Abstract
The process of oligodendrocyte differentiation is regulated by a dynamic interaction between a genetic and an epigenetic program. Recent studies, addressing nucleosomal histone modifications have considerably increased our knowledge regarding epigenetic regulation of gene expression during oligodendrocyte development and aging. These results have generated new hypotheses regarding the mechanisms underlying the decreased efficiency of endogenous remyelination in response to demyelinating injuries with increasing age.
In this review, we present an overview of the epigenetic mechanisms regulating gene expression at specific stages of oligodendrocyte differentiation and maturation as well as the changes that occur with aging.
Keywords: histone modification, oligodendrocyte differentiation, microRNA, Multiple Sclerosis
1. Introduction
Loss of myelin and insufficient compensation by endogenous remyelination contribute to the functional impairment that characterizes multiple sclerosis (MS). The restricted remyelination in MS has been associated with the limited myelinating capacity of endogenous oligodendrocyte precursor cells (OPCs) (Chari, 2007;Franklin and Ffrench-Constant, 2008). As with other regenerative processes, remyelination becomes even less efficient with age (Shields et al., 2000). This decline has been attributed to impaired OPC recruitment (Sim et al., 2002) and inefficient differentiation of OPC in old animals (Woodruff et al., 2004). To promote remyelination in the central nervous system either by endogenous or implanted remyelinating cells, it is essential to understand the molecular mechanisms underlying oligodendrocyte development and differentiation and to define the impact of aging on these processes. It is anticipated that insights in the epigenetic regulation of myelin formation and repair will provide novel targets for therapeutic intervention.
In the past three years, our knowledge regarding the multiple origins of OPCs during embryonic development has increased considerably. In the embryonic spinal cord, for instance, the first wave of OPC emerges from the pMN region in the ventral domain of the neural tube, followed by a second smaller wave of OPC formation in the dorsal part of the developing spinal cord (Battiste et al., 2007;Cai et al., 2005;Vallstedt et al., 2005). In the brain, three waves of OPC formation can be recognized: the earliest in the ventral regions of the telencephalon and diencephalon, followed by a second wave of OPCs from the lateral and/or caudal ganglionic eminences and, finally, a third wave of OPCs arising from dorsal cortical progenitors (Kessaris et al., 2006;Kessaris et al., 2008;Richardson et al., 2006). From these oligodendrogenic centers, OPCs migrate and populate the different parts of the developing brain and spinal cord.
Various transcription factors, active during one or all of the stages of oligodendrocyte formation, maturation and myelination, have been identified (Nicolay et al., 2007;Sher et al., 2008a;Wegner, 2008). Selective expression or silencing of genes at each of stage is ultimately controlled by epigenetic regulatory mechanisms which determine the accessibility of the transcriptional machinery to the chromosomal territories containing the specific genes. So the specific nuclear location of oligodendrocytic genes, for instance within heterochromatin or euchromatin, in the central or peripheral region of the nucleus, adjacent or distant from nuclear transcription domains, and near or far from nuclear splicing factor compartments, is of crucial importance. Chromatin condensation and decondensation can be mediated by various (hetero)chromosomal proteins like HP1 and ATP-dependent, SWI/SNF-like complexes, respectively, but the best characterized chromatin modifications take place at the level of the nucleosomes, the basic units of chromatin. A nucleosome consists of 150 bp DNA wrapped around histone octamers. Amino acids in the protruding tails of these histone proteins are targets for enzymatic modifications, such as histone methyltransferase- (HMT), histone acetyltransferase- (HAT) and histone deacetylase- (HDAC) activity. The resulting structural histone modifications have critical consequences for transcriptional availability of the DNA in any specific region. Besides at histones, methyltransferases are active at DNA level, leading to silencing of gene expression due to DNA methylation. Recruitment of DNA methyltransferases, histone deacetylases and other enzymes appears to be controlled by a special group of proteins, the Polycomb group (PcG) proteins (Liu et al., 2007;Sparmann and Lohuizen van M., 2006). The PcG proteins form multiple, histone-bound polycomb repressive complexes (PRCs), of which PRC1 and PRC2 are the best described and as yet considered the most important ones. They are considered major epigenetic regulators of gene expression programs controlling differentiation. Finally, the discovery of microRNAs has unraveled an additional epigenetic mechanism of post-transcriptional regulation of gene expression, since these small RNAs have specific mRNA transcripts as targets and can block their translation. The identification of microRNAs involved in oligodendrocyte differentiation has only just begun.
In this article, we review the current literature about the above mentioned epigenetic mechanisms regulating gene expression at specific stages of oligodendrocyte differentiation, and discuss their potential role in etiopathogenetic concepts of demyelinating diseases.
2. Chromatin architecture in oligodendrocytes
The 3D organization of chromatin within the nucleus is a crucial epigenetic factor in the regulation of gene expression, and the resulting nuclear location of a gene prominently determines its possibilities for transcription. During differentiation, positioning of chromosome territories may expose sites to the transcriptional machinery, thus facilitating efficient gene expression. Heterochromatin, highly condensed chromatin with transcriptionally inactive genes, is generally found in the peripheral region of the nucleus of most cell types. Constitutive heterochromatin, chromatin that is always condensed, and facultative heterochromatin, chromatin which may be decondensed in some circumstances or during developmental stages, also seem to have specific nuclear distribution patterns. Euchromatin, in general, consists of decondensed, gene-rich, transcriptionally active chromatin and appears to be associated with nuclear domains enriched in splicing factors (Bartova et al., 2008).
With the use of genomic in situ hybridization (GISH), the localization of a specific gene in a precise region of the nucleus has become feasible. As far as oligodendrocytes are concerned, GISH has been used to locate the genes encoding for proteolipid protein (PLP) and myelin basic protein (MBP) (Nielsen et al., 2002). PLP and MBP are both upregulated during the differentiation of OPCs into mature oligodendrocytes. Both in OPCs and in the more mature oligodendrocytes, the PLP gene appeared to be consistently located in the nuclear periphery and not to be co-localized with the MBP gene. Increased transcriptional activity of the PLP gene was found to correlate with the local accumulation of splicing factors and of the myelin transcription factor 1 (Myt1), which is known to bind to the PLP promotor (Nielsen et al., 2002). These data suggest that oligodendrocyte lineage genes and nuclear proteins might have specific and separate patterns of nuclear distribution and that activation of oligodendrocyte specific genes is established by changes in the distribution of local proteins regulating transcription rather than changes in clustering of coordinately regulated genes.
3. Histone modification in oligodendrocyte differentiation
Histone code
Post-translational modifications of nucleosomal histones consist of a growing list of alterations to amino acid residues on histone tails (figure 1a) that modulate gene expression independent of changes in the DNA sequence (Ptashne, 2007). The post-translational modifications of amino acids include acetylation/deacetylation, methylation (Shilatifard, 2006), phosphorylation (Dyson et al., 2005), sumoylation (Iniguez-Lluhi, 2006), ubiquitination (Weake and Workman, 2008) and citrullination (Mastronardi et al., 2006). These modifications establish a “histone code” that is cell type specific. A dynamic balance of these modifications dictates the transcriptional state of chromatin, as some histone changes confer a more open and transcriptionally competent state and others silence gene activation by chromatin compaction. Histone acetylation is functionally associated with transcriptionally competent chromatin, characterized by dispersed euchromatin. In contrast, histone deacetylation is functionally correlated with transcriptionally inactive chromatin (Bartova et al., 2008). Closely associated with the state of histone acetylation is histone methylation of lysine residues. However, in this case both the position of the amino acid to be methylated and the number of methyl groups added determine whether transcriptional capacity is enhanced or inhibited. Specifically, trimethylation of lysine 4 on histone H3 is associated with transcriptional activation, while trimethylation of lysine 9 or 27 on histone H3 is associated with transcriptional silencing (Shilatifard, 2006;Sims, III and Reinberg, 2008) (figure 1b). Repressive histone methylation on K9 is often coupled with histone deacetylation and has been associated with chromatin condensation and the formation of heterochromatin. Similar to acetyl groups on lysine residues, methyl groups can also be enzymatically removed from histones, due to the activity of specific demethylases. Besides lysine residues, methylation of arginine residues also leads to enhanced transcriptional activity. Asymmetric methylation of arginine 3 on histone H4 and of arginine 17 on histone H3 result in gene activation, while symmetric methylation of arginine 8 on histone H3 and of arginine 3 on histone H4 is associated with gene repression (Paik et al., 2007;Pal and Sif, 2007). Arginine methylation can be reversed by enzymatic conversion of arginine into citrulline residues, thereby further enhancing the complexity of the histone code.
Figure 1.
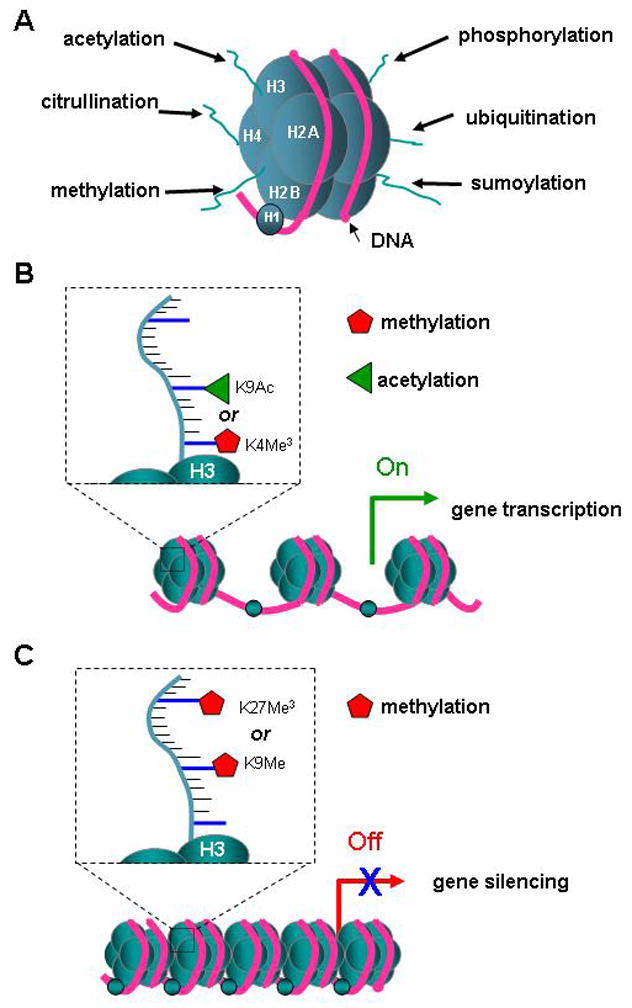
Histone modification. A. Model of the basic transcriptional unit, a nucleosome, shows the histone core octamer (2x H2A, H2B, H3, and H4, in blue) and the H1 linker protein, enwrapped 1.5 times by 150 bp of DNA (red). The histone tails can be modified by enzymes that catalyze methylation, acetylation, phosphorylation, citrullination, ubiquitination or sumoylation of specific amino acids. B. The scheme of 3 nucleosomes depicts a part of the chromatin with promotor and gene DNA that is open for transcription. The transcriptional active state is achieved by the acetylation or methylation of lysine 9 (K9) or lysine 4 (K4) respectively in the tail of histone 3 (H3). C. Transcriptional silencing can be caused by the histone modification resulting from the methylation of K9 or trimethylation of K27.
Histone acetyltransferases and oligodendrocyte differentiation
The rate and type of histone modifications in oligodendrocytes are controlled by a large group of enzymes and depend on the developmental stage, health and age of the cells. Histone modifications are clearly involved in cell fate decision and early stem cell differentiation, and this is also the case for late oligodendrocyte differentiation. Acetylation is essentially involved in the regulation of gene expression and is catalyzed by a family of histone acetyltransferases and reversed by histone deacetylase-mediated deacetylation. Members of the HAT family include GCN5, CBP and p300, each one being associated with transcriptional activation. Histone acetylases have been shown to play an important role in neural stem cell differentiation. Indeed, HAT p300 is recruited to the lineage specific gene glial fibrillary acidic protein (GFAP) during astrogliogenesis, controlling the amount of gene expression (Asklund et al., 2004;Song and Ghosh, 2004). In addition, removal of HDAC complexes from neurogenic marker genes, such as NeuroD, is necessary for neuronal differentiation to proceed (Hsieh et al., 2004). Whereas the acetylation of histones is necessary for transcriptional activation of several neuronal lineages, the role of HATs in oligodendrocyte differentiation is less pronounced. In fact, HDACs have been shown to play a more critical role in the progression of progenitor cells toward the oligodendrocyte lineage.
Histone deacetylases and oligodendrocyte differentiation
HDACs remove the acetyl groups from histone lysine residues and render the chromatin less accessible to transcription factors. HDACs have been classified into four broad classes, based on sequence homology to yeast histone deacetylases: class I (HDAC-1, -2, -3 and -8), class II (HDAC-4, -5, -6, -7, -9 and -10), class III (SIRT1-7), and class IV (HDAC11). Each class of HDAC has a unique expression pattern and is involved in different patterns of gene repression. Class I HDACs are generally localized to the nucleus and expressed in a ubiquitous manner in many tissues (Thiagalingam et al., 2003). They are often found as part of larger protein complexes that repress gene expression. Class II HDACs are generally localized in the cytoplasm and are shuttled to the nucleus when needed; the exception is HDAC10, which has been shown to be a constitutive nuclear protein. Moreover, class II HDACs are differentially expressed in human tissue, found more prominently in the heart, brain and skeletal muscle (Cress and Seto, 2000;Zhou et al., 2000). Class III HDACs are a family of NAD-dependent enzymes, also called “sirtuins,” due to their homology to the yeast Sir2 (Imai et al., 2000). They appear to play an important role in deacetylating histone H4 lysine 16 (Vaquero et al., 2007). However, Sirt1 has been shown to affect several additional proteins, including p53 (Cheng et al., 2003;Langley et al., 2002;Luo et al., 2001;Vaziri et al., 2001), FoxO (Brunet et al., 2004;Motta et al., 2004), p300/CBP (Motta et al., 2004), NFB (Yeung et al., 2004) and histone H1 (Vaquero et al., 2004). HDAC11, the only current member of class IV HDACs, shares properties of class I and class II. The size of the protein falls in line with class I HDACs but it is differentially expressed in human tissues, a characteristic of class II HDACs (Thiagalingam et al., 2003).
Unlike other neuronal lineages, oligodendrocyte differentiation of progenitor cells is initiated by a global histone deacetylation program (Marin-Husstege et al., 2002). During the early stages of oligodendrocyte development, HDACs mediate two critical processes: suppressing the choice of alternative lineages and repressing the expression of inhibitors of myelin specific genes (figure 2a). HDACs contribute to oligodendrocyte epigenetic identity by blocking critical transcription factors of alternative lineages, in particular Sox2 (Lyssiotis et al., 2007;Shen and Casaccia-Bonnefil, 2008). The importance of histone deacetylation in determining the direction of neural stem cell differentiation has been demonstrated in differentiation experiments in vitro with HDAC inhibitors. Differentiation of neural stem cells in the presence of HDAC inhibitors results in a noticeable reduction in oligodendrocytes on the account of a larger population of astrocytes and neurons (He et al., 2007b;Liu et al., 2007;Siebzehnrubl et al., 2007). Moreover, to initiate oligodendrocyte differentiation, HDAC1 and HDAC2 are recruited to the promoters of inhibitory molecules of myelin specific genes (He et al., 2007a;Shen and Casaccia-Bonnefil, 2008). Transcriptional inhibitors of oligodendrocyte differentiation include Hes5 (Kondo and Raff, 2000a;Liu et al., 2006), Sox11 and Tcf4 (He et al., 2007b), Id2 and Id4 (Kondo and Raff, 2000b;Marin-Husstege et al., 2006;Samanta and Kessler, 2004). Yin Yang 1 is the transcription factor that facilitates the recruitment of HDAC1 to the promoters of some of these transcriptional inhibitors, allowing for subsequent expression of myelin specific genes (He et al., 2007a). Thus, the global deacetylation program initiated by HDACs, especially specific isoforms of class I HDACs, enables expression of an oligodendrocyte transcriptional profile. SIRT2, a member of class III HDACs, is also enriched during oligodendrocyte development but it does not target its deacetylase capacity towards histones H3 or H4. Rather, SIRT2 targets acetylated α-tubulin, possibly effecting morphological differentiation of oligodendrocytes (Tang and Chua, 2008).
Figure 2.
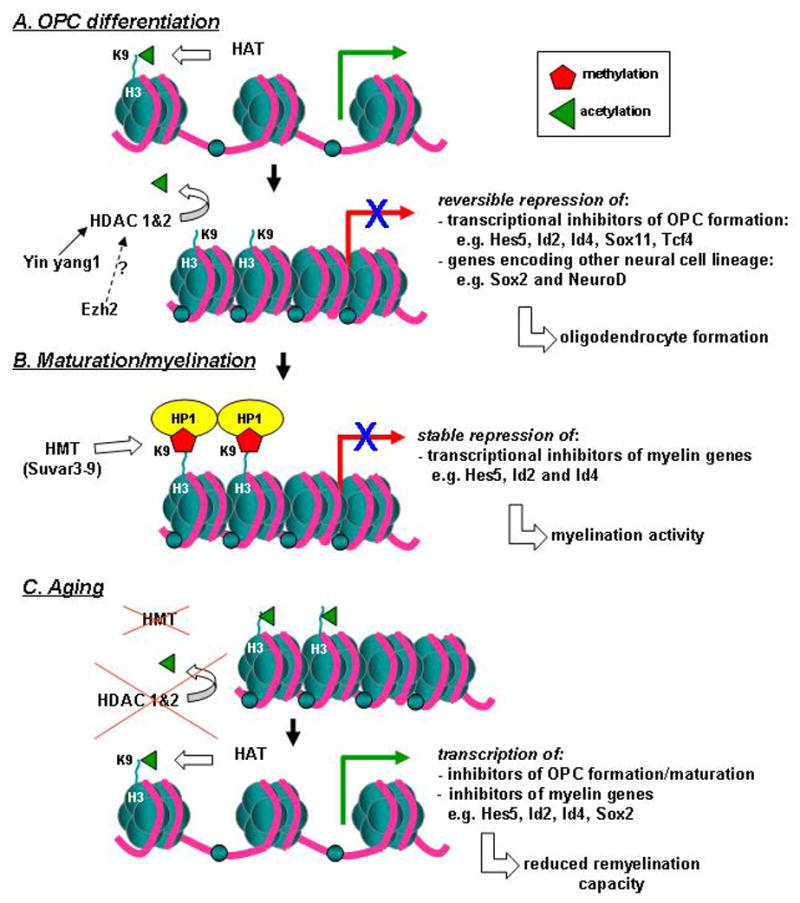
Histone modification and oligodendrocyte development. A. The differentiation of neural stem cells into oligodendrocyte precursor cells (OPCs) requires the recruitment of class I histone deacetylases, HDACs ( in particular 1 & 2) at genes encoding transcriptional inhibitors of OPC formation (e.g. Hes5, Id2, Id4, Sox11, and Tcf4) and those encoding the fate towards neurons or astrocytes. The HDAC activity at lysine 9 (K9) of histone protein 3 (H3) leads to the silencing of the expression of these genes, enabling oligodendrocyte formation. Yin Yang 1 and presumably Ezh2 are thought to facilitate the recruitment of the HDACs at these specific locations. The repression is still reversible, since some regulation is required to prevent premature myelination B. Essential stable repression of inhibitors of myelin genes at the mature stage of the oligodendrocyte, is established via the action of histone methyl transferases, HMTs (in particular of the Suvar3-9 family) resulting in the methylation of H3K9, followed by the binding of HP1 (histone protein 1) causing compaction of the nucleosome. The stable silencing of the myelin gene inhibitors promote myelination activity. C. The remyelination capacity of OPCs in aged animals is reduced due to diminished recruitment of HDACs and HMTs, resulting in histone acetyl transferase (HAT) activity to prevail causing an acetylated, transcriptional activated state of genes blocking proper OPC development and maturation and myelination.
Histone methyltransferases and oligodendrocyte differentiation
Histone methyltransferases (HMTases) are enzymes that catalyze the addition of methyl groups to various histone lysine residues. As noted above, histone methylation of specific residues is mediated by distinct enzymes. Methylation of lysine 4 on histone H3 (H3K4) is catalyzed by the mixed lineage leukemia (MLL) complex. Specifically, the SET domain of MLL is responsible for methylation of lysine K4 in the tail of histone H3. A number of HMTases are capable of methylating histone H3 at lysine 9 (H3K9), most notably the Su(var)3-9 family. However, the mechanism and the machinery responsible for methylation is dependent on the substrate and status of the cell, as dimethylases play a major role in euchromatin and trimethylases play a major role in heterochromatin (Shilatifard, 2006).
A prominent HMTase involved in methylation of lysine 27 on histone H3 (H3K27) is enhancer of zeste homologue 2 (Ezh2). Ezh2 is a member of the so-called Polycomb Group (PcG) proteins, which have been shown to play a key role in maintaining stem cell pluripotency (Boyer et al., 2006), cell proliferation (Erhardt et al., 2003), early embryogenesis (Varambally et al., 2002) and X chromosome inactivation (Plath et al., 2003). PcG proteins form polycomb repressive complexes like PRC1, which is comprised of the components Bmi1, HPC, EDR, Ring1/1a/1b/2 and YY1, and PRC2, which includes Ezh2, SUZ12 (suppressor of zeste 12) and EED (embryonic ectoderm development). The PRC2 complex, via Ezh2, initiates repressive activity by trimethylating histone H3 lysine 27 (H3K27) at target gene promoters; subsequently the methylated K27 serves as an anchor point for the recruitment of PRC1, which induces the transcriptional repressive state by recruiting HDACs. It has been shown that methylation at H3K27 by Ezh2 plays a role in the selection of the oligodendrocyte lineage. Neural stem cells differentiating into neurons or astrocytes down-regulate their expression of Ezh2, but the expression of Ezh2 in OPCs, even up to the stage of premyelinating immature oligodendrocytes, remains high. Forced increase in levels of Ezh2 in differentiating neural stem cells favored oligodendrocyte lineage choice, while Ezh2 silencing caused the opposite (Sher et al., 2008b). Ezh2 seemed to facilitate OPC formation by preventing a neuronal or astrocytic cell fate choice, as well as by stimulating OPC proliferation. Moreover, Ezh2 appeared to be involved in the differentiation of OPCs to immature oligodendrocytes; Ezh2 expression is down regulated in mature oligodendrocytes at the onset of myelination. This versatile effect of Ezh2 at different stages of oligodendrocyte formation and maturation must be ascribed to stage dependent changes in the target genes and/or the composition and activity of the PRC complexes involved. This is in line of what has been demonstrated before in other cell types and implies a far more dynamic role for Ezh2 than previously anticipated (Chopra and Mishra, 2005). The target genes and the varying composition of the PRC complexes at the various stages of oligodendrocyte formation still remain to be unraveled.
Histone demethylases and oligodendrocyte differentiation
While it was believed that histone methylation is a permanent modification, the discovery of lysine-specific demethylase enzymatic activities has revealed the dynamic process of histone methylation. Similar to methylation, demethylation is a differential process requiring distinct enzymes. LSD1 is a demethylase specific for mono- and di-methyl groups on lysine residues, while Jumonji domain-containing enzymes (JmjC) catalyze removal of tri-methyl groups from lysine residues (Benevolenskaya, 2007;Cheng and Zhang, 2007;Culhane and Cole, 2007). These histone demethylases are often coupled into a complex with other histone modifiers to enhance their individual activities, such as combining H3K4 trimethylation with H3K27 demethylation (Lan et al., 2008). While, so far no direct relevance of histone demethylation for oligodendrocyte differentiation has been found, involvement of H3K4 demethylase in the repression of neuronal genes in non-neuronal tissues has been shown (Tahiliani et al., 2007).
Although less studied, methylation of arginine residues provides another mode of dynamic gene regulation. In mammals, protein arginine methyltransferase 1 (PRMT1) and co-activator associated arginine methyltransferase 1 (CARM1) work in conjunction to catalyze methylation of arginine 3 on histone H4 (H4R3) and of arginines 2 and 17 on histone H3 (H3R2; H3R17), which results in gene activation. Meanwhile, PRMT5 catalyzes the methylation of arginine 8 on histone H3 (H3R8) and of arginine 3 on histone H4 (H4R3), which is associated with gene silencing (Paik et al., 2007;Pal and Sif, 2007). In both cases of arginine methylation, removal of the methyl group is not facilitated by any histone demethylases, as seen for lysine residues, but rather, the reaction is reversed by specific deiminases (Wang et al., 2004). Human peptidylarginine deiminase 2 (PAD2) and peptidylarginine deiminase 4 (PAD4) both convert arginine into citrulline, which removes the methyl group from the residue. Similar to histone demethylation, there is currently no direct role for arginine methylation in oligodendrocyte development. However, high levels of PAD2 and PAD4 enzyme, as well as subsequently high levels of citrullination, have been observed in myelin isolated from patients with multiple sclerosis (Wood et al., 2008) and experimental models of demyelination (Mastronardi et al., 2006).
4. Histone modification in oligodendrocyte maturation and aging
We have seen that class I HDACs play a crucial role in the actual generation and differentiation of early oligodendrocytes by preventing the lineage choice of neural stem cells leading to neuronal or astrocytic differentiation and by repressing the inhibitors of myelin gene expression. At the next stage of immature oligodendrocytes (after established lineage choice), up to the onset of myelination, the role of these HDACs is still committed to render myelin genes in a transcriptionally active state by forming repressive complexes with transcriptional inhibitors (Liu et al., 2006;Romm et al., 2005;Wei et al., 2005). The significance of HDAC activity in this specific period has been demonstrated in vivo by administration of valproic acid (VPA, a class 1 HDAC inhibitor) in postnatal rats (Shen et al., 2005). During the first 10 postnatal days, administration of VPA resulted in significant hypomyelination, with delayed expression of late differentiation markers and retained expression of progenitor markers. Administration of VPA after onset of myelination had no effect on myelin gene expression. During the time after the onset of myelination, a more stable repression of the transcriptional inhibitors of the myelin genes is effectuated by methylation at H3K9 (Shen et al., 2005) (figure 2b). This methylation is coupled with increased association of HP1, a protein that directs chromatin compaction by binding to adjacent H3K9 sites. Histone deacetylases such as HDAC11 and SIRT2 play another distinct role in oligodendrocytes. HDAC11 catalyzes deacetylation of lysine 9 and 14 on histone H3 (H3K9; H3K14) and it has been proposed to activate the expression of both myelin basic protein (MBP) and proteolipid protein (PLP) genes, possibly by modulating specific regulatory regions (Liu et al., 2009). Indeed, disruption of HDAC11 expression increased acetylation at the promoter region of both MBP and PLP and led to a subsequent decrease in their transcript levels, although it cannot be excluded that additional genes could have been affected by the silencing of HDAC11 (Liu et al., 2009). In a separate set of experiments, it was also shown that a cytosolic deacetylase, SIRT2, is recruited by the myelin protein PLP to the myelin compartment (Werner et al., 2007). In the cytosol, SIRT2 has been shown to deacetylate tubulin; however, it is likely that it may play an additional role in facilitating the morphological maturation of oligodendrocytes and myelin formation (Tang and Chua, 2008).
Histone modification in oligodendrocytes during aging and in diseases
The capacity of OPCs to acquire and maintain a high level of HDAC activity is necessary for oligodendrocyte differentiation. A similar epigenetic identity is also necessary for OPCs in the adult brain during cases of remyelination, as demonstrated in mice fed with a cuprizone-containing diet that induced demyelination. Removal of cuprizone led to spontaneous remyelination in young mice that recapitulated a gene expression pattern observed during initial oligodendrocyte development, characterized by increased activity of HDACs. In older mice, however, inefficient remyelination after cuprizone treatment was observed (Shen et al., 2008). Shen et al. detected decreased histone deacetylation and repressive methylation in oligodendrocytes of normal aging mice (Shen et al., 2008a) (figure 2c). The result of these epigenetic changes was an increased expression of inhibitory molecules, including Hes5, Ids and Sox2 (Shen et al., 2008a), thereby partially explaining the decreased remyelinating ability of the aging brain and the failure of replenishment therapies aimed at increasing the OPC population (Woodruff et al., 2004).
Apart from the epigenetic changes affecting remyelination potential, aberrant histone modification patterns in oligodendrocytes have been proposed to be part of the pathogenic process leading to demyelinating disorders such as MS (Casaccia-Bonnefil et al., 2008). High nuclear levels of the PAD4 (and PAD2) enzyme, as well as subsequently high levels of citrullination, have been observed in normal appearing white matter isolated from patients with multiple sclerosis (Wood et al., 2008) and experimental models of demyelination (Moscarello et al., 2007;Mastronardi et al., 2006). Under normal conditions, PAD4 is localized in the cytosol, but in the presence of an abnormally increased level of tumor necrosis factor α (TNFα), it is translocated to the nucleus, causing increased citrullination of histones. These molecular changes occur before the onset of symptoms in animal models of demyelination, indicating that TNFα-induced PAD4 nuclear localization and subsequent histone citrullination may be part of the etiopathogenesis of the disease. Increased levels of PAD2 must be ascribed to an increase in DNA demethylase activity, resulting in a hypomethylation of the PAD2 promoter (Casaccia-Bonnefil et al., 2008).
5. DNA methylation in oligodendrocytes
DNA methylation is an important epigenetic mechanism for regulating gene expression. It takes place at the CG sequences within the immediate 5′ promoter regions of genes. These regions have CpG contents greater than 60%, compared to 40% for the bulk DNA. Methylation of cytosine in CpG sequences turns off promoter activity and thereby decreases gene expression, since methylation affects both DNA structure and the binding of transcription factors. DNA methylation is regulated by the balance between DNA methyltransferases and DNA demethylases. As DNA methylation normally provides stable repression of genes, DNA methylation patterns are often cell-type specific, as seen in adult oligodendrocytes.
Interestingly, specific alterations in DNA methylation patterns found in brains from MS patients, have led to novel ideas about the pathogenesis of MS (Casaccia-Bonnefil et al., 2008). MS brains have a very specific two-fold higher DNA demethylase activity in comparison to normal brains and for instance brains of Alzheimer’s, Parkinson’s or Huntington’s patients (Mastronardi et al., 2007). As a consequence, DNA isolated from the white matter of MS brains contained only about 1/3 of the amount of methylated cytosine found in DNA from normal subjects or Alzheimer’s, Parkinson’s or Huntington’s patients (Mastronardi et al., 2007). The consequence of higher DNA demethylase activity in MS brains is that specific, normally repressed genes, particularly those with a high CpG content in their promoter, are reactivated. One of those genes is PAD2, which is involved in the deimination of myelin basic protein (MBP). While in normal brains only a very small proportion of MBP is citrullinated, the brains of patients with chronic MS are comprised of a larger percentage of citrullinated MBP, approximately 40% of the total MBP (Moscarello et al., 1986;Moscarello et al., 1994). The conversion of the majority of arginines in MBP into citrullines by increased levels of PAD2 resulted in aberrant interactions with other myelin components (Finch and Moscarello, 1972;Wood and Moscarello, 1989). Several studies have indicated the potential importance of abnormal citrullination of MBP in the sensitization of T-cells and the subsequent enhancement of the auto-immune response underlying MS pathogenesis (D’Souza et al., 2005;Musse et al., 2006;Tranquill et al., 2000;Zhou et al., 1995).
6. microRNA in oligodendrocytes
Whereas the previous epigenetic mechanisms regulating gene expression all operate at the transcriptional level, microRNAs (miRNA) are involved in mRNA translational regulation. miRNAs are endogenous small non-coding RNAs with a length of approximately ~22 nucleotides. They are encoded by DNA that resides between or within protein-coding genes. The long primary miRNA transcripts are processed into precursor miRNA stem-loops of ~60 nucleotides by the nuclear RNA polymerase type-III Drosha and then transported to the cytoplasm by exportin-5. After further trimming by Dicer, a cytoplasmic RNA polymerase type-III, they obtain their mature, short (~22 nucleotides), single-stranded form, which is bound to argonaute proteins. This silencing complex ultimately mediates the miRNA suppression of specific target mRNAs by imperfectly base pairing at the 3′ untranslated region (3′ UTR). This can either lead to the destabilization and degradation of the target mRNA or to its translational repression. In general, miRNAs are present at very high copy numbers per cell and they can regulate hundreds, thousands or even more mRNA targets, and so their impact on gene expression regulation must be considered profound. Apart from involvement of miRNAs in various cancers, specific microRNAs have been shown to modulate muscle differentiation (Chen et al., 2006), ES cell lineage commitment (Ivey et al., 2008;Tay et al., 2008;Krichevsky et al., 2006), haematopoietic lineage commitment (Johnnidis et al., 2008;Lu et al., 2008;Koralov et al., 2008), and cardiovascular development (Zhao et al., 2007), as well as synaptic development (Schratt et al., 2006).
In comparison to other organs, human and rodent brains contain many more types of miRNAs that are expressed at much higher levels, possibly reflecting their importance in the development and maintenance of the integrity of this complex organ. The expression of miRNAs is not homogenous over all brain regions and it alters during brain development. Although most of the functions of miRNAs in the brain still have to be discovered, recent studies in various animal models have implicated miRNAs as major players in neuroprotection and neurodegeneration (see review (Nelson et al., 2008). miRNA expression profiles of the separate neural populations, neurons, astrocytes and oligodendrocytes, have hardly been addressed and are mainly restricted to neurons. However, Lau et al. (2008) recently published a study in which they identified for the first time the miRNA expression profile (“microRNAome”) of developing oligodendrocytes isolated from postnatal (P7) rat brain (Lau et al., 2008). They were able to identify 98 miRNAs in developing rat oligodendrocytes, of which 43 changed their expression levels during the transition from oligodendrocyte progenitor cell (A2B5+/GalC−) to premyelinating oligodendrocyte (A2B5+/GalC+). Of the 98 identified miRNAs, 37 displayed a mRNA target bias; the expression level of the predicted target of 13 miRNAs appeared to be dynamically regulated during oligodendrocyte differentiation. So far, the most interesting example of that appeared to be miR-9, which is down-regulated during oligodendrocyte differentiation and inversely correlated with the expression of its predicted targets. One of these targets is peripheral myelin protein (PMP) 22, a protein produced by Schwann cells. PMP22 mRNA, but not protein, is present in oligodendrocytes. Lau et al. demonstrated that miR-9 interacts with the 3′ untranslated region of PMP22 and down-regulates its expression (Lau et al., 2008) in oligodendrocytes, whereas Schwann cells lack miR-9. These data show that miRNAs play an important role in the expression of oligodendrocytic genes, regulating the process of differentiation and maturation.
7. Conclusions
As in other cell types, the achievement of functional maturity of oligodendrocytes is the result of an intricate interplay between extrinsic signals and intrinsic, i.e. genetic and epigenetic, factors. While there is a substantial amount of information about essential oligodendrogenic transcription factors, oligodendrocyte differentiation genes and myelin genes, the regulation of their expression and/or silencing at specific stages during oligodendrocyte development and maturation by epigenetic factors is now just getting slowly unraveled. The identification of these epigenetic regulatory pathways has already led to novel ideas about etiopathogenic mechanisms underlying demyelinating diseases such as multiple sclerosis and may offer new therapeutic targets. Moreover, understanding the pathways leading to a stable specific differentiation of stem cells will benefit the production of pure, functional oligodendrocytes for cell replacement therapies.
Acknowledgments
PCB NIH R01-NS42925; NMSS RG3957; JLH was supported by a fellowship NIH T32 GM007280; FS is supported by a a grant of the Dutch Foundation MS Research, MS 04-554MS.
Reference List
- Asklund T, Appelskog IB, Ammerpohl O, Ekstrom TJ, Almqvist PM. Histone deacetylase inhibitor 4-phenylbutyrate modulates glial fibrillary acidic protein and connexin 43 expression, and enhances gap-junction communication, in human glioblastoma cells. Eur J Cancer. 2004;40:1073–1081. doi: 10.1016/j.ejca.2003.11.034. [DOI] [PubMed] [Google Scholar]
- Bartova E, Krejci J, Harnicarova A, Galiova G, Kozubek S. Histone modifications and nuclear architecture: a review. J Histochem Cytochem. 2008;56:711–721. doi: 10.1369/jhc.2008.951251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battiste J, Helms AW, Kim EJ, Savage TK, Lagace DC, Mandyam CD, Eisch AJ, Miyoshi G, Johnson JE. Ascl1 defines sequentially generated lineage-restricted neuronal and oligodendrocyte precursor cells in the spinal cord. Development. 2007;134:285–293. doi: 10.1242/dev.02727. [DOI] [PubMed] [Google Scholar]
- Benevolenskaya EV. Histone H3K4 demethylases are essential in development and differentiation. Biochem Cell Biol. 2007;85:435–443. doi: 10.1139/O07-057. [DOI] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature. 2006;441:349–353. doi: 10.1038/nature04733. [DOI] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- Cai J, Qi Y, Hu X, Tan M, Liu Z, Zhang J, Li Q, Sander M, Qiu M. Generation of oligodendrocyte precursor cells from mouse dorsal spinal cord independent of Nkx6 regulation and Shh signaling. Neuron. 2005;45:41–53. doi: 10.1016/j.neuron.2004.12.028. [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Pandozy G, Mastronardi F. Evaluating epigenetic landmarks in the brain of multiple sclerosis patients: a contribution to the current debate on disease pathogenesis. Prog Neurobiol. 2008;86:368–378. doi: 10.1016/j.pneurobio.2008.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chari DM. Remyelination in multiple sclerosis. Int Rev Neurobiol. 2007;79:589–620. doi: 10.1016/S0074-7742(07)79026-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JF, Mandel EM, Thomson JM, Wu Q, Callis TE, Hammond SM, Conlon FL, Wang DZ. The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38:228–233. doi: 10.1038/ng1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng HL, Mostoslavsky R, Saito S, Manis JP, Gu Y, Patel P, Bronson R, Appella E, Alt FW, Chua KF. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci U S A. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Zhang X. Structural dynamics of protein lysine methylation and demethylation. Mutat Res. 2007;618:102–115. doi: 10.1016/j.mrfmmm.2006.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra VS, Mishra RK. To SIR with Polycomb: linking silencing mechanisms. Bioessays. 2005;27:119–121. doi: 10.1002/bies.20191. [DOI] [PubMed] [Google Scholar]
- Cress WD, Seto E. Histone deacetylases, transcriptional control, and cancer. J Cell Physiol. 2000;184:1–16. doi: 10.1002/(SICI)1097-4652(200007)184:1<1::AID-JCP1>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Culhane JC, Cole PA. LSD1 and the chemistry of histone demethylation. Curr Opin Chem Biol. 2007;11:561–568. doi: 10.1016/j.cbpa.2007.07.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza CA, Wood DD, She YM, Moscarello MA. Autocatalytic cleavage of myelin basic protein: an alternative to molecular mimicry. Biochemistry. 2005;44:12905–12913. doi: 10.1021/bi051152f. [DOI] [PubMed] [Google Scholar]
- Dyson MH, Thomson S, Inagaki M, Goto H, Arthur SJ, Nightingale K, Iborra FJ, Mahadevan LC. MAP kinase-mediated phosphorylation of distinct pools of histone H3 at S10 or S28 via mitogen- and stress-activated kinase 1/2. J Cell Sci. 2005;118:2247–2259. doi: 10.1242/jcs.02373. [DOI] [PubMed] [Google Scholar]
- Erhardt S, Su IH, Schneider R, Barton S, Bannister AJ, Perez-Burgos L, Jenuwein T, Kouzarides T, Tarakhovsky A, Surani MA. Consequences of the depletion of zygotic and embryonic enhancer of zeste 2 during preimplantation mouse development. Development. 2003;130:4235–4248. doi: 10.1242/dev.00625. [DOI] [PubMed] [Google Scholar]
- Finch PR, Moscarello MA. A myelin protein fraction extracted with thioethanol. Brain Res. 1972;42:177–187. doi: 10.1016/0006-8993(72)90051-0. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Ffrench-Constant C. Remyelination in the CNS: from biology to therapy. Nat Rev Neurosci. 2008;9:839–855. doi: 10.1038/nrn2480. [DOI] [PubMed] [Google Scholar]
- He Y, Dupree J, Wang J, Sandoval J, Li J, Liu H, Shi Y, Nave KA, Casaccia-Bonnefil P. The transcription factor Yin Yang 1 is essential for oligodendrocyte progenitor differentiation. Neuron. 2007a;55:217–230. doi: 10.1016/j.neuron.2007.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Sandoval J, Casaccia-Bonnefil P. Events at the transition between cell cycle exit and oligodendrocyte progenitor differentiation: the role of HDAC and YY1. Neuron Glia Biol. 2007b;3:221–231. doi: 10.1017/S1740925X08000057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J, Nakashima K, Kuwabara T, Mejia E, Gage FH. Histone deacetylase inhibition-mediated neuronal differentiation of multipotent adult neural progenitor cells. Proc Natl Acad Sci U S A. 2004;101:16659–16664. doi: 10.1073/pnas.0407643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Iniguez-Lluhi JA. For a healthy histone code, a little SUMO in the tail keeps the acetyl away. ACS Chem Biol. 2006;1:204–206. doi: 10.1021/cb600188m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey KN, Muth A, Arnold J, King FW, Yeh RF, Fish JE, Hsiao EC, Schwartz RJ, Conklin BR, Bernstein HS, Srivastava D. MicroRNA regulation of cell lineages in mouse and human embryonic stem cells. Cell Stem Cell. 2008;2:219–229. doi: 10.1016/j.stem.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnnidis JB, Harris MH, Wheeler RT, Stehling-Sun S, Lam MH, Kirak O, Brummelkamp TR, Fleming MD, Camargo FD. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature. 2008;451:1125–1129. doi: 10.1038/nature06607. [DOI] [PubMed] [Google Scholar]
- Kessaris N, Fogarty M, Iannarelli P, Grist M, Wegner M, Richardson WD. Competing waves of oligodendrocytes in the forebrain and postnatal elimination of an embryonic lineage. Nat Neurosci. 2006;9:173–179. doi: 10.1038/nn1620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessaris N, Pringle N, Richardson WD. Specification of CNS glia from neural stem cells in the embryonic neuroepithelium. Philos Trans R Soc Lond B Biol Sci. 2008;363:71–85. doi: 10.1098/rstb.2006.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo T, Raff M. Basic helix-loop-helix proteins and the timing of oligodendrocyte differentiation. Development. 2000a;127:2989–2998. doi: 10.1242/dev.127.14.2989. [DOI] [PubMed] [Google Scholar]
- Kondo T, Raff M. The Id4 HLH protein and the timing of oligodendrocyte differentiation. EMBO J. 2000b;19:1998–2007. doi: 10.1093/emboj/19.9.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koralov SB, Muljo SA, Galler GR, Krek A, Chakraborty T, Kanellopoulou C, Jensen K, Cobb BS, Merkenschlager M, Rajewsky N, Rajewsky K. Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell. 2008;132:860–874. doi: 10.1016/j.cell.2008.02.020. [DOI] [PubMed] [Google Scholar]
- Krichevsky AM, Sonntag KC, Isacson O, Kosik KS. Specific microRNAs modulate embryonic stem cell-derived neurogenesis. Stem Cells. 2006;24:857–864. doi: 10.1634/stemcells.2005-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan F, Nottke AC, Shi Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr Opin Cell Biol. 2008;20:316–325. doi: 10.1016/j.ceb.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, Pelicci PG, Kouzarides T. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau P, Verrier JD, Nielsen JA, Johnson KR, Notterpek L, Hudson LD. Identification of dynamically regulated microRNA and mRNA networks in developing oligodendrocytes. J Neurosci. 2008;28:11720–11730. doi: 10.1523/JNEUROSCI.1932-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Han YR, Li J, Sun D, Ouyang M, Plummer MR, Casaccia-Bonnefil P. The glial or neuronal fate choice of oligodendrocyte progenitors is modulated by their ability to acquire an epigenetic memory. J Neurosci. 2007;27:7339–7343. doi: 10.1523/JNEUROSCI.1226-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Hu Q, D’ercole AJ, Ye P. Histone deacetylase 11 regulates oligodendrocyte-specific gene expression and cell development in OL-1 oligodendroglia cells. Glia. 2009;57:1–12. doi: 10.1002/glia.20729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Guo S, Ebert BL, Zhang H, Peng X, Bosco J, Pretz J, Schlanger R, Wang JY, Mak RH, Dombkowski DM, Preffer FI, Scadden DT, Golub TR. MicroRNA-mediated control of cell fate in megakaryocyte-erythrocyte progenitors. Dev Cell. 2008;14:843–853. doi: 10.1016/j.devcel.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Nikolaev AY, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- Lyssiotis CA, Walker J, Wu C, Kondo T, Schultz PG, Wu X. Inhibition of histone deacetylase activity induces developmental plasticity in oligodendrocyte precursor cells. Proc Natl Acad Sci U S A. 2007;104:14982–14987. doi: 10.1073/pnas.0707044104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marin-Husstege M, He Y, Li J, Kondo T, Sablitzky F, Casaccia-Bonnefil P. Multiple roles of Id4 in developmental myelination: predicted outcomes and unexpected findings. Glia. 2006;54:285–296. doi: 10.1002/glia.20385. [DOI] [PubMed] [Google Scholar]
- Marin-Husstege M, Muggironi M, Liu A, Casaccia-Bonnefil P. Histone deacetylase activity is necessary for oligodendrocyte lineage progression. J Neurosci. 2002;22:10333–10345. doi: 10.1523/JNEUROSCI.22-23-10333.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastronardi FG, Noor A, Wood DD, Paton T, Moscarello MA. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J Neurosci Res. 2007;85:2006–2016. doi: 10.1002/jnr.21329. [DOI] [PubMed] [Google Scholar]
- Mastronardi FG, Wood DD, Mei J, Raijmakers R, Tseveleki V, Dosch HM, Probert L, Casaccia-Bonnefil P, Moscarello MA. Increased citrullination of histone H3 in multiple sclerosis brain and animal models of demyelination: a role for tumor necrosis factor-induced peptidylarginine deiminase 4 translocation. J Neurosci. 2006;26:11387–11396. doi: 10.1523/JNEUROSCI.3349-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscarello MA, Brady GW, Fein DB, Wood DD, Cruz TF. The role of charge microheterogeneity of basic protein in the formation and maintenance of the multilayered structure of myelin: a possible role in multiple sclerosis. J Neurosci Res. 1986;15:87–99. doi: 10.1002/jnr.490150109. [DOI] [PubMed] [Google Scholar]
- Moscarello MA, Mastronardi FG, Wood DD. The role of citrullinated proteins suggests a novel mechanism in the pathogenesis of multiple sclerosis. Neurochem Res. 2007;32:251–256. doi: 10.1007/s11064-006-9144-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moscarello MA, Wood DD, Ackerley C, Boulias C. Myelin in multiple sclerosis is developmentally immature. J Clin Invest. 1994;94:146–154. doi: 10.1172/JCI117300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta MC, Divecha N, Lemieux M, Kamel C, Chen D, Gu W, Bultsma Y, McBurney M, Guarente L. Mammalian SIRT1 represses forkhead transcription factors. Cell. 2004;116:551–563. doi: 10.1016/s0092-8674(04)00126-6. [DOI] [PubMed] [Google Scholar]
- Musse AA, Boggs JM, Harauz G. Deimination of membrane-bound myelin basic protein in multiple sclerosis exposes an immunodominant epitope. Proc Natl Acad Sci U S A. 2006;103:4422–4427. doi: 10.1073/pnas.0509158103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Wang WX, Rajeev BW. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional control of oligodendrogenesis. Glia. 2007;55:1287–1299. doi: 10.1002/glia.20540. [DOI] [PubMed] [Google Scholar]
- Nielsen JA, Hudson LD, Armstrong RC. Nuclear organization in differentiating oligodendrocytes. J Cell Sci. 2002;115:4071–4079. doi: 10.1242/jcs.00103. [DOI] [PubMed] [Google Scholar]
- Paik WK, Paik DC, Kim S. Historical review: the field of protein methylation. Trends Biochem Sci. 2007;32:146–152. doi: 10.1016/j.tibs.2007.01.006. [DOI] [PubMed] [Google Scholar]
- Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003;300:131–135. doi: 10.1126/science.1084274. [DOI] [PubMed] [Google Scholar]
- Ptashne M. On the use of the word ‘epigenetic’. Curr Biol. 2007;17:R233–R236. doi: 10.1016/j.cub.2007.02.030. [DOI] [PubMed] [Google Scholar]
- Richardson WD, Kessaris N, Pringle N. Oligodendrocyte wars. Nat Rev Neurosci. 2006;7:11–18. doi: 10.1038/nrn1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romm E, Nielsen JA, Kim JG, Hudson LD. Myt1 family recruits histone deacetylase to regulate neural transcription. J Neurochem. 2005;93:1444–1453. doi: 10.1111/j.1471-4159.2005.03131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samanta J, Kessler JA. Interactions between ID and OLIG proteins mediate the inhibitory effects of BMP4 on oligodendroglial differentiation. Development. 2004;131:4131–4142. doi: 10.1242/dev.01273. [DOI] [PubMed] [Google Scholar]
- Schratt GM, Tuebing F, Nigh EA, Kane CG, Sabatini ME, Kiebler M, Greenberg ME. A brain-specific microRNA regulates dendritic spine development. Nature. 2006;439:283–289. doi: 10.1038/nature04367. [DOI] [PubMed] [Google Scholar]
- Shen S, Casaccia-Bonnefil P. Post-translational modifications of nucleosomal histones in oligodendrocyte lineage cells in development and disease. J Mol Neurosci. 2008;35:13–22. doi: 10.1007/s12031-007-9014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Li J, Casaccia-Bonnefil P. Histone modifications affect timing of oligodendrocyte progenitor differentiation in the developing rat brain. J Cell Biol. 2005;169:577–589. doi: 10.1083/jcb.200412101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen S, Sandoval J, Swiss VA, Li J, Dupree J, Franklin RJ, Casaccia-Bonnefil P. Age-dependent epigenetic control of differentiation inhibitors is critical for remyelination efficiency. Nat Neurosci. 2008;11:1024–1034. doi: 10.1038/nn.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sher F, Balasubramaniyan V, Boddeke E, Copray S. Oligodendrocyte differentiation and implantation: new insights for remyelinating cell therapy. Curr Opin Neurol. 2008a;21:607–614. doi: 10.1097/WCO.0b013e32830f1e50. [DOI] [PubMed] [Google Scholar]
- Sher F, Rossler R, Brouwer N, Balasubramaniyan V, Boddeke E, Copray S. Differentiation of Neural Stem Cells into Oligodendrocytes: Involvement of the Polycomb Group Protein Ezh2. Stem Cells. 2008b;26:2875–2883. doi: 10.1634/stemcells.2008-0121. [DOI] [PubMed] [Google Scholar]
- Shields S, Gilson J, Blakemore W, Franklin R. Remyelination occurs as extensively but more slowly in old rats compared to young rats following fliotoxin-induced CNS demyelination. Glia. 2000;28:77–83. doi: 10.1002/(sici)1098-1136(20000101)29:1<102::aid-glia12>3.0.co;2-1. [DOI] [PubMed] [Google Scholar]
- Shilatifard A. Chromatin modifications by methylation and ubiquitination: implications in the regulation of gene expression. Annu Rev Biochem. 2006;75:243–269. doi: 10.1146/annurev.biochem.75.103004.142422. [DOI] [PubMed] [Google Scholar]
- Siebzehnrubl FA, Buslei R, Eyupoglu IY, Seufert S, Hahnen E, Blumcke I. Histone deacetylase inhibitors increase neuronal differentiation in adult forebrain precursor cells. Exp Brain Res. 2007;176:672–678. doi: 10.1007/s00221-006-0831-x. [DOI] [PubMed] [Google Scholar]
- Sim FJ, Zhao C, Penderis J, Franklin RJ. The age-related decrease in CNS remyelination efficiency is attributable to an impairment of both oligodendrocyte progenitor recruitment and differentiation. J Neurosci. 2002;22:2451–2459. doi: 10.1523/JNEUROSCI.22-07-02451.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, III, Reinberg D. Is there a code embedded in proteins that is based on post-translational modifications? Nat Rev Mol Cell Biol. 2008;9:815–820. doi: 10.1038/nrm2502. [DOI] [PubMed] [Google Scholar]
- Song MR, Ghosh A. FGF2-induced chromatin remodeling regulates CNTF-mediated gene expression and astrocyte differentiation. Nat Neurosci. 2004;7:229–235. doi: 10.1038/nn1192. [DOI] [PubMed] [Google Scholar]
- Sparmann A, Lohuizen van M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Tahiliani M, Mei P, Fang R, Leonor T, Rutenberg M, Shimizu F, Li J, Rao A, Shi Y. The histone H3K4 demethylase SMCX links REST target genes to X-linked mental retardation. Nature. 2007;447:601–605. doi: 10.1038/nature05823. [DOI] [PubMed] [Google Scholar]
- Tang BL, Chua CE. SIRT2, tubulin deacetylation, and oligodendroglia differentiation. Cell Motil Cytoskeleton. 2008;65:179–182. doi: 10.1002/cm.20253. [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Thiagalingam S, Cheng KH, Lee HJ, Mineva N, Thiagalingam A, Ponte JF. Histone deacetylases: unique players in shaping the epigenetic histone code. Ann N Y Acad Sci. 2003;983:84–100. doi: 10.1111/j.1749-6632.2003.tb05964.x. [DOI] [PubMed] [Google Scholar]
- Tranquill LR, Cao L, Ling NC, Kalbacher H, Martin RM, Whitaker JN. Enhanced T cell responsiveness to citrulline-containing myelin basic protein in multiple sclerosis patients. Mult Scler. 2000;6:220–225. doi: 10.1177/135245850000600402. [DOI] [PubMed] [Google Scholar]
- Vallstedt A, Klos JM, Ericson J. Multiple dorsoventral origins of oligodendrocyte generation in the spinal cord and hindbrain. Neuron. 2005;45:55–67. doi: 10.1016/j.neuron.2004.12.026. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Scher M, Lee D, Erdjument-Bromage H, Tempst P, Reinberg D. Human SirT1 interacts with histone H1 and promotes formation of facultative heterochromatin. Mol Cell. 2004;16:93–105. doi: 10.1016/j.molcel.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Vaquero A, Sternglanz R, Reinberg D. NAD+-dependent deacetylation of H4 lysine 16 by class III HDACs. Oncogene. 2007;26:5505–5520. doi: 10.1038/sj.onc.1210617. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, Rubin MA, Chinnaiyan AM. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng EE, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Wysocka J, Sayegh J, Lee YH, Perlin JR, Leonelli L, Sonbuchner LS, McDonald CH, Cook RG, Dou Y, Roeder RG, Clarke S, Stallcup MR, Allis CD, Coonrod SA. Human PAD4 regulates histone arginine methylation levels via demethylimination. Science. 2004;306:279–283. doi: 10.1126/science.1101400. [DOI] [PubMed] [Google Scholar]
- Weake VM, Workman JL. Histone ubiquitination: triggering gene activity. Mol Cell. 2008;29:653–663. doi: 10.1016/j.molcel.2008.02.014. [DOI] [PubMed] [Google Scholar]
- Wegner M. A matter of identity: transcriptional control in oligodendrocytes. J Mol Neurosci. 2008;35:3–12. doi: 10.1007/s12031-007-9008-8. [DOI] [PubMed] [Google Scholar]
- Wei Q, Miskimins WK, Miskimins R. Stage-specific expression of myelin basic protein in oligodendrocytes involves Nkx2.2-mediated repression that is relieved by the Sp1 transcription factor. J Biol Chem. 2005;280:16284–16294. doi: 10.1074/jbc.M500491200. [DOI] [PubMed] [Google Scholar]
- Werner HB, Kuhlmann K, Shen S, Uecker M, Schardt A, Dimova K, Orfaniotou F, Dhaunchak A, Brinkmann BG, Mobius W, Guarente L, Casaccia-Bonnefil P, Jahn O, Nave KA. Proteolipid protein is required for transport of sirtuin 2 into CNS myelin. J Neurosci. 2007;27:7717–7730. doi: 10.1523/JNEUROSCI.1254-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DD, Ackerley CA, Brand B, Zhang L, Raijmakers R, Mastronardi FG, Moscarello MA. Myelin localization of peptidylarginine deiminases 2 and 4: comparison of PAD2 and PAD4 activities. Lab Invest. 2008;88:354–364. doi: 10.1038/labinvest.3700748. [DOI] [PubMed] [Google Scholar]
- Wood DD, Moscarello MA. The isolation, characterization, and lipid-aggregating properties of a citrulline containing myelin basic protein. J Biol Chem. 1989;264:5121–5127. [PubMed] [Google Scholar]
- Woodruff RH, Fruttiger M, Richardson WD, Franklin RJ. Platelet-derived growth factor regulates oligodendrocyte progenitor numbers in adult CNS and their response following CNS demyelination. Mol Cell Neurosci. 2004;25:252–262. doi: 10.1016/j.mcn.2003.10.014. [DOI] [PubMed] [Google Scholar]
- Yeung F, Hoberg JE, Ramsey CS, Keller MD, Jones DR, Frye RA, Mayo MW. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Ransom JF, Li A, Vedantham V, von DM, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNA-1-2. Cell. 2007;20(129):303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- Zhou SR, Moscarello MA, Whitaker JN. The effects of citrullination or variable amino-terminus acylation on the encephalitogenicity of human myelin basic protein in the PL/J mouse. J Neuroimmunol. 1995;62:147–152. doi: 10.1016/0165-5728(95)00112-3. [DOI] [PubMed] [Google Scholar]
- Zhou X, Richon VM, Rifkind RA, Marks PA. Identification of a transcriptional repressor related to the noncatalytic domain of histone deacetylases 4 and 5. Proc Natl Acad Sci U S A. 2000;97:1056–1061. doi: 10.1073/pnas.97.3.1056. [DOI] [PMC free article] [PubMed] [Google Scholar]


