Abstract
Following successful gene rearrangement at αβ T-cell receptor (TCR) loci, developing thymocytes express both CD4 and CD8 co-receptors and undergo a life-or-death selection event known as positive selection to identify cells expressing TCRs with potentially useful ligand specificities. Positively selected thymocytes must then decide whether to differentiate into CD4+ helper T cells or CD8+ cytotoxic T cells, a crucial decision known as CD4/CD8 lineage choice. This Review summarizes recent advances in our understanding of the cellular and molecular events involved in lineage fate decision and discusses them in the context of the major models of CD4/CD8 lineage choice.
Throughout development, bipotential cells use environmental cues to determine cell fate, and elucidating the mechanisms by which they do so continues to be a fascinating area of investigation. An immunologically relevant example of bipotential cell-fate determination is the differentiation of CD4+CD8+ (double positive, DP) thymocytes into either CD4+ helper T cells or CD8+ cytotoxic T cells. DP thymocytes are the first cells in the T-cell developmental pathway to express fully assembled αβ T-cell receptor (TCR) complexes on the cell surface (Fig. 1), and it is the ligand specificity of their TCR that determines their subsequent developmental fate. αβTCRs are somatically generated transmembrane receptors with clonally unique structures that allow for a hugely diverse repertoire of recognition specificities. However, most thymocytes express αβTCRs that are incapable of engaging self MHC molecules and are therefore not useful to the host immune system. To eliminate cells expressing TCRs that cannot engage self MHC molecules, DP thymocytes are subjected to strict selection pressures in which cells bearing potentially useful TCR are the only ones signalled to survive and to continue their differentiation into functionally mature T cells. The vast majority of DP thymocytes do not receive TCR survival signals and undergo ‘death by neglect’ because their TCR cannot engage self MHC molecules. This life-or-death TCR mediated signalling event in DP thymocytes is referred to as ‘positive selection’ and results in the survival and maturation of cells bearing potentially useful TCRs.
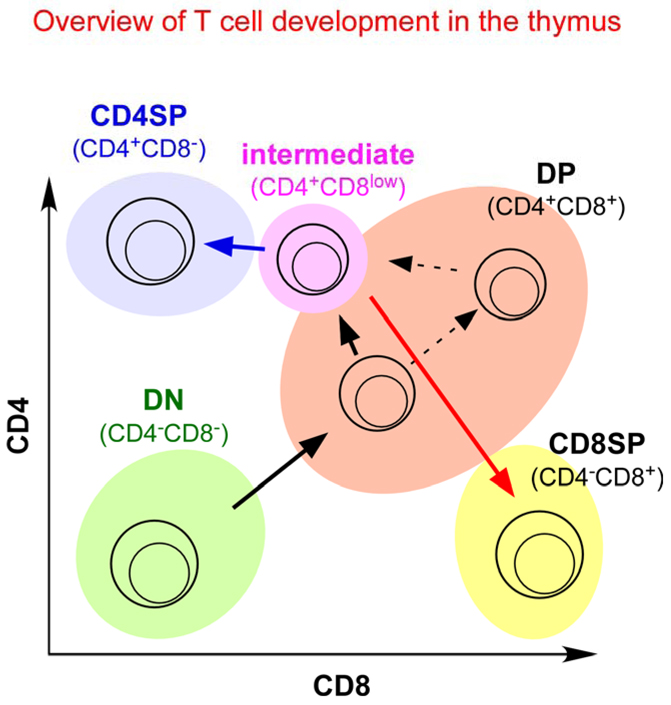
Figure 1. Overview of T-cell development in the thymus.
Thymocyte subpopulations can be identified by cell-surface co-receptor expression. Double negative (CD4−CD8−, DN) cells, which express neither CD4 nor CD8, are the most immature cells in the thymus. DN cells differentiate into double positive (CD4+CD8+, DP) thymocytes, which are the first cells to express a functional αβ T-cell receptor (TCR). DP thymocytes that express potentially useful TCR specificities are signalled by the TCR to undergo positive selection and to become intermediate (CD4+CD8low) cells, which then differentiate into either CD4 single positive (CD4+CD8−, CD4 SP) or CD8 SP (CD4−CD8+) mature thymocytes. Depending on the timing of their expression of a functional αβTCR, DP thymocytes can be signalled to undergo positive selection either when they express low levels of both co-receptors or when they express high levels of both co-receptors.
The success of positive selection in identifying potentially useful TCRs requires that DP thymocytes depend solely on signals downstream of TCR ligation for their survival, and that DP thymocytes be unresponsive to other survival signals. As a result, DP thymocytes are unique among T-lineage cells in that they are virtually refractory to the pro-survival cytokine interleukin-7 (IL-7), in part because DP thymocytes do not express receptors for IL-7 or most other pro-survival cytokines, and in part because DP thymocytes express high levels of SOCS1 (suppressor of cytokine signalling 1), a potent intracellular suppressor of cytokine signal transduction1, 2.
DP thymocytes are also unique among T-lineage cells in that they express both CD4 and CD8 co-receptors. CD4 and CD8 co-receptors are transmembrane proteins with extracellular domains that promote TCR engagement of MHC ligands and intracellular domains that enhance TCR signal transduction. As a result, CD4/CD8 co-receptors are molecules that promote signalling by MHC-restricted TCRs. The extracellular domains of CD4 and CD8 co-receptors bind specifically to invariant determinants on MHC class II and MHC class I molecules respectively, while their intracellular domains associate with the nonreceptor protein tyrosine kinase LCK, which initiates TCR signal transduction when enzymatically activated3–6. By binding to the same peptide–MHC complexes that have engaged the TCR, CD4 and CD8 co-receptors bring intracellular LCK into physical proximity with cytosolic domains of the engaged TCR to initiate signalling7, 8. And, by expressing both co-receptor molecules, DP thymocytes receive signals from both MHC-class-I- and MHC-class-II-restricted TCRs so that all potentially useful TCRs can generate positive selection signals and rescue DP thymocytes from cell death.
DP thymocytes that have been positively selected ultimately develop into either CD4+ or CD8+ single positive (SP) T cells, with their precise lineage fate determined by the MHC restriction specificity of their TCR (Box 1). With remarkable consistency, DP thymocytes signalled by MHC-class-II-restricted TCRs differentiate into CD4+ T cells, whereas DP thymocytes signalled by MHC-class-I-restricted TCRs differentiate into CD8+ T cells. The mechanism by which TCR specificity dictates CD4/CD8 lineage choice has been a difficult problem to unravel, and has been the subject of intense debate for the 20 years since TCR-transgenic mice first revealed that CD4/CD8 lineage choice was determined by the MHC-restriction specificity of the TCR9. Fortunately, the environmental cues, cellular signals, and transcription factors involved in lineage choice have now been significantly clarified. Although many aspects continue to be debated, a coherent and unified picture of positive selection and CD4/CD8 lineage choice is now emerging.
BOX 1 | Possible basis for thymic selection of an MHC-restricted TCR repertoire.
It is not understood why double-positive (DP) thymocytes bearing MHC-restricted T-cell receptors (TCRs) are the only DP thymocytes that are signalled to undergo positive selection, and why DP thymocytes bearing TCRs with the potential to engage non-MHC ligands that are expressed in the thymus are not also positively selected. Given that randomly generated TCRs have extensive diversity, it seems unlikely that all TCRs would display exclusive specificity for MHC ligands, although this is a formal possibility104, 105. A recently proposed solution106 to this problem is based on the fact that both CD4 and CD8 co-receptor molecules are expressed on individual DP thymocytes and that their content of LCK is limited107. So, most of the LCK that would be available to initiate TCR signalling in DP thymocytes associates with one or the other co-receptor molecule, with little ‘free’ LCK remaining108. So, productive TCR signalling in DP thymocytes requires co-engagement of TCR with co-receptors associated with LCK106. TCRs that engage MHC ligands do so together with CD4 or CD8 molecules, whereas TCRs that engage non-MHC ligands do so independently of co-receptor molecules. Consequently, DP thymocytes can be signalled by MHC-restricted TCRs to undergo positive selection, whereas DP thymocytes bearing MHC-independent TCRs cannot be signalled and die of neglect — even though their TCRs may have engaged an intrathymic ligand106. So, co-expression of CD4 and CD8 by DP thymocytes contributes in two ways to focusing the mature T-cell repertoire on MHC: by inhibiting positive selection signalling by TCRs that engage non-MHC ligands in the thymus106, and by promoting positive selection signalling by TCRs that engage MHC ligands.
The cellular and molecular mechanisms underlying CD4/CD8 lineage choice have been as much the subject of abstract model building as they have been the subject of experimental analyses. In trying to understand CD4/CD8 lineage choice, abstract models have provided the intellectual rationale for experiments that followed. This Review discusses the major models of CD4/CD8 lineage choice and uses them as prisms through which to understand the experimental findings that have enlightened our understanding of this biological puzzle.
Classical models of CD4/CD8 lineage choice
As DP thymocytes are bipotential cells that express both CD4 and CD8 co-receptors, CD4/CD8 lineage choice was classically considered to be the transcriptional termination of one or the other co-receptor gene as a consequence of the same TCR signalling event that mediates positive selection. All classical models of CD4/CD8 lineage choice incorporate this perspective and fall into two major categories as either ‘stochastic’ or ‘instructive’ which differ in whether co-receptor termination is random or instructed. Stochastic and instructive models of CD4/CD8 lineage choice were initially thought to represent two extremes that covered the full spectrum of logical possibilities10, but both types of classical models are actually based on a shared set of fundamental principles, namely: positive selection and lineage commitment are simultaneous events induced by the same TCR signals; TCR signals during positive selection can selectively terminate either Cd4 or Cd8 gene expression; and selective termination of either co-receptor gene is irreversible and indicative of commitment to the opposite co-receptor lineage.
The stochastic selection model
The stochastic selection model of CD4/CD8 lineage choice postulates that termination of co-receptor gene expression during positive selection of DP thymocytes occurs randomly11–16 and that a second TCR-dependent ‘rescue’ step occurs after positive selection, so that only SP thymocytes with matching TCR and co-receptors survive and differentiate into mature T cells (Fig 2A). Direct support for the stochastic selection model has mainly come from ‘co-receptor rescue’ experiments in which persistent expression of transgene-encoded co-receptors resulted in mature T cells bearing TCRs with MHC-restriction specificities that were inappropriate for their T-cell lineage11, 12, 15, 16. For example, expression of transgenic CD4 proteins resulted in mature CD8+ T cells bearing mismatched MHC-class-II-restricted TCRs, presumably because transgenically forced expression of CD4 permitted MHC-class-II-restricted TCRs to rescue short-lived SP thymocytes that had terminated endogenous Cd4 gene expression12, 17. Similarly, deletion of the endogenous control element responsible for silencing Cd4 transcription resulted in persistent CD4 expression by all thymocytes and resulted in mature CD8+ T cells bearing mismatched MHC-class-II-restricted TCRs14.
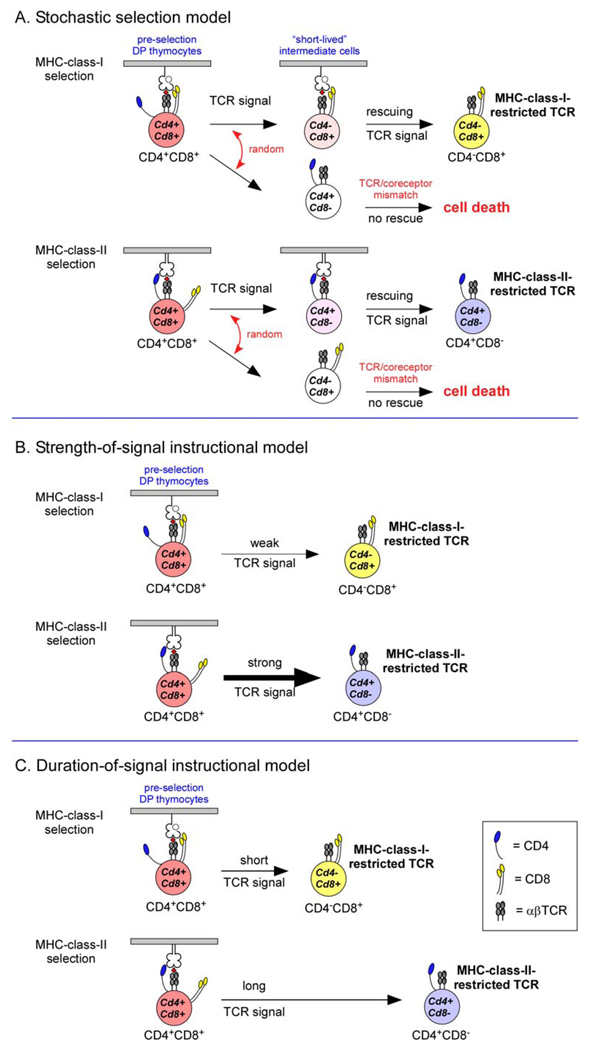
Figure 2. Classical models of CD4/CD8 lineage choice.
A. The stochastic selection model postulates that positive selecting T-cell receptor (TCR) signals randomly terminate expression of one or the other co-receptor molecule, resulting in the generation of “short-lived” intermediate cells, which will undergo programmed cell death unless rescued by a second TCR signal. Because the TCR-mediated rescue signal requires TCRs and co-receptors that are matched, 50% of positively selected thymocytes will fail to survive and mature.
B. The strength-of-signal instructional model postulates that strong TCR signals terminate Cd8 transcription whereas weak TCR signals terminate Cd4 transcription. Signalling by CD4 and MHC-class-II-restricted TCRs is strong resulting in mature CD4+ T cells, whereas signalling by CD8 and MHC-class-I-restricted TCRs is weak resulting in mature CD8+ T cells.
C. The duration-of-signal instructional model postulates that long and/or strong TCR signals terminate Cd8 transcription, whereas short and/or weak TCR signals terminate Cd4 transcription. MHC-class-I-restricted and MHC-class-II-restricted TCR signals are proposed to differ in both duration and intensity.
Because the stochastic selection model predicts that CD4/CD8 lineage choice occurs randomly in TCR-signalled DP thymocytes, lineage choice is predicted to be highly inefficient, with 50% of potentially useful TCRs lost because they were present on thymocytes that no longer expressed the matching co-receptor molecule. Yet, the numbers of T cells rescued by transgenic coreceptors never approached 50% of positively selected thymocytes as would be predicted for a random event. And experiments that measured the efficiency of repertoire selection of thymocytes bearing transgenic TCRs demonstrated that repertoire selection could approach 90% efficiency18, a level that cannot be reconciled with the central premise of the stochastic selection model.
The rescue step of the stochastic selection model requires that newly arising SP thymocytes be short-lived cells that die rapidly if their TCR and co-receptors do not have matching MHC specificities. However, this key requirement of the stochastic selection model has been contradicted by observations that SP thymocytes with mismatched TCR and co-receptors are not short-lived, but are sufficiently long-lived to differentiate into functionally mature T cells that emigrate into the periphery19, 20.
Thus, core principles of the stochastic selection model have been contradicted by experimental observation.
Strength-of-signal instructional model
Instructive models of CD4/CD8 lineage choice postulate that TCR signals direct DP thymocytes during positive selection to specifically terminate expression of the mismatching co-receptor molecule. Consequently, instructive models require that MHC-class-I- and MHC-class-II-restricted TCR signals be sufficiently distinct from one another to specify termination of the mismatching co-receptor molecule.
In the original instruction model, CD4 and CD8 co-receptors were hypothesized to transduce qualitatively different instructional signals21, but this idea was subsequently replaced by the proposal that DP thymocytes were instructed by differences in the strength of signals transduced by TCR and co-receptor co-engagements during positive selection22 (Fig. 2B). Because the cytosolic tail of CD4 binds significantly more intracellular LCK than the cytosolic tail of CD85, 23, TCR and CD4 co-engagement generates strong signals whereas TCR and CD8 co-engagement generates weak signals, and it is the strength of these signals that induces thymocytes to specifically terminate either Cd8 or Cd4 gene expression22. Formulation of the strength-of-signal instructional model was prompted by experiments that used chimeric co-receptor transgenes that encoded CD8α molecules with cytosolic tails engineered to express the cytosolic domain of CD422. In vivo expression of chimeric CD8–CD4 transgenic molecules resulted in the development of MHC-class-I-restricted CD4+ T cells, presumably because MHC-class-I-restricted DP thymocytes were directed to become CD4+ T cells. However, thymocytes bearing very low affinity MHC-class-I-restricted TCRs specific for the HY antigen were not directed to differentiate into CD4+ T cells22. Consequently, it was proposed that lineage choice was dictated by the overall strength of signals transduced by co-engaged TCR and co-receptor molecules, with strong signals promoting CD4-lineage choice and weak signals promoting CD8-lineage choice22. Since MHC-class-I- and MHC-class-II-restricted TCRs presumably had similar ligand affinities, it was quantitative differences in the intensity of signalling between CD4 and CD8 co-receptors that mainly determined CD4/CD8 lineage choice.
The strength-of-signal model provided a straightforward explanation for experiments that manipulated the activity in DP thymocytes of intracellular kinases such as LCK24, 25, CSK (C-terminal SRC kinase)26, TEC kinases27–29 and extracellular-signal-regulated kinases (ERKs)30–33. These experiments revealed that increased kinase activity favoured CD4+ T-cell differentiation, whereas decreased kinase activity favoured CD8+ T-cell differentiation. However, the crucial experiments for the strength-of-signal model assessed its core concept by directly altering the signalling intensity of the TCR or co-receptor molecules themselves. When these experiments were performed, it became clear that signal intensity did not determine CD4/CD8 lineage choice.
The effect of TCR signalling intensity on lineage choice was experimentally assessed by altering the number of immunoreceptor tyrosine-based activation motifs (ITAMs) contained within each TCR signalling complex34. Reductions in the number of TCR ITAMs reduced TCR signalling intensity and resulted in the generation of fewer SP T cells, but it did not alter CD4/CD8 lineage choice34. Regardless of the number of ITAMs, thymocytes expressing MHC-class-II-restricted TCRs still differentiated into CD4+ T cells and thymocytes expressing MHC-class-I-restricted TCRs still differentiated into CD8+ T cells, a finding that was recently confirmed with ITAM-deletion mutant mice35, contradicting the core concept of the strength-of-signal model.
The contribution of co-receptor ligation to TCR signalling intensity has also been carefully re-examined for its effect on CD4/CD8 lineage choice. Experiments to reassess the impact of chimeric CD8–CD4 transgenic co-receptor molecules on lineage choice by MHC-class-I-restricted thymocytes replicated the original experiments that prompted the strength-of-signal model22, but in CD8α-deficient mice36. Without endogenous CD8α molecules to complicate the experimental results, it could be seen that expression of stronger signalling chimeric CD8–CD4 co-receptors did not preferentially direct MHC-class-I-restricted thymocytes to differentiate into CD4+ T cells36, as had been originally thought22. Consequently, a definitive assessment of the role of co-receptor signal strength in determining lineage choice was carried out with mice whose endogenous Cd8a gene was engineered to encode the cytosolic tail of CD437. The engineered endogenous gene (named CD8.4) encoded stronger signalling CD8-CD4 chimeric co-receptor proteins whose effects on thymocyte development were independent of potential transgenic artifacts. The results were unequivocal: expression of stronger signalling CD8.4 co-receptors quantitatively increased thymic selection of MHC-class-I-restricted T cells but had no impact on CD4/CD8 lineage choice, as MHC-class-I-restricted T cells in CD8.4 mice were exclusively CD8+ T cells37. So, experiments with both endogenous and transgene encoded CD8-CD4 chimeric coreceptor molecules contradicted the premise of the strength-of-signal instructional model.
Although central requirements of the strength-of-signal model of CD4/CD8 lineage choice have been directly contradicted by experimental observation, experimental testing of the strength-of-signal model provided an answer to a different question — why CD4+ T cells outnumber CD8+ T cells in most mammalian species. Because the signalling intensity of CD4 co-receptors is greater than that of CD8 co-receptors, TCR co-engagement with CD4 induces quantitatively more DP thymocytes to undergo positive selection than TCR co-engagement with CD8, resulting in greater numbers of mature CD4+ than CD8+ T cells36, 37.
Duration-of-signal instructional model: a classical model with a twist
The duration-of-signal instructional model is an updated version of the original strength-of-signal model with the important twist that TCR signal duration, perhaps in addition to signal strength, determines CD4/CD8 lineage choice (Fig. 2C). In this model, TCR signals of long duration instruct DP thymocytes to terminate Cd8 gene expression and to differentiate into CD4+ T cells, whereas TCR signals of short duration instruct DP thymocytes to terminate Cd4 gene expression and to differentiate into CD8+ T cells38. While it was originally unclear why MHC-class-I- and MHC-class-II-restricted TCR signals would be of different duration38, an explanation adopted from the kinetic signaling model39, 40 (see below) provided a solution: all TCR-signalled DP thymocytes selectively reduce surface CD8 co-receptor expression which disrupts MHC-class-I-restricted TCR signaling but does not affect MHC-class-II-restricted TCR signaling40. Consequently, MHC-class-I-specific TCR signaling in CD4+CD8low thymocytes is of shorter duration than MHC-class-II-specific TCR signaling40.
There is now general acceptance that phenotypically CD4+CD8low thymocytes are lineage-uncommitted cells39 and are the precursors of both CD4+ and CD8+ mature T cells41, 42. However, the molecular basis by which surface CD8 expression is selectively reduced on TCR-signalled DP thymocytes so that they adopt an asymmetric CD4+CD8low phenotype is vigorously disputed39, 40, 43–45. The duration-of-signal instructional model 38, 43–45 postulates that TCR-mediated positive selection signalling in DP thymocytes has complex effects on cell-surface expression of both CD4 and CD8 coreceptor proteins that cause signalled DP thymocytes to ultimately appear CD4+CD8low, despite continued expression of both Cd4 and Cd8 genes46. Indeed, as a classical model, the duration-of-signal instructional model requires lineage choice to occur in thymocytes that are transcriptionally Cd4+Cd8+, despite their CD4+CD8low appearance45. However, the requirement that lineage-uncommitted CD4+CD8low thymocytes be transcriptionally Cd4+Cd8+ has not been experimentally verified38, 46, 47. In fact the opposite is true: in vivo experiments48 have documented that MHC-class-I-signalled DP thymocytes become CD4+CD8low intermediate cells by downregulating Cd8 transcription and becoming Cd4+Cd8−. In these experiments48, each DP thymocyte expressed two allelically distinct CD8 proteins whose expression was regulated by different transcriptional control elements, with one CD8 allele under the control of its endogenous cis-regulatory elements and the other under the control of heterologous transgenic elements. If MHC-class-I signalled DP thymocytes became CD4+CD8low cells by internalizing cell-surface CD8 proteins, then surface expression of both allelic CD8 proteins would be reduced. Alternatively, if MHC-class-I-signalled DP thymocytes became CD4+CD8low cells by terminating endogenous Cd8 gene expression, then surface expression of endogenously encoded cell-surface CD8 would alone be reduced. In fact, in vivo MHC-class-I-induced signalling in DP thymocytes only reduced surface expression of endogenously encoded CD8 proteins and not that of transgenically encoded CD8 proteins. So, the asymmetric loss of surface co-receptor expression on TCR-signalled DP thymocytes is due to downregulation of Cd8 gene expression48, and is not due to internalization of CD8 surface proteins48, 49.
Thus, the requirement of the duration-of-signal instructional model that makes it a classical model has been experimentally contradicted. However, the concept that TCR signal duration is a major determinant of CD4/CD8 lineage choice remains intact and is a central feature of the kinetic signalling model (see below).
The kinetic signalling model: a non-classical model of CD4/CD8 lineage choice
CD4/CD8 lineage choice seems to be best explained by the kinetic signalling model, which proposes that CD4/CD8 lineage choice is determined by TCR signal duration and proposes that cytokines of the common cytokine-receptor γ-chain (γc) family, such as IL-7, serve as ‘sensors’ which detect the duration of the TCR signal39, 40, 50 (Fig. 3). The kinetic signalling model is based on a different set of fundamental principles than those that underlie classical models, as it was prompted by experimental observations that could not be reconciled with the concepts on which classical models are based.
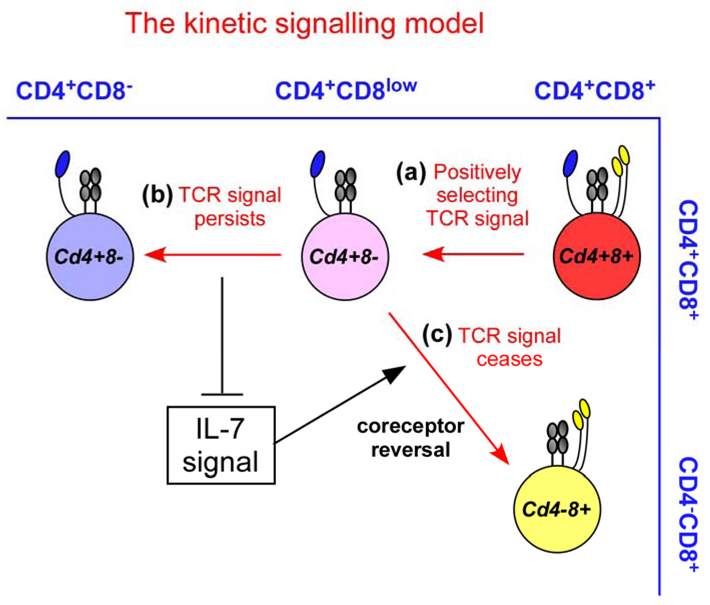
Figure 3. The kinetic signalling model of CD4/CD8 lineage choice.
Regardless of the specificity of their T-cell receptor (TCR), positively selecting TCR signals induce double-positive (DP) thymocytes that are transcriptionally Cd4+Cd8+ to terminate Cd8 gene expression and to convert into Cd4+Cd8−intermediate thymocytes. Because of absent Cd8 gene transcription, Cd4+Cd8− intermediate thymocytes appear phenotypically as CD4+CD8low cells (step a), and these are the cells in which lineage choice is made. Persistence of TCR signalling in Cd4+Cd8− intermediate thymocytes blocks interleukin-7 (IL-7) signalling and induces differentiation into mature CD4+ T cells (step b). Cessation or disruption of TCR signalling in Cd4+Cd8− permits IL-7 signalling, which induces Cd4+Cd8− intermediate thymocytes to undergo co-receptor reversal to become Cd4−Cd8+ and to differentiate into CD8+ T cells (step c).
Observations made both in vitro and in vivo indicated that TCR-signalled DP thymocytes terminated Cd8 transcription but not Cd4 transcription during positive selection, regardless of the MHC-restriction specificity of the TCR39. And most importantly, despite termination of Cd8 gene transcription, TCR-signalled thymocytes still remained lineage-uncommitted cells with the ability to differentiate into either CD4+ or CD8+ T cells39 (Fig. 3). These observations violate the fundamental principle underlying all classical models that co-receptor gene termination is irreversible and indicative of commitment to the opposite lineage.
Based on these observations, the kinetic signalling model proposed that TCR-signalled DP thymocytes first terminate Cd8 gene transcription and then assess the effect of absent Cd8 transcription on TCR signalling39, 40, 50. If TCR-mediated positive selection signals persist in the absence of Cd8 transcription, thymocytes differentiate into CD4+ T cells. If TCR-mediated positive selection signalling ceases in the absence of Cd8 transcription, thymocytes differentiate into CD8+ T cells. Such a simple assessment by TCR-signalled thymocytes can accurately identify their appropriate cell fate, as signalling by MHC-class-II-restricted TCRs is not dependent on CD8 and so would persist in the absence of Cd8 transcription (Fig. 4A), whereas signalling by MHC-class-I-restricted TCRs is dependent on CD8 and so would cease in the absence of Cd8 transcription (Fig. 4B). For the kinetic signalling model, it is axiomatic that positive selection and lineage choice are sequential, not simultaneous, events; that TCR-mediated positive selection signals terminate Cd8 transcription, converting signalled DP thymocytes into intermediate thymocytes with a transcriptionally Cd4+Cd8− phenotype that remain lineage-uncommitted cells; and that CD4/CD8 lineage choice occurs in Cd4+Cd8− intermediate thymocytes and is based on whether TCR signalling persists or ceases. The kinetic signalling model also proposes that the persistence or cessation of TCR signalling inversely regulates signalling induced by IL-7 and other γc cytokines, which consequently serve as sensors of TCR signal duration39, 40, 50.
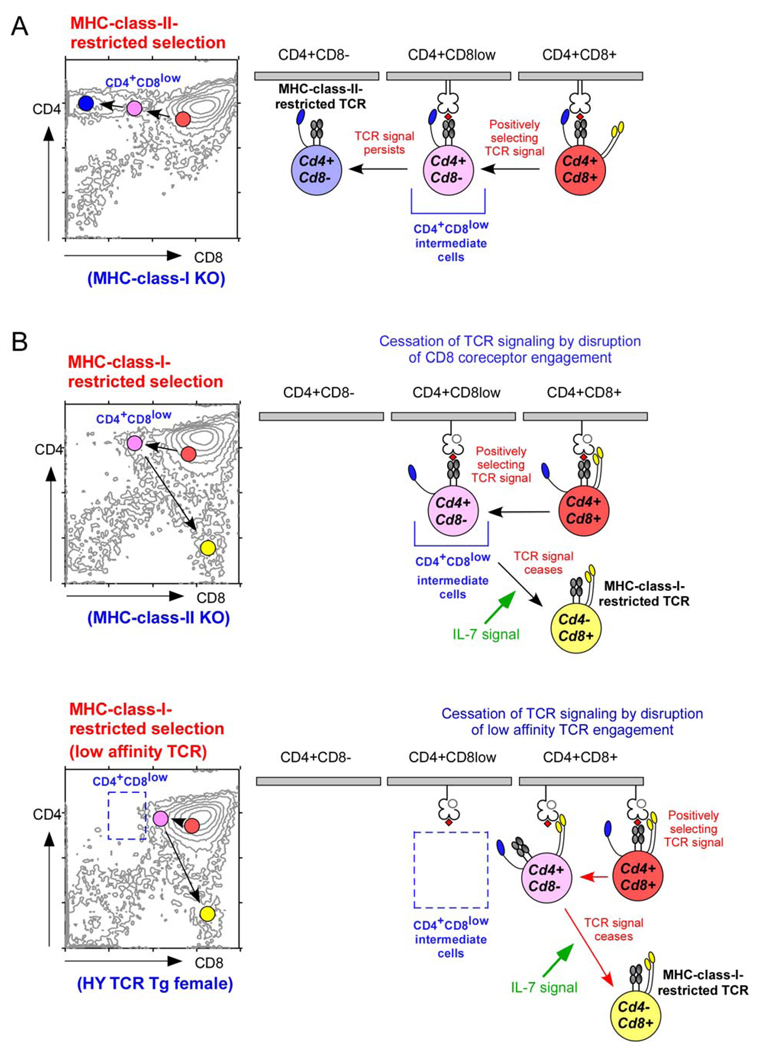
Figure 4. Lineage-fate mapping according to the kinetic signalling model of CD4/CD8 lineage choice.
Regardless of the MHC-restriction specificity of the T-cell receptor (TCR), positively selecting TCR signals convert double-positive (DP) thymocytes into Cd4+Cd8− intermediate thymocytes in which lineage choice is made. CD4/CD8 lineage direction is then dependent on whether positively selecting TCR signals persist or cease. A. MHC-class-II-restricted TCR signalling is independent of CD8 expression and, therefore, persists in Cd4+Cd8− intermediate thymocytes. Persistent TCR signalling induces intermediate thymocytes to differentiate into mature CD4+ T cells. B. MHC-class-I-restricted TCR signalling is dependent on CD8 expression and, therefore, ceases in Cd4+Cd8− intermediate thymocytes. Cessation of TCR signalling permits interleukin-7 (IL-7) signalling, which induces intermediate thymocytes to undergo co-receptor reversal and to differentiate into mature CD8+ T cells. The appearance of Cd4+Cd8− intermediate thymocytes at the point that TCR signalling ceases varies according to the ligand affinity of individual TCRs. Signalling by high affinity MHC-class-I-restricted TCRs (upper panel) can persist in Cd4+Cd8− intermediate thymocytes until intermediate thymocytes have lost sufficient CD8 to become CD4+CD8low cells. However, signalling by low affinity MHC-class-I-restricted TCRs (lower panel) is dependent on high CD8 expression and is disrupted by small reductions in cell-surface CD8 levels, when intermediate thymocytes still appear as CD4+CD8+ cells.
In the kinetic signalling model, TCR-signalled DP thymocytes undergo positive selection during which they terminate Cd8 gene expression and become Cd4+Cd8− intermediate thymocytes that remain bipotential despite absent Cd8 gene expression. Because they are no longer transcribing Cd8, Cd4+Cd8− intermediate thymocytes express steadily diminishing amounts of cell-surface CD8 so that most appear phenotypically as CD4+CD8low cells. The progressive decrease in cell-surface CD8 eventually disrupts CD8-dependent signalling by MHC-class-I-restricted TCRs (Fig. 4B). Consequently, most Cd4+Cd8− intermediate thymocytes appear phenotypically as CD4+CD8low cells, but, in TCR-transgenic mice, the appearance of Cd4+Cd8− intermediate thymocytes can cover a wide spectrum from CD4+CD8+ to CD4+CD8low to CD4+CD8−, depending on the ligand affinity and CD8 dependence of their transgenic TCR40. At one extreme, Cd4+Cd8− intermediate thymocytes bearing high affinity MHC-class-I-restricted TCRs might be CD4+CD8− because TCR signalling would not be disrupted until CD8 cell-surface levels fall to barely detectable levels. At the other extreme, Cd4+Cd8− intermediate thymocytes bearing low-affinity MHC-class-I-restricted TCRs (such as the HY transgenic TCR) might be CD4+CD8+ because TCR signalling would be disrupted by even slight reductions in CD8 cell-surface levels (Fig. 4C). Thus, intermediate thymocytes in which lineage choice occurs are best defined by their Cd4+Cd8− transcriptional phenotype, not their cell-surface phenotype which can vary with the ligand affinity of their TCR.
Cytokine signals and co-receptor reversal
For uncommitted Cd4+Cd8− intermediate thymocytes to differentiate into CD8+ T cells, they must terminate Cd4 transcription and reinitiate Cd8 transcription, molecular events collectively referred to as ‘co-receptor reversal’39 (Fig. 4C). Indeed, co-receptor reversal is the cornerstone of the kinetic signalling model and depends on signalling by IL-7 and possibly other intrathymic γc cytokines39, 51. As co-receptor reversal only occurs in thymocytes that are no longer receiving TCR signals, cytokines such as IL-7 are important for their survival51. In addition to providing survival signals, IL-7 and other γc cytokines have been shown in vitro to promote co-receptor reversal by enhancing Cd4 silencing and promoting re-initiation of Cd8 transcription51.
If IL-7 signals contribute to CD8-lineage fate, there must be a mechanism by which intermediate thymocytes subvert IL-7 signalling to differentiate into CD4-lineage T cells. Whereas persistent TCR signalling inhibits IL-7 signal transduction in mature T cells both in vitro52 and in vivo53, the effect of TCR signalling on IL-7 signal transduction in intermediate thymocytes has not yet been directly demonstrated. However, persistent TCR signalling has been shown in vitro to prevent intermediate thymocytes from undergoing co-receptor reversal in response to IL-739, 51.
The theory that differentiation into CD8+ T cells is IL-7 dependent whereas differentiation into CD4+ T cells is IL-7 independent is supported by several different observations. CD4 and CD8 SP thymocytes differ significantly in cell-surface expression of glucose transporter type 1 (GLUT1) which is quantitatively upregulated by IL-754, 55. Whereas GLUT1 expression by CD8 SP thymocytes is high, its expression by CD4 SP thymocytes is barely detectable51, indicating that IL-7 receptor (IL-7R) signalling occurs in thymocytes during CD8+ T-cell development but not during CD4+ T-cell development. The importance of γc-cytokine-induced signalling in CD8+ T-cell differentiation is further supported by reports that genetic ablation of SOCS1 results in preferential generation of CD8 SP thymocytes1, 2, 56; that blockade of IL-7R signalling by IL-7R- and γc-specific antibodies selectively abrogates the generation of CD8+ T cells39, 51; and that deletion of GFI1 (growth-factor independent 1), a negative regulator of IL-7R expression, selectively increases CD8+ T-cell differentiation57.
The proposed crosstalk between TCR and cytokine receptor signalling provides a mechanism whereby cytokine receptor signalling functions as a ‘sensor’ of TCR signal duration. Persistent TCR signalling impairs IL-7R signal transduction and drives differentiation of Cd4+Cd8− intermediate thymocytes into CD4+ T cells. However, if TCR signalling is disrupted, IL-7R signals initiate co-receptor reversal, which leads to differentiation into CD8+ T cells. Consequently, the kinetic signalling model proposes that lineage choice is determined by different types of signals: CD4+ T-cell differentiation is driven by TCR signals, whereas CD8+ T-cell differentiation is driven by cytokine receptor signals. Interestingly, the molecular mechanisms underlying these events are steadily becoming clarified (see below).
Co-receptor gene transcription and kinetic signalling
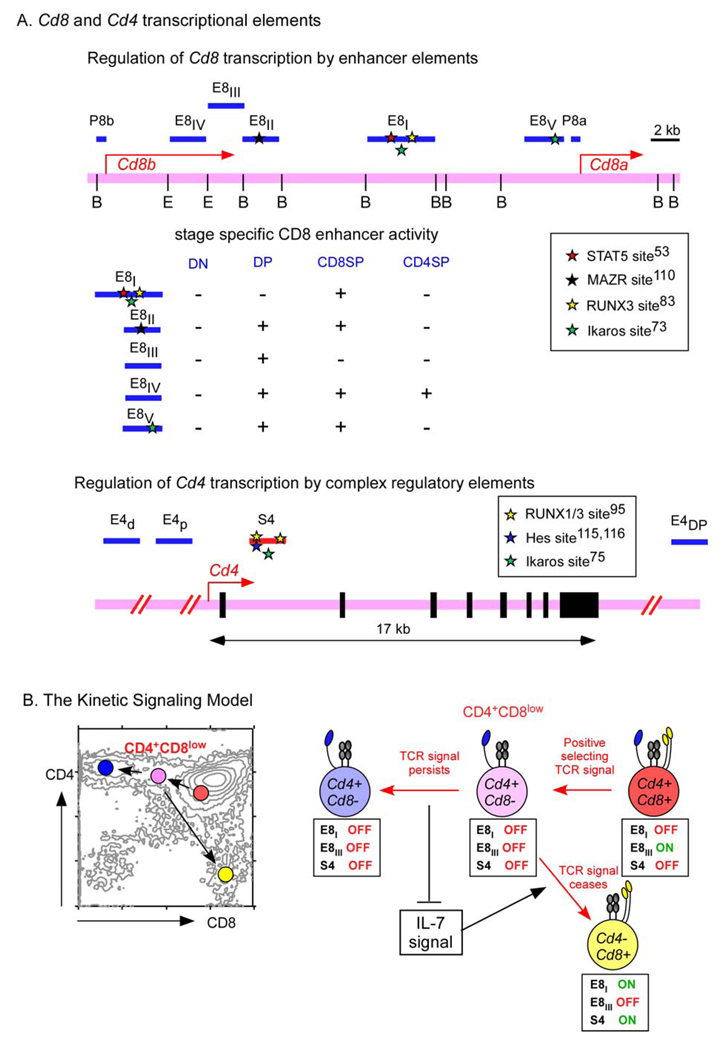
Cd4 and Cd8 transcription are regulated quite differently from one another58, 59 (Fig. 5A). Cell-specific expression of Cd4 results from the activity of a silencer element that abrogates Cd4 transcription in CD4− cells60, 61. By contrast, cell-specific expression of Cd8 is the result of stage-specific enhancer elements that actively induce its expression in CD8+ T cells62, 63. Five enhancer elements that regulate expression of the Cd8a gene have been identified (known as E8I–E8V)62–65, and two of these enhancer elements might be particularly relevant to understanding CD4/CD8 lineage choice, as the E8III enhancer is active only in DP thymocytes66 and the E8I enhancer is active in CD8 SP thymocytes and CD8+ T cells64, 67. It is possible to combine the concepts of the kinetic signalling model with the transcriptional control elements that regulate Cd4 and Cd8 gene expression to better understand lineage fate decisions (Fig. 5B). The kinetic signalling model predicts that E8III enhancer activity is suppressed by positive selecting TCR signals, since TCR signalling suppresses Cd8 expression in DP thymocytes. In fact, it has been shown that TCR signalling of DP thymocytes suppresses E8III enhancer activity20. It can also be predicted that E8I enhancer activity is responsive to the IL-7R signals that re-initiate Cd8 transcription in Cd4+Cd8− intermediate thymocytes undergoing co-receptor reversal. In fact, E8I enhancer activity, as well as Cd8a transcription, have both been shown to be increased by IL-7-induced STAT5 (signal transducer and activator of transcription 5) signals53. Interestingly, STAT5 deficiency in mice does not specifically abrogate CD8+ T-cell differentiation68, but this could be due to IL-7R signal transduction by other STAT molecules that substitute for STAT5 in STAT5-deficient thymocytes.
Figure 5. Regulation of Cd4 and Cd8 gene expression.
A. Cell-specific expression of the Cd8a gene is controlled by stage-specific enhancer elements, of which five are known (E8I-E8V). Nuclear factors that bind to these regions include Ikaros 73, RUNX1 (runt-related transcription factor 1)95, 109, RUNX383, 87, 95, MAZR (MAZ-related factor)110 and STAT5 (signal transducer and activator of transcription 5)53. In contrast to Cd8 gene expression, tissue specific expression of the Cd4 gene is not accomplished by Cd4 enhancer elements (E4)111–113, but is mainly controlled by activation of a silencer element (S4) that is located in the first intron60, 61, 109, 114. Nuclear factors that bind to the CD4 silencer element include RUNX, MYB and HES195, 109, 115, 116. B. Changes in co-receptor transcription during positive selection and lineage choice according to the kinetic signalling model. CD8 expression on pre-selection double-positive (DP) thymocytes is driven in part by the E8III Cd8 enhancer, which is turned off by T-cell receptor (TCR)-mediated positive selection signalling20, 62. In CD4+CD8low intermediate thymocytes, persistent TCR-mediated positive selection signalling inhibits Cd8 gene expression and inhibits Cd4 silencer activity, so that intermediate cells differentiate into CD4+ T cells. However, cessation of TCR-mediated positive selection signalling in intermediate thymocytes results in re-initiation of Cd8 gene expression, at least in part, by induction of E8I Cd8 enhancer activity, which is responsive to interleukin-7 receptor (IL-7R) signalling53.
In vivo assessments
A key concept of the kinetic signalling model that has been tested in vivo is that TCR-signal disruption invariably leads to CD8-lineage choice, even for thymocytes expressing MHC-class-II-restricted TCRs39, 51. One experimental model used to assess this prediction was carried out in mice in which expression of ζ-chain associated protein kinase of 70 kDa (ZAP70) was placed under the control of enhancer and promoter elements from the adenosine deaminase (ADA) gene so that ZAP70 expression would be halted during positive selection69. In ADA–ZAP70 transgenic mice (which lacked endogenous ZAP70 expression), ZAP70 was expressed in DP thymocytes but not in CD4+CD8low intermediate thymocytes. As ZAP70 is required for TCR-signal transduction70–72, TCR signalling ceased in all CD4+CD8low intermediate thymocytes69. As a result, all positively selected thymocytes in ADA–ZAP70 transgenic mice, including those expressing MHC-class-II-restricted TCRs, differentiated into CD8+ T cells, confirming that cessation of TCR signalling in intermediate thymocytes results exclusively in CD8-lineage choice.
A core concept of the kinetic signalling model is that TCR-mediated positive selection signals lead to the termination of Cd8 transcription, which eventually disrupts MHC-class-I-restricted TCR signalling and leads to CD8-lineage choice. However, if regulatory elements of the Cd8 gene also controlled Cd4 expression, one would predict that positive selection would be followed by a steady reduction in cell-surface CD4 expression that would eventually disrupt MHC-class-II-restricted TCR signalling and therefore lead to CD8-lineage choice, despite the MHC-class-II-restriction of the TCR. This prediction has been tested in an in vivo experimental model in which CD4 expression was placed under the control of the E8III enhancer (which is active in pre-selection DP thymocytes but is inactivated by TCR signalling)20. In E8III–CD4 transgenic mice that lack endogenous CD4 expression (referred to as 8DP4 mice), the expression of transgenic CD4 proteins was high on pre-selection DP thymocytes but steadily declined on intermediate thymocytes in parallel with endogenous CD8 protein expression. As a result, all positively selected thymocytes in 8DP4 mice, including those expressing MHC-class-II-restricted TCRs, differentiated exclusively into CD8+ T cells, which indicates that termination of co-receptor gene transcription during positive selection promotes CD8-lineage fate, regardless of the MHC restriction specificity of the TCR and regardless of which co-receptor protein is involved20.
Transcription factors involved in CD4/CD8 lineage choice
Our understanding of CD4/CD8 lineage choice has been significantly advanced by the identification of transcription and nuclear factors that influence CD4/CD8 lineage choice and thereby regulate Cd4 and Cd8 transcription. Some of these factors are involved in chromatin remodeling, including Ikaros73, 74, Mi-2β75, 76, and the SWI/SNF-like BAF chromatin remodeling complexes77, whereas other factors directly regulate the transcription of downstream effector genes78–86. The most relevant of the latter factors for this discussion are Th-POK (T-helper-inducing POZ/Kruppel-like factor; also known as cKROX and ZFP67)47, 81, 84, 85 and RUNX3 (runt-related transcription factor 3)80, 83, 84, 87 which, together with TOX (thymus high-mobility group box protein)78, 88, 89 and GATA3 (GATA-binding protein 3)82, 90, 91, contribute to a molecular understanding of CD4/CD8 lineage choice (Figure 5).
Th-POK
Th-POK is a zinc-finger protein that is encoded by the Zbtb7b gene81, 92. In an exciting series of experiments, two laboratories discovered that Th-POK was singularly important for CD4 lineage choice and CD4+ T-cell differentiation81, 85. Th-POK is expressed by CD4+ but not by CD8+ T cells81, 85 and was found to be the molecule that was mutated in helper-deficient (HD) mice that were unable to generate CD4+ T cells19, 93. In HD mice, MHC-class-II signalled thymocytes failed to differentiate into CD4+ T cells and instead differentiated into mature CD8+ T cells. These mice were found to have a point mutation in the second zinc finger domain of Th-POK that presumably disrupts DNA binding81. Thus, HD mice demonstrated that CD4+ T-cell differentiation requires a functional Th-POK molecule.
Reciprocal experiments revealed that expression of transgene-encoded Th-POK proteins throughout thymocyte development forced virtually all positively selected thymocytes to differentiate into CD4+ T cells, even those with MHC-class-I-restricted TCRs81, 85. As it seems to be both necessary and sufficient for CD4-lineage choice, it has been suggested that Th-POK is a master regulator of CD4-lineage choice and CD4+ T-cell differentiation45. Consistent with this perspective, retroviral transduction of mature CD8+ T cells with Th-POK led to reduced T-cell cytotoxicity and induced some CD4+ T-helper-cell characteristics, indicating that even mature CD8+ T cells are susceptible to the CD4-lineage-promoting effects of Th-POK94. Th-POK affects both Cd4 and Cd8 transcription, but in opposite ways: Th-POK maintains Cd4 transcription by preventing factors such as RUNX3 from silencing it95, and Th-POK reduces Cd8 expression by downregulating the enhancer activity of E8I94.
Th-POK is first expressed by TCR-signalled DP thymocytes that are CD4+CD8low cells47 — that is, by the cells in which CD4/CD8 lineage choice occurs. However, the level of Th-POK expression at this stage is too low to limit their bipotentiality. Interestingly, Th-POK expression in CD4+CD8low thymocytes is upregulated following persistent TCR signalling47, which is concordant with the kinetic signalling concept that CD4-lineage choice is induced in intermediate thymocytes by persistent TCR signalling.
RUNX proteins
RUNX proteins are members of the runt-domain family of transcription factors which have similar structural organizations and conserved DNA binding sites96. In the thymus, RUNX1 is expressed mostly by DN thymocytes and RUNX3 by post-selection CD8+ thymocytes95. The observation that RUNX proteins bind to the Cd4 silencer element and silence Cd4 expression indicated a role for RUNX proteins in CD4/CD8 lineage choice95.
RUNX1 and RUNX3 both bind to the Cd4 silencer element and silence Cd4 transcription in the cells in which they are expressed95. Indeed, RUNX deficiency results in Cd4 gene de-repression95, 97, whereas RUNX3 transgenic overexpression downregulates Cd4 transcription87, 98. During normal thymocyte development, RUNX3 is not expressed by pre-selection DP thymocytes and may first be expressed at low levels by CD4+CD8low intermediate thymocytes83, 99. Importantly, RUNX3 expression is upregulated during differentiation of CD4+CD8low thymocytes into CD8+ T cells, when RUNX3 provides two critical functions. First, RUNX3 binds to the Cd4 silencer element and silences Cd4 transcription95. Second, RUNX3 binds to the E8I Cd8 enhancer element and re-initiates Cd8 transcription83. Thus, RUNX3 may be the transcriptional mediator of co-receptor reversal by silencing Cd4 and reinitiating Cd8 gene expression during the differentiation of Cd4+Cd8− intermediate thymocytes into Cd4−Cd8+ mature T cells.
Recently, it was shown that RUNX proteins also bind to a sequence in the gene encoding Th-POK (Zbtb7b) and extinguish Th-POK expression84. Thus, the silencing of Zbtb7b gene expression is another mechanism by which RUNX3 promotes CD8+ T-cell differentiation. Importantly, however, the intrathymic signals that upregulate RUNX3 expression during CD8+ T-cell differentiation have not yet been identified. Nonetheless, the kinetic signalling model predicts that RUNX3 expression by intermediate thymocytes would be upregulated, either directly or indirectly, by IL-7 and possibly other cytokine signals.
TOX
High mobility group (HMG) box proteins are DNA-binding proteins that regulate gene expression by modulating local chromatin structure and recruiting other nuclear factors100, 101. TOX is an HMG box protein that was first discovered because it was upregulated in TCR-signalled DP thymocytes and so was postulated to have a role in positive selection and/or lineage choice89. Although transgenic overexpression of TOX has complex effects that are not yet fully understood89, recent experiments in TOX-deficient mice revealed that positively selected thymocytes do not become CD4+CD8low cells, but instead become CD4lowCD8low cells which fail to differentiate into CD4+ T cells78. Reversal of TOX deficiency by introduction of a TOX transgene restores both the appearance of positively selected thymocytes as CD4+CD8low cells and their ability to differentiate into CD4-lineage cells78. Thus, TOX seems to be important for maintaining or upregulating CD4 expression in positively selected DP thymocytes78, 102. This perspective explains the importance of TOX for CD4+ T-cell differentiation, as CD4 co-receptor expression is required for persistent MHC-class-II-restricted TCR signalling in intermediate thymocytes.
GATA3
GATA3 is an enhancer-binding zinc finger protein that functions as a lineage specific transcription factor in T cells at various stages of development82, 90. GATA3 is expressed in the earliest progenitor T cells and is required for thymocytes to differentiate beyond the DN stage of development90. GATA3 also plays an important role in CD4 lineage choice based on observations that GATA3 is preferentially expressed in CD4+ T cells90; that GATA3 expression is upregulated by TCR-signalling in DP thymocytes82; and that sustained expression of GATA3 blocks generation of CD8+ T cells91. In addition, conditional deletion of Gata3 in DP thymocytes markedly decreased CD4 T cell numbers without affecting CD8 T cell generation103, indicating a critical role for GATA3 in the survival and/or differentiation of positively selected thymocytes into CD4-lineage T cells. But, unlike Th-POK, GATA3 does not seem to be a CD4-lineage-specifying factor because forced expression of GATA3 does not re-direct MHC-class-I-restricted thymocytes to differentiate into CD4+ T cells82. However, because it is expressed in positively selected thymocytes earlier than either Th-POK or RUNX3, GATA3 may be upstream of these other factors so that its specific role in CD4 lineage choice may be more difficult to discern.
A synthesis
Although our knowledge is still far from complete, it is possible to integrate what we currently understand about the transcriptional activities of Th-POK, RUNX3, TOX, and GATA3 into a coherent view of CD4/CD8 lineage choice (Figure 5). TCR-mediated positive selection signals terminate Cd8 gene expression (in part by suppressing the activity of the E8III Cd8 enhancer) and upregulate TOX which maintains or increases Cd4 transcription so that positively selected DP thymocytes convert into Cd4+Cd8− intermediate cells (which appear as CD4+CD8low thymocytes). If positively selecting TCR signals are MHC-class-II-restricted, TCR signalling in CD4+CD8low thymocytes persists and upregulates both GATA3 and Th-POK, preventing Cd4 gene silencing and promoting CD4+ T-cell differentiation. Alternatively, if positively selecting TCR signals are MHC-class-I-restricted, TCR signalling in CD4+CD8low intermediate thymocytes is disrupted or ceases, which results in loss of GATA3 and Th-POK expression, IL-7R signalling, and upregulation of RUNX3. RUNX3 then silences both Zbtb7b and Cd4 transcription, leading to termination of CD4-lineage potential and reinitiation of Cd8 gene expression (in part by activating the E8I Cd8 enhancer). In the presence of IL-7 and other cytokines, CD8 lineage thymocytes then proceed to differentiate into mature CD8+ T cells.
In this way, cell-surface TCR and co-receptor signalling can be integrated with the transcriptional factors involved in CD4/CD8 lineage choice to reveal an increasingly detailed picture of how lineage fate decisions occur in the thymus.
Concluding remarks
Understanding the basis for CD4/CD8 lineage choice in the thymus is central to our understanding of thymocyte development. Consequently, CD4/CD8 lineage choice remains one of the most intensively studied and debated lineage decisions in immunology. Model building has had an indispensable role in determining the logic by which DP thymocytes ascertain their appropriate lineage fate. However, the rules of logic stipulate that models can never be proven but only disproven, so model testing will continue to provide the driving force behind many of the most informative experiments in this field. Rigorous testing of the kinetic signalling model will hopefully lead to new and deeper insights, as the proposed role of cytokines in CD8-lineage choice opens new avenues of investigation. The recent identification of nuclear factors involved in CD4/CD8 lineage choice promises to provide the circuitry102 that links signalling events at the cell membrane with changes in gene expression patterns during thymocyte selection. However, a great deal still remains to be explained and understood, including a molecular definition of ‘lineage commitment’ and a greater understanding of how CD4/CD8 lineage choice results in distinct helper and cytotoxic cellular functions.
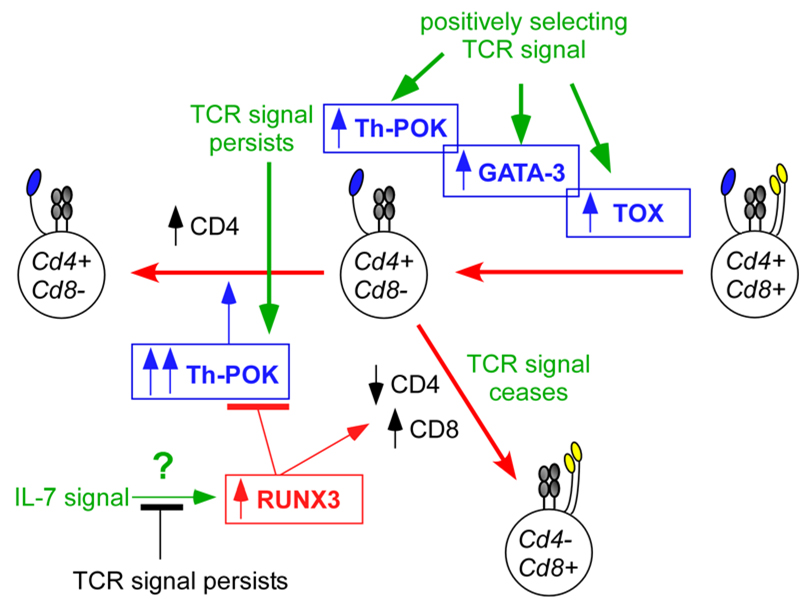
Figure 6. Environmental cues and nuclear factors that influence the CD4/CD8 decision.
Environmental cues that influence CD4/CD8 lineage choice must ultimately be translated by developing thymocytes into molecular events mediated by nuclear factors that differentially affect co-receptor gene expression. Here, we consider the interactions among four different transcription factors: Th-POK (T-helper-inducing POZ/Kruppel-like factor), RUNX3 (runt-related transcription factor 3), TOX (thymus high-mobility group box protein) and GATA3 (GATA-binding protein 3). Three of these factors are important for CD4+ T-cell differentiation (Th-POK, TOX and GATA3), and only one (RUNX3) is known to be important for CD8+ T-cell differentiation. During positive selection, T-cell receptor (TCR) signals upregulate TOX, GATA3 and Th-POK. TOX upregulation is necessary for TCR-signalled DP thymocytes to phenotypically become CD4+CD8low intermediate thymocytes78. GATA3 upregulation is important for the differentiation of CD4+CD8low thymocytes into CD4+ T cells82, 103. And Th-POK expression in TCR-signalled thymocytes, which is significantly upregulated in CD4+CD8low intermediate thymocytes by persistent TCR signalling47, is required for CD4-lineage commitment81, 85, 93 and for preventing Cd4 gene silencing by RUNX proteins117. It is not yet known what environmental signal upregulates RUNX3 expression, but it is hypothesized that its expression may be upregulated by interleukin-7 receptor (IL-7R) signalling. In any event, RUNX3 performs three important functions that promote the differentiation of intermediate thymocytes into CD8+ T cells: first, RUNX3 binds to the Cd4 silencer element and silences Cd4 gene expression95; second, RUNX3 binds to the E8I Cd8 enhancer element and re-initiates Cd8 gene expression83, and third RUNX3 silences Th-POK gene expression84.
Acknowledgements
Most importantly, we would like to recognize our many colleagues who made important contributions to our current understanding of CD4/CD8 lineage fate decisions in the thymus that we were unable to adequately acknowledge and reference in this review. We are grateful to Drs. Batu Erman, Sophia Sarafova, Remy Bosselut and Naomi Taylor for many helpful discussions and for their critical reading of the manuscript. This research was supported by the Intramural Research Program of the NIH, NCI, Center for Cancer Research.
Glossary
- programmed cell death
A common form of cell death, which is also known as apoptosis. Many physiological and developmental stimuli cause apoptosis, and this mechanism is frequently used to delete unwanted, superfluous or potentially harmful cells. Apoptosis involves cell shrinkage, chromatin condensation, plasma-membrane blebbing and DNA fragmentation. Eventually, the cell breaks up into many membrane-bound 'apoptotic bodies', which are phagocytosed by neighbouring cells.
- immunoreceptor tyrosine-based activation motif
(ITAM). A short peptide motif containing tyrosine residues that is found in the cytoplasmic tail of several signalling adaptor proteins and that is necessary to recruit proteins that are involved in triggering activating signalling proteins. The consensus sequence is Tyr-X-X-(Leu/Ile)-X6–8-Tyr-X-X-(Leu/Ile), where X denotes any amino acid.
- common cytokine-receptor γ-chain
(γc). A shared cytokine receptor chain for a group of short-chained, four-helical bundle interleukins, including interleukin-2 (IL-2), IL-4, IL-7, IL-9, IL-15 and IL-21.
- HY transgenic TCR
An MHC-class-I-restricted TCR that recognizes an antigenic complex composed of H-2Db and the Y chromosome-encoded male antigen (H-Y). This was the first TCR-transgenic mouse ever constructed. In female mice, thymocytes expressing this clonotypic TCR transgene are positively selected and differentiate into CD8+ T cells, whereas in male mice thymocytes expressing the same TCR are negatively selected.
Biographies
Alfred Singer: Alfred Singer is Chief of the Experimental Immunology Branch of the National Cancer Institute. His research focuses on many different of aspects of thymocyte development, repertoire selection, and coreceptor function.
Stanley Adoro: Stanley Adoro is currently a graduate student in Immunology at the University of Pennsylvania. He is pursuing his dissertation research in the laboratory of Alfred Singer where he is investigating the regulation of coreceptor gene transcription and CD4/CD8 lineage choice in the thymus.
Jung-Hyun Park: Jung-Hyun Park received his Ph.D. in Immunology from the University of Wurzburg in Germany, and his postdoctoral training at the Korea Research Institute in Bioscience and Biotechnology, Korea. After joining the Experimental Immunology Branch of the National Cancer Institute as a research fellow, he studied the role of IL-7 receptor signaling in T cell development and homeostasis, which continues to be his major scientific interest.
Reference
- 1.Chong MM, et al. Suppressor of cytokine signaling-1 is a critical regulator of interleukin-7-dependent CD8+ T cell differentiation. Immunity. 2003;18:475–487. doi: 10.1016/s1074-7613(03)00078-5. [DOI] [PubMed] [Google Scholar]
- 2.Yu Q, et al. Cytokine signal transduction is suppressed in preselection double-positive thymocytes and restored by positive selection. J Exp Med. 2006;203:165–175. doi: 10.1084/jem.20051836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doyle C, Strominger JL. Interaction between CD4 and class II MHC molecules mediates cell adhesion. Nature. 1987;330:256–259. doi: 10.1038/330256a0. [DOI] [PubMed] [Google Scholar]
- 4.Norment AM, Salter RD, Parham P, Engelhard VH, Littman DR. Cell-cell adhesion mediated by CD8 and MHC class I molecules. Nature. 1988;336:79–81. doi: 10.1038/336079a0. [DOI] [PubMed] [Google Scholar]
- 5.Shaw AS, et al. The lck tyrosine protein kinase interacts with the cytoplasmic tail of the CD4 glycoprotein through its unique amino-terminal domain. Cell. 1989;59:627–636. doi: 10.1016/0092-8674(89)90008-1. [DOI] [PubMed] [Google Scholar]
- 6.Turner JM, et al. Interaction of the unique N-terminal region of tyrosine kinase p56lck with cytoplasmic domains of CD4 and CD8 is mediated by cysteine motifs. Cell. 1990;60:755–765. doi: 10.1016/0092-8674(90)90090-2. [DOI] [PubMed] [Google Scholar]
- 7.Veillette A, Bookman MA, Horak EM, Samelson LE, Bolen JB. Signal transduction through the CD4 receptor involves the activation of the internal membrane tyrosine-protein kinase p56lck. Nature. 1989;338:257–259. doi: 10.1038/338257a0. [DOI] [PubMed] [Google Scholar]
- 8.Veillette A, Zuniga-Pflucker JC, Bolen JB, Kruisbeek AM. Engagement of CD4 and CD8 expressed on immature thymocytes induces activation of intracellular tyrosine phosphorylation pathways. J Exp Med. 1989;170:1671–1680. doi: 10.1084/jem.170.5.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Teh HS, et al. Thymic major histocompatibility complex antigens and the alpha beta T-cell receptor determine the CD4/CD8 phenotype of T cells. Nature. 1988;335:229–233. doi: 10.1038/335229a0. This is the first report of a TCR transgenic mouse and demonstrates that CD4/CD8 lineage choice in the thymus is dictated by the MHC-restriction-specificity of the positively selecting TCR.
- 10.Janeway CA., Jr T-cell development. Accessories or coreceptors? Nature. 1988;335:208–210. doi: 10.1038/335208a0. [DOI] [PubMed] [Google Scholar]
- 11.Chan SH, Cosgrove D, Waltzinger C, Benoist C, Mathis D. Another view of the selective model of thymocyte selection. Cell. 1993;73:225–236. doi: 10.1016/0092-8674(93)90225-f. [DOI] [PubMed] [Google Scholar]
- 12. Davis CB, Killeen N, Crooks ME, Raulet D, Littman DR. Evidence for a stochastic mechanism in the differentiation of mature subsets of T lymphocytes. Cell. 1993;73:237–247. doi: 10.1016/0092-8674(93)90226-g. Published back-to-back with reference 11, these studies provided strong support for the stochastic selection model of CD4/CD8 lineage choice. In reference 11, MHC-II deficient mice were shown to contain CD4+CD8low thymocytes which were thought to be MHC-I-restricted cells that were shortlived and that had randomly made an incorrect CD4 lineage choice. Reference 12 was the first ‘co-receptor rescue experiment’ in which constitutive expression of transgenic coreceptor proteins promoted the differentiation of positively selected thymocytes into mature T cells of the inappropriate lineage.
- 13.Itano A, Kioussis D, Robey E. Stochastic component to development of class I major histocompatibility complex-specific T cells. Proc Natl Acad Sci U S A. 1994;91:220–224. doi: 10.1073/pnas.91.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leung RK, et al. Deletion of the CD4 silencer element supports a stochastic mechanism of thymocyte lineage commitment. Nat Immunol. 2001;2:1167–1173. doi: 10.1038/ni733. [DOI] [PubMed] [Google Scholar]
- 15.Robey E, Itano A, Fanslow WC, Fowlkes BJ. Constitutive CD8 expression allows inefficient maturation of CD4+ helper T cells in class II major histocompatibility complex mutant mice. J Exp Med. 1994;179:1997–2004. doi: 10.1084/jem.179.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan SH, Waltzinger C, Baron A, Benoist C, Mathis D. Role of coreceptors in positive selection and lineage commitment. Embo J. 1994;13:4482–4489. doi: 10.1002/j.1460-2075.1994.tb06770.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baron A, Hafen K, von Boehmer H. A human CD4 transgene rescues CD4-CD8+ cells in beta 2-microglobulin-deficient mice. Eur J Immunol. 1994;24:1933–1936. doi: 10.1002/eji.1830240834. [DOI] [PubMed] [Google Scholar]
- 18.Itano A, Robey E. Highly efficient selection of CD4 and CD8 lineage thymocytes supports an instructive model of lineage commitment. Immunity. 2000;12:383–389. doi: 10.1016/s1074-7613(00)80190-9. [DOI] [PubMed] [Google Scholar]
- 19. Keefe R, Dave V, Allman D, Wiest D, Kappes DJ. Regulation of lineage commitment distinct from positive selection. Science. 1999;286:1149–1153. doi: 10.1126/science.286.5442.1149. This study characterizes a spontaneous mutant mouse strain that lacks helper T cells and so is referred to as ‘helper deficient’ (HD; helper deficient). This study provides the first evidence that positive selection and CD4/CD8 lineage choice are sequential, temporally distinct events.
- 20. Sarafova SD, et al. Modulation of coreceptor transcription during positive selection dictates lineage fate independently of TCR/coreceptor specificity. Immunity. 2005;23:75–87. doi: 10.1016/j.immuni.2005.05.011. This study provides strong in vivo experimental support for the kinetic signaling model of CD4/CD8 lineage choice. Using an elaborate in vivo model in which CD4 coreceptor expression is regulated by Cd8 transcriptional control elements, this study shows that lineage commitment is determined by the transcriptional kinetics of coreceptor expression during positive selection signaling, and not by the identity or signaling strength of the coreceptor proteins themselves.
- 21.Seong RH, Chamberlain JW, Parnes JR. Signal for T-cell differentiation to a CD4 cell lineage is delivered by CD4 transmembrane region and/or cytoplasmic tail. Nature. 1992;356:718–720. doi: 10.1038/356718a0. [DOI] [PubMed] [Google Scholar]
- 22. Itano A, et al. The cytoplasmic domain of CD4 promotes the development of CD4 lineage T cells. J Exp Med. 1996;183:731–741. doi: 10.1084/jem.183.3.731. This study proposes the ‘strength-of-signal instructional model’ and provides the first experimental support.
- 23.Wiest DL, et al. Regulation of T cell receptor expression in immature CD4+CD8+ thymocytes by p56lck tyrosine kinase: basis for differential signaling by CD4 and CD8 in immature thymocytes expressing both coreceptor molecules. J Exp Med. 1993;178:1701–1712. doi: 10.1084/jem.178.5.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez-Hoyos G, Sohn SJ, Rothenberg EV, Alberola-Ila J. Lck activity controls CD4/CD8 T cell lineage commitment. Immunity. 2000;12:313–322. doi: 10.1016/s1074-7613(00)80184-3. [DOI] [PubMed] [Google Scholar]
- 25.Sohn SJ, Forbush KA, Pan XC, Perlmutter RM. Activated p56lck directs maturation of both CD4 and CD8 single-positive thymocytes. J Immunol. 2001;166:2209–2217. doi: 10.4049/jimmunol.166.4.2209. [DOI] [PubMed] [Google Scholar]
- 26.Schmedt C, Tarakhovsky A. Autonomous maturation of alpha/beta T lineage cells in the absence of COOH-terminal Src kinase (Csk) J Exp Med. 2001;193:815–826. doi: 10.1084/jem.193.7.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Broussard C, et al. Altered development of CD8+ T cell lineages in mice deficient for the Tec kinases Itk and Rlk. Immunity. 2006;25:93–104. doi: 10.1016/j.immuni.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 28.Liao XC, Littman DR. Altered T cell receptor signaling and disrupted T cell development in mice lacking Itk. Immunity. 1995;3:757–769. doi: 10.1016/1074-7613(95)90065-9. [DOI] [PubMed] [Google Scholar]
- 29.Schaeffer EM, et al. Tec family kinases modulate thresholds for thymocyte development and selection. J Exp Med. 2000;192:987–1000. doi: 10.1084/jem.192.7.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bommhardt U, Basson MA, Krummrei U, Zamoyska R. Activation of the extracellular signal-related kinase/mitogen-activated protein kinase pathway discriminates CD4 versus CD8 lineage commitment in the thymus. J Immunol. 1999;163:715–722. [PubMed] [Google Scholar]
- 31.Fischer AM, Katayama CD, Pages G, Pouyssegur J, Hedrick SM. The role of erk1 and erk2 in multiple stages of T cell development. Immunity. 2005;23:431–443. doi: 10.1016/j.immuni.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 32.Sharp LL, Schwarz DA, Bott CM, Marshall CJ, Hedrick SM. The influence of the MAPK pathway on T cell lineage commitment. Immunity. 1997;7:609–618. doi: 10.1016/s1074-7613(00)80382-9. [DOI] [PubMed] [Google Scholar]
- 33.Wilkinson B, Kaye J. Requirement for sustained MAPK signaling in both CD4 and CD8 lineage commitment: a threshold model. Cell Immunol. 2001;211:86–95. doi: 10.1006/cimm.2001.1827. [DOI] [PubMed] [Google Scholar]
- 34.Love PE, Lee J, Shores EW. Critical relationship between TCR signaling potential and TCR affinity during thymocyte selection. J Immunol. 2000;165:3080–3087. doi: 10.4049/jimmunol.165.6.3080. [DOI] [PubMed] [Google Scholar]
- 35.Holst J, et al. Scalable signaling mediated by T cell antigen receptor-CD3 ITAMs ensures effective negative selection and prevents autoimmunity. Nat Immunol. 2008;9:658–666. doi: 10.1038/ni.1611. [DOI] [PubMed] [Google Scholar]
- 36.Bosselut R, Feigenbaum L, Sharrow SO, Singer A. Strength of signaling by CD4 and CD8 coreceptor tails determines the number but not the lineage direction of positively selected thymocytes. Immunity. 2001;14:483–494. doi: 10.1016/s1074-7613(01)00128-5. [DOI] [PubMed] [Google Scholar]
- 37. Erman B, et al. Coreceptor signal strength regulates positive selection but does not determine CD4/CD8 lineage choice in a physiologic in vivo model. J Immunol. 2006;177:6613–6625. doi: 10.4049/jimmunol.177.10.6613. To experimentally test the strength-of-signal instructional model, this report uses gene knock-in technology to engineer the endogenous CD8α gene so that it encodes stronger signaling CD8-CD4 chimeric coreceptor proteins. In contradiction to the strength-of-signal model, the results obtained demonstrate that the strength of coreceptor signaling does not alter CD4/CD8 lineage choice. Instead, the results demonstrate that the strength of coreceptor signaling has a quantitative effect on the number of DP thymocytes that are positively selected to differentiate into mature T cells.
- 38.Yasutomo K, Doyle C, Miele L, Fuchs C, Germain RN. The duration of antigen receptor signalling determines CD4+ versus CD8+ T-cell lineage fate. Nature. 2000;404:506–510. doi: 10.1038/35006664. [DOI] [PubMed] [Google Scholar]
- 39. Brugnera E, et al. Coreceptor reversal in the thymus: signaled CD4+8+ thymocytes initially terminate CD8 transcription even when differentiating into CD8+ T cells. Immunity. 2000;13:59–71. doi: 10.1016/s1074-7613(00)00008-x. This is the original study to introduce the kinetic signaling model and to describe results which contradicted classical presumptions of CD4/CD8 lineage choice.
- 40.Singer A. New perspectives on a developmental dilemma: the kinetic signaling model and the importance of signal duration for the CD4/CD8 lineage decision. Curr Opin Immunol. 2002;14:207–215. doi: 10.1016/s0952-7915(02)00323-0. [DOI] [PubMed] [Google Scholar]
- 41.Lundberg K, Heath W, Kontgen F, Carbone FR, Shortman K. Intermediate steps in positive selection: differentiation of CD4+8int TCRint thymocytes into CD4-8+TCRhi thymocytes. J Exp Med. 1995;181:1643–1651. doi: 10.1084/jem.181.5.1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Suzuki H, Punt JA, Granger LG, Singer A. Asymmetric signaling requirements for thymocyte commitment to the CD4+ versus CD8+ T cell lineages: a new perspective on thymic commitment and selection. Immunity. 1995;2:413–425. doi: 10.1016/1074-7613(95)90149-3. Published simultaneously with reference 41, these reports identify CD4+CD8low thymocytes as precursors of both CD4+ and CD8+ T cells.
- 43.Aliahmad P, Kaye J. Commitment issues: linking positive selection signals and lineage diversification in the thymus. Immunol Rev. 2006;209:253–273. doi: 10.1111/j.0105-2896.2006.00345.x. [DOI] [PubMed] [Google Scholar]
- 44.Kappes DJ, He X, He X. CD4-CD8 lineage commitment: an inside view. Nat Immunol. 2005;6:761–766. doi: 10.1038/ni1230. [DOI] [PubMed] [Google Scholar]
- 45.Kappes DJ, He X, He X. Role of the transcription factor Th-POK in CD4:CD8 lineage commitment. Immunol Rev. 2006;209:237–252. doi: 10.1111/j.0105-2896.2006.00344.x. [DOI] [PubMed] [Google Scholar]
- 46.Lucas B, Germain RN. Unexpectedly complex regulation of CD4/CD8 coreceptor expression supports a revised model for CD4+CD8+ thymocyte differentiation. Immunity. 1996;5:461–477. doi: 10.1016/s1074-7613(00)80502-6. [DOI] [PubMed] [Google Scholar]
- 47.He X, et al. CD4-CD8 lineage commitment is regulated by a silencer element at the ThPOK transcription-factor locus. Immunity. 2008;28:346–358. doi: 10.1016/j.immuni.2008.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Bosselut R, Guinter TI, Sharrow SO, Singer A. Unraveling a revealing paradox: Why major histocompatibility complex I-signaled thymocytes "paradoxically" appear as CD4+8lo transitional cells during positive selection of CD8+ T cells. J Exp Med. 2003;197:1709–1719. doi: 10.1084/jem.20030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Barthlott T, Kohler H, Eichmann K. Asynchronous coreceptor downregulation after positive thymic selection: prolonged maintenance of the double positive state in CD8 lineage differentiation due to sustained biosynthesis of the CD4 coreceptor. J Exp Med. 1997;185:357–362. doi: 10.1084/jem.185.2.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singer A, Bosselut R. CD4/CD8 coreceptors in thymocyte development, selection, and lineage commitment: analysis of the CD4/CD8 lineage decision. Adv Immunol. 2004;83:91–131. doi: 10.1016/S0065-2776(04)83003-7. [DOI] [PubMed] [Google Scholar]
- 51.Yu Q, Erman B, Bhandoola A, Sharrow SO, Singer A. In vitro evidence that cytokine receptor signals are required for differentiation of double positive thymocytes into functionally mature CD8+ T cells. J Exp Med. 2003;197:475–487. doi: 10.1084/jem.20021765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Noguchi M, et al. Functional cleavage of the common cytokine receptor gamma chain (gammac) by calpain. Proc Natl Acad Sci U S A. 1997;94:11534–11539. doi: 10.1073/pnas.94.21.11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Park JH, et al. 'Coreceptor tuning': cytokine signals transcriptionally tailor CD8 coreceptor expression to the self-specificity of the TCR. Nat Immunol. 2007;8:1049–1059. doi: 10.1038/ni1512. [DOI] [PubMed] [Google Scholar]
- 54.Rathmell JC, Vander Heiden MG, Harris MH, Frauwirth KA, Thompson CB. In the absence of extrinsic signals, nutrient utilization by lymphocytes is insufficient to maintain either cell size or viability. Mol Cell. 2000;6:683–692. doi: 10.1016/s1097-2765(00)00066-6. [DOI] [PubMed] [Google Scholar]
- 55.Wofford JA, Wieman HL, Jacobs SR, Zhao Y, Rathmell JC. IL-7 promotes Glut1 trafficking and glucose uptake via STAT5-mediated activation of Akt to support T-cell survival. Blood. 2008;111:2101–2111. doi: 10.1182/blood-2007-06-096297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Catlett IM, Hedrick SM. Suppressor of cytokine signaling 1 is required for the differentiation of CD4+ T cells. Nat Immunol. 2005;6:715–721. doi: 10.1038/ni1211. [DOI] [PubMed] [Google Scholar]
- 57.Yucel R, Karsunky H, Klein-Hitpass L, Moroy T. The transcriptional repressor Gfi1 affects development of early, uncommitted c-Kit+ T cell progenitors and CD4/CD8 lineage decision in the thymus. J Exp Med. 2003;197:831–844. doi: 10.1084/jem.20021417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ellmeier W, Sawada S, Littman DR. The regulation of CD4 and CD8 coreceptor gene expression during T cell development. Annu Rev Immunol. 1999;17:523–554. doi: 10.1146/annurev.immunol.17.1.523. [DOI] [PubMed] [Google Scholar]
- 59.Kioussis D, Ellmeier W. Chromatin and CD4, CD8A and CD8B gene expression during thymic differentiation. Nat Rev Immunol. 2002;2:909–919. doi: 10.1038/nri952. [DOI] [PubMed] [Google Scholar]
- 60.Sawada S, Scarborough JD, Killeen N, Littman DR. A lineage-specific transcriptional silencer regulates CD4 gene expression during T lymphocyte development. Cell. 1994;77:917–929. doi: 10.1016/0092-8674(94)90140-6. [DOI] [PubMed] [Google Scholar]
- 61. Siu G, Wurster AL, Duncan DD, Soliman TM, Hedrick SM. A transcriptional silencer controls the developmental expression of the CD4 gene. Embo J. 1994;13:3570–3579. doi: 10.1002/j.1460-2075.1994.tb06664.x. Together with reference 60, these reports describe the identification of a Cd4 silencer element that transcriptionally suppresses CD4 coreceptor expression in CD4− thymocytes.
- 62.Ellmeier W, Sunshine MJ, Losos K, Littman DR. Multiple developmental stage-specific enhancers regulate CD8 expression in developing thymocytes and in thymus-independent T cells. Immunity. 1998;9:485–496. doi: 10.1016/s1074-7613(00)80632-9. [DOI] [PubMed] [Google Scholar]
- 63. Hostert A, et al. Hierarchical interactions of control elements determine CD8alpha gene expression in subsets of thymocytes and peripheral T cells. Immunity. 1998;9:497–508. doi: 10.1016/s1074-7613(00)80633-0. Together with reference 62, these reports identify individual enhancer elements responsible for the developmental and stage-specific transcription of the Cd8a gene during T cell development.
- 64.Ellmeier W, Sunshine MJ, Losos K, Hatam F, Littman DR. An enhancer that directs lineage-specific expression of CD8 in positively selected thymocytes and mature T cells. Immunity. 1997;7:537–547. doi: 10.1016/s1074-7613(00)80375-1. [DOI] [PubMed] [Google Scholar]
- 65.Hostert A, et al. A CD8 genomic fragment that directs subset-specific expression of CD8 in transgenic mice. J Immunol. 1997;158:4270–4281. [PubMed] [Google Scholar]
- 66.Feik N, et al. Functional and molecular analysis of the double-positive stage-specific CD8 enhancer E8III during thymocyte development. J Immunol. 2005;174:1513–1524. doi: 10.4049/jimmunol.174.3.1513. [DOI] [PubMed] [Google Scholar]
- 67.Hostert A, et al. A region in the CD8 gene locus that directs expression to the mature CD8 T cell subset in transgenic mice. Immunity. 1997;7:525–536. doi: 10.1016/s1074-7613(00)80374-x. [DOI] [PubMed] [Google Scholar]
- 68.Yao Z, et al. Stat5a/b are essential for normal lymphoid development and differentiation. Proc Natl Acad Sci U S A. 2006;103:1000–1005. doi: 10.1073/pnas.0507350103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liu X, Bosselut R. Duration of TCR signaling controls CD4-CD8 lineage differentiation in vivo. Nat Immunol. 2004;5:280–288. doi: 10.1038/ni1040. [DOI] [PubMed] [Google Scholar]
- 70.Kadlecek TA, et al. Differential requirements for ZAP-70 in TCR signaling and T cell development. J Immunol. 1998;161:4688–4694. [PubMed] [Google Scholar]
- 71.Liu X, et al. Restricting Zap70 expression to CD4+CD8+ thymocytes reveals a T cell receptor-dependent proofreading mechanism controlling the completion of positive selection. J Exp Med. 2003;197:363–373. doi: 10.1084/jem.20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Negishi I, et al. Essential role for ZAP-70 in both positive and negative selection of thymocytes. Nature. 1995;376:435–438. doi: 10.1038/376435a0. [DOI] [PubMed] [Google Scholar]
- 73.Harker N, et al. The CD8alpha gene locus is regulated by the Ikaros family of proteins. Mol Cell. 2002;10:1403–1415. doi: 10.1016/s1097-2765(02)00711-6. [DOI] [PubMed] [Google Scholar]
- 74.Urban JA, Winandy S. Ikaros null mice display defects in T cell selection and CD4 versus CD8 lineage decisions. J Immunol. 2004;173:4470–4478. doi: 10.4049/jimmunol.173.7.4470. [DOI] [PubMed] [Google Scholar]
- 75.Naito T, Gomez-Del Arco P, Williams CJ, Georgopoulos K. Antagonistic interactions between Ikaros and the chromatin remodeler Mi-2beta determine silencer activity and Cd4 gene expression. Immunity. 2007;27:723–734. doi: 10.1016/j.immuni.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 76.Williams CJ, et al. The chromatin remodeler Mi-2beta is required for CD4 expression and T cell development. Immunity. 2004;20:719–733. doi: 10.1016/j.immuni.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 77.Chi TH, et al. Reciprocal regulation of CD4/CD8 expression by SWI/SNF-like BAF complexes. Nature. 2002;418:195–199. doi: 10.1038/nature00876. [DOI] [PubMed] [Google Scholar]
- 78. Aliahmad P, Kaye J. Development of all CD4 T lineages requires nuclear factor TOX. J Exp Med. 2008;205:245–256. doi: 10.1084/jem.20071944. This study documents that, in the absence of the HMG-box protein TOX, development of CD4 lineage T cells is blocked, identifying TOX as one the nuclear factors required for CD4 lineage choice.
- 79.Bosselut R. CD4/CD8-lineage differentiation in the thymus: from nuclear effectors to membrane signals. Nat Rev Immunol. 2004;4:529–540. doi: 10.1038/nri1392. [DOI] [PubMed] [Google Scholar]
- 80.Egawa T, Tillman RE, Naoe Y, Taniuchi I, Littman DR. The role of the Runx transcription factors in thymocyte differentiation and in homeostasis of naive T cells. J Exp Med. 2007;204:1945–1957. doi: 10.1084/jem.20070133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. He X, et al. The zinc finger transcription factor Th-POK regulates CD4 versus CD8 T-cell lineage commitment. Nature. 2005;433:826–833. doi: 10.1038/nature03338. Together with reference 85, this study identifies Th-POK as a CD4 lineage-determining factor and as the putative CD4 ‘master gene’. Using positional cloning, this study identifies a point mutation in the zinc finger protein Th-POK as responsible for the absence of CD4+ T cells in HD (helper deficient) mice. Reciprocally, transgenic Th-POK proteins are shown to redirect the differentiation of MHC-I selected thymocytes cells into CD4+ T cells.
- 82.Hernandez-Hoyos G, Anderson MK, Wang C, Rothenberg EV, Alberola-Ila J. GATA-3 expression is controlled by TCR signals and regulates CD4/CD8 differentiation. Immunity. 2003;19:83–94. doi: 10.1016/s1074-7613(03)00176-6. [DOI] [PubMed] [Google Scholar]
- 83.Sato T, et al. Dual functions of Runx proteins for reactivating CD8 and silencing CD4 at the commitment process into CD8 thymocytes. Immunity. 2005;22:317–328. doi: 10.1016/j.immuni.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 84.Setoguchi R, et al. Repression of the transcription factor Th-POK by Runx complexes in cytotoxic T cell development. Science. 2008;319:822–825. doi: 10.1126/science.1151844. [DOI] [PubMed] [Google Scholar]
- 85. Sun G, et al. The zinc finger protein cKrox directs CD4 lineage differentiation during intrathymic T cell positive selection. Nat Immunol. 2005;6:373–381. doi: 10.1038/ni1183. Together with reference 81, this study identifies Th-POK as a CD4 lineage-determining factor and as the putative CD4 ‘master gene’. Using the technique of gene microarrays, this study identifies the zinc-finger protein Th-POK as a transcription factor directing positively selected thymocytes to differentiate into CD4+ T cells, regardless of the MHC-restriction-specificity of their TCR.
- 86.Taniuchi I, Ellmeier W, Littman DR. The CD4/CD8 lineage choice: new insights into epigenetic regulation during T cell development. Adv Immunol. 2004;83:55–89. doi: 10.1016/S0065-2776(04)83002-5. [DOI] [PubMed] [Google Scholar]
- 87.Grueter B, et al. Runx3 regulates integrin alpha E/CD103 and CD4 expression during development of CD4-/CD8+ T cells. J Immunol. 2005;175:1694–1705. doi: 10.4049/jimmunol.175.3.1694. [DOI] [PubMed] [Google Scholar]
- 88.Aliahmad P, et al. TOX provides a link between calcineurin activation and CD8 lineage commitment. J Exp Med. 2004;199:1089–1099. doi: 10.1084/jem.20040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wilkinson B, et al. TOX: an HMG box protein implicated in the regulation of thymocyte selection. Nat Immunol. 2002;3:272–280. doi: 10.1038/ni767. [DOI] [PubMed] [Google Scholar]
- 90.Hendriks RW, et al. Expression of the transcription factor GATA-3 is required for the development of the earliest T cell progenitors and correlates with stages of cellular proliferation in the thymus. Eur J Immunol. 1999;29:1912–1918. doi: 10.1002/(SICI)1521-4141(199906)29:06<1912::AID-IMMU1912>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 91.Nawijn MC, et al. Enforced expression of GATA-3 during T cell development inhibits maturation of CD8 single-positive cells and induces thymic lymphoma in transgenic mice. J Immunol. 2001;167:715–723. doi: 10.4049/jimmunol.167.2.715. [DOI] [PubMed] [Google Scholar]
- 92.Galera P, Musso M, Ducy P, Karsenty G. c-Krox, a transcriptional regulator of type I collagen gene expression, is preferentially expressed in skin. Proc Natl Acad Sci U S A. 1994;91:9372–9376. doi: 10.1073/pnas.91.20.9372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dave VP, Allman D, Keefe R, Hardy RR, Kappes DJ. HD mice: a novel mouse mutant with a specific defect in the generation of CD4(+) T cells. Proc Natl Acad Sci U S A. 1998;95:8187–8192. doi: 10.1073/pnas.95.14.8187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Jenkinson SR, et al. Expression of the transcription factor cKrox in peripheral CD8 T cells reveals substantial postthymic plasticity in CD4-CD8 lineage differentiation. J Exp Med. 2007;204:267–272. doi: 10.1084/jem.20061982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Taniuchi I, et al. Differential requirements for Runx proteins in CD4 repression and epigenetic silencing during T lymphocyte development. Cell. 2002;111:621–633. doi: 10.1016/s0092-8674(02)01111-x. This report identifies RUNX family transcription factors as critical mediators of CD4 repression.
- 96.Levanon D, Groner Y. Structure and regulated expression of mammalian RUNX genes. Oncogene. 2004;23:4211–4219. doi: 10.1038/sj.onc.1207670. [DOI] [PubMed] [Google Scholar]
- 97.Woolf E, et al. Runx3 and Runx1 are required for CD8 T cell development during thymopoiesis. Proc Natl Acad Sci U S A. 2003;100:7731–7736. doi: 10.1073/pnas.1232420100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kohu K, et al. Overexpression of the Runx3 transcription factor increases the proportion of mature thymocytes of the CD8 single-positive lineage. J Immunol. 2005;174:2627–2636. doi: 10.4049/jimmunol.174.5.2627. [DOI] [PubMed] [Google Scholar]
- 99.Liu X, Taylor BJ, Sun G, Bosselut R. Analyzing expression of perforin, Runx3, and Thpok genes during positive selection reveals activation of CD8-differentiation programs by MHC II-signaled thymocytes. J Immunol. 2005;175:4465–4474. doi: 10.4049/jimmunol.175.7.4465. [DOI] [PubMed] [Google Scholar]
- 100.Schilham MW, Clevers H. HMG box containing transcription factors in lymphocyte differentiation. Semin Immunol. 1998;10:127–132. doi: 10.1006/smim.1998.0114. [DOI] [PubMed] [Google Scholar]
- 101.Stros M, Launholt D, Grasser KD. The HMG-box: a versatile protein domain occurring in a wide variety of DNA-binding proteins. Cell Mol Life Sci. 2007;64:2590–2606. doi: 10.1007/s00018-007-7162-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hedrick SM. Thymus lineage commitment: a single switch. Immunity. 2008;28:297–299. doi: 10.1016/j.immuni.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 103.Pai SY, et al. Critical roles for transcription factor GATA-3 in thymocyte development. Immunity. 2003;19:863–875. doi: 10.1016/s1074-7613(03)00328-5. [DOI] [PubMed] [Google Scholar]
- 104.Huseby ES, et al. How the T cell repertoire becomes peptide and MHC specific. Cell. 2005;122:247–260. doi: 10.1016/j.cell.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 105.Zerrahn J, Held W, Raulet DH. The MHC reactivity of the T cell repertoire prior to positive and negative selection. Cell. 1997;88:627–636. doi: 10.1016/s0092-8674(00)81905-4. [DOI] [PubMed] [Google Scholar]
- 106. Van Laethem F, et al. Deletion of CD4 and CD8 coreceptors permits generation of alphabetaT cells that recognize antigens independently of the MHC. Immunity. 2007;27:735–750. doi: 10.1016/j.immuni.2007.10.007. This study offers a novel explanation for thymic selection of an exclusively MHC-restricted TCR repertoire. The authors propose that CD4 and CD8 coreceptors on DP thymocytes bind intracellular LCK and consequently diminish the amount of free LCK available to the TCR. As a result, TCR signaling in DP thymocytes is exclusively co-receptor-dependent, requiring co-engagement of TCR and co-receptors which occurs only with MHC dependent ligands. TCR that engage non-MHC ligands in the thymus cannot access LCK and so are unable to transduce intracellular signals that would rescue thymocytes from death by neglect. The net effect is that only MHC-specific TCR can signal DP thymocytes to undergo positive selection.
- 107.Wiest DL, Ashe JM, Abe R, Bolen JB, Singer A. TCR activation of ZAP70 is impaired in CD4+CD8+ thymocytes as a consequence of intrathymic interactions that diminish available p56lck. Immunity. 1996;4:495–504. doi: 10.1016/s1074-7613(00)80415-x. [DOI] [PubMed] [Google Scholar]
- 108.Haughn L, et al. Association of tyrosine kinase p56lck with CD4 inhibits the induction of growth through the alpha beta T-cell receptor. Nature. 1992;358:328–331. doi: 10.1038/358328a0. [DOI] [PubMed] [Google Scholar]
- 109.Yu M, et al. Nucleoprotein structure of the CD4 locus: implications for the mechanisms underlying CD4 regulation during T cell development. Proc Natl Acad Sci U S A. 2008;105:3873–3878. doi: 10.1073/pnas.0800810105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Bilic I, et al. Negative regulation of CD8 expression via Cd8 enhancer-mediated recruitment of the zinc finger protein MAZR. Nat Immunol. 2006;7:392–400. doi: 10.1038/ni1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Adlam M, Duncan DD, Ng DK, Siu G. Positive selection induces CD4 promoter and enhancer function. Int Immunol. 1997;9:877–887. doi: 10.1093/intimm/9.6.877. [DOI] [PubMed] [Google Scholar]
- 112.Uematsu Y, Donda A, De Libero G. Thymocytes control the CD4 gene differently from mature T lymphocytes. Int Immunol. 1997;9:179–187. doi: 10.1093/intimm/9.1.179. [DOI] [PubMed] [Google Scholar]
- 113.Wurster AL, Siu G, Leiden JM, Hedrick SM. Elf-1 binds to a critical element in a second CD4 enhancer. Mol Cell Biol. 1994;14:6452–6463. doi: 10.1128/mcb.14.10.6452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zou YR, et al. Epigenetic silencing of CD4 in T cells committed to the cytotoxic lineage. Nat Genet. 2001;29:332–336. doi: 10.1038/ng750. [DOI] [PubMed] [Google Scholar]
- 115.Allen RD, 3rd, Kim HK, Sarafova SD, Siu G. Negative regulation of CD4 gene expression by a HES-1-c-Myb complex. Mol Cell Biol. 2001;21:3071–3082. doi: 10.1128/MCB.21.9.3071-3082.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kim HK, Siu G. The notch pathway intermediate HES-1 silences CD4 gene expression. Mol Cell Biol. 1998;18:7166–7175. doi: 10.1128/mcb.18.12.7166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Wildt KF, et al. The transcription factor Zbtb7b promotes CD4 expression by antagonizing Runx-mediated activation of the CD4 silencer. J Immunol. 2007;179:4405–4414. doi: 10.4049/jimmunol.179.7.4405. [DOI] [PubMed] [Google Scholar]