Abstract
The ATP-binding cassette (ABC) superfamily consists of both importers and exporters. These transporters have, by tradition, been classified according to the ATP hydrolyzing constituents, which are monophyletic. The evolutionary origins of the transmembrane porter proteins/domains are not known. Using five distinct computer programs, we here provide convincing statistical data suggesting that the transmembrane domains of ABC exporters are polyphyletic, having arisen at least three times independently. ABC1 porters arose by intragenic triplication of a primordial two-transmembrane segment (TMS)-encoding genetic element, yielding six TMS proteins. ABC2 porters arose by intragenic duplication of a dissimilar primordial three-TMS-encoding genetic element, yielding a distinctive protein family, nonhomologous to the ABC1 proteins. ABC3 porters arose by duplication of a primordial four-TMS-encoding genetic element, yielding either eight- or 10-TMS proteins. We assign each of 48 of the 50 currently recognized families of ABC exporters to one of the three evolutionarily distinct ABC types. Currently available high-resolution structural data for ABC porters are fully consistent with our findings. These results provide guides for future structural and mechanistic studies of these important transport systems.
Keywords: ATP-binding cassette (ABC) transporter, Evolutionary origin, Polyphyletic, Protein structure, Transport mechanism
Introduction
Over the past 20 years, our laboratory has identified more than 600 families of established and putative transport systems (Busch and Saier 2002; Saier 2000), which are described and tabulated in the Transporter Classification Database (TCDB http://www.tcdb.org) (Saier et al. 2006; Saier et al. 2009). These families have been defined on the basis of homology of the transmembrane constituents that form the transport channels and therefore comprise the actual porters.
The ATP-binding cassette (ABC) superfamily (TC 3.A.1) is considered to be one of the two largest superfamilies of transmembrane transporters found in nature, the other being the major facilitator superfamily (MFS, TC 2.A.1) (Higgins 1992, 2007; Pao et al. 1998). These two superfamilies have about equal numbers of functionally characterized systems, described in the scientific literature and TCDB (Busch and Saier 2002; Saier 2000). However, by tradition, the ABC superfamily is defined on the basis of its energy coupling proteins, the monophyletic ATP hydrolyzing (ABC) domains, rather than the integral membrane porter domains (Higgins 1992, 2007; Saurin, Hofnung and Dassa 1999).
We have long wondered if the membrane constituents, like the ATP-hydrolyzing (ABC) constituents, are monophyletic. However, previous efforts in our laboratory and in others have not provided an answer to this important question (Higgins 1992, 2007; Khwaja et al. 2005). Dissimilar three-dimensional (3-D) structures, as noted for certain ABC uptake (Hvorup et al. 2007) vs. export (Aller et al. 2009; Dawson and Locher 2006; Ward et al. 2007) transporters, although suggestive, do not prove independent origin. For those that are homologous, a single transport mechanism and common structural features can be predicted. However, if ABC porters are polyphyletic, there is no basis for extrapolating findings made with one phylogenetic group of proteins to another.
To establish that different membrane proteins are polyphyletic (arose independently of each other), it is necessary to establish distinct routes of evolutionary appearance. This has become possible due to (1) the availability of more sensitive software (Yen et al. 2009b; Zhai and Saier 2002; Zhou et al. 2003), (2) the availability of larger numbers of homologues resulting from genome sequencing, and (3) application of the superfamily principle (Doolittle 1981; Saier 1994).
To establish homology between repeat elements in the transmembrane domains of ABC exporters, we used the superfamily principle to extend the significant internal homology decision to other evolutionarily related proteins (e.g., derived from a common ancestor) (Doolittle 1981; Saier 1994). This principle, established in 1981 (Doolittle 1981), has been used to establish homology for distantly related members of extensive superfamilies (Saier 2003; Tran and Saier 2004; Yen et al. 2009a, b). The key word here is “related.” That is, we first must establish homology throughout most of their lengths for all of the proteins belonging to a single family. This was done for members of each of the three families or topological types of transmembrane domains/proteins, which we designated ABC1, ABC2 and ABC3. Then, homology was established for internal repeat elements in representative transmembrane domains using statistical approaches based on five distinct computer programs. Once this was done, it was not necessary to establish homology for every repeat element in every member of the family (Doolittle 1981; Saier 1994; Saier et al. 2009) because all of these proteins are homologous to each other. Thus, all ABC1 porters possess the internal triplication where the primordial precursor protein was a simple two-transmembrane segment (TMS) hairpin structure. The same procedures and arguments showed that all ABC2 proteins, which we demonstrated are homologous to each other, contain an internal duplication of three TMSs. The presence of four TMS repeat units in ABC3 proteins was established previously (Khwaja et al. 2005).
Many families of integral membrane transport proteins evolved independently of each other following different evolutionary pathways (Saier 2003). They most often did so by intragenic multiplication events where the primordial genes encoded channel-forming peptides with one, two, three or four α-helical TMSs (Saier 2003). They duplicated, triplicated or quadruplicated—sometimes in a single step, sometimes in more than one step (Nelson et al. 1999; Saier 2003).
The results reported here clearly suggest that the membrane constituents of ABC transporters are polyphyletic, having arisen independently at least three times, following these three distinct evolutionary pathways. One of these three topological types, identified in ABC export pumps, ABC2, appears to have been the precursor of at least some ABC uptake porters (unpublished observations).
Methods
To establish homology (common ancestry), either between two proteins or between two internal segments in a set of homologous proteins, the IC and GAP programs were initially used (Devereux et al. 1984; Zhai and Saier 2002; Zhou et al. 2003). For establishing homology among putative full-length homologues or repeat sequences of greater than 60 amino acyl residues, a value of 10 standard deviations (sd) is considered sufficient (Saier 1994; Saier et al. 2009). According to Dayhoff et al. (1983), this value corresponds to a probability of 10−24 that this degree of similarity arose by chance. The two proteins (or sets of domains) to be compared were subjected to PSI-BLAST searches of the NCBI nonredundant protein database with a single iteration (Yen et al. 2009b) (criteria as described below). The query sequences were the membrane domains of the SmbG protein (TC 3.A.1.111.4) and the KpsM protein (TC 3.A.1.101.1) for the ABC1 and ABC2 sets of proteins, respectively. Characterization of prokaryotic ABC3 homologues is described in Khwaja et al. (2005), although the occurrence of the eukaryotic members of this family (3.A.1.207) is first reported here.
We have found that a single iteration with a cut-off value of e−4 for the initial BLAST search and a cut-off value of e−5 for the iteration reliably retrieves homologues with very few false positives. Nevertheless, all proteins giving e−8 or larger were tested for homology using the GAP program with default settings, requiring a comparison score of at least 10 sd in order to conclude that these proteins share a common origin. All hits that satisfied these criteria were put through a modified CDHit program with 90% cut-off (Li and Godzik 2006; Yen et al. 2009b) to eliminate redundancies, fragmentary sequences and sequences with similarities of >90% identity. A multiple alignment was generated with the Clustal X program (Thompson et al. 1997), and homology of all aligned sequences throughout the relevant transmembrane domains was established using the IC and GAP programs (Devereux et al. 1984; Zhai and Saier 2002). Internal regions to be examined for repeats were excised from the full-length protein sequences based on the multiple alignment as described by Zhou et al. (2003), and dissimilar segments were compared with potentially homologous regions of the same proteins using the IC and GAP programs with default settings and 500 random shuffles. The ATP hydrolyzing (ABC) domains of these systems were excluded, and only the transmembrane domains were used in the analyses. Additional programs used to provide further evidence for homology were the LALIGN, GGSEARCH and GLSEARCH programs (found at http://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml) as well as the PairwiseStatSig program (found at http://brendelgroup.org/bioinformatics2go/PairwiseStatSig.php). The data generated with these four programs are presented in Supplemental Tables S1–S4 on our Web site (http://biology.ucsd.edu/~msaier/supmat/ABC123), and a summary of these results is presented here in Table 3.
Table 3.
Confirmation of the presence of homologous repeat units in ABC1 and ABC2 proteins using four different programs
| ABC type | TMSsa | Protein 1 (organism) | Protein accessionb | TMSsa | Protein 2 (organism) | Protein accessionb | E valuec (LALIGN) | E valuec (GGSEARCH) | E valuec (GLSEARCH) | E valuec (PairwiseStatSig) | E valuec (average) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ABC-1 | 1 + 2 | Clostridium leptum DSM 753 | ZP_02078939 | 3 + 4 | Colwellia psychrerythraea 34H | YP_267051 | 5.9 × e−6 | 2.5 × e−13 | 1.6 × e−14 | 5.2 × e−6 | 6.0 × e−10 |
| ABC-1 | 1 + 2 | Caulobacter crescentus CB15 | NP_419501 | 5 + 6 | Rhodoferax ferrireducens T118 | YP_523966 | 2.6 × e−6 | 5.4 × e−15 | 4.9 × e−15 | 7.9 × e−7 | 9.0 × e−11 |
| ABC-1 | 3 + 4 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 | YP_855895 | 5 + 6 | Unknown homologue | ZP_01811308 | 1.4 × e−6 | 5.5 × e−13 | 2.7 × e−10 | 1.7 × e−6 | 4.0 × e−9 |
| ABC-2 | 1–3 | Streptomyces avermitilis MA-4680 | NP_825831 | 4–6 | Burkholderia sp. 383 | YP_368972 | 5.8 × e−6 | 2.0 × e−14 | 4.5 × e−12 | 2.0 × e−6 | 1.0 × e−9 |
aTMSs, the transmembrane α-helical segments compared
bProtein accession number in the NCBI protein database
cE values reflect the probability that the alignment occurred by chance
Average hydropathy, amphipathicity and similarity plots for sets of homologues were generated with the AveHAS program (Zhai and Saier 2001b), while Web-based hydropathy, amphipathicity and predicted topology for an individual protein were estimated using the WHAT program (Zhai and Saier 2001a). These programs were updated as described by Yen et al. (2009a, b). Sequences were spliced for statistical analyses as described by Zhou et al. (2003).
Results
Fifty families of phylogenetically distinct ABC exporters from bacteria, archaea and eukaryotes are currently recognized in the TCDB (Table 1) (Saier et al. 2006, 2009). The most common topological type for the membrane domains of exporters has six TMSs, but some have four, eight, 10 or 12 TMSs (Higgins 1992; Khwaja et al. 2005; Tusnady et al. 1997). We show that all three of the ABC efflux porter topological types identified here (ABC1, ABC2 and ABC3) are present in all three domains of living organisms, although one of them (ABC3) has been found only in lower, not in higher, eukaryotes.
Table 1.
Type assignment for all 50 families of ABC efflux systems identified and listed in TCDBa
| TC number | Family name | Known substrate range | ABC type |
|---|---|---|---|
| 3.A.1.101 | Capsular polysaccharide exporter (CPSE) | CPS | 2 |
| 3.A.1.102 | Lipooligosaccharide exporter (LOSE) | LOS | 2 |
| 3.A.1.103 | Lipopolysaccharide exporter (LPSE) | LPS | 2 |
| 3.A.1.104 | Techoic acid exporter (TAE) | TA | 2 |
| 3.A.1.105 | Drug exporter-1 (DrugE1) | Drugs | 2 |
| 3.A.1.106 | Lipid exporter (LipidE) | PL, LPS, lipid A, drugs, peptides | 1 |
| 3.A.1.107 | Putative heme exporter (HemeE) | Heme, cytochrome c | 2 |
| 3.A.1.108 | ß-Glucan exporter (GlucanE) | Polysaccharides | 1 |
| 3.A.1.109 | Protein-1 exporter (Prot1E) | Proteins | 1 |
| 3.A.1.110 | Protein-2 exporter (Prot2E) | Proteins | 1 |
| 3.A.1.111 | Peptide-1 exporter (Pep1E) | Bacteriocin, peptides | 1 |
| 3.A.1.112 | Peptide-2 exporter (Pep2E) | Other peptides | 1 |
| 3.A.1.113 | Peptide-3 exporter (Pep3E) | Antibiotics, siderophores | 1 |
| 3.A.1.114 | Probable glycolipid exporter (DevE) | Glycolipids | 3 |
| 3.A.1.115 | Na+ exporter (NatE) | Sodium | 2 |
| 3.A.1.116 | Microcin B17 exporter (McbE) | Bacteriocins, peptides | 2 |
| 3.A.1.117 | Drug exporter-2 (DrugE2) | Drugs, lipids, dyes | 1 |
| 3.A.1.118 | Microcin J25 exporter (McjD) | Peptides, antibiotics | 1 |
| 3.A.1.119 | Drug/siderophore exporter-3 (DrugE3) | Drugs, siderophores | 1 |
| 3.A.1.120 | (Putative) Drug resistance ATPase-1 (DrugRA1) | Drugs | b |
| 3.A.1.121 | (Putative) Drug resistance ATPase-2 (DrugRA2) | Drugs, antibiotics | b |
| 3.A.1.122 | Macrolide exporter (MacB) | Macrolides, heme | 3 |
| 3.A.1.123 | Peptide-4 exporter (Pep4E) | Drugs, peptides | 1 |
| 3.A.1.124 | 3-Component peptide-5 exporter (Pep5E) | Bacteriocins | 2 |
| 3.A.1.125 | Lipoprotein translocase (LPT) | O.M. Lipoproteins | 3 |
| 3.A.1.126 | ß-Exotoxin I exporter (ß-ETE) | Proteins | 2 |
| 3.A.1.127 | AmfS peptide exporter (AmfS-E) | Peptides, morphogens | 2 |
| 3.A.1.128 | SkfA peptide exporter (SkfA-E) | Modified peptides | 2 |
| 3.A.1.129 | CydDC cysteine exporter (CydDC-E) | Cysteine | 1 |
| 3.A.1.130 | Multidrug/hemolysin exporter (MHE) | Drugs, hemolysins | 2 |
| 3.A.1.131 | Bacitracin resistance (Bcr) | Bacteriocins | 2 |
| 3.A.1.132 | Gliding motility ABC transporter (Gld) | Polysaccharides, copper ions | 2 |
| 3.A.1.133 | Peptide-6 exporter (Pep6E) | Peptides | 2 |
| 3.A.1.134 | Peptide-7 exporter (Pep7E) | Peptides, bacteriocins | 3 |
| 3.A.1.135 | Drug exporter-4 (DrugE4) | Drugs | 1 |
| 3.A.1.136 | Uncharacterized ABC3-type (U-ABC3-1) | Unknown | 3 |
| 3.A.1.137 | Uncharacterized ABC3-type (U-ABC3-2) | Unknown | 3 |
| 3.A.1.138 | Unknown ABC2-type (ABC2-1) | Unknown | 2 |
| 3.A.1.201 | Multidrug resistance exporter (MDR) | Drugs, fatty acids, lipids | 1 |
| 3.A.1.202 | Cystic fibrosis transmembrane exporter (CFTR) | Chloride | 1 |
| 3.A.1.203 | Peroxysomal fatty acyl CoA transporter (P-FAT) | Long chain fatty acids | 1 |
| 3.A.1.204 | Eye pigment precursor transporter (EPP) | Pigments, drugs, hemes | 2 |
| 3.A.1.205 | Pleiotropic drug resistance (PDR) | Drugs, steroids, nucleotides, acids | 2 |
| 3.A.1.206 | a-Factor sex pheromone exporter (STE) | Peptides | 1 |
| 3.A.1.207 | Eukaryotic ABC3 (E-ABC3) | Unknown | 3 |
| 3.A.1.208 | Drug conjugate transporter (DCT) | Drugs, conjugates, anions, peptides, folates | 1 |
| 3.A.1.209 | MHC peptide transporter (TAP) | Peptides | 1 |
| 3.A.1.210 | Heavy metal transporter (HMT) | Drugs, metal conjugates, heme | 1 |
| 3.A.1.211 | Cholesterol/phospholipid/retinal (CPR) flippase | Drugs, sterols, lipids, retinal, surfactants, proteins, peptides, xenobiotics | 2 |
| 3.A.1.212 | Mitochondrial peptide exporter (MPE) | Peptides | 1 |
aTC number within the ABC superfamily (3.A.1) is provided in the first column. The family name is given in the second column, with the family abbreviation presented in parentheses. Column 3 indicates the substrates known to be transported, while column 4 indicates to which topological type family members belong
bThe membrane constituents of these two families of putative ABC drug export systems have not been identified
Families of ABC exporters, classified under TC 3.A.1, are listed in Table 1. TC numbers 3.A.1.1–27 (with one possible exception, see below) are families of uptake porters, each forming a distinct phylogenetic cluster with unique functional characteristics. All but one, the chloroplast lipid uptake system (TC 3.A.1.27.1), are from prokaryotes. These uptake families are not presented in Table 1. TC numbers 3.A.1.101–138 are largely prokaryotic exporters, where again each family consists of members that are phylogenetically coherent. Members of a family usually transport related types of substrates (Table 1). TC numbers 3.A.1.201–212 are largely eukaryotic exporters and frequently exhibit broad substrate specificities (Busch and Saier 2002). These 50 families are listed in Table 1 according to their TC numbers, together with their names, abbreviations, substrate ranges and topological types, as revealed in this study.
When identifying internal repeats for the six TMS ABC membrane proteins, each internal membrane protein or domain (lacking the ATP-hydrolyzing domain) was divided into segments of either two or three TMSs. Thus, all proteins were divided into thirds and halves with two TMSs and three TMSs per segment, respectively. Then, for the thirds, all first segments were compared with the second and third segments, and the second segments were compared with the third segments. For the halves, all first halves were compared with second halves. For ABC1 proteins, only comparisons between the thirds gave scores above 5 sd, while for ABC2 proteins, only comparisons between the two halves gave scores above 5 sd. In fact, the highest scores obtained for the “correct” alignments were 10 sd, as reported in Table 2. This value is 5 sd higher than that obtained for the alternative comparisons. In all cases, the alignments giving rise to the top scores were checked to ensure that the correct TMSs, corresponding to the proposed repeat elements as displayed in Fig. 1, were aligned and that they aligned throughout these segments.
Table 2.
Statistical data, obtained with the GAP and IC programs, establishing homology between the repeat units within ABC porters of the three currently recognized topological types, ABC1–3
| ABC type | TMSsa | Protein 1 (organism) | Protein 1 accessionb | TMSsa | Protein 2 (organism) | Protein 2 accessionb | C.S.c (sd) | %Id | %Se | #Gf | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| A | ABC1 | 1 + 2 | Clostridium leptum DSM 753 | ZP_02078939 | 3 + 4 | Colwellia psychrerythraea 34H | YP_267051 | 10.0 | 30.4 | 39.1 | 1 |
| B | ABC1 | 1 + 2 | Caulobacter crescentus CB15 | NP_419501 | 5 + 6 | Rhodoferax ferrireducens T118 | YP_523966 | 10.1 | 31.3 | 40.3 | 2 |
| C | ABC1 | 3 + 4 | Aeromonas hydrophila subsp. hydrophila ATCC 7966 | YP_855895 | 5 + 6 | Unknown | ZP_01811308 | 10.1 | 38.5 | 47.7 | 3 |
| D | ABC2 | 1–3 | Streptomyces avermitilis MA-4680 | NP_825831 | 4–6 | Burkholderia sp. 383 | YP_368972 | 10.2 | 39.6 | 47.3 | 5 |
| E | ABC3 | 1–4 | Streptomyces avermitilis MA-4680 g | NP_828260 | 7–10 | Streptomyces avermitilis MA-4680 g | NP_828260 | 38.0g | 31.5g | 41.0g | 16g |
aTMSs, the transmembrane α-helical segments compared
bProtein accession number in the NCBI protein database
cC.S., comparison score
d%I, percent identity
e%S, percent similarity
f#G, number of gaps (see Figs. 2 and 3)
gThe alignment and statistical analysis for ABC3 family members, upon which the values reported here were obtained, can be found in Khwaja et al. (2005)
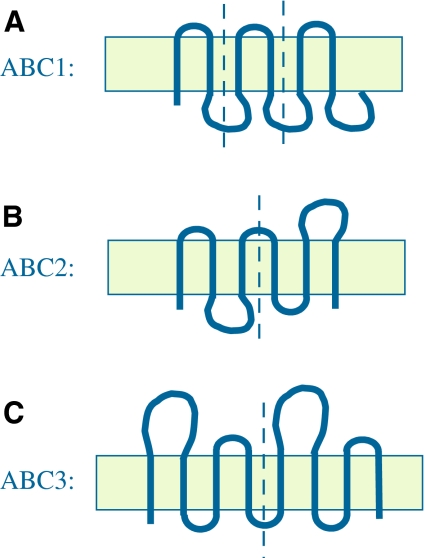
Fig. 1.
Three topological types of ABC exporters, illustrating the types of internal repeats present in each one. In all cases, vertical dashed lines separate the repeat units. a ABC1: a six-TMS topology resulting from intragenic triplication of a primordial two-TMS-encoding genetic element. The three hairpin repeats have the same orientation in the membrane. b ABC2: a six-TMS topology resulting from intragenic duplication of a primordial three-TMS-encoding genetic element. The two three-TMS repeats have opposite orientation in the membrane. c ABC3: an eight-TMS topology resulting from intragenic duplication of a primordial four-TMS-encoding genetic element (Khwaja et al. 2005). The two four-TMS repeats have the same orientation in the membrane
This treatment relies on the superfamily principle (Doolittle 1981; Saier 1994), which states that if A is homologous to B and B is homologous to C, then A is homologous to C, an obvious precept that nevertheless is sometimes denied. Note that homology is an absolute term, meaning “sharing a common evolutionary origin.” Thus, if it can be shown that A and B share a common origin and B and C share a common origin, then it follows that A and C must also share a common origin (Doolittle 1981).
While there are degrees of similarity or identity, there are no degrees of homology. Relevant to the studies reported here, if proteins A and B are homologous throughout their lengths and the first third of A is homologous to the second third of B, then the first thirds are homologous to the second thirds in both A and B. Further, if members of a set of these proteins are homologous throughout their lengths, then the first thirds of all of them must be homologous to the second thirds of all of them. Similarly, if the first half of one protein in a set of homologues is homologous to the second half of another protein within this set, then all halves of the homologous proteins within this set are homologous to each other (Doolittle 1981; Saier 1994).
Homology is considered established (Saier et al. 2009; Yen et al. 2009b; Zhai and Saier 2002) when two sequences of at least 60 residues (an average-sized protein domain) give a comparison score of 10 sd or greater, corresponding to a probability of 10−24 that the degree of similarity observed occurred by chance (Dayhoff et al. 1983). These criteria for homology are more rigorous than those used by most other investigators, and in no instance have these values been shown to incorrectly predict homology (Saier 2003).
When the transmembrane domains of the proteins comprising 48 of the 50 families of efflux pumps were compared, all of these proteins proved to be homologous to one of the three sets of proteins which we designated ABC1, ABC2 and ABC3 (Table 1). ABC families 3.A.1.120 and 3.A.1.121 could not be assigned as the transmembrane domains of these putative multidrug resistance systems have not been identified. In no case, however, did a protein in one of these three sets (ABC1, ABC2 and ABC3) prove to be homologous to a protein in another set. Thus, the basis for assigning these proteins to the three sets was both homology (or lack thereof) and establishment of independent pathways of evolution.
Statistical analyses leading to the conclusion of homology using the GAP and IC programs were confirmed using four other programs. These programs are LALIGN, GGSEARCH, GLSEARCH and PairwiseStatSig, each based on a different set of assumptions (see “Methods”). The raw data obtained with each of these programs are presented in Supplementary Tables S1–S4, and a summary of the results obtained using these four programs is presented in Table 3. When the results from the four programs were averaged, the values for the comparisons reported were about e−10, strongly indicative of homology (Table 3, last column).
The most widespread (and therefore possibly the oldest) topological type of demonstrably homologous ABC exporters is ABC1. These ubiquitous proteins most commonly have six TMSs with their N and C termini inside (Dawson and Locher 2006; Higgins 1992, 2007; Ward et al. 2007). Twenty-one (13 from prokaryotes and eight from eukaryotes) of the 50 recognized efflux families as well as one putative uptake family (3.A.1.23) could be shown to be homologous to members of the ABC1 family (Table 1). Fifty percent of the ABC exporters in the TCDB are of this type. These proteins proved to have arisen from a two-TMS hairpin precursor by intragenic triplication, as shown schematically in Fig. 1a and demonstrated statistically, first using the IC and GAP programs (Table 2) (Zhai and Saier 2002; Zhou et al. 2003) and subsequently using four additional programs (Table 3).
The sequence similarities of the three hairpin repeat elements in these six TMS proteins are demonstrated in Fig. 2a–c and Table 2 (rows A–C) where the superfamily principle (Doolittle 1981) is invoked to establish homology of the repeat units in all members of this ABC1 family. These membrane domains are often fused to ABC domains to form “half-sized” ABC transport proteins with the membrane (M) and cytoplasmic (C) (ATP hydrolyzing) domains fused, most frequently in the order M–C. Particularly (but not exclusively) in eukaryotes, these can be duplicated to yield full-length ABC transport systems, with a total of 12 TMSs, in an MCMC arrangement in a single, large, polypeptide chain. Additional domains can be fused to this basic structure, as occurs frequently in eukaryotes but seldom in prokaryotes (Higgins 1992, 2007). However, in all of our studies, only the six-TMS transmembrane domains were analyzed. These porters actively expel all kinds of substrates, from simple ions and sugars to drugs and macromolecules (Table 1). These substrates are exported from the cell cytoplasm into the extracellular milieu or into intracellular organelles of eukaryotes.
Fig. 2.
Alignments of three two-TMS hairpin repeats in ABC1-type transmembrane porters. All of the porters included in this study are homologous throughout their lengths. a Comparison of TMSs 1 and 2 (ZP_02078939) with TMSs 3 and 4 (YP_267051) in ABC1 proteins. This alignment gave a comparison score of 10.0 sd. b Comparison of TMSs 1 and 2 (NP_419501) with TMSs 5 and 6 (YP_523966) in ABC1 proteins. This alignment gave a comparison score of 10.1 sd. c Comparison of TMSs 3 and 4 (YP_855895) with TMSs 5 and 6 (ZP_01811308) in ABC1 proteins. This alignment gave a comparison score of 10.1 sd. In these figures, as well as in Fig. 3, shaded areas represent predicted α-helical TMSs based on the WHAT program (Zhai and Saier 2001a). A vertical line indicates an identity, a colon denotes a close similarity and a dot indicates a more distant similarity, all as defined by the GAP program (Devereux et al. 1984)
ABC exporters of the second type are designated ABC2 (Reizer et al. 1992). These proteins also have six TMSs per polypeptide chain with N and C termini inside. They include 20 recognized exporter families, 17 of prokaryotes and three of eukaryotes (Table 1). Thirty-four percent of the exporters in the TCDB are of this type. All of the membrane domains in porters of these 20 families are demonstrably homologous to each other using the statistical criteria presented here (≥10 sd for segments of at least 60 aas using the GAP and IC programs; see “Methods”).
Each of the ABC2 proteins proved to contain a three-TMS repeat unit, always duplicated in tandem, so that the two homologous halves are of opposite orientation in the membrane (Fig. 3 and Table 2, row D) (Saier 2003). The alignment shown in Fig. 3 gave a comparison score of 10.2 sd, which by our criteria is sufficient to establish homology (Saier 1994; Saier et al. 2009). This conclusion of homology was substantiated using four additional programs as presented in Supplementary Tables S1–S4 and summarized in Table 3 (see above and “Methods”). Therefore, these proteins evidently arose by intragenic duplication of a primordial three-TMS element (Fig. 1b). None of these proteins exhibits statistically significant sequence similarity with ABC1 or ABC3 homologues. The 20 known ABC2 families of exporters, represented in many organismal kingdoms, are seldom duplicated or fused to other domains. However, exceptions are the members of family 128 where the six-TMS unit is duplicated to give proteins of 12 TMSs with two fused ABC2 domains (M–M). ABC2 porters export complex carbohydrates (capsular polysaccharides, lipo-oligosaccharides, lipopolysaccharides, teichoic acids), drugs, peptides and Na+ (Table 1).
Fig. 3.
Comparison of TMSs 1, 2 and 3 with TMSs 4, 5 and 6 in ABC2-type transmembrane porters. This alignment gave a comparison score of 10.2 sd
Members of the third type of homologous ABC exporter, ABC3 (Khwaja et al. 2005), have a basic four-TMS topology (Fig. 1c). These four-TMS proteins have a large extracellular loop between TMSs 1 and 2 and can be (but need not be) fused N-terminally to an ABC domain (order C–M, see Fig. 4a). When duplicated to the eight- or 10-TMS proteins, they are never fused to ABC domains, which are encoded as distinct but adjacent cistrons, usually within single operons in prokaryotes (Table 2, row E) (Khwaja et al. 2005). The extra two nonhomologous putative TMSs in the 10-TMS proteins separate the two four-TMS repeat units (Fig. 4b). The very high scores obtained when these four-TMS repeat units are compared suggests that ABC3 duplications occurred much later than did those giving rise to ABC1 and ABC2 proteins. This suggestion is in agreement with the facts that ABC3 transporters can have two such units encoded by two distinct genes (Khwaja et al. 2005) and that both the intragenically duplicated and the unduplicated genes can be found in large numbers.
Fig. 4.
Three topological types identified for ABC3 porters. a The basic four-TMS unit may or may not be fused N-terminally to an ABC (ATP-hydrolyzing) domain, but the eight- and 10-TMS proteins are never fused to ABC domains (Khwaja et al. 2005). b Two putative topological types with eight and 10 TMSs, respectively, resulting from intragenic duplication of the four-TMS unit shown in a. The two extra TMSs (in white) in the 10-TMS proteins are centrally located between the two repeat units. (Reproduced from Khwaja et al. 2005, with permission.)
Seven homologous TC families (six from prokaryotes and one from unicellular eukaryotes) exhibit the ABC3 topological arrangements (Table 1). They comprise 12% of all ABC exporters tabulated in the TCDB. They transport peptides such as bacteriocins, heme and macrolide antibiotics; and they function in glycolipid and small lipoprotein export to the outer membranes of gram-negative bacteria (Table 1). Finally, two families (3.A.1.120 and 121) include known ATP hydrolyzing (C) domains which confer drug resistance to certain bacteria, but the transmembrane domains (M) have not been identified. Four percent of the ABC superfamily entries in the TCDB are of this type.
Uptake systems can have their ABC receptors and membrane domains fused to each other and duplicated in various arrangements (see TCDB; Higgins 1992, 2007; Saier et al. 2009). Each of the 27 recognized phylogenetic families of these porters exhibits a characteristic range of substrates and certain size/domain characteristics (see TCDB). Family assignments are thus made on the basis of the phylogeny of all three constituents as well as function which correlate. These porters can take up all kinds of small biological molecules and occasionally macromolecules (Busch and Saier 2002; see TCDB). At least some of these porters are of the ABC2 topological type with demonstrable homology with ABC2 exporters and a recognizable internal three-TMS repeat element. In some of these proteins, the C-terminal TMS has been lost, yielding a five-TMS topology; and in a few, the five-TMS element is found duplicated, so there are 10 TMSs per polypeptide chain (E. I. Sun, W. H. Zheng & M. H. Saier, unpublished observations). Analyses of these uptake systems will be described elsewhere.
Discussion
We have conducted statistical analyses using five distinct programs that use five different algorithms, each based on a different set of assumptions, to provide convincing evidence for the independent evolutionary origins of three distinct families of membrane domains of ABC exporters. Indeed, the available X-ray structures published for three ABC exporters suggest a unified fold, in agreement with our conclusion that the membrane domains of all three of the solved exporters prove to be of the ABC1 type (two from TC family 3.A.1.106 and one from family 3.A.1.201; see Table 1) (Aller et al. 2009; Dawson and Locher 2006; Ward et al. 2007). Our studies allow us to predict not only that these porters exhibit similar folds (even though one is from a gram-negative bacterium, one is from a gram-positive bacterium and one is from a human) but also that all ABC1 family proteins will exhibit this fold because they all share a common origin.
The five uptake systems for which high-resolution X-ray structures are available are the vitamin B12 porter of Escerichia coli (BtuCDF;, TC 3.A.1.13.5; Hvorup et al. 2007), the probable metal chelate uptake system of Haemophilus influenzae (HI1471, TC 3.A.1.14.11; Pinkett et al. 2007), the methionine transporter of E. coli (MetNI, TC 3.A.1.24.1; Kadaba et al. 2008), the maltose porter of E. coli (MalEFGK, TC 3.A.1.1.1; Oldham et al. 2007) and the molybdate porter of Methanosarcina acetivorans (ModABC, TC 3.A.1.8.1; Gerber et al. 2008). The membrane domains of all of these porters exhibit the same fold, strongly suggestive of a common ancestry. However, they do not show 3-D similarity to the exporters in their transmembrane domains. This is exactly as we would predict since our analyses indicate that uptake porters are of the ABC2 type, which have evolved independently of the ABC1-type exporters. All exporters for which 3-D data are available are of the ABC1 type, as noted above. Thus, the published 3-D structural data currently available are in full agreement with our conclusion that the ABC1 and ABC2 transmembrane protein domains evolved independently of each other. No high-resolution 3-D structure has yet been presented for one of the ABC3 family proteins or for an ABC2 exporter.
The work reported here and by Khwaja et al. (2005) provides convincing evidence for not just two but at least three independently arising topological types of ABC exporters. One of these topological types (ABC2), and possibly only this one, occurs in uptake porters (unpublished results). The origin(s) and relationships of remaining importers have yet to be established. We propose that the ABC2 exporters, being ubiquitous and therefore probably more ancient than the uptake systems, which are restricted to prokaryotes and chloroplasts, gave rise to these importers rather than the other way around. Further studies will be required to establish this postulate.
The establishment of independent origin is not possible by solving 3-D protein structures and is achievable only using bioinformatic approaches as described here (Saier 2003). The results reveal that the ABC superfamily, like the bacterial phosphoenolpyruvate-dependent phosphotransferase system (PTS) superfamily (TC 4.A.1–6) (Saier et al. 2005), consists of a mosaic of independently evolving porters. Both the ABC and PTS porters are therefore “functional superfamilies.” In each of these two functional superfamilies, three distinct, independently evolving, unrelated families of porters have now been identified. However, the PTS also uniquely includes an independently evolving family of nontransporting PTS proteins, the phosphoenolpyruvate-dependent dihydroxyacetone phosphorylating kinases (Gutknecht et al. 2001; Saier et al. 2005). Future efforts will be devoted to defining more precisely the constituents of these independently evolving superfamilies, identifying still other topological types if they exist and determining the evolutionary origins of all currently recognized ABC uptake porter families.
Acknowledgements
We thank Dr. Yu-feng Tsai, Dr. Ming Ren Yen and Dorjee Tamang for software development; Dorjee Tamang for assistance with figure preparation and TCDB maintenance; Van Duong and Jeeni Criscenzo for help with manuscript preparation; Dr. Geoff Chang for valuable discussions; and the National Institutes of Health (GM077402) for financial support.
Open Access This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G (2009) Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science 323:1718–1722 [DOI] [PMC free article] [PubMed]
- Busch W, Saier MH Jr (2002) The transporter classification (TC) system, 2002. Crit Rev Biochem Mol Biol 37:287–337 [DOI] [PubMed]
- Dawson RJ, Locher KP (2006) Structure of a bacterial multidrug ABC transporter. Nature 443:180–185 [DOI] [PubMed]
- Dayhoff MO, Barker WC, Hunt LT (1983) Establishing homologies in protein sequences. Methods Enzymol 91:524–545 [DOI] [PubMed]
- Devereux J, Haeberli P, Smithies O (1984) A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res 12:387–395 [DOI] [PMC free article] [PubMed]
- Doolittle RF (1981) Similar amino acid sequences: chance or common ancestry? Science 214:149–159 [DOI] [PubMed]
- Gerber S, Comellas-Bigler M, Goetz BA, Locher KP (2008) Structural basis of trans-inhibition in a molybdate/tungstate ABC transporter. Science 321:246–250 [DOI] [PubMed]
- Gutknecht R, Beutler R, Garcia-Alles LF, Baumann U, Erni B (2001) The dihydroxyacetone kinase of Escherichia coli utilizes a phosphoprotein instead of ATP as phosphoryl donor. EMBO J 20:2480–2486 [DOI] [PMC free article] [PubMed]
- Higgins CF (1992) ABC transporters: from microorganisms to man. Annu Rev Cell Biol 8:67–113 [DOI] [PubMed]
- Higgins CF (2007) Multiple molecular mechanisms for multidrug resistance transporters. Nature 446:749–757 [DOI] [PubMed]
- Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP (2007) Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science 317:1387–1390 [DOI] [PubMed]
- Kadaba NS, Kaiser JT, Johnson E, Lee A, Rees DC (2008) The high-affinity E. coli methionine ABC transporter: structure and allosteric regulation. Science 321:250–253 [DOI] [PMC free article] [PubMed]
- Khwaja M, Ma Q, Saier MH Jr (2005) Topological analysis of integral membrane constituents of prokaryotic ABC efflux systems. Res Microbiol 156:270–277 [DOI] [PubMed]
- Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659 [DOI] [PubMed]
- Nelson RD, Kuan G, Saier MH Jr, Montal M (1999) Modular assembly of voltage-gated channel proteins: a sequence analysis and phylogenetic study. J Mol Microbiol Biotechnol 1:281–287 [PubMed]
- Oldham ML, Khare D, Quiocho FA, Davidson AL, Chen J (2007) Crystal structure of a catalytic intermediate of the maltose transporter. Nature 450:515–521 [DOI] [PubMed]
- Pao SS, Paulsen IT, Saier MH Jr (1998) Major facilitator superfamily. Microbiol Mol Biol Rev 62:1–34 [DOI] [PMC free article] [PubMed]
- Pinkett HW, Lee AT, Lum P, Locher KP, Rees DC (2007) An inward-facing conformation of a putative metal-chelate-type ABC transporter. Science 315:373–377 [DOI] [PubMed]
- Reizer J, Reizer A, Saier MH Jr (1992) A new subfamily of bacterial ABC-type transport systems catalyzing export of drugs and carbohydrates. Protein Sci 1:1326–1332 [DOI] [PMC free article] [PubMed]
- Saier MH Jr (1994) Computer-aided analyses of transport protein sequences: gleaning evidence concerning function, structure, biogenesis, and evolution. Microbiol Rev 58:71–93 [DOI] [PMC free article] [PubMed]
- Saier MH Jr (2000) A functional-phylogenetic classification system for transmembrane solute transporters. Microbiol Mol Biol Rev 64:354–411 [DOI] [PMC free article] [PubMed]
- Saier MH Jr (2003) Tracing pathways of transport protein evolution. Mol Microbiol 48:1145–1156 [DOI] [PubMed]
- Saier MH Jr, Hvorup RN, Barabote RD (2005) Evolution of the bacterial phosphotransferase system: from carriers and enzymes to group translocators. Biochem Soc Trans 33:220–224 [DOI] [PubMed]
- Saier MH Jr, Tran CV, Barabote RD (2006) TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res 34:D181–D186 [DOI] [PMC free article] [PubMed]
- Saier MH Jr, Yen MR, Noto K, Tamang DG, Elkan C (2009) The Transporter Classification Database: recent advances. Nucleic Acids Res 37:D274–D278 [DOI] [PMC free article] [PubMed]
- Saurin W, Hofnung M, Dassa E (1999) Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48:22–41 [DOI] [PubMed]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882 [DOI] [PMC free article] [PubMed]
- Tran CV, Saier MH Jr (2004) The principal chloroquine resistance protein of Plasmodium falciparum is a member of the drug/metabolite transporter superfamily. Microbiology 150:1–3 [DOI] [PubMed]
- Tusnady GE, Bakos E, Varadi A, Sarkadi B (1997) Membrane topology distinguishes a subfamily of the ATP-binding cassette (ABC) transporters. FEBS Lett 402:1–3 [DOI] [PubMed]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G (2007) Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci USA 104:19005–19010 [DOI] [PMC free article] [PubMed]
- Yen MR, Chen J, Marquez JL, Sun EI, Saier MH Jr (2009a) Multi drug resistance: polgenetic characterization of superfamilies of secondary carriers that include drug exporters. In: Yan Q (ed) Membrane transporters in drug discovery and development: methods and protocols (in press)
- Yen MR, Choi J, Saier MH Jr (2009b) Bioinformatic analyses of transmembrane transport: novel software for deducing protein phylogeny, topology, and evolution. J Mol Microbiol Biotechnol 17:163–176 [DOI] [PMC free article] [PubMed]
- Zhai Y, Saier MH Jr (2001a) A Web-based program (WHAT) for the simultaneous prediction of hydropathy, amphipathicity, secondary structure and transmembrane topology for a single protein sequence. J Mol Microbiol Biotechnol 3:501–502 [PubMed]
- Zhai Y, Saier MH Jr (2001b) A Web-based program for the prediction of average hydropathy, average amphipathicity and average similarity of multiply aligned homologous proteins. J Mol Microbiol Biotechnol 3:285–286 [PubMed]
- Zhai Y, Saier MH Jr (2002) A simple sensitive program for detecting internal repeats in sets of multiply aligned homologous proteins. J Mol Microbiol Biotechnol 4:375–377 [PubMed]
- Zhou X, Yang NM, Tran CV, Hvorup RN, Saier MH Jr (2003) Web-based programs for the display and analysis of transmembrane α-helices in aligned protein sequences. J Mol Microbiol Biotechnol 5:1–6 [DOI] [PubMed]