Abstract
Phosphorylation of bacterial ribosomal proteins has been known for decades; however, there is still very limited information available on specific locations of the phosphorylation sites in ribosomal proteins and the role they might play in protein synthesis. In this study, we have mapped the specific phosphorylation sites in twenty-four E. coli ribosomal proteins by tandem mass spectrometry. Specific detection of phosphorylation was achieved by either phosphorylation specific visualization techniques, ProQ staining and antibodies for phospho-Ser, Thr, and Tyr, or by mass spectrometry equipped with a capability to detect addition and the loss of the phosphate moiety. Enrichment by immobilized metal affinity and/or strong cation exchange chromatography was used to improve the success of detection of the low abundance phosphopeptides. We found the small subunit (30S) proteins S3, S4, S5, S7, S11, S12, S13, S18, and S21 and the large subunit (50S) proteins L1, L2, L3, L5, L6, L7/L12, L13, L14, L16, L18, L19, L21, L22, L28, L31 to be phosphorylated at one or more residues. Potential roles for each specific site in ribosome function were deduced through careful evaluation of the given site of the phosphorylation in 3D-crystal structure models of ribosomes and the previous mutational studies of E. coli ribosomal proteins.
Introduction
Ribosomes are large ribonucleoprotein particles consisting of two subunits in all species. In bacteria, the subunits are designated as the small subunit (30S) and the large subunit (50S), which together make up the 70S ribosome. The 30S subunit is responsible for binding and decoding mRNAs in addition to assisting the 50S subunit in translocation.1 The large subunit (50S) possesses the peptidyl transferase activity which is buried within the 23S ribosomal RNA (rRNA).2,3 Earlier biochemical and mutational studies of ribosomal proteins have revealed the importance of many of these proteins in mRNA and tRNA recognition, decoding, formation of binding sites for translation factors, peptidyl transferase activity, formation of the peptide exit tunnel, and finally in helicase activity of the ribosomes.4-8 The recent progress in the structural and biochemical studies of bacterial ribosomes has provided an enormous amount of information on the overall rRNA architecture and the details of protein-RNA interactions in each subunit, as well as the details of the interaction of the ribosome with the initiation and elongation factors, mRNA, and tRNA.2,5,7,9-12 Surprisingly, there is limited information available on regulation of ribosome function by post-translational modifications (PTM) of these proteins, especially by phosphorylation. Phosphorylation of Ser, Thr, and Tyr residues is a common type of PTM seen in signal transduction pathways. In addition to regulation of protein function, phosphorylation could also serve as a molecular switch for conformational changes during protein synthesis on the ribosome. In two earlier in vitro phosphorylation studies on E. coli and Streptomyces collinus ribosomes, the ribosomal proteins phosphorylated by a protein kinase from rabbit skeletal muscle and a protein kinase associated with the ribosomes were identified, respectively.13,14 E. coli ribosomal proteins S4, S9, S18, S19, L2, L3, L5, L7/L12, L10, and L33 were found to be phosphorylated by the protein kinase from rabbit skeletal muscle at their Ser and Thr residues13 and S. collinus ribosomal proteins S3, S4, S12, S13, S14, S18, L2, L7/L12, L16, L17, and L23 were determined to be Ser and Thr phosphorylated by the protein kinase associated with the ribosomes.15 The in vitro phosphorylation of ribosomal proteins was found to change the conformation or substrate binding sites in the ribosome and to lead to a 50% loss of activity in protein synthesis.13-15 In a recent high-throughput analysis of the steady-state phosphoproteome of E. coli, ribosomal proteins S7, L7/L12, L16, L19, and L28 were reported to be phosphorylated.16 Moreover, phosphorylation of L7/L12 and L18 in Thermus thermophilus and Bacillus stearothermophilus was reported previously.17,18 However, the knowledge of specific sites of phosphorylation of ribosomal proteins is still very incomplete, which significantly limits our ability to understand the regulation of prokaryotic protein synthesis by phosphorylation. In this study, we conducted a comprehensive study to map the sites of phosphorylation in E. coli ribosomal proteins through utilization of a combination of biochemical techniques and tandem mass spectrometry. Of the fifty-four proteins in the 70S ribosome, we found twenty-four to be steady-state phosphorylated. These ribosomal proteins are S3, S4, S5, S7, S11, S12, S13, S18, S21 in the small subunit and L1, L2, L3, L5, L6, L7/L12, L13, L14, L16, L18, L19, L21, L22, L28, L31 in the large subunit. Our findings should now allow ribosome researchers to investigate the role of specific phosphorylation sites in regulation of protein synthesis by mutational analysis of ribosomal proteins.
Materials and Methods
Preparation of E. coli ribosomes
E. coli ribosomes from W strain were prepared following a previously described method. 19,20 Briefly, E. coli cells were grown to an optical density of 0.5 at A600. After grinding the cells (usually 1.5-2 g/L) using alumina, the cell paste was resuspended in ribosomal Buffer A (20 mM Tris-HCl pH 7.6, 10 mM MgCl2, 1 mM DTT). Crude ribosomes were sedimented at 48,000 rpm in a Beckman 50.2 Ti rotor for 3.5 h from the cell suspension. The crude ribosomal pellet was then resuspended in Ribosomal Buffer B (50 mM Tris-HCl pH 7.6, 20 mM MgCl2, 1 mM DTT, 1 M NH4Cl) and centrifuged at 48,000 rpm in a Beckman 50.2 Ti rotor for 5 h at 4 °C to remove the ribosome associated translational factors. Finally, the high-salt washed ribosomes were resuspended in Ribosomal Buffer C (10 mM Tris-HCl pH 7.6, 10 mM MgCl2, 1 mM DTT, and 50 mM NH4Cl) and 70S ribosomes were isolated from 10-30% sucrose linear gradients prepared in Buffer C. Phosphatase (2 mM imizadole, 1 mM sodium orthovanadate, 1.15 mM sodium molybdate, 1 mM sodium fluoride, and 4 mM sodium tartrate dehydrate) and protease (0.1 mM PMSF) inhibitors were also added to the ribosome preparations that are intended to be used in the proteomic analyses.
Immunoblot and 2D-gel analyses
Approximately, 15 A260 units of E. coli ribosomes were treated with RNase A, acetone precipitated, and the resulting pellet was resuspended in a lysis buffer consisting of 9.8M urea, 2% (w/v) NP-40, 2% ampholytes pI 5-7 and 3-10, and 100 mM DTT. The samples were loaded on NEPHGE (non-equilibrium pH gradient electrophoresis) tube gels and equilibrated in buffer containing 60 mM Tris-HCl pH 6.8, 2% SDS, 100 mM DTT, and 10% glycerol. 21 The first dimension NEPHGE gels of ribosomal proteins were loaded on 20% second dimension SDS-PAGE gels. Then, the gels were either stained with Coomassie Blue or transferred to PVDF membranes for immunoblotting analysis to probe with the following primary antibodies (Sigma-Aldrich, Inc.) for: phosphoserine at 1:20,000, phosphothreonine at 1:10,000, and phosphotyrosine at 1:5,000 and the secondary streptavidin HRP antibody (Amersham Biosciences Inc.) at 1:30,000 dilutions. The membranes were developed using the SuperSignal West Femto Max Sensitivity Substrate (Pierce Biochemicals Inc.) according to the protocol provided by the manufacturer.
ProQ staining and in vivo 32P-ortho phosphate labeling
Approximately, 1 A260 unit of purified E. coli ribosomes were separated on a 16% SDS-PAGE gel and stained with ProQ Diamond Phosphoprotein Gel Stain (Molecular Probes Inc.). Phosphorylated proteins were visualized by a laser scanner at an excitation wavelength of 532 nm. For in vivo 32P-ortho phosphate labeling of ribosomal proteins, bacteria were grown to an early log phase at 37°C in LB medium. The culture spun down and the pellet was resuspended in 20 ml of phosphate-free M9 medium. Isotope, 10 μCi/ml of 32P-orthophosphate, was added and followed by 1h incubation at 37°C. The cell pellet was resuspended in ribosomal Buffer A and sonicated before the sedimentation of crude ribosomes as described above. Then, the purified ribosomal proteins were subjected to SDS-PAGE and autoradiography.
Enrichment of phosphopeptides by immobilized metal affinity chromatography (IMAC)
In order to maximize detection of phosphorylated peptides/proteins in our in-solution digestion of 70S ribosome samples, IMAC/SwellGel phosphopeptide enrichment was performed. The E. coli 70S ribosome sample (7.5 A260 units) was incubated with 10 μg of RNase A at 37 °C for 1 h and denatured in 6 M Guanidine HCl, injected onto C4 –like polymeric reverse-phase trap cartridge (Michrom Bioresources, Inc.) for desalting, concentration, and removal of the degraded RNA. The eluate was dried in a SpeedVac, resuspended in 30 μL of 25 mM NH4HCO3, and digested by trypsin (Promega Corporation) at 1:50 ratio at 37 °C overnight. Phosphorylated tryptic peptides in the resulting in-solution digest were enriched by using a IMAC/SwellGel gallium-chelated resin (Phosphopeptide Isolation Kit, Pierce Biotechnology, Inc.) according to the protocol provided by the manufacturer. The samples were analyzed by LC-MS/MS phosphorylation detection and mapping.
Enrichment of phosphopeptides by strong cation exchange (SCX) chromatography
In-solution tryptic digests of the 70S ribosomal proteins were injected on a BioBasic SCX column (2.1×250 mm, 5 micron, 300A, ThermoFinnigan Co.). Peptides were eluted with a gradient of 5 mM phosphate (pH 2.7), 25% acetonitrile (Mobile phase A) and 5 mM phosphate (pH 2.7), 25% acetonitrile, 350 mM KCl (Mobile phase B). The gradient started with a hold at 0% B for 5 min. It was ramped to 15% B in 20 min, then quickly to 80% B in 5 min, and held there for 5 min. The column was equilibrated at 0% B for 15 min prior to next injection. Absorbance over the range of 200 to 300 nm was measured and absorbance at 210 nm was plotted. Fractions (600 μL) were collected throughout for 30 minutes. Early fractions, which are expected to be enriched in phosphopeptides, were reduced to 20 μL using a SpeedVac. The SCX fractions were online desalted and analyzed by LC-MS/MS.
Protein identification and phosphorylation site mapping by tandem mass spectrometry
The spots corresponding to phosphorylated proteins detected by immunoblot analysis were excised from a Coomassie stained gel and in-gel digestions with trypsin or Lys-C were performed. 22 For protein identification and phosphorylation mapping by tandem mass spectrometry, tryptic digests were analyzed by capillary liquid chromatography- nanoelectrospray ionization - tandem mass spectrometry (LC-MS/MS). Tandem MS spectra obtained by fragmenting a peptide by collision-induced dissociation (CID) were acquired using a capillary liquid chromatography - nanoelectrospray ionization - tandem mass spectrometry (LC-MS/MS) system that consisted of a Surveyor HPLC pump, a Surveyor Micro AS autosampler, and an LTQ linear ion trap mass spectrometer (ThermoFinnigan). A portion (3-5 μL) of each fraction was injected and loaded onto a peptide trap (Michrom peptide CapTrap, C8 like resin, 0.3×1mm, 5μ) over 3 min at 10 μL/min for on-line desalting and concentration. Utilizing the six-port switching valve of the LTQ mass spectrometer, the peptide trap was then placed in line with the analytical column, a PicoFrit column (0.075 × 150 mm) packed in-house with Wide Bore C18 reverse phase resin (Supelco Co., 5 μ, 300 Å). The column was eluted at ∼250 μL/min (obtained by a flow splitting of 150 μL/min flow using a Valco T) by a gradient that consisted of 0.1% formic acid (Solvent A) and 0.1% formic acid in acetonitrile (Solvent B). The peptides were eluted by ramping the solvent B to 40% over either 30 min or 120 min depending on the complexity of the protein mixture in a given fraction. Spray voltage was optimized routinely for sensitivity at around 2 kV. Capillary temperature was set at 180 °C. Tube lens was at 70V. Tandem MS collision-induced dissociation (CID) spectra were acquired for ions above a predetermined intensity threshold using the automated data-dependent acquisition feature. For phosphopeptide mapping, MS3 was triggered by neutral loss detection in the CID MS/MS spectra due to the neutral loss of phosphate moiety in phosphopeptides. This loss of phosphate moiety is detected as a dominant peak in MS/MS spectrum at an m/z value 98, 49, or 32.7 lower than that of the phosphopeptide for singly-, doubly-, or triply -charged peptides, respectively.
Both the MS/MS and MS3 spectra acquired were processed by Xcalibur 2.0 and searched against an in-house generated 70S and Swiss-Prot protein sequence database using the search engines Turbo Sequest embedded in Bioworks 3.2 (ThermoFinnigan Inc) and Mascot 2.2 (Matrix Science), which are licensed and maintained locally. Database searches were performed with cysteine carbamidomethylation as fixed modification when alkylation by iodoacetamide was performed. The variable modifications were methionine oxidation (+16 Da) and phosphorylation (+80 Da) of Ser, Thr, and Tyr residues and loss of water (-18 Da) from Ser and Thr due to β-elimination during loss of the phosphate moiety. Up to 2 missed cleavages were allowed for the protease of choice. Peptide mass tolerance and fragment mass tolerance were set to 3 and 2 Da, respectively. Tandem MS spectra matched to phosphopeptides with a Mascot scores of ≥40 were further evaluated manually at the raw data level with the consideration of overall data quality, signal-to-noise of matched peaks, and the presence of dominant peaks that did not match to any theoretical m/z value. The spectra obtained from the LC-MS/MS analyses were also searched against the Swiss-Prot database using the Turbo Sequest search engine using setting similar to those used with the Mascot engine. Search results were reported as correlation coefficient (XCorr) and that is the cross-correlation value between the observed peptide fragment mass spectrum and the one theoretically predicted. XCorr cut-off values were >1.5, 2.0, and 2.5 for +1, +2, and +3 charged peptides, respectively.
Results and Discussion
Structural and functional information obtained from recent studies of ribosomes have focused our attentions to rRNA; however, many of the biological functions are also contributed by the ribosomal proteins in the ribosome.23,24 In general, proteins of the large and small subunit are located on the outer surface of the subunits and the ones with long peptide tails permeate into ribosomal RNA (rRNA) and shape the rRNA into the correct tertiary structure.25,26 As summarized in Table I, many functions of the ribosomal proteins appear to be essential for ribosome structure and protein synthesis in bacteria.24 The large subunit proteins that are surrounding the peptidyl transferase active site and the elongation factor binding sites are functional proteins of the 50S subunit, while the proteins located in the mRNA binding path and decoding center are the functional sites of the small subunit (Table I).11,25,26 Therefore, phosphorylation of these ribosomal proteins can be one of the possible regulatory mechanism(s) employed in regulation of their functions in bacterial translation.
Table I. Phosphorylated Proteins of E.coli Ribosomes Detected by Immunoblotting and LC-MS/MS Analysis.
| Protein | pI | MW | Phosphosites | Detection | Function* |
|---|---|---|---|---|---|
| S3 | 10.27 | 25.9 | Ser, Thr, Tyr | IB/MS | mRNA binding to ribosomes |
| S4 | 10.05 | 23.5 | Ser, Thr, Tyr | IB/MS | mRNA binding /Functional mutations |
| S5 | 10.11 | 17.5 | Ser, Thr, Tyr | IB/MS | mRNA binding /Functional mutations |
| S7 | 10.30 | 17.6 | Ser, Thr, Tyr | IB/MS | mRNA and tRNA binding at the E site |
| S9 | 10.94 | 14.9 | Ser, Thr, Tyr | IB/MS | Interaction with P site tRNA |
| S11 | 11.33 | 13.8 | Ser, Thr | IB/MS | mRNA and tRNA binding at the E site |
| S12 | 10.88 | 13.7 | Ser, Thr, Tyr | IB/MS | tRNA decoding at the A site |
| S13 | 10.78 | 13.1 | Ser, Thr | MS | Subunit joining |
| S18 | 10.60 | 8.9 | Ser, Thr, Tyr | MS | mRNA binding to ribosomes |
| S21 | 11.15 | 8.5 | Tyr | IB/MS | mRNA binding to ribosomes |
| L2 | 10.93 | 29.9 | Ser,Thr | IB/MS | Required for peptidyl transferase |
| L3 | 9.90 | 22.2 | Ser,Thr | IB/MS | Required for peptidyl transferase |
| L4 | 9.72 | 22.1 | Ser,Thr,Tyr | IB/MS | Lines peptide exit tunnel |
| L5 | 9.49 | 20.3 | Tyr | IB/MS | Interaction with P site tRNA |
| L7/L12 | 4.60 | 12.2 | Ser, Thr | MS | Factor-binding stalk |
| L9 | 6.17 | 15.8 | Ser,Thr,Tyr | IB/MS | |
| L10 | |||||
| L11 | 9.64 | 14.9 | Ser,Thr | IB/MS | Factor binding |
| L13 | 9.91 | 16.0 | Thr,Tyr | IB/MS | |
| L15 | 11.18 | 15.0 | Tyr | IB/MS | |
| L16 | 11.22 | 15.3 | Tyr | IB/MS | A site tRNA binding |
| L19 | 10.57 | 13.3 | Ser,Thr,Tyr | IB/MS | Subunit joining |
| L20 | 11.47 | 13.5 | Ser, Tyr | IB/MS | |
| L22 | 10.23 | 12.2 | Ser, Thr, Tyr | IB/MS | Lines peptide tunnel |
| L23 | 9.94 | 11.2 | Ser, Thr | IB/MS | At tunnel exit, interacts with chaperones and SRP |
| L24 | 10.21 | 11.3 | Ser, Thr | IB/MS | At tunnel exit, interacts with chaperones and SRP |
Adapted from Brodersen and Nissen (24). IB;immunobloting, MS; mass spectrometry.
In this study, we determined the phosphorylation status of ribosomal proteins in E. coli 70S ribosomes at steady-state. First, separation of E. coli ribosomal proteins on 2D-NEPHGE gels was performed and the gels were transferred to PVDF membranes for immunoblotting. A total of fourteen phosphorylated E. coli ribosomal proteins were initially identified in this analysis. Fig. 1B, C, and D show blots probed with phosphotyrosine, phosphoserine, and phosphothreonine, respectively. Ribosomal proteins S4, S7, S9, S12, S21, L2, L9, L15, L16, L19, and L20 were detected as Tyr phosphorylated, whereas S4, S9, S11, S12, L2, L9, L11, L17, L19, and L20 were Ser and/or Thr phosphorylated (Table I). As detected in the Coomassiee blue stained 2D-NEPHGE gels, majority of the ribosomal proteins are between 10-20 kDa and extremely basic (Fig. 1A and Table I). Even though identification of ribosomal proteins in crowded regions of the Coomassie gel was not a setback, to obtain a better separation and confirm the type of ribosomal protein phosphorylation(s) on 2D-NEPHGE gels, separation conditions were optimized (Fig. 2A). This resulted in the identification of additional phosphorylated ribosomal proteins S3, S5, L3, L4, L5, L13, L22, and L24. Many of these proteins were found either in multiple spots or forming a streaky pattern verifying the phosphorylation of these proteins and to be phosphorylated mainly at Ser and Thr residues detected by immunoblotting. In addition to the analysis of phosphorylated ribosomal proteins, immunoblot analysis of dephosphorylated ribosomes was also performed with the phosphotyrosine antibody. After preincubation of ribosomes with alkaline phosphatase, a decreased signal from the Tyr phosphorylated ribosomal proteins was observed (data not shown).
Fig. 1. Two-dimensional gel analysis of phosphorylated E. coli ribosomal proteins.
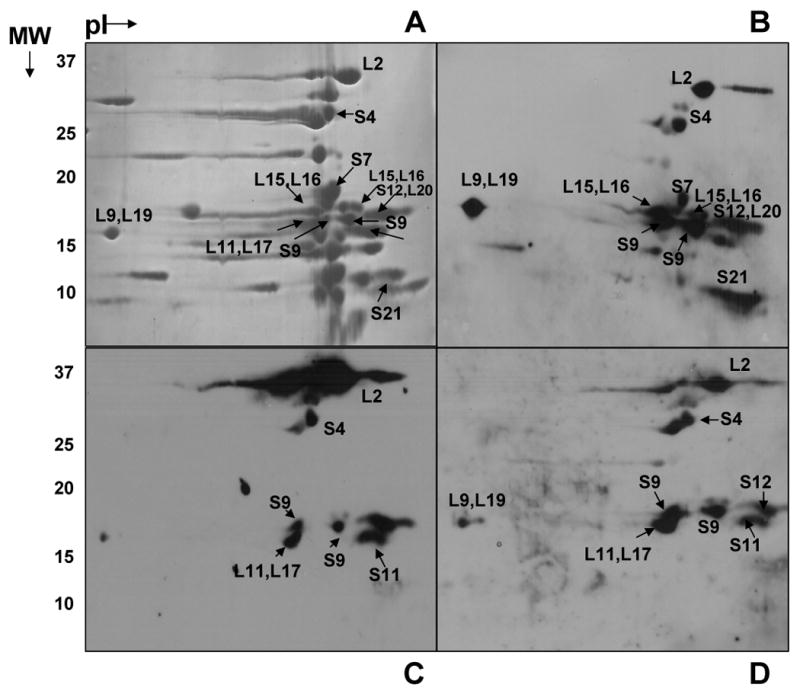
(A) 15 A260 units of E. coli ribosomes were separated on NEPHGE gels using pI 5-7 and 3-10 ampholytes. The gel was stained with Coomassie Blue. (B, C, D) Immunoblotting analysis of E. coli ribosomal proteins using phosphotyrosine, phosphoserine, and phosphothreonine antibodies, respectively. The regions labeled correspond to phosphorylated proteins identified by LC-MS/MS analysis from the tryptic digests of the gel pieces. Not all of the proteins that are found to be phosphorylated are marked in the figures.
Fig. 2. Further analysis of phosphorylated E. coli ribosomal proteins by NEPHGE gels.
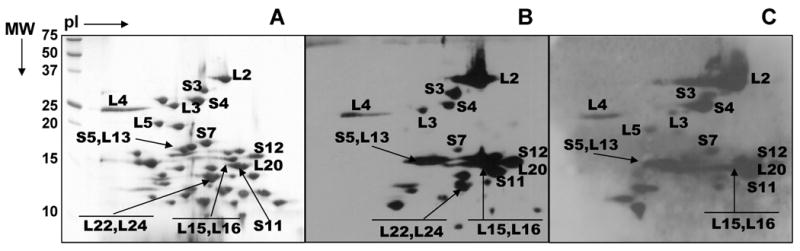
(A) 15 A260 units of E. coli ribosomes were better separated on NEPHGE gels using pI 8-10 and 3-10 ampholytes. The gel was stained with Coomassie Blue. (B and C) Immunoblotting analysis of E. coli ribosomal proteins using phosphoserine and phosphotyrosine antibodies, respectively. The regions labeled correspond to phosphorylated proteins identified by LC-MS/MS analysis from the tryptic and Lys-C digests of the gel pieces. Not all of the proteins that are found to be phosphorylated are marked in the figures.
In addition to immunoblotting analysis, 32P-orthophosphate was added to E. coli cell culture to promote incorporation of 32P phosphate into phosphorylated ribosomal proteins. Due to addition of limited amount of 32P ortho phosphate, most of the labeled phosphate was incorporated into rRNA and a very small amount of 32P-labeled the phosphoproteins of 70S ribosomes; therefore, RNaseA treatment of the ribosomes caused smearing of the phosphorylated proteins rather than obtaining well separated phosphorylated ribosomal protein bands (Fig. 3B). We also stained the ribosomal proteins separated on SDS-PAGE gels using phosphorylation specific ProQ stain (Fig. 3C). As seen in Fig. 3B and 3C, the majority of ribosomal proteins around 15kDa were highly phosphorylated in both 32P-labeling and ProQ staining of the ribosomal proteins.
Fig. 3. Detection of phosphorylated E. coli ribosomal proteins by [32P]-orthophosphate labeling and ProQ phospho staining.
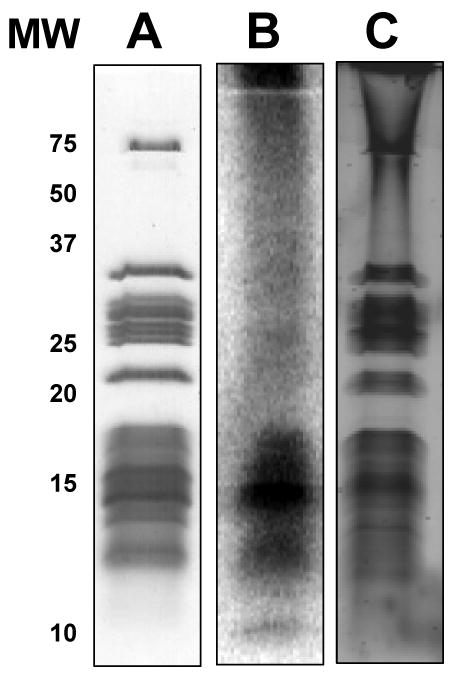
(A) Coomassie Blue stained gel of E. coli ribosomes. (B) E. coli ribosomes visualized by phosphor-imaging after labeling with [32P]-orthophosphate. (C) ProQ Diamond phosphoprotein stained gel of E. coli ribosomes visualized by a laser scanner at 532 nm.
Following the detection of the phosphorylated proteins of E. coli ribosomes detected by phosphorylation specific antibodies and the ProQ staining, mapping of the specific sites of phosphorylation was conducted using variety of proteomics approaches. These approaches employed various techniques to separate the proteins and enrich the phosphopeptides prior to tandem mass spectrometry (MS/MS). Phosphopeptide analysis by mass spectrometry presents unique challenges mainly due to varying amounts of phosphorylated peptides and also some difficulties faced during chromatography and even tandem mass spectrometry. In one approach, the ribosomal proteins were separated on 2D-gels prepared at two different ampholyte conditions and each spot detected by Coomassie Blue staining was digested in-gel by a protease (Fig. 1A and 2A). Although the coverage from the tryptic in-gel digestion of ribosomal proteins was usually above 50%, it was difficult to map some of the phosphorylation sites from the tryptic digests due to the basic character of the ribosomal proteins. For example, it was not possible to map any phosphorylation sites for S21 after trypsin and Lys-C digestions of the gel spots. Since some potential phosphorylation sites are in the lysine-rich regions of the basic protein, tryptic peptides from these regions were below the lower limit of the mass analyzer (∼ m/z 400) and were therefore not analyzed by the mass spectrometer.
As reported by many structural studies, conformational changes during translation is associated with many different functional states of ribosomes and these different functional states are essential for protein synthesis.27-30 For this reason, different stoichiometries of phosphorylated proteins or even specific phosphorylation sites within the same protein is possible due to different functional stages of ribosomes prepared at steady-state conditions. For example stoichiometry for phosphorylation of L18 is reported as 1:1 ratio; however, if the phosphorylation is regulatory, not structural, only a small fraction of the protein would be phosphorylated because of the different stages of translation at steady-state. 18 To overcome the low abundancy issue for some of the phosphorylation sites and map additional phosphorylation sites, two other proteomic approaches were employed with in-solution tryptic digestions of ribosomal proteins followed by two commonly used phosphopeptide enrichment methods, IMAC or SCX. In addition to two different 2D-gel conditions, these methods allowed us identify additional phosphopeptides (Table II). Basically, these separation/enrichment techniques helped us to simplify the sample as much as possible to facilitate the detection of the phosphopeptides within the time window available to the mass spectrometer. Samples enriched by IMAC using Galium-chelated resin also contained significant number of acidic peptides as expected in addition to the phosphopeptides. However, we chose not to convert the acidic moieties to their methyl esters due to significant loss of sample experienced during this process. The SCX chromatography proved to be very efficient and effective in both fractionation of the peptide mixtures and the enrichment of the phosphopeptides. At low pH (∼2.7), positive charges (2+ and higher for most tryptic peptides) on phosphopeptides are reduced due to presence of negatively charged phosphate groups and therefore result in convenient early elution of phosphopeptides in cation exchange chromatography. The early fractions were concentrated and easily desalted online since they all had less than ∼20 mM KCl as they eluted from the column. MS/MS analysis indeed produced more phosphopeptides in these early SCX fractions compared to the later fractions, which are expected to contain mostly non-phosphorylated peptides.
Table II. Phosphorylated Peptides of E. coli Ribosomal Proteins Detected by LC-MS/MS Analysis.
| Start - End | Sequence | Phosphosite | XCorr | m/z | Mr(expt) | Mr(calc) | Delta |
|---|---|---|---|---|---|---|---|
| S3- 30S | |||||||
| 27-37 | EFADNLDSˆDFK | S34 | 2.04 | 647.67 | 1283.34 | 1284.26 | -0.92 |
| 114-125 | LVADSˆITSQLER | S118 (T120, S121) | 3.06 | 658.02 | 1314.04 | 1314.49 | 0.44 |
| S4- 30S | |||||||
| 26-43 | AIDT#KCKIEQAPGQHGAR | T29 | 2.66 | 668.37 | 2003.11 | 2004.15 | 1.04 |
| 128-145 | VVNIAS#YQVSPNDVVSIR | S133 (Y134, S137) | 3.62 | 1020.69 | 2040.38 | 2041.19 | 0.81 |
| 128-145 | VVNIA#SY#QVSPNDVVSIR | S133, Y134 (S137) | 2.72 | 1061.00 | 2121.00 | 2121.17 | 0.17 |
| 165-176 | EKPT#WLEVDAGK | T169 | 2.70 | 727.99 | 1454.96 | 1453.52 | -1.44 |
| 188-205 | S#DLSADINEHLIVELY#SK | S188, Y203 (S204) | 2.98 | 735.51 | 2204.51 | 2207.21 | 2.70 |
| S5- 30S | |||||||
| 29-44 | IFS#FTALTVVGDGNGR | S31 (T33) | 2.06 | 867.88 | 1734.75 | 1734.83 | 0.08 |
| 29-51 | IFS#FT#ALTVVGDGNGRVGFGY#GK | S31, T33 (T36), Y49 | 2.84 | 868.45 | 2603.35 | 2603.60 | 0.25 |
| 93-111 | VFMQPAS#EGTGIIAGGAM*R | S99 | 1.30 | 995.66 | 1989.31 | 1990.19 | -0.13 |
| S7-30S | |||||||
| 53-75 | SˆGKS#ELEAFEVALENVRPTVEVK | S53, S56 | 3.52 | 865.26 | 2594.78 | 2594.81 | 0.03 |
| 56-75 | S#ELEAFEVALENVRPTVEVK | S56 | 3.29 | 780.39 | 2339.15 | 2340.51 | 1.36 |
| 79-94 | VGGS#T#Y#QVPVEVR | S82, T83, Y84 | 2.35 | 816.51 | 1632.02 | 1631.49 | -0.53 |
| 79-97 | VGGS#T#Y#QVPVEVRPVR | S82, T83, Y84 | 3.19 | 662.46 | 1985.37 | 1983.93 | -1.44 |
| 111-136 | GDKS#M*ALRLANELSDAAENKGTAVK | S114 (S124) | 3.92 | 896.60 | 2687.78 | 2686.88 | -0.90 |
| 143-150 | MAEANKAFAHY#R | Y153 | 2.43 | 745.95 | 1490.88 | 1489.58 | 1.30 |
| S11-30S | |||||||
| 14-36 | QVS#DGVAHIHASFNNTIVTITDR | S16 (S25) | 2.78 | 859.29 | 2575.86 | 2576.70 | 0.83 |
| 53-68 | GS#RKS#T#PFAAQVAAER | S54, S57, T58 | 1.92 | 639.19 | 1915.56 | 1916.80 | 1.23 |
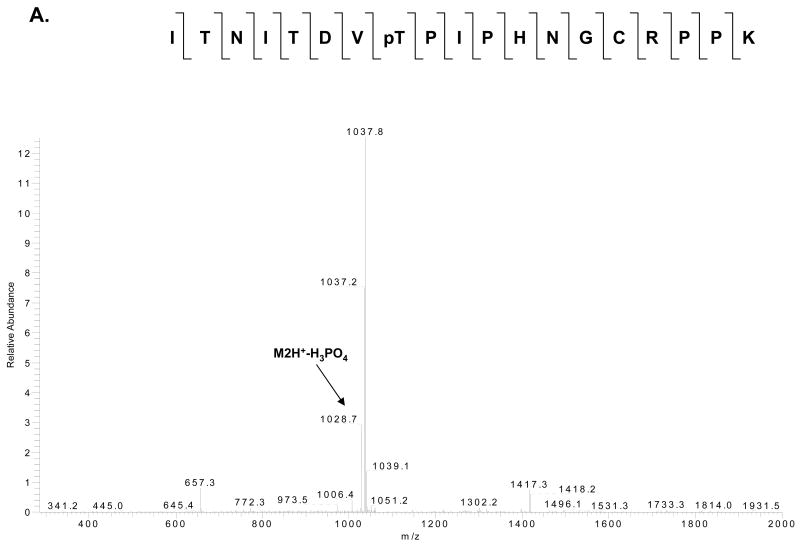
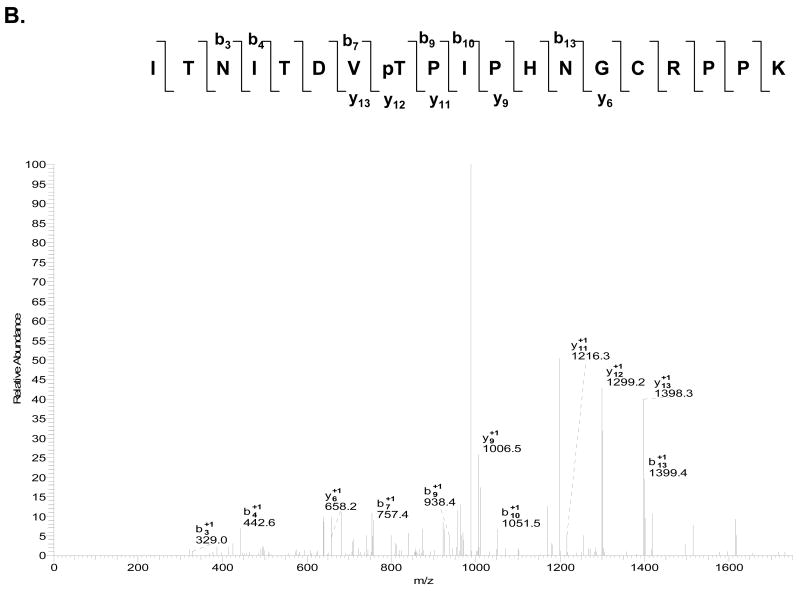
| 106-125 | ITNITDVT#PIPHNGCRPPK | T113 (T110) | 2.75 | 718.95 | 2154.83 | 2154.37 | 0.46 |
| 106-125 | ITNITDVTˆPIPHNGCRPPK | T113 (T110) | 2.69 | 1028.71 | 2056.37 | 2056.39 | -0.02 |
| S12- 30S | |||||||
| 36-50 | VY#TT#TPKKPNS#ALRK | Y37, T39 (T38), S45 | 2.18 | 972.55 | 1944.09 | 1944.93 | 0.85 |
| S13-30S | |||||||
| 44-56 | IS#ELSEGQIDTLR | S45 | 2.18 | 770.59 | 1540.18 | 1541.58 | 1.40 |
| S18- 30S | |||||||
| 30-42 | NY#IT#ESˆGKIVPSR | Y31, T33, S35 | 2.42 | 803.66 | 1606.31 | 1606.61 | 0.29 |
| L1-50S | |||||||
| 37-53 | FVES#VDVAVNLGIDARK | S40 | 2.65 | 956.54 | 1912.07 | 1913.06 | 0.99 |
| L2- 50S | |||||||
| 183-202 | VEADCRAT#LGEVGNAEHM*LR | T190 | 3.41 | 757.56 | 2270.67 | 2268.41 | -2.26 |
| L3- 50S | |||||||
| 14-33 | IFT#EDGVSIPVTVIEVEANR | T16 (S21, T25) | 3.65 | 757.04 | 2270.67 | 2269.43 | 0.33 |
| 14-33 | IFTˆEDGVS#IPVTVIEVEANR | T16 (S21), S21 (T25) | 3.96 | 751.41 | 2252.22 | 2251.43 | -0.79 |
| L5- 50S | |||||||
| 15-29 | LMT#EFNYNSVM*QVPR | T17 | 3.66 | 963.12 | 1925.24 | 1926.10 | 0.86 |
| 15-29 | LMT#EFNY#NSVM*QVPR | T17, Y21 (S23) | 3.38 | 669.19 | 2005.56 | 2006.08 | 0.52 |
| 125-147 | GNY#SM*GVREQIIFPEIDYDKVDR | Y127 or S128 | 2.58 | 948.57 | 2843.68 | 2842.03 | -1.65 |
| L6- 50S | |||||||
| 69-84 | ALLNS#MVIGVTEGFT#K | S73,T83 (T79) | 2.52 | 921.47 | 1841.93 | 1840.95 | -0.98 |
| L10-50S | |||||||
| 125-150 | LATLPTYEEAIARLM*ATMKEAS#AGK | S147 | 3.33 | 922.11 | 2764.31 | 2764.11 | -0.20 |
| L13- 50S | |||||||
| 14-23 | DWY#VVDATGK | Y16 | 2.06 | 616.83 | 1232.64 | 1234.23 | 1.59 |
| 39-60 | HKAEY#TˆPHVDTGDYIIVLNADK | Y44, T45 (T50) | 4.40 | 854.70 | 2562.08 | 2562.73 | 0.65 |
| 41-60 | AEY#TPHVDT#GDY#IIVLNADK | Y43 (T44), T49, Y53 | 3.01 | 825.71 | 2475.12 | 2475.37 | 0.25 |
| L14- 50S | |||||||
| 1-17 | M*IQEQTMLNVADNS#GAR | S14 (T6) | 2.73 | 664.62 | 1991.85 | 1991.09 | -0.76 |
| 109-122 | S#EKFMKIISLAPEVL | S109 (S117) | 2.58 | 596.68 | 1788.02 | 1786.06 | -1.96 |
| L16- 50S | |||||||
| 18-33 | GLAQGT#DVSFGSFGLK | T24 (S27) | 2.27 | 831.93 | 1662.85 | 1664.73 | 1.88 |
| 100-113 | VLY#EMDGVPEELAR | Y103 | 2.09 | 850.74 | 1700.48 | 1701.82 | 1.34 |
| 128-136 | TTFVT#KTVM | T132 (T134) | 1.53 | 554.77 | 1108.54 | 1108.23 | -0.31 |
| L18- 50S | |||||||
| 34-56 | HIYAQVIAPNGSˆEVLVAASTVEK | S45 (S52, T53) | 4.78 | 794.12 | 2380.37 | 2379.71 | -0.66 |
| L19- 50S | |||||||
| 14-28 | QDVPSˆFRPGDTVEVK | S18 | 3.09 | 829.02 | 1657.04 | 1656.84 | -0.20 |
| L21- 50S | |||||||
| 1-10 | M*Y#AVFQSGGK | Y2 | 2.01 | 592.77 | 1184.53 | 1184.24 | -0.29 |
| L22- 50S | |||||||
| 29-41 | VSQALDILT#Y#T#NK | T37, Y38, T39 | 2.39 | 854.25 | 1707.48 | 1706.60 | -0.88 |
| 100-110 | T#S#HIT#VVVSDR | T100, S101, T104 | 2.24 | 728.06 | 1455.10 | 1454.29 | -0.81 |
| L28- 50S | |||||||
| 4-18 | VCQVT#GKRPVTGNNR | T8 | 2.40 | 855.26 | 1709.51 1 | 709.8500 | 0.34 |
| L31- 50S | |||||||
| 9-23 | YEEITAS#CS#CGNVMK | S15 (T13), S17 (S15) | 2.91 | 899.22 | 1797.43 | 1795.83 | -1.60 |
| L31-B2- 50S | |||||||
| 34-52 | VIELDGVTYPYVTIDVS#SK | S50 (S51) | 2.47 | 1090.55 | 2178.60 | 2179.35 | -0.74 |
(ˆ) and (#) show the phosphorylated Ser(S), Thr (T), and Tyr(Y) residues. Residues in parenthesis show the other possible phosphorylation site(s) detected. (*) indicates the methionine oxidation. Mass spectrometry data was searched against the E. coli protein sequences in SwissProt database using Bioworks 3.2.
The mass spectrometry data obtained from phosphorylated peptides either by 2D-gel and phosphopeptide specific enrichment techniques is listed in Table II and each spectrum was included in the supplementary Fig. S1. The spectral data were processed and searched against the Swiss-Prot database using both Mascot and Sequest search engines; however, only the Sequest scores are provided in the tables. In the case of uncertainty about the exact location of the phosphorylation site for phosphopeptides with more than one possible site, other possible sites were given in parenthesis in Tables II. Moreover, protein sequences of phosphorylated proteins of E. coli ribosomes were aligned with homologous ribosomal proteins from other bacterial species and human mitochondria to evaluate the conservation of phosphorylation sites in these proteins (Supplemental Fig. S2). In the following paragraphs, phosphorylated ribosomal proteins were grouped according to their possible roles in ribosome function and potential roles of phosphorylation is discussed.
Phosphorylated proteins located around the mRNA binding path
Phosphorylated proteins S3, S4, S5, S7, S11, S18 and S21 are located between the head, shoulder and platform of the 30S subunit, which forms the part of the mRNA binding path interacting with the 3′ and 5′-ends of mRNAs during translation.31-33 Closing of this path around the mRNA has been suggested to have a role in processivity and directionality of mRNA movement.33,34 The mRNA is surrounded by S3 at the top, S4 at the bottom, and S5 at the lower left (Fig. 4A). S7 and S11, S18 and S21 are located in the head and platform regions of the small subunit right across from each other respectively, and both interact with mRNA and tRNA at different conformational stages of ribosomes during translation.25,26,33,35
Fig. 4. 3D-model of the 30S subunit from E. coli indicating the location of each phosphorylated ribosomal protein.
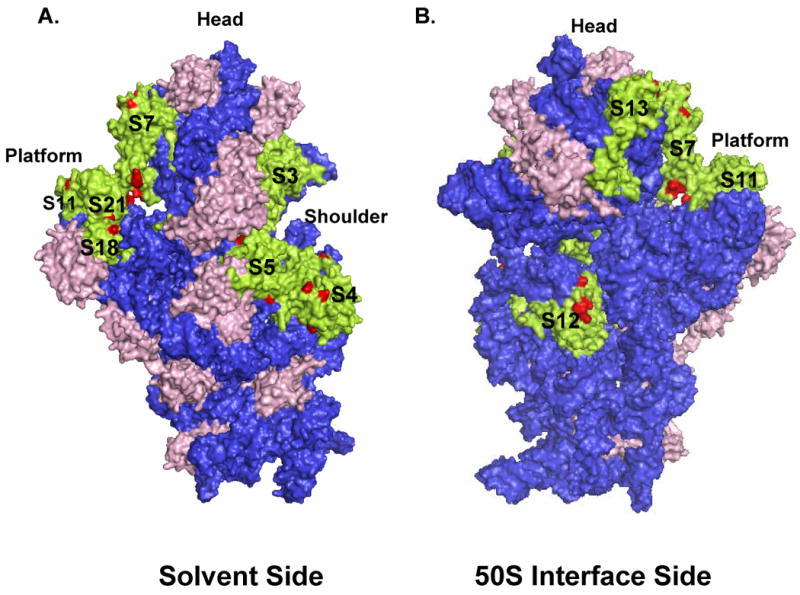
The phosphorylated ribosomal proteins in the 30S subunit are represented by green color, while the phosphorylated residues mapped by mass spectrometry and unphosphorylated proteins are colored red and pink, respectively. Coordinates of the E. coli 30S subunit were obtained from the Protein Data Bank (Acc. # 2AW7). (A) Depicting the 30S subunit from the solvent side, while (B) gives a view of the small subunit from the 50S interface.
In the immunoblotting analyses, phosphorylation of S3 and S4 was clearly detected by all three antibodies, (Fig. 1 and 2). Phosphorylation sites found in S3 are Ser34 and Ser118 (Thr120 or Ser121) (Table II). Some of these residues, specifically, Ser34, appear to be involved in intermolecular interactions to provide possible conformational changes and the other phosphorylated residue, Ser118 (Thr120 or Ser121), is located in the α-helix and loop region of the domain II extending towards the mRNA-binding path. In the MS/MS analysis, phosphorylation sites in S4 were found at Thr29, Ser133, Tyr134, (Ser137), Thr169, and Ser188 (Ser204) residues (Table II). S5 is a 17.5 kDa protein phosphorylated at Ser, Thr, and Tyr residues as detected in both immunoblotting of 2D-gels and mapping by tandem mass spectrometry (Fig. 2, Table II). The phosphorylated Ser, Thr, and Tyr residues mapped in these proteins are distributed mainly at the solvent accessible sites (Fig. 4A).
Ribosomal S7 is a primary rRNA binding protein responsible for arranging the 3′ major domain of the 16S rRNA to fold properly enabling other proteins to bind.36,37 Structural studies clearly showed that the head and platform of the 30S subunit undergo conformational changes during subunit association, binding of aminoacyl-tRNA or factors, and translocation.1,5,26,38,39 In our studies, S7 was found to be phosphorylated in immunoblotting analyses performed with phospho amino acid specific antibodies and in MS/MS analyses at residues Ser53, Ser56, Ser82, Thr83, Tyr84, Ser114(Ser124) and Tyr149 (Table II). Phosphorylation sites at Ser53, Ser56, and Ser114 (Ser124) are in close proximity to each other and located at the top of the S7 protein. In addition, a phosphopeptide containing Ser82, Thr83, and Tyr84 residues was also repeatedly detected in the analysis (Table II). Another phosphorylated Tyr residue, Tyr153, was found in a peptide located near the mRNA binding path. Interestingly, these residues are at the tip of the flexible arm of the S7 towards the shoulder region, where it interacts with S11 and phosphorylation of these residues will hinder mRNA binding in this region (Fig. 4B). Therefore, phosphorylation of S7 at these residues might be important in release of mRNA and/or tRNA from the ribosome.
S11 is a tertiary RNA binding protein located in the platform region of the small subunit with highly conserved Ser and Thr residues. An interaction at the E site was found between S7 and S11, which most likely is involved in the 30S conformational changes that take place during protein synthesis.35,40 When either S7 or S11 mutated, translational errors such as codon misreading and frameshifting were increased.41 Phosphorylation of highly conserved Ser and Thr residues may play an important function in conformational changes of the 30S subunit and/or translocation to ensure proper readthrough of the mRNAs. In fact, S11 is mainly phosphorylated at Ser and Thr residues as determined from immunoblotting analyses of 2D-gels and by mass spectrometry at residues Ser16 (Ser25), Ser54, Ser57, Thr58, and Thr113 (Thr110) (Table II). All these residues are located in two N-terminal β-sheet structures where S11 interacts with the 16S rRNA. Phosphorylated residues, Ser54, Ser57, and Thr58, are all positioned in a loop region between the N-terminal tail and the helix1 where S11 interacts with S7 at the mRNA binding path. Especially, Thr58 is a very highly conserved residue among the bacterial, archaeal, and mitochondrial S11 homologs (Fig 5C). The other phosphorylated residue(s), Thr113 or Thr110, is located in protein-protein interfaces; specifically Thr113 is located near the positively charged residues the C-terminal ends of S18 and S21 proteins (Fig. 4B).11 Phosphorylation of these residues in S11 may cause a major rearrangement of S11, S18, and S21 in the platform region of the 30S subunit during mRNA decoding and translocation.
Fig. 5. MS/MS and MS3 spectra of a phosphorylated tryptic peptide from E. coli ribosomal S11.

(A) CID MS/MS spectrum for doubly–charged phosphorylated peptide ITNITDVpTPIPHNGCRPPK (m/z 1077.5) with a neutral loss of 49 Da. (B) MS3 fragmentation spectrum of the dephosphorylated peptide (m/z 1028.7). (C) The primary sequence alignment of the tryptic peptide from E. coli S11 with the peptides from the same region of several bacterial and the human mitochondrial S11 sequences. Abbreviations for bacterial species: ECOLI; E. coli, HAEIN; Haemophilus Influenzae, MYCTU; Mycobacterium tuberculosis, BACSU; Bacillus subtilis; and THET8; Thermus thermophilus. The alignment was processed with the CLUSTALW program in Biology Workbench and the results are displayed in BOXSHADE. The two asterisks indicate possible phosphorylated residues as detected in the LC-MS/MS analysis of the phosphorylated tryptic peptide.
S18 and S21 are located in the platform region of the 30S subunit that interacts with 5′ ends and the Shine Dalgarno helix in bacterial mRNAs. Specific interaction of this helix and S18 is important for the initiation complex formation.42-45 In the crystal structure of the initiation complex solved by Yusupova et al., the highly flexible and basic N-terminal end of the S18 protein is in close contact with the Shine Dalgarno helix in the initiation complex whereas in the post-initiation complex this helix is shifted and rotated to interact with S2 rather than S18.46 The S18 protein was determined to be phosphorylated at residues Tyr31, Thr33, and Ser35 (Table II). On the other hand, S21 is found to be phosphorylated only at Tyr residues in the immunoblotting experiments (Fig. 1B). Interestingly, there are two highly conserved Tyr residues in bacterial S21 homologs; however, no phosphorylated Tyr residue was detected and mapped in our LC-MS/MS analysis due to the size of the tryptic peptides obtained from S21 spot. Possibly, the phosphorylated residues in both S18 and S21 might be involved in either protein-protein or protein-mRNA interactions in the platform region exposed to mRNA binding surface of the protein (Fig. 4B).
Phosphorylated proteins at the interface region of the small and large subunits
S12 and L2 are the two proteins located at the interface between the small and large subunits of the ribosome near the tRNA-binding sites.35 In the LC-MS/MS analysis of 2D-gel spots, ribosomal proteins S12 and L20 were identified in the same spot. This S12 and/or L20 spot appears to be Ser, Thr, and Tyr phosphorylated in the immunoblotting experiments (Fig. 1 and 2). Due to high Lys and Arg content of these proteins, only phosphorylated tryptic peptide detected was from S12. For this reason, we did not include L20 as a phosphorylated ribosomal protein in Table I.
Phosphorylation of S12 at Ser and Thr residues was also demonstrated by a ribosome associated kinase in S. collinus. 15 S12 is important for tRNA decoding at the A site specifically in relation to the wobble base (the third position), where coordination of a magnesium ion occurs. 24 For this reason, it is conceivable to suggest that the phosphorylation of S12 might be important in the A site tRNA binding, especially after locating the phosphorylated residues in close proximity to the hinge region of the 16S rRNA around position 912 and H44 in the crystal structure (Fig. 4B).
L2 is another phosphorylated protein located right at the interface between the subunits and this protein absolutely required for subunit association as a primary rRNA binding protein and important for peptidyl transferase activity. 47-49 Evidently, L2 is the second largest 50S protein consisting of 273 residues and it is one of the most highly conserved ribosomal proteins with numerous functions in protein synthesis. Our data shows that L2 is a highly phosphorylated at Ser, Thr, and Tyr residues (Fig. 1 and 2, Table I). Previously, in vitro phosphorylation of this E. coli ribosomal protein by a protein kinase from rabbit skeletal muscle was reported.13 Even though L2 was found to be phosphorylated at Ser, Thr and Tyr residues due to high occurrence of tryptic Lys and Arg sites, we were not able to detected many of the highly conserved and predicted phosphorylation sites in L2. Only one phosphorylated peptide was identified in the mass spectrometric analysis of the 2D-gel spot (Fig. 1A and 2A). The only phosphorylated Thr residue is located at position 190 and in the globular domain of the L2 protein (Table II). In the event of Thr190 phosphorylation, the globular domain of L2 may be expanded due to charge repulsion in the rRNA-protein interface and this expansion might be important in subunit association/dissociation (Fig. 6A).
Fig. 6. 3D-model of the E. coli 50S subunit showing the position of each phosphorylated ribosomal protein.
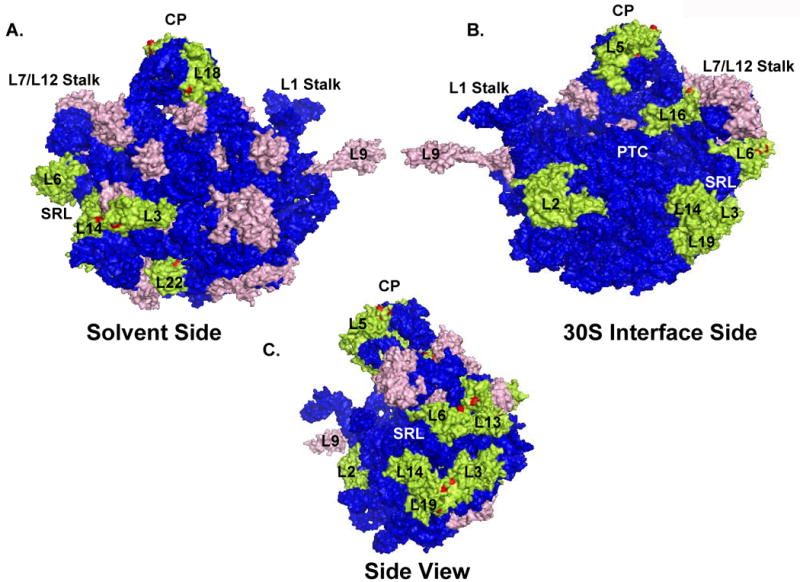
The color coding is the same as in Fig.4. Coordinates of the E. coli 50S subunit were obtained from the Protein Data Bank (Acc. # 2AW4). (A) Depicting the 50S subunit from the solvent side, (B) gives a view of the large subunit from the 30S interface, and (C) is the side view of the large subunit showing the phosphorylated proteins around the SRL. Specific regions, peptidyl transferase center (PTC), central protuberance (CP), sarcin-ricin loop (SRL), L1 and L7/L12 stalks and the 50S ribosomal proteins were labeled in the model generated by PyMol software (DeLano Scientific LLC).
Phosphorylated proteins located at the central protuberance and 30S head regions
L5 is the only protein located at the interface region of the central protuberance of the 50S subunit, where it interacts with the small subunit proteins S13 and S19 in the head of the 30S subunit. Ribosomal L5 is a 20 kDa protein phosphorylated at Ser, Thr, and Tyr residues as detected by immunoblotting and tandem MS analyses (Fig.2, Table I). Some of these phosphorylation sites were found as one or two phosphorylations per peptide; however, it was not possible to determine the exact phosphorylation sites in some of the peptides (Table II). Many of the phosphorylation sites in L5 are probably involved in conformational rearrangements of L5 and 23S rRNA during translocation in the central protuberance. The phosphorylated peptide containing the phosphorylated Tyr127 has another very highly conserved Tyr residue at position 142; however, the score for phosphorylation of Tyr142 is low compared to the Tyr127 phosphorylation. On the other hand, Tyr142 is more likely to be phosphorylated according to the NetPhos predictions. Moreover, Tyr142 is located in the extended β4-β5 loop and might be significant in regulating the role of L5 in subunit association and ratchet-like movement of EF-G bound ribosomes during translocation and subunit dissociation.39,50,51 In the E. coli structure, the side chain of Tyr142 is surrounded by hydrophobic residues (Ile135, Ile140, Val145, and Val148). 11,39 Phosphorylation of this Tyr residue gets phosphorylated at any stage of translation would cause rejection from the hydrophobic pocket and cause conformational changes in interactions between the two flexible loops of L5 and S13 and S19 proteins at the inter-subunit bridges B1a, B1b, and B1c.
In the small subunit, S13 is located right across from the L5 and only one phosphorylated peptide was detected from S13 containing Ser45, Ser48 and Thr54. Even though the highest phosphorylation score was obtained for Ser45, Ser48 is more conserved among the bacterial S13 proteins and it is predicted to be phosphorylated (Fig. S2). All of these possible phosphorylation sites either Ser45 or Ser48 residues are on the surface of S13 at the rRNA-protein interface.
Phosphorylated proteins surrounding the SRL region
Ribosomal proteins L3, L6, L13, L14 and L19 are positioned below the L7/L12 stalk organizing the sarcin-ricin loop (SRL) domain, mainly domain VI of 23S rRNA, for factor binding (Fig. 6A). 26,52 Phosphorylation of L3, L6, L13, and 19 were detected on 2D-gels and identified in the LC-MS/MS analyses; however, due to the high degree of phosphorylation and differences in pIs, it was difficult to determine specific phosphorylation status of some of these proteins in the immunoblotting analyses (Fig. 1 and 2, Table I). Therefore, most of the phosphorylated peptides for these proteins were identified in repeated analyses of IMAC and SCX from the in-solution digestions of ribosomal proteins.
L3 is one of the proteins that might have an effect on peptidyl transferase activity of ribosomes due to its close proximity to the active center.53,54 In immunoblotting and mass spectrometric analyses, L3 is mainly phosphorylated at Ser and Thr residues at positions Thr16, Ser21, and Thr25 (Fig. 1 and 2, Table II). These phosphorylated residues reside at the N-terminal end of the protein at the L3-L14 protein interface to possibly increase the strength of protein-protein interactions; however, Ser21 is completely solvent accessible and may be involved directly in factor binding.
L6 is located right below the L7/L12 stalk and extended towards the back of the large subunit. 11 Similar to proteins surrounding the SRL region, the surface of L6 and L13 proteins provide interaction sites for elongation factors binding to the ribosomes. 52,55 The peptide identified for L6 revealed phosphorylation sites at residues Ser73 (Thr79) and Thr83 (Table II). It is conceivable to propose a role for all these phosphorylation sites in regulation of factor binding to L6 due to their location below the SRL region (Fig. 6B, C). Especially, residues Thr79 and Thr83 are highly conserved in L6 homologs from many bacteria, and Thr83 is replaced by a Glu residue in Bacillus possibly implying the requirement for a negatively charged residue in that position (Fig. S2).
Ribosomal L13 was one of the eight ribosomal proteins that were found in active sub-ribosomal particles containing both 23S and 5S rRNA.56 C-terminal end of the L13 is closely positioned at the factor binding site between L3 and L6 proteins, towards the back of the 50S subunit and the other end interacts with ribosomal proteins L20 and L21 (Fig. 6A).52 Almost all of the phosphorylated residues found at Tyr16, Tyr44, Thr45, Thr50, and Tyr53 positions are solvent accessible towards the back of the 50S subunit with possible roles in binding of ribosome interacting proteins. The other phosphorylated protein that is interacting with L13 is L21 and the only phosphorylated residue detected in our analysis is located at Tyr2. This residue is at the N-terminal end of L21 that interacts with L20 and L13.11
L14 and L19 form the parts of the intersubunit bridges between the small and large subunits; therefore they may have significant roles in subunit association.26,57 The ribosomal L14 protein is phosphorylated at Thr6 and Ser109 residues. Even though L19 and L9 proteins were not clearly separable due to their molecular masses and pIs, the 2D-gel spot appeared to be phosphorylated at Ser, Thr and Tyr residues (Fig. 1, Table I). We were able to map a Ser phosphorylation site in L19 protein but could not detect the peptide covering the highly conserved Tyr98 and Tyr99 residues. The phosphorylated L19 peptide obtained from tryptic digests of 2D-gel spot revealed the phosphorylation of Ser18 and phosphorylation of this residue was also determined to be phosphorylated in a recent high throughput phospoproteomics analysis of the E. coli proteome.16 From a study using reverse genetics, it was shown that L19 is essential for cell viability.58 The mutations in L19 resulted in increased sensitivity to aminoglycosides active at the A-site, indicating a perturbation of the decoding step; however, this mutation was compensated by a mutation in S12 at Lys42.59 We have detected two phosphorylation sites, Ser18 and Ser82 (Ser84), in L19 and interestingly, these residues are in the same axis as Gln40 in the crystal structure of 50S subunit at 3.5 Å.11 Therefore, the phosphorylated form of L19 may be the active form allowing for the assembly and decoding processes to occur.
Phosphorylated L7/L12 stalk proteins
L7/L12 stalk is located on the opposite side of the L1 stalk in the 50S subunit and is essential for the function of elongation factors EF-Tu and EF-G, initiation factor IF-2, and release factor RF3 on the ribosome.38,60-62 The stalk is formed by L10, L11 and multiple copies of L7/L12 and due to flexibility of this stalk only L11 was mapped in the crystal structure of the ribosome11. Among these three proteins only L11 was detected as phosphorylated in a spot overlapping with L17 in the 2D-immunoblotting analysis of the ribosome preparations (Fig. 1). We were not able to map any specific phosphorylation sites on L11 while it was possible to detect phosphorylated peptides in-solution digestions of ribosome preparations for phosphorylated L7/L12 by MS/MS (Table II).
In early cross-linking studies, L7/L12 was determined to be the only ribosomal protein that is present in multiple copies, four copies in E. coli and six copies in thermophiles.17,63 Based on structural investigations by cryo-EM and NMR, the protein is determined to be composed of two distinct globular domains connected by a linker called the “hinge region”. The N-terminal domain has a role in dimerization and anchoring L7/L12 to the ribosome, while the C-terminal domain interacts with the factors during translation. Finally, the hinge region, including residues 33 to 52, of L7/L12 is an alanine rich region responsible for large conformational changes of the C-terminal domain of the protein during translocation.30,64-66 Recently, requirement of L7/L12 phosphorylation and acetylation for ribosome association was demonstrated by mass spectrometry; however, the specific sites for phosphorylation were not determined.17,67 In a recent phosphoproteome analysis of E. coli, L7/L12 was also found to be phosphorylated at Ser89.16 We determined three additional phosphorylation sites that are located at the Ser15, Ser33 and Thr52 (Table II). Clearly, phosphorylation of residues located at the beginning (Ser33) and end (Thr52) of the linker or hinge region could possibly play a role in the conformational changes of the C- and N-terminal domains of L7/L12 during translation.
Other phosphorylated proteins of the 70S ribosome
Ribosomal protein L16 is an essential component of the bacterial ribosome that organizes the tRNA interacting site in the 50S subunit.68 It is also one of the only few ribosomal proteins come close to the peptidyl transferase cleft, and therefore, a preferred target for antibiotic binding to the ribosome (Fig. 6B). In the 2D-gel systems used, L16 was found in multiple spots along with L15 (Fig. 2); however, the mass spectrometric analysis of these spots revealed the presence of phosphorylated peptides only from L16. We have found three phosphorylated peptides with phosphorylation sites located at residues Thr24 (Ser27), Tyr103, and Thr132 (Thr134) (Table II) and many of these phosphorylated residues are located in the globular part of the L16 away from the cleft possibly involved in protein binding to the L7/L12 stalk base (Fig. 6B).
Another phosphorylated protein that we detected is L22 and it is located at a L22 protein consist of a globular domain exposed to the rear surface of the large subunit and an extension penetrating into the 23S rRNA forming the lining of the peptide exit tunnel.52,69 The extensions contributing to the lining of the peptide exit tunnel have been implicated in a gating mechanism that might regulate the exit of nascent peptides and mutations in these extensions are also involved in antibiotic sensitivity.70-72 In the 2D-gel system, L22 was detected in multiple spots (Fig. 2) along with L24 and only two phosphorylated peptides were detected in the mass spectrometric analysis of the tryptic digests of these spots. Both peptides had multiple phosphorylation sites at Thr37, Tyr38, Thr39 and Thr100, Ser101, Thr104 residues. The phosphorylated peptide containing Thr37, Tyr38 and Thr39 residues are located in the globular domain near the L32 interface and the other phosphorylated residues Thr100, Ser101 and Thr104 are at the peptide exit tunnel.
L18 was only identified in LC-MS/MS analysis of in- solution digestions of 70S ribosomes with over 60% coverage. In fact, Bloemink and Moore have shown that the phosphorylation of L18 is necessary for 5S rRNA binding to 23S rRNA and the phosphorylated Ser residue was predicted to be at position 57 in B. stearothermophilus L18.18 In the mass spectrometric analysis of peptides from L18, we determined that Ser45 is the most likely residue to be phosphorylated in E. coli rather than Ser52 that corresponds to Ser57 in B. stearothermophilus. However, the phosphorylated peptide containing the Ser45 residue was also predicted to be phosphorylated at Ser52 or Thr53 with a lower score (Table II). All these residues that are located in a peptide extends at the rRNA-protein interface may be involved in L18 binding to 5S rRNA. In addition, L18 is near the end of the signal recognition particle (SRP) binding path and phosphorylated Ser45 can also be important for SRP-large subunit interactions.73
Conclusions
Data presented here and previous studies on phosphorylated proteins of bacterial ribosomes demonstrate the significance of phosphorylation in function and regulation of protein synthesis. 13-16 Therefore, in order to better understand the role of essential ribosomal proteins in protein synthesis, post-translational modification analysis of these proteins was a mandatory task for proteomics researchers. We accomplished this task partially by identifying the phosphorylated ribosomal proteins and specific phosphorylation sites at steady-state levels in E. coli. Mapping of the phosphorylated residues in these ribosomal proteins will provide structurally relevant insights into the role of phosphorylation in ribosome function and regulation. As modeled in the crystal structures of E. coli subunits, the phosphorylated residues are mostly on the surface of ribosomal proteins with solvent accessible -OH side chains where functions of ribosomal proteins could be regulated by this modification. In general, phosphorylation patterns seen in the small and large subunit, especially around the mRNA binding path and SRL region, imply the significance of this modification in protein synthesis. On the other hand, phosphorylated ribosomal proteins with no assigned function might participate in an unknown function or conformational change which is yet to be discovered.
Supplementary Material
Fig. S1 – MS/MS and MS3 spectra of a phosphorylated tryptic peptides from E. coli ribosomal proteins. CID MS/MS and MS3 spectra for phosphorylated peptides indicating either a neutral loss of 98 Da from the phosphorylated peptide or addition of 80 Da to the unphosphorylated peptide shown by ˆ and #, respectively. (*) indicates the methionine oxidation. Mass spectrometry data was searched against the E. coli protein sequences in SwissProt database using Bioworks 3.2.
Fig. S2 – The primary sequence alignments of E. coli ribosomal proteins in several bacterial species and the human mitochondrial sequence. Abbreviations for bacterial species: ECOLI; E. coli, HAEIN; Haemophilus Influenzae, MYCTU; Mycobacterium tuberculosis, BACSU; Bacillus subtilis; and THET8; Thermus thermophilus. Alignments were processed with the CLUSTALW program in Biology Workbench and the results are displayed in BOXSHADE. Asterisks indicate possible phosphorylated residues as detected in the LC-MS/MS analysis of the phosphorylated tryptic peptide.
Acknowledgments
The authors thank Dr. Katsuhiko Murakami for his valuable contribution in modeling the ribosome structure by PyMol software. This work was supported by the National Institute of Health grants R01 GM071034-01A1 and EB005197-01 to E.C.K.
References
- 1.Carter AP, Clemons J, Brodersen DE, Morgan-Warren RJ, Hartsch T, Wimberly BT, Ramakrishnan V. Crystal Structure of an Initiation Factor Bound to the 30S Ribosomal Subunit. Science. 2001;291:498–501. doi: 10.1126/science.1057766. [DOI] [PubMed] [Google Scholar]
- 2.Nissen P, Hansen J, Ban N, Moore PB, Steitz TA. The Structural Basis of Ribosome Activity in Peptide Bond Synthesis. Science. 2000;289:920–930. doi: 10.1126/science.289.5481.920. [DOI] [PubMed] [Google Scholar]
- 3.Bacher JM, de Crecy-Lagard V, Schimmel PR. Inhibited cell growth and protein functional changes from an editing-defective tRNA synthetase. Proc Natl Acad Sci USA. 2005;102:1697–1701. doi: 10.1073/pnas.0409064102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker AM, Draper DE. Messenger RNA recognition by fragments of ribosomal protein S4. J Biol Chem. 1995;270:22939–22945. doi: 10.1074/jbc.270.39.22939. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal RK, Spahn CM, Penczek P, Grassucci RA, Nierhaus KH, Frank J. Visualization of tRNA movements on the Escherichia coli 70S ribosome during the elongation cycle. J Cell Biol. 2000;150:447–460. doi: 10.1083/jcb.150.3.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodnina MV, Wintermeyer W. Ribosome fidelity: tRNA discrimination, proofreading and induced fit. Trends Biochem Sci. 2001;26:124–130. doi: 10.1016/s0968-0004(00)01737-0. [DOI] [PubMed] [Google Scholar]
- 7.Dallas A, Noller HF. Interaction of Translation Initiation Factor 3 with the 30S Ribosomal Subunit. Mol Cell. 2001;8:855–864. doi: 10.1016/s1097-2765(01)00356-2. [DOI] [PubMed] [Google Scholar]
- 8.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 9.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 Å resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 10.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 11.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 Å resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 12.Selmer M, Dunham CM, Murphy FVT, Weixlbaumer A, Petry S, Kelley AC, Weir JR, Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 13.Traugh JA, Traut RR. Phosphorylation of ribosomal proteins of Escherichia coli by protein kinase from rabbit skeletal muscle. Biochemistry. 1972;11:2503–2509. doi: 10.1021/bi00763a019. [DOI] [PubMed] [Google Scholar]
- 14.Mikulik K, Suchan P, Bobek J. Changes in ribosome function induced by protein kinase associated with ribosomes of Streptomyces collinus producing kirromycin. Biochem Biophys Res Commun. 2001;289:434–443. doi: 10.1006/bbrc.2001.6017. [DOI] [PubMed] [Google Scholar]
- 15.Mikulik K, Janda I. Protein kinase associated with ribosomes phosphorylates ribosomal proteins of Streptomyces collinus. Biochem Biophys Res Commun. 1997;238:370–376. doi: 10.1006/bbrc.1997.7297. [DOI] [PubMed] [Google Scholar]
- 16.Macek B, Gnad F, Soufi B, Kumar C, Olsen JV, Mijakovic I, Mann M. Phosphoproteome analysis of E. coli reveals evolutionary conservation of bacterial Ser/Thr/Tyr phosphorylation. Mol Cell Proteomics. 2008;7:299–307. doi: 10.1074/mcp.M700311-MCP200. [DOI] [PubMed] [Google Scholar]
- 17.Ilag LL, Videler H, McKay AR, Sobott F, Fucini P, Nierhaus KH, Robinson CV. Heptameric (L12)6/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc Natl Acad Sci USA. 2005;102:8192–8197. doi: 10.1073/pnas.0502193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bloemink MJ, Moore PB. Phosphorylation of ribosomal protein L18 is required for its folding and binding to 5S rRNA. Biochem. 1999;38:13385–13390. doi: 10.1021/bi9914816. [DOI] [PubMed] [Google Scholar]
- 19.Remold-O'Donnell E, Thach RE. A new method for the purification of initiation factor F2 in high yield, and an estimation of stoichiometry in the binding reaction. J Biol Chem. 1970;245:5737–5742. [PubMed] [Google Scholar]
- 20.Graves M, Breitenberger C, Spremulli LL. Euglena gracilis chloroplast ribosomes: Improved isolation procedure and comparison of elongation factor specificity with prokaryotic and eukaryotic ribosomes. Arch Biochem Biophys. 1980;204:444–454. doi: 10.1016/0003-9861(80)90055-7. [DOI] [PubMed] [Google Scholar]
- 21.Cahill A, Baio D, Cunningham C. Isolation and characterization of rat liver mitochondrial ribosomes. Anal Biochem. 1995;232:47–55. doi: 10.1006/abio.1995.9962. [DOI] [PubMed] [Google Scholar]
- 22.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 23.Draper D, Reynaldo LP. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999;27:381–388. doi: 10.1093/nar/27.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brodersen DE, Nissen P. The social life of ribosomal proteins. FEBS J. 2005;272:2098–2108. doi: 10.1111/j.1742-4658.2005.04651.x. [DOI] [PubMed] [Google Scholar]
- 25.Cate JH, Yusupov MM, Yusupova GZ, Earnest TN, Noller HF. X-ray crystal structures of 70S ribosome functional complexes. Science. 1999;285:2095–2104. doi: 10.1126/science.285.5436.2095. [DOI] [PubMed] [Google Scholar]
- 26.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JHD, Noller HF. Crystal Structure of the Ribosome at 5.5 A Resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 27.Simonetti A, Marzid S, Jenner L, Myasnikov A, Romby P, Yusupov G, Klaholz BP, Yusupov M. A structural view of translation initiation in bacteria. Cell Mol Life Sci. 2008 doi: 10.1007/s00018-008-8416-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vila-Sanjurjo A, Schuwirth BS, Hau CW, Cate JH. Structural basis for the control of translation initiation during stress. Nat Struct Mol Biol. 2004;11:1054–1059. doi: 10.1038/nsmb850. [DOI] [PubMed] [Google Scholar]
- 29.Berk V, Zhang W, Pai RD, Cate JH. Structural basis for mRNA and tRNA positioning on the ribosome. Proc Natl Acad Sci USA. 2006;103:15830–15834. doi: 10.1073/pnas.0607541103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Datta PP, Sharma MR, Qi L, Frank J, Agrawal RK. Interaction of the G′ domain of elongation factor G and the C-terminal domain of ribosomal protein L7/L12 during translocation as revealed by cryo-EM. Mol Cell. 2005;20:723–731. doi: 10.1016/j.molcel.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Dontsova OA, Rosen KV, Bogdanova SL, Skripkin EA, Kopylov AM, Bogdanov AA. Identification of the Escherichia coli 30S ribosomal subunit protein neighboring mRNA during initiation of translation. Biochimie. 1992;74:363–371. doi: 10.1016/0300-9084(92)90114-t. [DOI] [PubMed] [Google Scholar]
- 32.Frank J, Zhu J, Penczek P, Li Y, Srivastava S, Verschoor A, Radermacher M, Grassuccl R, Lata R, Agrawal R. A model of protein synthesis based on cryo-electron microscopy of the E. coli ribosome. Nature. 1995;376:441–444. doi: 10.1038/376441a0. [DOI] [PubMed] [Google Scholar]
- 33.Yusupova GZ, Yusupov MM, Cate JH, Noller HF. The Path of messenger RNA through the Ribosome. Cell. 2001;106:233–241. doi: 10.1016/s0092-8674(01)00435-4. [DOI] [PubMed] [Google Scholar]
- 34.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 35.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 36.Green R, Noller H. Ribosomes and translation. Ann Rev Biochem. 1997;66:679–716. doi: 10.1146/annurev.biochem.66.1.679. [DOI] [PubMed] [Google Scholar]
- 37.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J Mol Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 38.Agrawal RK, Heagle AB, Penczek P, Grassucci RA, Frank J. EF-G-dependent GTP hydrolysis induces translocation accompanied by large conformational changes in the 70S ribosome. Nat Struct Biol. 1999;6:643–647. doi: 10.1038/10695. [DOI] [PubMed] [Google Scholar]
- 39.Valle M, Zavialov A, Sengupta J, Rawat U, Ehrenberg M, Frank J. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–134. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 40.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J Biol Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 41.Robert F, Brakier-Gingras L. A functional interaction between ribosomal proteins S7 and S11 within the bacterial ribosome. J Biol Chem. 2003;278:44913–44920. doi: 10.1074/jbc.M306534200. [DOI] [PubMed] [Google Scholar]
- 42.Brandt R, Gualerzi CO. Ribosomal localization of the mRNA in the 30S initiation complex as revealed by UV crosslinking. FEBS Lett. 1992;311:199–202. doi: 10.1016/0014-5793(92)81101-q. [DOI] [PubMed] [Google Scholar]
- 43.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 44.Stade K, Rinke-Appel J, Brimacombe R. Site-directed cross-linking of mRNA analogues to the Escherichia coli ribosome; identification of 30S ribosomal components that can be cross- linked to the mRNA at various points 5′ with respect to the decoding site. Nuc Acids Res. 1989;17:9889–9908. doi: 10.1093/nar/17.23.9889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jagannathan I, Culver GM. Assembly of the central domain of the 30S ribosomal subunit: roles for the primary binding ribosomal proteins S15 and S8. J Mol Biol. 2003;330:373–383. doi: 10.1016/s0022-2836(03)00586-2. [DOI] [PubMed] [Google Scholar]
- 46.Yusupova G, Jenner L, Rees B, Moras D, Yusupov M. Structural basis for messenger RNA movement on the ribosome. Nature. 2006;444:391–394. doi: 10.1038/nature05281. [DOI] [PubMed] [Google Scholar]
- 47.Marquardt O, Roth HE, Wystup G, Nierhaus KH. Binding of Escherichia coli ribosomal proteins to 23S RNA under reconstitution conditions for the 50S subunit. Nuc Acids Res. 1979;6:3641–3650. doi: 10.1093/nar/6.11.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willumeit R, Forthmann S, Beckmann J, Diedrich G, Ratering R, Stuhrmann HB, Nierhaus KH. Localization of the protein L2 in the 50 S subunit and the 70 S E. coli ribosome. J Mol Biol. 2001;305:167–177. doi: 10.1006/jmbi.2000.4289. [DOI] [PubMed] [Google Scholar]
- 49.Rohl R, Nierhaus KH. Assembly map of the large subunit (50S) of Escherichia coli ribosomes. Proc Natl Acad Sci USA. 1982;79:729–733. doi: 10.1073/pnas.79.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nakashima T, Yao M, Kawamura S, Iwasaki K, Kimura M, Tanaka I. Ribosomal protein L5 has a highly twisted concave surface and flexible arms responsible for rRNA binding. RNA. 2001;7:692–701. doi: 10.1017/s1355838201002345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Perederina A, Nevskaya N, Nikonov O, Nikulin A, Dumas P, Yao M, Tanaka I, Garber M, Gongadze G, Nikonov S. Detailed analysis of RNA-protein interactions within the bacterial ribosomal protein L5/5S rRNA complex. RNA. 2002;8:1548–1557. [PMC free article] [PubMed] [Google Scholar]
- 52.Ban N, Nissen P, Hansen J, Moore PB, Steitz TA. The Complete Atomic Structure of the Large Ribosomal Subunit at 2.4 Å Resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- 53.Diedrich G, Spahn CM, Stelzl U, Schafer MA, Wooten T, Bochkariov DE, Cooperman BS, Traut RR, Nierhaus KH. Ribosomal protein L2 is involved in the association of the ribosomal subunits, tRNA binding to A and P sites and peptidyl transfer. EMBO J. 2000;19:5241–5250. doi: 10.1093/emboj/19.19.5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bischof O, Urlaub H, Kruft V, Wittmann-Liebold B. Peptide environment of the peptidyl transferase center from Escherichia coli 70 S ribosomes as determined by thermoaffinity labeling with dihydrospiramycin. J Biol Chem. 1995;270:23060–23064. doi: 10.1074/jbc.270.39.23060. [DOI] [PubMed] [Google Scholar]
- 55.Agrawal R, Penczek P, Grassucci R, Frank J. Visualization of elongation factor G on the Escherichia coli 70S ribosome: the mechanism of translocation. Proc Nat Acad Sci USA. 1998;95:6134–6138. doi: 10.1073/pnas.95.11.6134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khaitovich P, Mankin AS, Green R, Lancaster L, Noller HF. Characterization of functionally active subribosomal particles from Thermus aquaticus. Proc Natl Acad Sci USA. 1999;96:85–90. doi: 10.1073/pnas.96.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gao H, Sengupta J, Valle M, Korostelev A, Eswar N, Stagg SM, Van Roey P, Agrawal RK, Harvey SC, Sali A, et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell. 2003;113:789–801. doi: 10.1016/s0092-8674(03)00427-6. [DOI] [PubMed] [Google Scholar]
- 58.Persson BC, Bylund GO, Berg DE, Wikstrom PM. Functional analysis of the ffh-trmD region of the Escherichia coli chromosome by using reverse genetics. J Bacteriol. 1995;177:5554–5560. doi: 10.1128/jb.177.19.5554-5560.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Maisnier-Patin S, Paulander W, Pennhag A, Andersson DI. Compensatory Evolution Reveals Functional Interactions between Ribosomal Proteins S12, L14 and L19. J Mol Biol. 2006 doi: 10.1016/j.jmb.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 60.Savelsbergh A, Mohr D, Kothe U, Wintermeyer W, Rodnina MV. Control of phosphate release from elongation factor G by ribosomal protein L7/12. EMBO J. 2005;24:4316–4323. doi: 10.1038/sj.emboj.7600884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stark H, Rodnina MV, Wieden HJ, van Heel M, Wintermeyer W. Large-scale movement of elongation factor G and extensive conformational change of the ribosome during translocation. Cell. 2000;100:301–309. doi: 10.1016/s0092-8674(00)80666-2. [DOI] [PubMed] [Google Scholar]
- 62.Wahl MC, Moller W. Structure and function of the acidic ribosomal stalk proteins. Curr Protein Pept Sci. 2002;3:93–106. doi: 10.2174/1389203023380756. [DOI] [PubMed] [Google Scholar]
- 63.Traut RR, Lambert JM, Kenny JW. Ribosomal protein L7/L12 cross-links to proteins in separate regions of the 50 S ribosomal subunit of Escherichia coli. J Biol Chem. 1983;258:14592–14598. [PubMed] [Google Scholar]
- 64.Bocharov EV, Sobol AG, Pavlov KV, Korzhnev DM, Jaravine VA, Gudkov AT, Arseniev AS. From structure and dynamics of protein L7/L12 to molecular switching in ribosome. J Biol Chem. 2004;279:17697–17706. doi: 10.1074/jbc.M313384200. [DOI] [PubMed] [Google Scholar]
- 65.Bocharov EV, Gudkov AT, Budovskaya EV, Arseniev AS. Conformational independence of N- and C-domains in ribosomal protein L7/L12 and in the complex with protein L10. FEBS Lett. 1998;423:347–350. doi: 10.1016/s0014-5793(98)00121-5. [DOI] [PubMed] [Google Scholar]
- 66.Bubunenko MG, Chuikov SV, Gudkov AT. The length of the interdomain region of the L7/L12 protein is important for its function. FEBS Lett. 1992;313:232–234. doi: 10.1016/0014-5793(92)81198-u. [DOI] [PubMed] [Google Scholar]
- 67.Gordiyenko Y, Deroo S, Zhou M, Videler H, Robinson CV. Acetylation of L12 increases interactions in the Escherichia coli ribosomal stalk complex. J Mol Biol. 2008;380:404–414. doi: 10.1016/j.jmb.2008.04.067. [DOI] [PubMed] [Google Scholar]
- 68.Nishimura M, Yoshida T, Shirouzu M, Terada T, Kuramitsu S, Yokoyama S, Ohkubo T, Kobayashi Y. Solution structure of ribosomal protein L16 from Thermus thermophilus HB8. J Mol Biol. 2004;344:1369–1383. doi: 10.1016/j.jmb.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 69.Ban N, Nissen P, Hansen J, Capel M, Moore PB, Steitz TA. Placement of protein and RNA structures into a 5 Å -resolution map of the 50S ribosomal subunit. Nature. 1999;400:841–847. doi: 10.1038/23641. [DOI] [PubMed] [Google Scholar]
- 70.O'Connor M, Gregory ST, Dahlberg AE. Multiple defects in translation associated with altered ribosomal protein L4. Nucleic Acids Res. 2004;32:5750–5756. doi: 10.1093/nar/gkh913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zengel JM, Jerauld A, Walker A, Wahl MC, Lindahl L. The extended loops of ribosomal proteins L4 and L22 are not required for ribosome assembly or L4-mediated autogenous control. RNA. 2003;9:1188–1197. doi: 10.1261/rna.5400703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlunzen F, Harms JM, Franceschi F, Hansen HA, Bartels H, Zarivach R, Yonath A. Structural basis for the antibiotic activity of ketolides and azalides. Structure. 2003;11:329–338. doi: 10.1016/s0969-2126(03)00022-4. [DOI] [PubMed] [Google Scholar]
- 73.Schaffitzel C, Oswald M, Berger I, Ishikawa T, Abrahams JP, Koerten HK, Koning RI, Ban N. Structure of the E. coli signal recognition particle bound to a translating ribosome. Nature. 2006;444:503–506. doi: 10.1038/nature05182. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 – MS/MS and MS3 spectra of a phosphorylated tryptic peptides from E. coli ribosomal proteins. CID MS/MS and MS3 spectra for phosphorylated peptides indicating either a neutral loss of 98 Da from the phosphorylated peptide or addition of 80 Da to the unphosphorylated peptide shown by ˆ and #, respectively. (*) indicates the methionine oxidation. Mass spectrometry data was searched against the E. coli protein sequences in SwissProt database using Bioworks 3.2.
Fig. S2 – The primary sequence alignments of E. coli ribosomal proteins in several bacterial species and the human mitochondrial sequence. Abbreviations for bacterial species: ECOLI; E. coli, HAEIN; Haemophilus Influenzae, MYCTU; Mycobacterium tuberculosis, BACSU; Bacillus subtilis; and THET8; Thermus thermophilus. Alignments were processed with the CLUSTALW program in Biology Workbench and the results are displayed in BOXSHADE. Asterisks indicate possible phosphorylated residues as detected in the LC-MS/MS analysis of the phosphorylated tryptic peptide.