Abstract
Phenomics is an emerging transdiscipline dedicated to the systematic study of phenotypes on a genome-wide scale. New methods for high-throughput genotyping have changed the priority for biomedical research to phenotyping, but the human phenome is vast and its dimensionality remains unknown. Phenomics research strategies capable of linking genetic variation to public health concerns need to prioritize development of mechanistic frameworks that relate neural systems functioning to human behavior. New approaches to phenotype definition will benefit from crossing neuropsychiatric syndromal boundaries, and defining phenotypic features across multiple levels of expression from proteome to syndrome. The demand for high throughput phenotyping may stimulate a migration from conventional laboratory to web-based assessment of behavior, and this offers the promise of dynamic phenotyping –the iterative refinement of phenotype assays based on prior genotype-phenotype associations. Phenotypes that can be studied across species may provide greatest traction, particularly given rapid development in transgenic modeling. Phenomics research demands vertically integrated research teams, novel analytic strategies and informatics infrastructure to help manage complexity. The Consortium for Neuropsychiatric Phenomics at UCLA has been supported by the NIH Roadmap Initiative to illustrate these principles, and is developing applications that may help investigators assemble, visualize, and ultimately test multi-level phenomics hypotheses. As the transdiscipline of phenomics matures, and work is extended to large-scale international collaborations, there is promise that systematic new knowledgebases will help fulfill the promise of personalized medicine and the rational diagnosis and treatment of neuropsychiatric syndromes.
Keywords: phenotype, genetics, genomics, informatics, cognition, psychiatry
Introduction: Phenomics as an Emergent Transdiscipline
The nominal completion of the human genome project has fostered enormous enthusiasm and carried with it the promise of biomedical breakthroughs and a new era of personalized medicine in which genetic profiles serve as bases for rational diagnosis and treatment. Almost a decade into the post-genomic era, there is an emerging consensus that some of the problems are even harder than originally hoped, and nowhere is this sense of sobriety clearer than in neuropsychiatric research. David Goldstein recently remarked: “There is absolutely no question that for the whole hope of personalized medicine, the news has been just about as bleak as it could be” (Wade, 2008). Others have expressed more optimism (Maher et al., 2008) based on recent reports of positive findings from genome-wide association studies (GWAS) of schizophrenia and bipolar disorder (Ferreira et al., 2008, O’Donovan et al., 2008). But there remain obvious challenges both in identifying relevant genetic variants, and once these variants are identified, determining what roles these variants may play in neuropsychiatric illness.
Freimer and Sabatti (2003) suggested that “The Human Phenome Project” is now an imperative to follow-on and fulfill the promise of the human genome project. Given the continued decreases in cost and increasing availability of high-throughput genotyping platforms, it is today even clearer that phenotyping comprises the key rate- and cost-limiting factor in human genetics. Beyond the time and cost needed for phenotyping, the field faces an even grander challenge: What phenotypes should we be studying? The human “phenome”, which Freimer and Sabatti referred to as the “…manifold human phenotypes from molecule to mind…”, is a big place. The human genome, with only three billion bases, selected from a pool of only four nucleic acids, organized in a neat one-dimensional sequence, pales in comparison to the human phenome, which contains an unknown number of elements, many of which are characterized by enormous inter-individual variation that is at best only partially understood, and for which the dimensionality remains unknown. Phenomics – operationally defined as the systematic study of phenotypes on a genome-wide scale – is critically important to provide traction for biomedical advances in the post-genomic era (Bilder, 2008).
“Systematic study” implies not only the assessment of phenotypes in a well operationalized fashion, but further that the strategy for studying phenotypes is defined in relevant biological contexts, with appreciation that any given study’s capacity to sample the human phenome is limited to a minute fraction of the phenomic state-space (all the possible kinds of phenotypes and their individual variants). “On a genome-wide scale” implies that examination of any specific phenotype involves considering the likely complex genetic contributions to that manifestation from the entire genome, and acknowledges that many of the putative “candidate genes” suggested by prior research may turn out to be false-positive signals.
In practical terms, phenomics demands broad scientific expertise, including genetics, molecular biology, cell biology, systems biology, and higher levels of phenotypic expression, and these experts must be capable of communicating effectively and collaboratively designing and executing translational research projects on a large scale. For neuropsychiatric phenomics, the systems-level experts must include experts with knowledge about neural systems, cognitive systems, and neuropsychiatric symptoms and syndromes. Further expertise in mathematical modeling, statistics, and information sciences is needed to confront the many novel data analytic challenges, and enable the definition, visualization, and testing of complex multi-level hypotheses. Phenomics is therefore best defined as a “transdiscipline”, as it engages contributing disciplines interactively and synthetically to generate a new, emergent discipline.
Neuropsychiatric Phenomics: Reframing Models of Structure-Function Relations
Discerning the genetic contributions to any complex human illness is fraught with challenges and demands broad phenomics scope, but neuropsychiatric genetics confronts unique obstacles. Among the most intriguing of the distinctions involves the mind-brain problem itself. While we might consider the emergent properties of any organ to involve qualitatively similar unknowns, the emergence of complex human behavior from brain function remains the epitome of scientific challenges. For example, while cardiovascular function involves myriad unresolved mysteries, already multi-scale bioengineering models have been developed for the heart at molecular genetic, cellular, tissue, and whole organ levels, enabling prediction of whole organ consequences of genetic variation (Crampin et al., 2004). Similar efforts have helped model pulmonary function, kidney function and more (see for example, the International Union of Physiological Sciences Physiome Project). In contrast, there remain major gaps in explanations of how psychological processes relate to brain function, and this comprises a non-trivial limitation to modeling the biological bases of neuropsychiatric syndromes. Contemporary theorists have written extensively about the likely form of this relation, and increasingly call attention to the emergent properties of mental states that are not simply reducible to the interactions of more elementary component processes (Tononi and Edelman, 1998, Edelman, 2003, Seth et al., 2006). Kendler has written eloquently about the need for iterative evolution of our concepts about structure-function relations, and highlights that our desideratum is more likely an implementation rather than a reductionist replacement solution (Kendler, 2008). While these theoretical models are appealing, mechanistic models ultimately will require more detail about exactly what emerges from what, and will likely require some unexpected reframing of current concepts about psychological functions, to effectively advance phenomics research.
It is further critical to recognize that previous methods for carving brain function “at its joints” are likely to have generated divisions that align poorly with those that can be genetically determined. Much of what we have learned about brain-behavior relations comes from models that are of questionable value to understanding the genetic roots of brain function and dysfunction. To take an extreme example, the discipline of clinical neuropsychology has learned much from the study of individuals with brain injury, stroke, or neoplasm, but it is obvious that genetic variation is extremely unlikely to have similar effects on brain function. Thus the conventional catalogs of neuropsychological “domains” (i.e., language, memory, executive functions and so on) should not be expected to align well with the cognitive functions affected by genetic variation.
A more subtle point is that there is not a compelling reason to expect that decomposition of behavior, based on analyses within the behavioral level alone, will provide a useful target for genetic analysis. It might be hoped, for example, that the “normal” factorial structure of cognitive abilities revealed through decades of cognitive testing might be reflected in genetic variants. We can certainly seek genetic correlates of such constructs as “intelligence” or “memory” (see Sabb et al., this issue), and since most of the constructs have reasonably high heritability, it is clear that these are related to genetic variation (or epigenetic variation). It remains unclear to what degree any of the relations identified will be specific. The “generalist gene hypothesis” is based on findings that a single set of genes affects most cognitive abilities. Indeed the genetic correlations (i.e., the extent to which genetic effects on one trait are correlated with genetic effects on another trait) are approximately .80 among diverse verbal, spatial, memory, mathematical and reading abilities (Butcher et al., 2006, Kovas and Plomin, 2006). But how many genes must be involved to explain the heritability of these traits, which is generally estimated to be approximately 40%. Initial work using multi-stage genome-wide scanning revealed four candidate single nucleotide polymorphisms (SNPs), each of which explained only ~0.2% of phenotypic variance, and together these four SNPs accounted for only ~0.8% of variance in the cognitive phenotypes (Butcher et al., 2005). Given that these specific analyses compared extreme groups (those high and low in mental ability, thus perhaps not reflecting fully the quantitative trait distribution), and used what are now outmoded genotyping methods (they used DNA pooling and only examined 10,000 SNP’s), these estimates of shared variance might be considered pessimistic. But so far there is little evidence from more recent GWAS of substantially more robust genetic associations with similar phenotypes. This might suggest that at least several hundred genes will ultimately be associated with cognitive ability in general, and that the majority of these will account for less than 1% of phenotypic variance.
Similar high genetic correlations may be found for a range of other neuropsychiatric syndromes (Kendler et al., 2003), and help to explain the high “co-morbidity” of these syndromes. The challenge this poses for gene discovery at the syndromal level may be further exacerbated by the application of a taxonomic classification system that has reified invalid distinctions. It is already recognized that few of the diagnostic categories in the DSM-IV represent valid classes even when examined in terms of the symptoms used to define the syndromes (Haslam and Kim, 2002, Haslam, 2003), and it is hoped that the DSM-V may include indices of the continuous trait dimensions that will better reflect symptomatic variation (Kraemer, 2007). Even if some syndromal classes are verified to represent true “taxa” or “latent classes” based on the covariance structure of the symptoms used to define the syndromes, this is no guarantee that these syndromes will have a clearer genetic basis. For example, there is evidence that a “melancholic” subtype of depression may represent a valid taxon distinct from other forms of depression and from healthy groups (Haslam and Beck, 1994, Ambrosini et al., 2002), yet more detailed epidemiological data offer less support for the distinctiveness of this subgroup (Kessing, 2007), and assessment of familial risk reveals a pattern more consistent with a quantitative trait than genetically distinctive subgroup (Kendler, 1997). Thus empirical evidence suggesting discrete syndromal classes may be helpful, but is not sufficient to assert a clearer genetic basis.
The high genetic correlations for behavioral phenotypes may be an expected consequence of attempting to identify relations across biological scales that involve emergent functional consequences. Given that the level of gene effects must be translated through gene expression of proteins, to the functional roles played by those proteins in cellular systems and signaling pathways, to the functioning of those cells and signaling pathways in integrated neural circuits, and then ultimately make the brain-to-behavior traversal, there is no shortage of opportunities for specific effects to be obscured. Fisher has commented eloquently that “…the ability to undertake genetic analyses while employing only the most basic abstract concept of ‘the gene’, and without any understanding of molecular pathways, has become both a blessing and a curse, particularly in studies of the brain” (Fisher, 2006). The phenomics strategy explicitly calls for efforts to redefine phenotypes as multi-level combinations of measures that may offer more realistic constraints on the mechanistic paths leading from genome to syndrome.
Phenotype Prioritization for Phenomics Research
The phenomics strategy can be seen as an extension of the endophenotype approach that embraces multi-level modeling. This strategy acknowledges that many of the putative endophenotypes or intermediate phenotypes being investigated in biological psychiatry today may not possess much simpler genetic architecture than do the highest level syndromal phenotypes (Flint and Munafo, 2007). While this strategy does not replace many elements of previously suggested qualities important for phenotype prioritization (Gottesman and Gould, 2003, Bearden and Freimer, 2006), it shifts the emphases (see Table 1).
Table 1.
Considerations for Phenotype Selection in Phenomics Research
| Genetic Validity |
| a) moderately high heritability; association with related disorders in family members |
| b) relate to known common functional genetic variation |
| Measurement Properties |
| a) reliability (nomothetic validity, internal consistency); test-retest reliability (at least within a particular state, and preferably across states in illnesses which have an episodic pattern); and other desirable psychometric properties (e.g., discriminating power across a broad range of individual differences) |
| b) have good concurrent validity (convergent and divergent validity) with respect to hypothesized endophenotypes |
| Relevance for Human Applications |
| a) related to higher level constructs in humans, enabling links to clinical effectiveness and outcomes |
| b) suitable for application in clinical trials |
| c) associated with neuropsychiatric morbidity |
| d) cross-disorders relevance – the phenotype construct has been identified as important in more than one diagnostic syndrome |
| Relevance for Translational Investigation |
| a) homologies of expression across species enabling both basic and clinical investigation |
| b) latent endophenotype constructs relate to neural systems models sufficiently well studied that physiologically plausible manipulations can be effected and measured using currently available techniques |
| c) implicated neural systems already are targets of known partially effective treatments for existing disorders, or these systems are the targets of new chemical entities |
| Knowledge of Environmental Effects |
| a) known environmental effects on phenotype expression can usefully constrain modeling of phenotypic variance explained by genotypic variance, and may be particularly valuable in model organisms where environments can be strictly controlled (epigenetic mechanisms are seen as an area of rapid expansion in knowledge and systems in which these can be studied effectively may be prioritized |
For example, heritability remains important, but the suggestion that a valuable endophenotype should associate with illness and show familial cosegregation with illness may be less critical, depending on how “illness” is defined. The critical point is that the most valuable phenotypes for phenomics research may well be those that cross boundaries of the current diagnostic taxonomy. Indeed, if it is true that substantial genetic correlations exist for neuropsychiatric syndromes, it may be most fruitful to study phenotypes that are shared across multiple diagnostic syndromes, and that by limiting research to more narrowly defined diagnostic groups, the genetic signals from the strongest genetic contributions may be lost amidst other less important diagnosis-specific “noise.” It is further unclear how helpful conventional heritability statistics are to prioritizing phenotypes, given that many well defined behavioral traits possess heritabilities exceeding 40% (Sabb et al., 2008), and higher heritability does not seem to assure a simpler genetic architecture. Since the principal value of identifying high heritability is to assure that the phenotypic trait is likely to be meaningfully related to genetic variation (i.e., that it possesses some evidence of genetic validity), it may also be valuable in phenotype selection to have evidence that the phenotype is significantly associated with known genetic variants. Further, to leverage the power of GWAS using quantitative trait loci, it is helpful if the genetic variant is common in human populations (although it should be recognized that with the advent of ever-increased capacity for genotyping, it may soon be possible for phenotypes associated with rare variants to be detected). It is also helpful if the genetic variant is associated with known functional effects at translational or transcriptional levels, in order to foster greater traction in molecular biology research.
Among the desiderata for phenotype selection, it is difficult to overemphasize the importance of fundamental measurement properties. Given that the upper limit on validity will be imposed by reliability, it is critical for research to focus on those measures that show adequate internal consistency (helping assure that a meaningfully coherent construct is being measured in the first place). If that is true, it is further important to identify those that are relatively stable over time and organismic states, or if there is some fluctuation that this fluctuation itself is a part of the phenotypic assay. The importance of phenotypic stability has been emphasized by others; in brief it is logical to propose that our fixed genetic inheritance will be most easily associated with features that are stable. But there also may be important phenotypic features that have instability as their signature, in which case it is precisely this variability that needs to be assessed. Finally, it is valuable to assure that the sensitivity of phenotype measurement is strong across widely varying levels of phenotype expression. For example, a cognitive phenotype is likely best measured by a test that shows strong measurement properties at both higher and lower levels of that ability, and if the measurement is biased towards either higher or lower levels of ability, then meaningful genetic effects may never be detected not because these do not exist but rather because the test is too insensitive.
A critical aspect of understanding the measurement properties of specific phenotypes is determining how amenable these are to high throughput phenotyping. Given the changing finances of genomics research, it has become clear that the rate-limiting step in advancing knowledge about human disease has shifted dramatically from genetics to phenomics. Phenotyping now far exceeds the time and financial costs of genotyping. Until “high throughput phenotyping” methods are developed, accrual of knowledge in phenomics will be thwarted. Development of detailed costing models is advocated to identify the most cost effective methods for addressing key phenomic targets. High throughput collection of behavioral data is virtually impossible using traditional laboratory based methods. For example, a “comprehensive” examination of cognitive abilities may require ten to twenty hours, and briefer assessments routinely fail to provide broad phenotypic coverage, or fail to specify adequately the phenotypes of interest, or both.
There are two major hopes in advancing high throughput phenotyping. One hope is based on leveraging advances in modern psychometric theory. Using item response theory and computerized adaptive testing algorithms, it is now routinely possible to increase efficiency in psychological assessment by a factor of two. Thus one can either specify the construct with double the precision, or decrease testing time for the same measurement precision in half the time of the original test. There are multiple challenges in using these methods, including: (a) the construct itself needs to be well operationalized in advance; and (b) relatively large samples (i.e., 500 or more with complete response sets) are suggested to apply these methods. A second hope is based on the incredible growth in use of the internet, with more than 100 million people in the United States using the web daily. If we can provide well structured phenotyping tools, enabling widespread access for individuals to provide meaningful data about themselves, the results could rapidly revolutionize behavioral genomics. A paradigm shift that could accelerate discovery would involve dynamic phenotyping, by which we mean the iterative refinement of phenotype assays based on prior genotype-phenotype associations. There remains considerable skepticism among scientists about both sampling biases and validity of data collected using web-based rather than conventional laboratory-based methods, despite demonstrations to the contrary (Krantz et al., 1997, Buchanan and Smith, 1999, Krantz and Dalal, 2000, Andersson et al., 2006, Chiasson et al., 2006, Cunningham et al., 2006, Graham et al., 2006). While much work remains to address these concerns in a compelling manner, the potential increases in throughput may dramatically outweigh, and even provide solutions to the sampling and quality control issues. Given that conventional laboratory studies running for 5 years often have difficulty ascertaining and examining a few thousand individuals, the opportunities for sub-sampling and data cleaning with several hundred thousand individuals may appear increasingly attractive, particularly as we increasingly recognize the value of samples with tens of thousands of individuals.
There are some features of phenotypes that may be less important to advance genetics research, but may be prioritized because of their relevance to other human applications. Biomedical research ultimately seeks to alleviate human suffering, and thus those phenotypes that can be meaningfully related to clinical morbidity, or the clinical effectiveness and outcomes of treatments, may be important targets for study. Those phenotypes that are suitable for application in clinical trials may be particularly valuable given the possible opportunity to link phenotypic variation to putative effects at the levels of cellular systems or signaling pathways. Consistent with and extending the points made earlier about the value of cross-disorder phenotypes, those phenotypes that are relevant across multiple categories of neuropsychiatric illness may have even broader public health significance than those that do not. For example, finding the genetic bases of “normal” variation in memory or attentional control may be of greater value than identifying genetic associations with Alzheimer’s disease or ADHD.
The phenomics strategy also prioritizes phenotypes that are relevant for translational investigation, and thus those phenotypes that cross species boundaries. This is not intended to discount the importance of uniquely human phenotypes, and focusing on these is an alternate approach with great potential but different goals. For example, it may be possible to gain traction on the genetic bases of language by examining differences in gene expression between human and chimpanzee (Oldham and Geschwind, 2006). But the phenomics approach suggests that the “low hanging fruit” in neuropsychiatric genetics may be picked most readily by focusing on phenotypes that are generally well conserved phylogenetically. The primary rationale for this is that by examining phylogenetically conserved phenotypes, we increase the opportunity to gain traction on intervening biology using basic science models.
Of particular value today are transgenic models that can help illuminate the cellular systems and signaling pathways affected by genetic variants, and therefore a priority may be given to phenotypes that can be studied in both mouse and human. For phenotypes that have reasonable homologies from mouse to man, there is a high likelihood that there will be even more robust homologies in larger rodents and non-human primates, enabling more detailed analysis of higher level neuropsychopharmacological effects and neural systems level phenotypes.
Linked to prioritization of cross-species phenotypes is the idea that phenotypes selected for study may be most useful if embedded in plausible mechanistic hypotheses. The better fleshed out the biological mechanisms, the higher the likelihood that meaningful connections will be established with other biological knowledge. Thus, other factors being equal, a phenotype for which there exists already a set of mechanistic models may be preferable to one that does not possess similar evidence. While all such mechanistic models are so far incomplete, working within a framework that includes relevant empirical science may help link findings to existing evidence and either further develop more effective models, or help prune these and better specify superior models for the future.
A critical point to consider in any phenotype prioritization effort is that examining “criteria” does not enable generation of some figure-of-merit for phenotype selection. In the CNP, we had started with the aim of generating a phenotype selection algorithm, and on more careful consideration recognized that at best it might be possible to generate a phenotype “profiling tool” capable of characterizing the strengths and weaknesses of a given phenotype. Some of the features noted above are likely to conflict directly with others (for example, validation with respect to molecular targets may run counter to validation with respect to outcomes).
There is further a substantial risk of phenotype reification that can actively interfere with discovery. For example, a phenotype prioritized because it is influenced reliably by a known molecular entity (a drug) might be seen as valuable by virtue of links to signaling pathways in a mechanistic model, but this might be misleading if the mechanistic model is not well understood. A case in point is the phenotype involving induction of catalepsy in rodents, used for decades as a screening test for antipsychotic drug development. This may have canalized drug discovery towards agents more likely to produce extrapyramidal symptoms than antipsychotic efficacy.
Finally, the phenomics strategy emphasizes that phenotype selection should weigh cautiously evidence from candidate gene strategies. There may be value in research that uses a ‘bottom-up’ approach, starting with functional genetic variation, through identification of proteins affected, through identification of signaling and other cellular processes affected, through neural systems function to behavior. But there should also be appropriate concern that this approach can undermine discovery of novel gene-phene correlations. It may be an impediment to discovery to be tied to known genomic ‘hot spots’, many of which were identified using phenotypes that are acknowledged to be suboptimal, and also may be subject to various other methodological problems (including population stratification and linkage disequilibrium) that may have led to false positive identification of regions that are distant from the driving functional genomic regions of greater interest.
Managing Complexity in the Human Phenome
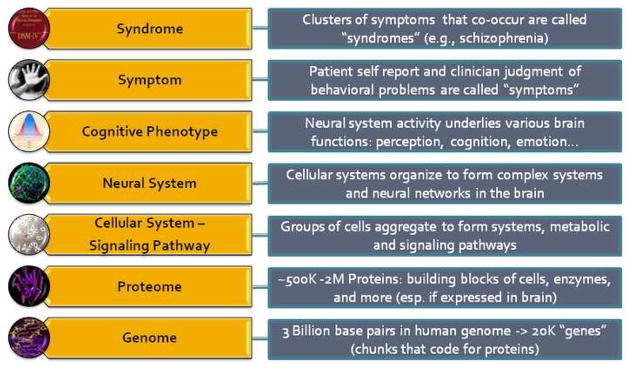
To facilitate this work, we have adopted a simplified seven-layer schema to reflect some of the key traversals across levels of inquiry and biological scales that are important in the complete representation of a phenomics hypothesis from genome to syndrome (Figure 1). While experts representing a specific disciplinary perspective may argue coherently about the validity of these “levels”, we have found this helpful to foster transdisciplinary communication. The bottom few layers reflect basic biological “dogma” that is generic to all phenomics sub-disciplines (i.e., that genes “code” for proteins, that proteins exert their biological effects via incorporation in cellular systems and signaling pathways). Above this level, the schema is customized for neuropsychiatric research, by focusing on the assembly of cellular systems and signaling systems into neural systems, suggesting that activity within neural systems leads to the emergence of cognitive functions (which we define broadly to encompass a complete range of mental faculties and operations, including mood, affective, mnestic, linguistic, and thinking abilities), and that these cognitive phenotypes form the basis of clinically observable symptoms, which in turn are used to define neuropsychiatric syndromes.
Figure 1.
Seven-level schematic illustrating relevant components of mechanistic “genome to syndrome” hypotheses for phenomics research
Given this simple framework, some crude calculations may offer perspective on the volume of the phenomics search space. Acknowledging that this schema is oversimplified reinforces the assertion that resulting computations are probably underestimates. But assuming only seven transformations across levels are needed to generate a plausible genome-to-syndrome hypothesis, we can estimate the impact of both pleiotropy and polygenicity on phenomics research programs. Consider the case of a modest five-fold expansion across levels (i.e., a given gene influences 5 proteins, a given protein influences 5 cellular systems and signaling pathways, and so on). Ascending through seven levels would generate 15,625 effects at the “syndromal” level. A ten-fold expansion at each level would generate more than a million effects after promulgation across seven levels. Similarly, if we imagine the spectrum of plausible polygenic contributions to high level phenotypic traits (i.e., that a syndrome is identified by multiple symptoms, that each symptom reflects variation in multiple cognitive phenotypes, which are in turn the product of action across multiple neural systems, and so on), it is easy to imagine that hundreds if not thousands of genetic variations contribute to complex neuropsychiatric syndromal phenotypes.
Comparable complexity is revealed by posing the question slightly differently. Imagine our goal is to determine how much variance is shared between observations at each level of analysis. Assume further that the amount of variance shared is constant across levels. For a single gene to account for 25% of variance in a complex syndrome would require that a feature on each level must share at least 80% variance with a feature on the next level. This scenario is likely unrealistic, given existing data showing that this exceeds the reliability of virtually all of the higher phenotypic features, and that genotype may explain only ~20% of variance at the level of the transcript (Flint and Munafo, 2007). Still optimistic but possibly realistic is identifying between-level associations equivalent to 50% shared variance. Through seven levels, this would result in 1.6% shared variance between a specific genotype and syndromal phenotype (i.e., among the strongest results so far obtained in GWAS research). Best supported by data are relations across levels that show shared variance of approximately 20% (consistent both with the Flint and Munafo review, and further with typical correlations among various phenotypes in neuropsychiatric research in the range of .4 to .5). In this scenario, a genetic variant would share only .01% with a phenotypic variant, or in other words, some 5000 genetic variants would be needed to explain the overall heritability of approximately 50% that have been identified through family-based studies of high level personality traits and neuropsychiatric diagnostic phenotypes.
There are so far insufficient empirical data to draw firm conclusions on the actual complexity we face in neuropsychiatric genetics, but if the results of recent GWAS in both neuropsychiatry and other complex disorders are a useful guide, it appears that even the strongest signals being detected are explaining less than 1% of phenotypic variance, and thus small contributions from many genetic variants appears more the rule than the exception. As noted above, the findings of Plomin and his group that 4 genetic variants together explained only 0.8% variance in intelligence, suggests that at least hundreds of genes are likely involved to explain heritability of 40%. Such estimates ignore the possibility that epigenetic factors account for some currently unknown proportion of this variance.
The promise of phenomics rests in part on the concept that mechanistic biological hypotheses linking genome to syndrome can help set constraints on these massive webs of associations. There is hope that elements of convergence, following self-organizing principles that have been discerned in other disciplines as diverse as chemistry and economics may be applicable also to the emergence of multiple aspects of gene expression and higher level phenotypes (Kauffman, 1993, Nykter et al., 2008). Considerable work already has advanced the idea that the emergent properties of cognition from neural systems activity may depend on self-organizing neural networks (Tononi et al., 1994, 1999, Sporns et al., 2005). Similar progress is being made applying information science and network principles to the study of gene networks and other biological systems (Sridhar et al., 2007, Zheng et al., 2007, Centler et al., 2008).
A major question remains: at what level of a multi-level mechanistic network is it most fruitful to attempt identifying such phenotypic convergence? Is it most fruitful to focus on the transcriptome, the proteome, the signalome, or elsewhere?
Narrowing the Search Space for Phenomics Research: Examples from the CNP
Within the Consortium for Neuropsychiatric Phenomics (CNP), which is one of nine Interdisciplinary Research Consortia supported by the NIH Roadmap Initiative starting in 2008 (http://nihroadmap.nih.gov/interdisciplinary/index.asp), we have prioritized investigation of two cognitive phenotypes for translational research. We refer to these phenotypes as response inhibition mechanisms, and memory mechanisms. Phenotype selection benefited from considering the criteria outlined above, together with input from two teams that considered catalogs of phenotypes worthy of investigation both across psychiatric disorders (cross-disorders workgroup) and across different species (cross-species workgroup). In the course of considering a diversity of possible phenotypic targets, one important question emerged: At what level of phenotypic expression would it be best to focus attention?
The need to narrow focus in phenomics research is prompted by the sheer magnitude of possible associations as summarized above. A key tenet of the phenomics strategy is to maximize agnosticism in recognition of the many unknowns we face (including the possibility that many “candidate genes” reflect false positive findings, and that many phenotypes are ill-defined). But given extensive pleiotropy together with the likelihood that neuropsychiatric traits are polygenic – perhaps massively polygenic – it is clearly implausible to conduct research that will provide an unbiased sample of the human phenome (i.e., phenome-wide association).
We determined that for the purposes of neuropsychiatric phenomics (i.e., establishing a research program dedicated to linking genomic to neuropsychiatric syndromal levels of description), a focus on cognitive phenotypes would be most fruitful. The key rationale for focusing on this level of analysis is that cognitive phenotypes may be meaningfully related to higher level symptoms and syndromes on the one hand, and to underlying neural systems activity on the other hand. In this way, cognitive phenotypes were seen as offering a potential mediating link from the study of behavioral phenotypes to their brain bases, and interposing a bottleneck to facilitate transdisciplinary research capable of marrying clinical to basic research in neuropsychiatry. Many cognitive phenotypes further manifest many of the desirable properties alluded to previously, including: (a) genetic validity (with heritability of most well-defined cognitive phenotypes approximating 50%; (b) reasonable measurement properties (with internal consistency indices approximating .90 and test-retest reliability of many measures approximating .80); (c) relevance for translational research (since for at least some cognitive phenotypes, there exist non-human homologs or analogs).
Among the critical linkages, cognitive phenotypes may offer valuable tools for bridging from human research to basic research via the neural systems level. Current neuroimaging methods particularly have enabled interrogation of neural systems activation effects associated with cognitive manipulations, thus forging a relational link that bridges observable behavior to underlying biology. Some imaging methods even permit insight into cellular and signaling correlates of cognitive phenomena. The capacity to elicit neural systems responses to cognitive probes in humans can be actively complemented by the study of similar cognitive processes in lower-level biological systems in non-human species. Thus we conceptualize the vertical research strategy as operating both top-down in humans, from the level of syndromes through symptoms and cognitive phenotypes to neural systems (complemented by GWAS), and bottom-up in mice, given the facility with which we can create new transgenic models and determine the effects of the genetic manipulations on molecular expression, cellular systems and signaling pathways, and neural systems.
The CNP’s selection of response inhibition and memory mechanisms as focal phenotypes was based on the criteria noted above, and our capacity to examine these in integrated top-down and bottom-up research, given the specific areas of expertise possessed by members of an extensive group of collaborators. The research team of the Consortium for Neuropsychiatric Phenomics includes more than 50 investigators representing dozens of disciplines including genetics, statistical genetics, molecular biology, neurobiology, systems neuroscience, cognitive neuroscience, neuropsychopharmacology, clinical neurosciences (including psychiatry, neurology, psychology, and neuropsychology), public health, statistics, psychometrics, and computer science. We have established research teams to address both the response inhibition and memory mechanisms themes using the top down approaches (human GWAS studies with approximately 10 hours of cognitive phenotyping per individual, and additional neuroimaging investigations in a subgroup). These are complemented by bottom-up studies in which we are creating transgenic models based on candidate genes of putative relevance to these themes. For example, one set of studies in the response inhibition theme focuses on a bacterial artificial chromosome transgenic model that permits cell-specific alteration in expression of the G protein-coupled receptor 6 gene Gpr6 within medium spiny neurons in the striatum. These studies are following up on exciting findings that Gpr6 −/− mice show increased initiation of responses under instrumental conditioning schedules, but decreased capacity to withhold the same responses under leaner schedules (Lobo et al., 2007). In the memory mechanisms theme, ongoing work is following up on findings examining transgenic manipulations of Disc1 and Dysbindin, which have shown a range of cognitive, neurophysiological, and neuroanatomic deficits paralleling those observed in patients with neuropsychiatric illnesses such as schizophrenia and bipolar disorder (Cannon et al., 2005, Li et al., 2007).
Another unique feature of this work, dictated by the phenomics strategy, is that the GWAS is principally targeting a group of community volunteers rather than specific diagnostic groups. This approach is based on the idea that quantitative traits representing these phenotypes are likely to be most powerfully identified in broad population samples, rather than in groups selected for diagnostic phenotypes, which are biased towards the extremes of the relevant phenotype distributions. To the extent that individuals with neuropsychiatric disorders represent extreme values on these quantitative traits, the relevant phenotyping assays will show restricted range and thus reduced power to detect associations. The CNP is therefore focusing its GWAS assessments of response inhibition and memory phenotypes on 2000 community individuals (recruited from the Los Angeles metropolitan region, and thus referred to as the “LA2K” study), and will also study smaller samples of 100 individuals in each of three diagnostic groups (including people with diagnoses of Schizophrenia, Bipolar I Disorder, and Attention Deficit/Hyperactivity Disorder or ADHD). The 300 patients are included primarily to characterize their performance on response inhibition and memory phenotype assays, rather than to help identify novel genetic associations. If genetic associations with the cognitive phenotypes are identified in the LA2K sample, we can then determine the extent to which these genetic variants occur in the patient groups and if these appear useful in explaining deviant scores. This strategy is based on the premise that the genetic bases for quantitative traits such as memory and response inhibition are likely to show strong overlap between individuals who have psychiatric diagnoses and those who do not. If this is true, findings of genetic association in the non-diagnosed group will be informative for the diagnosed group. If this premise is wrong, and the phenotypic deviation shown among diagnosed individuals is due to “syndrome-specific” genetic factors, then the information derived from the LA2K will be a helpful start on the path to determining the unique genetic sources of these effects.
Data Analytic Strategies for Phenomics Research
The sheer scope of phenomics research in the GWAS era raises data analytic challenges on an unprecedented scale. It has become difficult even to represent, much less comprehend the hypotheses that we are beginning to interrogate. This complexity places a new burden on the field to develop data mining and informatics strategies that are capable of identifying meaningful associations across multiple levels, and involving hundreds of thousands of variables.
Given that analysis of genome-wide datasets has only been widespread over the last few years, there have been rapid advances in development of novel analytic methods. Most effort so far, however, has involved the analysis of an individual phenotype, usually a high level diagnostic phenotype, in a “case-control” study design. In these designs, the simplest possible analytic model involves direct comparison of allelic frequencies at each one of several hundred thousand (or more) single-nucleotide polymorphisms (SNPs), possibly supplemented by fine mapping studies or other approaches to replication and/or verification of candidate genetic signals. Given that this approach involves hundreds of thousands of statistical tests, control for false positive results is of paramount importance, and a variety of methods to determine false discovery rates have been suggested (Kang and Zuo, 2007, Wakefield, 2008). The basic analytic strategy, however, has widely remained one of conducting many independent tests, while acknowledging that the degree of independence remains uncertain. The importance of moving beyond the “massively univariate” perspective to consider gene X gene and gene X environment interaction effects is difficult to overestimate, as is highlighted in recent work (Burdick et al., 2008). Furthermore, recent reports suggest that important phenotypic variation may be related more strongly to either copy number variations or rare variants that may be missed using currently standard SNP arrays (Lencz et al., 2007, Sebat et al., 2007).
Efforts have been made to reduce dimensionality on the genomic level through identification of haplotypes, singular value decomposition and independent component analysis of gene expression data, and other novel analytic strategies (Li et al., 2005, Yuan and Li, 2007, Biswas et al., 2008, Madi et al., 2008). Advances in these methods may particularly aid in analysis of gene X gene interaction effects, which may be of great importance for complex polygenic phenotypes (Musani et al., 2007). Similar efforts have been made to reduce dimensionality at the phenotype level, again using singular value decomposition methods, clustering methods, or both (e.g., factor mixture modeling)(Muthen et al., 2006). The proliferation of new methods to advance dimension reduction at genotypic and phenotypic levels is exciting and will likely be helpful to advance phenomics research. Additional methods will likely be important, however, that are capable of both multi-level modeling of phenotypic constructs (in contrast to most dimension reduction strategies that are applied only within a single level of phenotypic observation), and that are capable of identifying the best combinations of genotype and phenotype that relate to each other.
“Genomic convergence” is a term coined to describe a multifactorial, multistep approach combining gene expression and genomic data to identify and prioritize targets (Hauser et al., 2003). We suggest that extending this strategy to include higher level phenomic data is a logical, but not trivial, next step. Similar conclusions have been drawn, and some interesting new approaches are emerging to enable superior multivariate, multilevel analysis and visualization of genotypic and phenotypic data together (Beyene et al., 2007). Given the challenges of reducing scope to aid the pragmatics of both comprehension and computation, methods are needed to efficiently prune multilevel models. Sparse canonical correlation analysis, so far applied to genomic and gene expression data (Parkhomenko et al., 2007), is appealing in its aim to maximize shared variance between genotype and phenotype, rather than maximize variance explained within each level considered separately and then examine associations between the factors that emerge, which has been the dominant model. Major questions remain about the most effective ways to prune such models; ultimately we believe biological hypotheses will enable rational constraints to be applied without jeopardizing discovery.
Implicit in this prioritization of multivariate gene-phene association is the likelihood that the traditional concepts of heritability may be usefully revised. Adopting the conventional definition of broad-sense heritability (H2) as the ratio of genetic variance (G2) to phenotypic variance (P2), Wikipedia suggests that: “Estimating heritability is not a simple process, since only P can be observed or measured directly” (accessed 10/29/08). While questions remain about how much genotypic variance can be observed or measured directly, current studies assessing hundreds of thousands of markers already may provide some estimates, and future methods will surely come closer. A component of the phenomics research program is to advance development of models that will estimate heritability directly from genomic data rather than from phenotype inheritance patterns.
*Omics Need Informatics
Informatics strategies are helping address the new and profound challenge of identifying meaningful signals from GWAS. Many aspects of informatics development require ontologies that provide sufficient identification of the concepts under study, and their possible inter-relations. The Gene Ontology project (http://www.geneontology.org) is serving a useful role already by providing a coherent framework with which investigators can identify gene products by cellular component, biological process and molecular function. The “Entrez” system provided by the National Center for Bioinformatics already contains, in addition to the widely used literature repository (PubMed) and a literature-based database containing annotations specifically relevant for functional genomics (Online Mendelian Inheritance in Man), a rich and steadily growing series of major databases covering Nucleotides (15 databases), Proteins (6 databases); Structure (3 databases); complete Genomes (5 databases), Expression (3 databases), and Chemicals (3 databases). Publications now routinely utilize these bioinformatics resources in creative ways to either constrain analysis, identify replication signals, or interpret results. Despite the rapid growth and utility of these knowledge resources, representation of higher-level phenomic information remains at best incomplete.
The lack of informatics resources for higher-level phenotype data currently poses a limit on phenomics research. To address one segment of this large gap, we have been working on developing ontologies and annotation databases for neuropsychiatric phenomics research, and particularly for the domain of cognitive phenotypes, which has many unique challenges (Bilder et al., submitted). This has involved the building of controlled vocabularies for cognitive concepts and tasks, symptoms, and syndromes. This work already has revealed some striking idiosyncrasies in patterns of term use in the biomedical literature. For example, a recent study aimed to identify published literature on heritability for the concept “cognitive control”, which has enjoyed dramatic increase in use over the last decade (Sabb et al., 2008). In identifying published work on “cognitive control” it was found, however, that this new construct was actually measured using exactly the same tests that had previously been used to index other constructs (“working memory”, “task switching/set shifting”, “response inhibition”, or “response selection”). This instability in the use of cognitive concept labels poses a challenge that needs to be addressed by anchoring cognitive concept use to the measurement level, much as the labeling of SNPs has benefited from adoption of the Reference SNP ID system (rs-numbers) that can be related to physical genomic sequence data. In contrast, gene naming conventions remain elusive and creativity abounds (Seringhaus et al., 2008).
Efforts to develop a useful taxonomy of cognitive tasks face challenges. Given that the goal of developing a task taxonomy is better anchoring and specification of the more fluid cognitive concepts, organizing cognitive concepts by “function” would involve circular logic and could be counterproductive, possibly leading to reification of inaccurate concepts. An alternative that may be less subject to bias is development of task taxonomies that are more like the familiar phylogenetic taxonomies of species. The analogies between task development and evolution of species may have additional merits. Most published tasks are clearly “descended” from “ancestors”, and often involve either (attempted) direct replication or subtle modification of the parent task. Less frequently, a task paradigm is changed substantially; we suggested that the term “task speciation” may help identify those events in task evolution marking differences significant enough that the results of the new task cannot be (or should not be) “mated” with those of the previously existing task to produce a “viable” meta-analytic result. Determinations about whether data should be pooled or not remain somewhat subjective, and different decisions may reflect differing goals of different investigators. But this kind of specification would nevertheless mark a significant advance over the current lack of conventions, and could facilitate development of guidelines on those features of cognitive test paradigms which when changed should prompt consideration of task speciation. Cognitive ontology development is considered in greater detail elsewhere (Bilder et al., submitted), and is the primary aim of an ongoing project, which we hope will benefit from collaborative network inputs (see www.cognitiveatlas.org).
Assuming that it is possible to develop ontologies representing higher phenomic levels, considerable additional effort is necessary to develop the methods for representing the complex, multi-level hypotheses capable of relating genome to syndrome. The CNP includes a project – Hypothesis Web Development for Phenomics Research (RL1LM009833) – that aims to develop tools to help develop, visualize, and share multilevel phenomics hypotheses. Currently scientific hypotheses tend to be articulated using a string of assertions that take the form: “X is associated with Y” [citation]; “Y is associated with Z” [citation]; “suggesting that X may influence Z, mediated by Y”. In addition to the logical leaps and failures to prove causality in such assertions, it is further conspicuous that citations are used to assert the “truth” of evidentiary components in these hypotheses, as though these were binary, when instead we are aware that the reported relations are only partially true, typically having exceeded some arbitrary threshold of significance. Given that the threshold for significance is usually based on disconfirming the null hypothesis of zero association, interpreting such findings as “truths” (implying 100% confidence) is at least partially misleading.
This highlights the potential value of conditioning assertions in terms of quantitative effect sizes. When two alternative hypotheses are presented as chains of assertions with binary “truth” values assigned to every link, there is no way to judge the relative merits of the hypotheses. In contrast, if links are supported by quantitative estimates documenting the strength of each association, it is possible to identify the stronger chain. Extending this rationale to even more complex, partially overlapping networks of associations, the effect size statistics can be used further to select (and prune) branches that will maximize overall hypothesis strength. It is hoped that the “Background & Significance Sections of the Future” may replace current practice with graphical “hypothesis webs” supported by such quantitative, empirical annotation. These efforts may benefit from newly developed strategies for Meta-Analytic Structural Equation Modeling, which enables analyses to be pooled at the level of study results, rather than at the individual subject level (Furlow and Beretvas, 2005, Riley et al., 2007, Cheung, 2008). While the same caveats apply to data pooling in these analyses as they do to any meta-analytic pooling of findings, the challenges are magnified as the complexity of models increases. Developing these methods offers the promise, however, that one day literature searching can be converted to knowledge mining with dynamic extraction of concept maps based on strength of association, and for discovery to be realized based on exploratory interrogation of maps with which investigators may previously have been unfamiliar.
Some tools are already freely available on-line to advance the hypothesis web concept (see Table 2). PubGraph (www.pubgraph.org) was developed to generate graphical concept webs where nodes in the graphs are concepts defined by PubMed queries, and edges connecting the nodes are annotated with Jaccard coefficients revealing the strength of association between concept pairs. PubAtlas (www.pubatlas.org) has further developed this literature querying facility to generate heat maps of concept co-occurrence matrices, and enable the historical changes in these maps to be plotted or played as movies, enabling researchers to visualize the dynamic growth of concept relations over time, and then select the relevant literature that underlies trends. PubAtlas further contains a series of pre-defined lexica (e.g., cognitive concepts, cognitive tasks, neuroanatomy), organized as series of PubMed queries, so that it is possible to “blast” any arbitrary search term against these lexica to find intersections with any term in the pre-defined set. PubBrain (www.pubbrain.org) takes concept-queries, intersects these with a modified version of the NeuroNames lexicon (neuroanatomic terms from the Foundational Model of Anatomy), and then projects the intersections on a three-dimensional probabilistic atlas of the brain (Shattuck et al., 2008). The Phenowiki (www.phenowiki.org) was developed to enable collaborative quantitative annotation of cognitive phenotype concepts (Sabb et al., 2008). The Phenowiki combines some features of a wiki (text annotation about cognitive concepts and tasks), with additional features of a relational database. These include quantitative data fields for empirical information about group difference effect sizes, effect sizes for associations between pairs of variables, study sample sizes and some demographic characteristics, along with psychometric properties for psychological test variables. Each of these applications offers partial solutions to a very large problem; it is hoped that further development and integration of these and similar tools will one day enable integrated development, visualization, and testing of complex multilevel hypotheses.
Table 2.
Web-applications developed to advance neuropsychiatric phenomics research
| Application | URL | Description |
|---|---|---|
| PubGraph | www.pubgraph.org | Creates graphs of PubMed query results, where nodes are concepts defined by each query and edges are annotated with association statistics |
| PubBrain | www.pubbrain.org | Generates co-occurrence statistics of PubMed query with neuroanatomic lexicon, and projects weighted results on a 3D probabilistic atlas of the human brain |
| PubAtlas | www.pubatlas.org | Generates co-occurrence statistics among PubMed queries, generates heat maps and history plots, provides pre-defined lexica for “blast” queries relevant to neuropsychiatric phenomics research |
| Phenowiki | www.phenowiki.org | Provides wiki-like text annotation for cognitive concepts and tests, and collaborative database entry of quantitative effect size, heritability, psychometric, and sample statistics |
| Cognitive Atlas | www.cognitiveatlas.org | Provides collaborative annotation for cognitive concepts and tests, including specification of assertions with evidence for and evidence against, and confidence voting (under development) |
Conclusions and Future Directions
Phenomics is an emerging transdiscipline that aims to leverage breakthroughs in genome-wide genotyping, burgeoning knowledge in the clinical and cognitive neurosciences, and unimagined developments in information and computer sciences, to yield traction on biomedical problems of enormous complexity. The phenomics perspective suggests that systematic study of multiple phenotypes, across multiple biological scales, will be important even if ever-larger sample sizes succeed in revealing robust genetic associations with high level syndromal phenotypes. The need for this perspective seems particularly compelling in its application to neuropsychiatric syndromes. As we move into the “post-GWAS era”, it is likely that declining costs and new methods will enable ever-finer mapping of genetic sequences, detecting rare variants and copy number variations not revealed by most current platforms. While so far it remains unclear to what extent epigenetic factors may help account for phenotypic variance not explained by genomic data, the theoretical potential is vast (Mehler, 2008), and some major initiatives are aiming to develop epigenomics more fully (http://nihroadmap.nih.gov/epigenomics/initiatives.asp), making it possible that epigenome-wide analysis may become a staple of biological characterization in the future. There remain major challenges in identifying and effectively studying human population samples sufficiently large, and capable of addressing challenges of genomic and phenomic diversity. Given the limited throughput of existing lab-based phenotyping methods, web-based ascertainment and phenotyping strategies may represent the most rational way forward, but considerable work is needed to translate what is now technically feasible into pragmatic deployments. While still at early stages in the Human Phenome Project, there remains great promise that phenomics can help us realize the vision of personalized medicine and rational neuropsychiatric diagnosis and treatment.
Acknowledgments
This work was supported by the Consortium for Neuropsychiatric Phenomics and its component awards (UL1DE019580, RL1MH083268, RL1MH083269, RL1DA024853, RL1MH083270, RL1LM009833, PL1MH083271, PL1NS062410), and The Cognitive Atlas (R01MH082795).
Abbreviations
- ADHD
Attention Deficit/Hyperactivity Disorder
- CNP
Consortium for Neuropsychiatric Phenomics
- DSM-IV
Diagnostic and Statistical Manual for Mental Disorders, 4th Edition
- DSM-V
Diagnostic and Statistical Manual for Mental Disorders, 5th Edition
- E2
environmental variance
- G2
genetic variance
- Gpr6
G protein-coupled receptor 6 gene
- GWAS
genome wide association studies
- H2
broad sense heritability
- LA2K
Study of 2000 community volunteers in Los Angeles
- NIH
National Institutes of Health
- rs
reference SNP
- SNP
single nucleotide polymorphism
- UCLA
University of California, Los Angeles
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ambrosini PJ, Bennett DS, Cleland CM, Haslam N. Taxonicity of adolescent melancholia: a categorical or dimensional construct? Journal of psychiatric research. 2002;36:247–256. doi: 10.1016/s0022-3956(02)00011-0. [DOI] [PubMed] [Google Scholar]
- Andersson G, Westoo J, Johansson L, Carlbring P. Cognitive bias via the Internet: a comparison of Web-based and standard emotional Stroop tasks in social phobia. Cogn Behav Ther. 2006;35:55–62. doi: 10.1080/16506070500372469. [DOI] [PubMed] [Google Scholar]
- Bearden C, Freimer N. Endophenotypes for psychiatric disorders: Ready for primetime? Trends in Genetics. 2006 doi: 10.1016/j.tig.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Beyene J, Tritchler D, Bull SB, Cartier KC, Jonasdottir G, Kraja AT, Li N, Nock NL, Parkhomenko E, Rao JS, Stein CM, Sutradhar R, Waaijenborg S, Wang KS, Wang Y, Wolkow P. Multivariate analysis of complex gene expression and clinical phenotypes with genetic marker data. Genet Epidemiol. 2007;31(Suppl 1):S103–109. doi: 10.1002/gepi.20286. [DOI] [PubMed] [Google Scholar]
- Bilder RM. Phenomics: building scaffolds for biological hypotheses in the post-genomic era. Biological psychiatry. 2008;63:439–440. doi: 10.1016/j.biopsych.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilder RM, Sabb FW, Parker DS, Kalar D, Chu WW, Fox J, Poldrack RA. Cognitive Ontologies for Neuropsychiatric Phenomics Research. Cognitive Neuropsychiatry. doi: 10.1080/13546800902787180. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas S, Storey JD, Akey JM. Mapping gene expression quantitative trait loci by singular value decomposition and independent component analysis. BMC Bioinformatics. 2008;9:244. doi: 10.1186/1471-2105-9-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan T, Smith JL. Research on the Internet: validation of a World-Wide Web mediated personality scale. Behav Res Methods Instrum Comput. 1999;31:565–571. doi: 10.3758/bf03200736. [DOI] [PubMed] [Google Scholar]
- Burdick KE, Kamiya A, Hodgkinson CA, Lencz T, DeRosse P, Ishizuka K, Elashvili S, Arai H, Goldman D, Sawa A, Malhotra AK. Elucidating the relationship between DISC1, NDEL1 and NDE1 and the risk for schizophrenia: evidence of epistasis and competitive binding. Human molecular genetics. 2008;17:2462–2473. doi: 10.1093/hmg/ddn146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butcher LM, Kennedy JK, Plomin R. Generalist genes and cognitive neuroscience. Curr Opin Neurobiol. 2006;16:145–151. doi: 10.1016/j.conb.2006.03.004. [DOI] [PubMed] [Google Scholar]
- Butcher LM, Meaburn E, Knight J, Sham PC, Schalkwyk LC, Craig IW, Plomin R. SNPs, microarrays and pooled DNA: identification of four loci associated with mild mental impairment in a sample of 6000 children. Human molecular genetics. 2005;14:1315–1325. doi: 10.1093/hmg/ddi142. [DOI] [PubMed] [Google Scholar]
- Cannon TD, Hennah W, van Erp TGM, Thompson PM, Lonnqvist J, Huttunen M, Gasperoni T, Tuulio-Henriksson A, Pirkola T, Toga AW, Kaprio J, Mazziotta J, Peltonen L. DISC1/TRAX Haplotypes Associate with Schizophrenia, Reduced Prefrontal Gray Matter, and Impaired Short- and Long-Term Memory. Archives of general psychiatry. 2005;62:1205–1213. doi: 10.1001/archpsyc.62.11.1205. [DOI] [PubMed] [Google Scholar]
- Centler F, Kaleta C, di Fenizio PS, Dittrich P. Computing chemical organizations in biological networks. Bioinformatics. 2008;24:1611–1618. doi: 10.1093/bioinformatics/btn228. [DOI] [PubMed] [Google Scholar]
- Cheung MW. A model for integrating fixed-, random-, and mixed-effects meta-analyses into structural equation modeling. Psychol Methods. 2008;13:182–202. doi: 10.1037/a0013163. [DOI] [PubMed] [Google Scholar]
- Chiasson MA, Parsons JT, Tesoriero JM, Carballo-Dieguez A, Hirshfield S, Remien RH. HIV behavioral research online. J Urban Health. 2006;83:73–85. doi: 10.1007/s11524-005-9008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crampin EJ, Halstead M, Hunter P, Nielsen P, Noble D, Smith N, Tawhai M. Computational physiology and the physiome project. Exp Physiol. 2004;89:1–26. doi: 10.1113/expphysiol.2003.026740. [DOI] [PubMed] [Google Scholar]
- Cunningham JA, Selby P, van Mierlo T. Integrated online services for smokers and drinkers? Use of the check your drinking assessment screener by participants of the Stop Smoking Center. Nicotine Tob Res. 2006;8(Suppl 1):S21–25. doi: 10.1080/14622200601039865. [DOI] [PubMed] [Google Scholar]
- Edelman GM. Naturalizing consciousness: a theoretical framework. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:5520–5524. doi: 10.1073/pnas.0931349100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O’Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, Fan J, Kirov G, Perlis RH, Green EK, Smoller JW, Grozeva D, Stone J, Nikolov I, Chambert K, Hamshere ML, Nimgaonkar VL, Moskvina V, Thase ME, Caesar S, Sachs GS, Franklin J, Gordon-Smith K, Ardlie KG, Gabriel SB, Fraser C, Blumenstiel B, Defelice M, Breen G, Gill M, Morris DW, Elkin A, Muir WJ, McGhee KA, Williamson R, Macintyre DJ, Maclean AW, St Clair D, Robinson M, Van Beck M, Pereira AC, Kandaswamy R, McQuillin A, Collier DA, Bass NJ, Young AH, Lawrence J, Nicol Ferrier I, Anjorin A, Farmer A, Curtis D, Scolnick EM, McGuffin P, Daly MJ, Corvin AP, Holmans PA, Blackwood DH, Gurling HM, Owen MJ, Purcell SM, Sklar P, Craddock N. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008 doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher SE. Tangled webs: tracing the connections between genes and cognition. Cognition. 2006;101:270–297. doi: 10.1016/j.cognition.2006.04.004. [DOI] [PubMed] [Google Scholar]
- Flint J, Munafo MR. The endophenotype concept in psychiatric genetics. Psychol Med. 2007;37:163–180. doi: 10.1017/S0033291706008750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freimer N, Sabatti C. The human phenome project. NatGenet. 2003;34:15–21. doi: 10.1038/ng0503-15. [DOI] [PubMed] [Google Scholar]
- Furlow CF, Beretvas SN. Meta-analytic methods of pooling correlation matrices for structural equation modeling under different patterns of missing data. Psychol Methods. 2005;10:227–254. doi: 10.1037/1082-989X.10.2.227. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. AmJ Psychiatry. 2003;160:636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Graham AL, Bock BC, Cobb NK, Niaura R, Abrams DB. Characteristics of smokers reached and recruited to an internet smoking cessation trial: a case of denominators. Nicotine Tob Res. 2006;8(Suppl 1):S43–48. doi: 10.1080/14622200601042521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haslam N. Categorical versus dimensional models of mental disorder: the taxometric evidence. Australian and New Zealand Journal of Psychiatry. 2003;37:696–704. doi: 10.1080/j.1440-1614.2003.01258.x. [DOI] [PubMed] [Google Scholar]
- Haslam N, Beck AT. Subtyping major depression: a taxometric analysis. J Abnorm Psychol. 1994;103:686–692. doi: 10.1037//0021-843x.103.4.686. [DOI] [PubMed] [Google Scholar]
- Haslam N, Kim HC. Categories and continua: A review of taxometric research. Genetic Social and General Psychology Monographs. 2002;128:271–320. [PubMed] [Google Scholar]
- Kang G, Zuo Y. Entropy-based joint analysis for two-stage genome-wide association studies. J Hum Genet. 2007;52:747–756. doi: 10.1007/s10038-007-0177-7. [DOI] [PubMed] [Google Scholar]
- Kauffman SA. The Origins of Order: Self Organization and Selection in Evolution. New York: Oxford University Press; 1993. [Google Scholar]
- Kendler KS. The diagnostic validity of melancholic major depression in a population-based sample of female twins. Archives of general psychiatry. 1997;54:299–304. doi: 10.1001/archpsyc.1997.01830160013002. [DOI] [PubMed] [Google Scholar]
- Kendler KS. Explanatory models for psychiatric illness. Am J Psychiatry. 2008;165:695–702. doi: 10.1176/appi.ajp.2008.07071061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendler KS, Prescott CA, Myers J, Neale MC. The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Archives of general psychiatry. 2003;60:929–937. doi: 10.1001/archpsyc.60.9.929. [DOI] [PubMed] [Google Scholar]
- Kessing LV. Epidemiology of subtypes of depression. Acta Psychiatr Scand Suppl. 2007:85–89. doi: 10.1111/j.1600-0447.2007.00966.x. [DOI] [PubMed] [Google Scholar]
- Kovas Y, Plomin R. Generalist genes: implications for the cognitive sciences. Trends Cogn Sci. 2006;10:198–203. doi: 10.1016/j.tics.2006.03.001. [DOI] [PubMed] [Google Scholar]
- Kraemer HC. DSM categories and dimensions in clinical and research contexts. International Journal of Methods in Psychiatric Research. 2007;16:S8–S15. doi: 10.1002/mpr.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krantz JH, Ballard J, Scher J. Comparing the results of laboratory and World-Wide Web samples on the determinants of female attractiveness. Behavioral Research Methods, Instruments & Computers. 1997;29:264–269. [Google Scholar]
- Krantz JH, Dalal R. Validity of Web-Based Psychological Research. In: Birnbaum MH, editor. Psychological experiments on the internet. New York: Academic Press; 2000. pp. 35–60. [Google Scholar]
- Lencz T, Morgan TV, Athanasiou M, Dain B, Reed CR, Kane JM, Kucherlapati R, Malhotra AK. Converging evidence for a pseudoautosomal cytokine receptor gene locus in schizophrenia. Molecular psychiatry. 2007;12:572–580. doi: 10.1038/sj.mp.4001983. [DOI] [PubMed] [Google Scholar]
- Li L, Jiang W, Li X, Moser KL, Guo Z, Du L, Wang Q, Topol EJ, Rao S. A robust hybrid between genetic algorithm and support vector machine for extracting an optimal feature gene subset. Genomics. 2005;85:16–23. doi: 10.1016/j.ygeno.2004.09.007. [DOI] [PubMed] [Google Scholar]
- Li W, Zhou Y, Jentsch JD, Brown RA, Tian X, Ehninger D, Hennah W, Peltonen L, Lonnqvist J, Huttunen MO, Kaprio J, Trachtenberg JT, Silva AJ, Cannon TD. Specific developmental disruption of disrupted-in-schizophrenia-1 function results in schizophrenia-related phenotypes in mice. Proc Natl Acad Sci U S A. 2007;104:18280–18285. doi: 10.1073/pnas.0706900104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobo MK, Cui Y, Ostlund SB, Balleine BW, Yang XW. Genetic control of instrumental conditioning by striatopallidal neuron-specific S1P receptor Gpr6. Nature neuroscience. 2007;10:1395–1397. doi: 10.1038/nn1987. [DOI] [PubMed] [Google Scholar]
- Madi A, Friedman Y, Roth D, Regev T, Bransburg-Zabary S, Ben Jacob E. Genome holography: deciphering function-form motifs from gene expression data. PLoS ONE. 2008;3:e2708. doi: 10.1371/journal.pone.0002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher BS, Riley BP, Kendler KS. Psychiatric genetics gets a boost. Nat Genet. 2008;40:1042–1044. doi: 10.1038/ng0908-1042. [DOI] [PubMed] [Google Scholar]
- Mehler MF. Epigenetic principles and mechanisms underlying nervous system functions in health and disease. Prog Neurobiol. 2008 doi: 10.1016/j.pneurobio.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musani SK, Shriner D, Liu N, Feng R, Coffey CS, Yi N, Tiwari HK, Allison DB. Detection of Gene × Gene Interactions in Genome-Wide Association Studies of Human Population Data. Human Heredity. 2007;63:67–84. doi: 10.1159/000099179. [DOI] [PubMed] [Google Scholar]
- Muthen B, Asparouhov T, Rebollo I. Advances in behavioral genetics modeling using Mplus: applications of factor mixture modeling to twin data. Twin Res Hum Genet. 2006;9:313–324. doi: 10.1375/183242706777591317. [DOI] [PubMed] [Google Scholar]
- Nykter M, Price ND, Aldana M, Ramsey SA, Kauffman SA, Hood LE, Yli-Harja O, Shmulevich I. Gene expression dynamics in the macrophage exhibit criticality. Proceedings of the National Academy of Sciences. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Donovan MC, Craddock N, Norton N, Williams H, Peirce T, Moskvina V, Nikolov I, Hamshere M, Carroll L, Georgieva L, Dwyer S, Holmans P, Marchini JL, Spencer CC, Howie B, Leung HT, Hartmann AM, Moller HJ, Morris DW, Shi Y, Feng G, Hoffmann P, Propping P, Vasilescu C, Maier W, Rietschel M, Zammit S, Schumacher J, Quinn EM, Schulze TG, Williams NM, Giegling I, Iwata N, Ikeda M, Darvasi A, Shifman S, He L, Duan J, Sanders AR, Levinson DF, Gejman PV, Buccola NG, Mowry BJ, Freedman R, Amin F, Black DW, Silverman JM, Byerley WF, Cloninger CR, Cichon S, Nothen MM, Gill M, Corvin A, Rujescu D, Kirov G, Owen MJ. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nat Genet. 2008 doi: 10.1038/ng.201. [DOI] [PubMed] [Google Scholar]
- Oldham MC, Geschwind DH. Deconstructing language by comparative gene expression: from neurobiology to microarray. Genes Brain Behav 5 Suppl. 2006;1:54–63. doi: 10.1111/j.1601-183X.2006.00195.x. [DOI] [PubMed] [Google Scholar]
- Parkhomenko E, Tritchler D, Beyene J. Genome-wide sparse canonical correlation of gene expression with genotypes. BMC Proceedings. 2007;1:S119. doi: 10.1186/1753-6561-1-s1-s119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley RD, Abrams KR, Sutton AJ, Lambert PC, Thompson JR. Bivariate random-effects meta-analysis and the estimation of between-study correlation. BMC Med Res Methodol. 2007;7:3. doi: 10.1186/1471-2288-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabb FW, Bearden CE, Glahn DC, Parker DS, Freimer N, Bilder RM. A collaborative knowledge base for cognitive phenomics. Molecular psychiatry. 2008;13:350–360. doi: 10.1038/sj.mp.4002124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebat J, Lakshmi B, Malhotra D, Troge J, Lese-Martin C, Walsh T, Yamrom B, Yoon S, Krasnitz A, Kendall J, Leotta A, Pai D, Zhang R, Lee YH, Hicks J, Spence SJ, Lee AT, Puura K, Lehtimaki T, Ledbetter D, Gregersen PK, Bregman J, Sutcliffe JS, Jobanputra V, Chung W, Warburton D, King MC, Skuse D, Geschwind DH, Gilliam TC, Ye K, Wigler M. Strong association of de novo copy number mutations with autism. Science. 2007;316:445–449. doi: 10.1126/science.1138659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seringhaus MR, Cayting PD, Gerstein MB. Uncovering trends in gene naming. Genome Biol. 2008;9:401. doi: 10.1186/gb-2008-9-1-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth AK, Izhikevich E, Reeke GN, Edelman GM. Theories and measures of consciousness: an extended framework. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:10799–10804. doi: 10.1073/pnas.0604347103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shattuck DW, Mirza M, Adisetiyo V, Hojatkashani C, Salamon G, Narr KL, Poldrack RA, Bilder RM, Toga AW. Construction of a 3D probabilistic atlas of human cortical structures. Neuroimage. 2008;39:1064–1080. doi: 10.1016/j.neuroimage.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, Tononi G, Kotter R. The human connectome: A structural description of the human brain. PLoS Comput Biol. 2005;1:e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar S, Dhamdhere K, Blelloch G, Halperin E, Ravi R, Schwartz R. Algorithms for efficient near-perfect phylogenetic tree reconstruction in theory and practice. IEEE/ACM Trans Comput Biol Bioinform. 2007;4:561–571. doi: 10.1109/TCBB.2007.1070. [DOI] [PubMed] [Google Scholar]
- Tononi G, Edelman GM. Consciousness and complexity. Science. 1998;282:1846–1851. doi: 10.1126/science.282.5395.1846. [DOI] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM. A measure for brain complexity: relating functional segregation and integration in the nervous system. Proceedings of the National Academy of Sciences of the United States of America. 1994;91:5033–5037. doi: 10.1073/pnas.91.11.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tononi G, Sporns O, Edelman GM. Measures of degeneracy and redundancy in biological networks. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:3257–3262. doi: 10.1073/pnas.96.6.3257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade N. New York Times. New York: New York Times; 2008. A Dissenting Voice as the Genome Is Sifted to Fight Disease. [Google Scholar]
- Wakefield J. Reporting and interpretation in genome-wide association studies. Int J Epidemiol. 2008;37:641–653. doi: 10.1093/ije/dym257. [DOI] [PubMed] [Google Scholar]
- Yuan S, Li KC. Context-dependent clustering for dynamic cellular state modeling of microarray gene expression. Bioinformatics. 2007;23:3039–3047. doi: 10.1093/bioinformatics/btm457. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Yeh CW, Yang CD, Jang SS, Chu IM. On the local optimal solutions of metabolic regulatory networks using information guided genetic algorithm approach and clustering analysis. J Biotechnol. 2007;131:159–167. doi: 10.1016/j.jbiotec.2007.06.019. [DOI] [PubMed] [Google Scholar]