 degradation rather than Mps1 message or protein levels. This is underscored by our observations with MCF7, 16N and 21NT cells, where neither the levels of the Mps1 message nor whole-cell levels of the Mps1 protein correlated with misregulation of Mps1 degradation. Accordingly, a simple analysis of Msp1 levels in archival tumor samples is unlikely to be informative. Instead it will be necessary to examine the level of Mps1 at centrosomes, or the response of Mps1 to roscovitine. While we might achieve the former in archival samples with difficulty, the latter will require the analysis of precious primary tissue samples. It should be possible to analyze Mps1 in homogenized primary tissue, but an assay protocol will first have to be worked out using normal tissue. Moreover, because multiple mechanisms can account for the presence of extra centrosomes (Doxsey 2002; Nigg 2002), it may also be necessary to directly assess centrosome duplication, which would also require live tissue. Tumors are complex heterogeneous environments consisting of stroma and tumor, and any assay will have to account for each compartment. Regardless, the data presented here demonstrate that Mps1 is misregulated in two tumor-derived cell lines through two distinct mechanisms. Together with our previous data demonstrating that preventing the degradation of Mps1 at centrosomes is sufficient to cause centrosome re-duplication in human cells (Kasbek et al. 2007), the data presented here support the suggestion that Mps1 might contribute to the production of extra centrosomes in human tumors. Thus, Mps1 represents a potential link between extra centrosomes and genomic instability.
Materials and Methods
Cells and Cell Culture
HeLa T-REx and 293 T-REx cells expressing the Native Tetracycline (Tet) Repressor were obtained from Invitrogen (Carlsbad, CA). MCF7, 16N, and 21NT cells were obtained from Dr. Heide Ford of the University of Colorado Health Sciences Center (Denver, CO). HeLa, HeLa T-REx, 293, 293 T-REx, MCF7, and U2OS cells were grown in Glutamax DMEM; RPE1 cells were grown in F12:DMEM (50:50); 16N and 21T cells were grown in MEM. All media was supplemented with 10% Fetal Bovine Serum (Hyclone, Logan, UT), 50 units/ml penicillin G, and 50 µg/ml streptomycin; in addition to this 16N and 21NT cells were supplemented with 12.5 ng/ml Epidermal Growth Factor (Sigma, St. Louis, MO), 1 µg/ml insulin (Sigma), and 1 µg/ml hydrocortisone (Sigma), while HeLa T-REx and 293 T-REx cells were supplemented with 5 µg/ml blasticidin. Cells were cultured at 37°C in a humidified chamber in the presence of 5% CO2, and all media, supplements, and antibiotics were from Invitrogen unless otherwise indicated. S-phase arrest was achieved using a 24 hr treatment with 4 mM hydroxyurea (HU; Sigma) as previously described (Fisk et al. 2003; Fisk and Winey 2001; Kasbek et al. 2007). For the generation of stable cell lines, 293 T-REx or HeLa T-REx cells were transfected with the Tet-regulated expression plasmids pHF168 or pHF169, and pH173 or pHF174, respectively (see below). At 48 hours after transfection G418 was added to 400 µg/ml. Cells were fed every three days, and G418 resistant clones were isolated after two weeks using cloning discs soaked in trypsin. Stable cell lines were maintained in the presence of blasticidin and G418, and expression of native or GFP-tagged Mps1 or Mps1Δ12/13 was induced by the addition of 1 µg/ml Doxycycline (Dox).
Plasmids
Previously described expression plasmids used for this study express GFP-tagged constructs from the SV40 promoter in the pECE vector backbone; pHF7 (GFP) (Fisk and Winey 2001); pHF36 (GFP-Mps1) and pHF80 (GFP-Cetn2) (Fisk et al. 2003); pHF60 (GFP-Mps1Δ12/13), pHF87 (GFP-cyclin A1), pHF97 (GFP-Mps1AAA) (Kasbek et al. 2007). Plasmids pHF168 (Tet-Mps1), pHF169 (Tet-Mps1Δ12/13), pHF173 (Tet-GFP-Mps1), pHF174 (Tet-GFP-Mps1Δ12/13) that express native or GFP-tagged Mps1 or Mps1Δ12/13 from the Tet operator were created using the Gateway® recombination system (Invitrogen); PCR products were cloned into pENTR/D/SD-TOPO, and open reading frames were transferred from the resulting entry clones into the pT-REx-DEST30 vector using LR Clonase. Primer sequences are available upon request. Note that while the resulting vectors require Tet for their expression in cells that express the native Tet Repressor, e.g. 293 T-REx or HeLa T-REx, they are constitutively expressed from the CMV promoter in cells that lack the native Tet Repressor. All transfections were performed using Effectine (Qiagen, Valencia CA). For transient expression experiments, cells were analyzed at 24 or 48 hours after transfection.
Cytology
Antibodies for Indirect immunofluorescence (IIF) were GTU-88 mouse anti-γ-Tubulin (Sigma) and rabbit anti-Ninein (Abcam, Cambridge, MA). Secondary antibodies for IIF were Alexa 488- and Alexa 594-conjugated donkey anti-rabbit and donkey anti-mouse (Molecular Probes, Eugene OR), and DNA was stained with Hoechst 33342 (Sigma). IIF was performed as described previously (Fisk et al. 2003; Fisk and Winey 2001; Kasbek et al. 2007). All images were acquired at ambient temperature using an Olympus IX-81 microscope, with a 60x Plan Apo oil immersion objective (1.4 NA) and a QCAM Retiga Exi FAST 1394 camera, and analyzed using the Slidebook software package (Intelligent Imaging Innovations, Denver CO). For centrosome re-duplication assays, cells were arrested in S-phase with a 24 hr HU treatment, medium was replaced with medium containing fresh HU, and centrosome number was determined after an additional 48 hr of S-phase arrest using γ-Tubulin staining as previously described (Fisk et al. 2003; Fisk and Winey 2001; Kasbek et al. 2007). Values in bar graphs represent the mean ± standard deviation of duplicate samples from three independent experiments. Between 50 and 100 cells were counted for each duplicate sample. For all experiments centrosome number was verified using an antibody against centrin to count centrioles.
Centrosomal accumulation of GFP-Mps1 and GFP-Mps1AAA was analyzed as previously described (Kasbek et al. 2007). Briefly, transfected cells were arrested in S-phase with a 24 hr HU treatment, and stained with Hoechst and an antibody against γ-Tubulin. Multi-plane images were captured and projected along the Z-axis after No-Neighbors deconvolution, and the fluorescence intensity of the GFP and γ-Tubulin signals along a line drawn through the center of the two centrosomes was determined using Slidebook Software. As previously described (Kasbek et al. 2007), the analysis was limited to cells with two centrosomes, and centrosomal localization was judged according to the following criteria: the maximum GFP signal must fall within two pixels of one of the two γ-Tubulin peaks, and all GFP maxima within 80% of the maximum GFP signal must fall within the boundary of one of the two γ-Tubulin peaks (boundaries defined by γ-Tubulin signal in excess of 90% of the peak value at either centrosome).
Immunoblot Analysis
Antibodies for Immunoblot analysis were C-19 rabbit anti-hMps1 (SCB540; Santa Cruz Biotechnology, Santa Cruz CA), N1 mouse anti-Mps1 (Invitrogen), mouse anti-GFP (Invitrogen), mouse anti-Rb (BD Biosciences Pharmingen, San Jose, CA), and DM1A mouse anti α-Tubulin (Sigma). The MDS rabbit antibody against Mps1 amino acids 420–507 was generated by injection of a fusion of this region of Mps1 to Glutathione-S-Transferase into rabbits (Lampire Biological Laboratories, Pipersville, PA), and purification of the resulting serum against a similar fusion to the Maltose Binding Protein. Secondary antibodies were Alexa 680-conjugated donkey anti-mouse (Molecular Probes) and IRDye800-conjugated donkey anti-rabbit (Rockland, Gilbertsville, PA). For analysis of Mps1 levels, cells were arrested in S-phase with a 24 hr HU treatment, incubated for an additional 24 hr in the presence of HU and 50µM roscovitine (EMD Biosciences CalBiochem, Gibstown, NJ) or the equivalent amount of DMSO, then harvested and analyzed by dual-color quantitative immunoblot analysis on the Odyssey imaging system (Li-Cor, Lincoln NE) as previously described (Kasbek et al. 2007).
RNA isolation and RT-PCR
Total RNA was isolated from U2OS cells using the RNeasy Mini Kit (Qiagen, Valencia, CA). Full-length cDNA was generated using the First Choice® RLM-RACE kit (Ambion, Foster City, CA). Briefly, using kit components total RNA was dephosphorylated, mRNAs were decapped, and a 5’ RACE adaptor was ligated to the 5’ phosphates now present only on mRNAs. First strand cDNA was synthesized using MMLV Reverse Transcriptase (RT) and a gene-specific primer (binding site 48 nt 3’ of the Mps1 stop codon). This cDNA was then amplified by nested PCR; an outer PCR reaction consisting of the RT primer and a primer complementary to the 5’-RACE adaptor, followed by several inner PCR reactions consisting of various Mps1-specific primers. The binding sites for the inner PCR primers are as follows; forward primers, 1, nt 997–1017; 2, nt 1115–1135; 3; nt 1243–1264; reverse primers, a, nt 1607-1587; b, nt 1698-1678. Primer sequences are available upon request. Mps1 RT and inner PCR primers are diagramed in Figure 5.
Acknowledgments
We would like to thank Thomas G. Paulson (Fred Hutchinson Cancer Research Center, Seattle, WA) and Adlina Mohd-Yusof (The Ohio State University) for critical reading of this manuscript. This work was supported by an NIH grant (GM77311) to H.A. F., and by seed grants from the Ohio State University Comprehensive Cancer Center Institutional ACS fund, the ACS, Ohio Division, and The Ohio Cancer Research Associates to H. A. F.
References
- Abrieu A, Magnaghi-Jaulin L, Kahana JA, Peter M, Castro A, Vigneron S, Lorca T, Cleveland DW, Labbe JC. Mps1 is a kinetochore-associated kinase essential for the vertebrate mitotic checkpoint. Cell. 2001;106(1):83–93. doi: 10.1016/s0092-8674(01)00410-x. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120(Pt 13):2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- Band V, Sager R. Tumor Progression in Breast Cancer. In: Rhim JS, Dritschilo A, editors. Neoplastic Transformation in Human Cell Systems in vitro. New Jersey: Humana Press; 1991. pp. 169–178. [Google Scholar]
- Band V, Zajchowski D, Swisshelm K, Trask D, Kulesa V, Cohen C, Connolly J, Sager R. Tumor progression in four mammary epithelial cell lines derived from the same patient. Cancer Res. 1990;50(22):7351–7357. [PubMed] [Google Scholar]
- Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Muller-Tidow C, Golub TR, Kawakamia K, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc Natl Acad Sci U S A. 2004;101(17):6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol Cell. 2002;10(3):439–440. doi: 10.1016/s1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- Duensing A, Liu Y, Perdreau SA, Kleylein-Sohn J, Nigg EA, Duensing S. Centriole overduplication through the concurrent formation of multiple daughter centrioles at single maternal templates. Oncogene. 2007;26(43):6280–6288. doi: 10.1038/sj.onc.1210456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellsworth DL, Ellsworth RE, Liebman MN, Hooke JA, Shriver CD. Genomic instability in histologically normal breast tissues: implications for carcinogenesis. Lancet Oncol. 2004a;5(12):753–758. doi: 10.1016/S1470-2045(04)01653-5. [DOI] [PubMed] [Google Scholar]
- Ellsworth DL, Ellsworth RE, Love B, Deyarmin B, Lubert SM, Mittal V, Shriver CD. Genomic patterns of allelic imbalance in disease free tissue adjacent to primary breast carcinomas. Breast Cancer Res Treat. 2004b;88(2):131–139. doi: 10.1007/s10549-004-1424-7. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Centrosomes and tumour suppressors. Curr Opin Cell Biol. 2002;14(6):700–705. doi: 10.1016/s0955-0674(02)00385-x. [DOI] [PubMed] [Google Scholar]
- Fisk HA, Mattison CP, Winey M. Human Mps1 protein kinase is required for centrosome duplication and normal mitotic progression. Proc Natl Acad Sci U S A. 2003;100(25):14875–14880. doi: 10.1073/pnas.2434156100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisk HA, Winey M. The mouse mps1p-like kinase regulates centrosome duplication. Cell. 2001;106(1):95–104. doi: 10.1016/s0092-8674(01)00411-1. [DOI] [PubMed] [Google Scholar]
- Ford HL, Landesman-Bollag E, Dacwag CS, Stukenberg PT, Pardee AB, Seldin DC. Cell cycle-regulated phosphorylation of the human SIX1 homeodomain protein. J Biol Chem. 2000;275(29):22245–22254. doi: 10.1074/jbc.M002446200. [DOI] [PubMed] [Google Scholar]
- Fukasawa K. Oncogenes and tumour suppressors take on centrosomes. Nat Rev Cancer. 2007;7(12):911–924. doi: 10.1038/nrc2249. [DOI] [PubMed] [Google Scholar]
- Guarguaglini G, Duncan PI, Stierhof YD, Holmstrom T, Duensing S, Nigg EA. The forkhead-associated domain protein Cep170 interacts with Polo-like kinase 1 and serves as a marker for mature centrioles. Mol Biol Cell. 2005;16(3):1095–1107. doi: 10.1091/mbc.E04-10-0939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick K, Weiss E, Luca FC, Winey M, Murray A. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption. Science. 1996;273:953–956. doi: 10.1126/science.273.5277.953. [DOI] [PubMed] [Google Scholar]
- Kanai M, Ma Z, Izumi H, Kim SH, Mattison CP, Winey M, Fukasawa K. Physical and functional interaction between mortalin and Mps1 kinase. Genes Cells. 2007;12(6):797–810. doi: 10.1111/j.1365-2443.2007.01091.x. [DOI] [PubMed] [Google Scholar]
- Kasbek C, Yang CH, Yusof AM, Chapman HM, Winey M, Fisk HA. Preventing the degradation of mps1 at centrosomes is sufficient to cause centrosome reduplication in human cells. Mol Biol Cell. 2007;18(11):4457–4469. doi: 10.1091/mbc.E07-03-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13(2):190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- La Terra S, English CN, Hergert P, McEwen BF, Sluder G, Khodjakov A. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J Cell Biol. 2005;168(5):713–722. doi: 10.1083/jcb.200411126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- Lingle WL, Barrett SL, Negron VC, D'Assoro AB, Boeneman K, Liu W, Whitehead CM, Reynolds C, Salisbury JL. Centrosome amplification drives chromosomal instability in breast tumor development. Proc Natl Acad Sci U S A. 2002;99(4):1978–1983. doi: 10.1073/pnas.032479999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc Natl Acad Sci U S A. 1998;95(6):2950–2955. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. Altered centrosome structure is associated with abnormal mitoses in human breast tumors. Am J Pathol. 1999;155(6):1941–1951. doi: 10.1016/S0002-9440(10)65513-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle WL, Salisbury JL. The role of the centrosome in the development of malignant tumors. Curr Top Dev Biol. 2000;49:313–329. doi: 10.1016/s0070-2153(99)49015-5. [DOI] [PubMed] [Google Scholar]
- Liu ST, Chan GK, Hittle JC, Fujii G, Lees E, Yen TJ. Human MPS1 Kinase Is Required for Mitotic Arrest Induced by the Loss of CENP-E from Kinetochores. Mol Biol Cell. 2003;14(4):1638–1651. doi: 10.1091/mbc.02-05-0074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Vucica Y, Rosenbaum JL. Kinetics and regulation of de novo centriole assembly. Implications for the mechanism of centriole duplication. Curr Biol. 2001;11(5):308–317. doi: 10.1016/s0960-9822(01)00094-x. [DOI] [PubMed] [Google Scholar]
- Milliken EL, Lozada KL, Johnson E, Landis MD, Seachrist DD, Whitten I, Sutton AL, Abdul-Karim FW, Keri RA. Ovarian hyperstimulation induces centrosome amplification and aneuploid mammary tumors independently of alterations in p53 in a transgenic mouse model of breast cancer. Oncogene. 2008;27(12):1759–1766. doi: 10.1038/sj.onc.1210815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2(11):815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17(5):215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- Ou YY, Mack GJ, Zhang M, Rattner JB. CEP110 and ninein are located in a specific domain of the centrosome associated with centrosome maturation. J Cell Sci. 2002;115(Pt 9):1825–1835. doi: 10.1242/jcs.115.9.1825. [DOI] [PubMed] [Google Scholar]
- Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149(2):317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pihan GA, Purohit A, Wallace J, Malhotra R, Liotta L, Doxsey SJ. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 2001;61(5):2212–2219. [PubMed] [Google Scholar]
- Pihan GA, Wallace J, Zhou Y, Doxsey SJ. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 2003;63(6):1398–1404. [PubMed] [Google Scholar]
- Salisbury JL, Whitehead CM, Lingle WL, Barrett SL. Centrosomes and cancer. Biol Cell. 1999;91(6):451–460. [PubMed] [Google Scholar]
- Schutz AR, Winey M. New alleles of the yeast MPS1 gene reveal multiple requirements in spindle pole body duplication. Mol Biol Cell. 1998;9(4):759–774. doi: 10.1091/mbc.9.4.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stucke VM, Sillje HH, Arnaud L, Nigg EA. Human Mps1 kinase is required for the spindle assembly checkpoint but not for centrosome duplication. Embo J. 2002;21(7):1723–1732. doi: 10.1093/emboj/21.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsikitis VL, Chung MA. Biology of ductal carcinoma in situ classification based on biologic potential. Am J Clin Oncol. 2006;29(3):305–310. doi: 10.1097/01.coc.0000198740.33617.2f. [DOI] [PubMed] [Google Scholar]
- Tsou MF, Stearns T. Controlling centrosome number: licenses and blocks. Curr Opin Cell Biol. 2006;18(1):74–78. doi: 10.1016/j.ceb.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Weiss E, Winey M. The Saccharomyces cerevisiae spindle pole body duplication gene MPS1 is part of a mitotic checkpoint. J. Cell Biol. 1996;132(1 & 2):111–123. doi: 10.1083/jcb.132.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittaker SR, Walton MI, Garrett MD, Workman P. The Cyclin-dependent kinase inhibitor CYC202 (R-roscovitine) inhibits retinoblastoma protein phosphorylation, causes loss of Cyclin D1, and activates the mitogen-activated protein kinase pathway. Cancer Res. 2004;64(1):262–272. doi: 10.1158/0008-5472.can-03-0110. [DOI] [PubMed] [Google Scholar]
- Winey M, Goetsch L, Baum P, Byers B. MPS1 and MPS2: Novel yeast genes defining distinct steps of spindle pole body duplication. J. of Cell Biol. 1991;114(4):745–754. doi: 10.1083/jcb.114.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C-H, Kasbek C, Fisk HA. The use of infrared fluorescent dyes in quantitative immunoblotting. In: Walker JM, editor. Protein Protocols Handbook. Third Edition ed. London: Humana Press; 2008. [Google Scholar]
- Yuan B, Xu Y, Woo JH, Wang Y, Bae YK, Yoon DS, Wersto RP, Tully E, Wilsbach K, Gabrielson E. Increased expression of mitotic checkpoint genes in breast cancer cells with chromosomal instability. Clin Cancer Res. 2006;12(2):405–410. doi: 10.1158/1078-0432.CCR-05-0903. [DOI] [PubMed] [Google Scholar]