Abstract
Chromatin remodeling enzymes use the energy of ATP hydrolysis to alter histone–DNA contacts and regulate DNA-based processes in eukaryotes. Whether different subfamilies of remodeling complexes generate distinct products remains uncertain. We have developed a protocol to analyze nucleosome remodeling on individual products formed in vitro. We used a DNA methyltransferase to examine DNA accessibility throughout nucleosomes that had been remodeled by the ISWI and SWI/SNF families of enzymes. We confirmed that ISWI-family enzymes mainly created patterns of accessibility consistent with canonical nucleosomes. In contrast, SWI/SNF-family enzymes generated widespread DNA accessibility. The protection patterns created by these enzymes were usually located at the extreme ends of the DNA and showed no evidence for stable loop formation on individual molecules. Instead, SWI/SNF family proteins created extensive accessibility by generating heterogeneous products that had fewer histone–DNA contacts than a canonical nucleosome, consistent with models in which a canonical histone octamer has been ‘pushed’ off of the end of the DNA.
INTRODUCTION
Chromatin packages nuclear DNA and thereby controls functions of the eukaryotic genome. Packaging of the DNA into nucleosome arrays, which can be folded into higher-order structures, provides the cell with a way to organize its genome in the nucleus. These structures also provide a means to tightly control access to the DNA sequence. Many sequence-specific DNA-binding factors and general transcription factors cannot bind to nucleosomal DNA (1,2). Consequently, they require enzymatic activities to expose their binding site to achieve their functions.
Different types of activities operate on the chromatin template and produce changes in DNA accessibility. Various enzymes covalently modify the histones or the DNA (3,4) while others use ATP hydrolysis to alter histone–DNA contacts. The latter proteins are termed chromatin or nucleosome remodeling factors. These enzymes have been implicated in virtually all DNA-based processes including replication, DNA damage repair and transcription. Consequently, chromatin remodeling is required for cell viability, homeostasis and proper metazoan development (5,6).
All the ATP-dependent chromatin remodelers studied so far belong to the SNF2-superfamily of ATPases. They have been divided into subfamilies based on their primary protein sequence (7,8). Extensive work from numerous laboratories suggests that these subfamilies show similarities as well as differences in their nucleosome binding and remodeling properties (6,9). The ISWI- and SWI/SNF-type of enzymes are two abundant classes of remodeling factors showing such differences. For example, most members of the Imitation Switch (ISWI)-family can regularly space nucleosome arrays in contrast to members of the Switch/Sucrose-Non-fermenting (SWI/SNF)-family (10,11). Additionally, ISWI-family remodeling complexes require adjacent naked DNA to remodel efficiently while SWI/SNF complexes do not. However, both families of remodeling complexes are similar in their ability to move nucleosomes translationally (a mechanism referred to as ‘sliding’).
Whether SWI/SNF enzymes create products of a different nature than ISWI-based factors to achieve their in vivo regulatory functions is still a matter of debate. Unlike ISWI-related enzymes, SWI/SNF-types of factors are efficient at exposing the entire nucleosomal DNA to nucleases, facilitating transcription factor binding to nucleosomal DNA in an ATP-dependent manner (12–18). SWI/SNF complexes form altered topologies, unusual dinucleosomal species, and can transfer octamers (19–22), properties that have led to proposals that they significantly disrupt histone–DNA contacts, perhaps via different mechanisms than seen with other remodeling complexes.
There are two prominent hypotheses to explain why SWI/SNF remodelers can perform functions that others cannot. The first proposes that SWI/SNF remodelers have an especially potent ability to move nucleosomes, allowing them to move nucleosomes off of the end of a DNA fragment or into an adjacent nucleosome (21,23–27). The second hypothesis proposes that SWI/SNF family remodelers induce DNA loops (28–31) and this looping allows access to sites internal to the starting nucleosome position (20,32–34)
These mechanistic issues have not been resolved, in part because of technical limitations. Indeed, most assays to study nucleosome remodeling rely on nucleases and/or changes in nucleosome electrophoretic mobility (6,35) and are thereby limited to looking at populations of nucleosomes. Remodeling factors rarely produce a single remodeled product, but rather create a panel of remodeled nucleosomes, and thus these technologies look at the average distribution across the spectrum of products. To circumvent these issues, optical or magnetic traps have been used to measure the energetics of remodeling on single molecules. These analyses offered insight into many of the physical characteristics of the remodeling process such as the force required to disrupt nucleosomes, the involvement and processivity of DNA tracking and the formation of DNA loops (36). In another study, Wang and colleagues probed histone-DNA contacts on single nucleosomes that were remodeled by ySWI/SNF by unzipping their DNA double helix and compared their ‘disruption signature’ to that of the nucleosome substrate. However, this approach could only monitor about half of a nucleosome at a time as nucleosomes became destabilized by the analysis (37). Thus to date, although very valuable, previous studies did not provide information about the accessibility of single DNA molecules throughout the entire length of remodeled nucleosomes.
To address the nature of the remodeled products more directly, we have developed an in vitro method that measures histone–DNA contacts on individual molecules after nucleosome remodeling. We have compared hundreds of individual remodeled products created by either ISWI or SWI/SNF family remodeling complexes. Data obtained using this approach are consistent with previous studies which showed that in bulk populations SNF2H, the human ISWI homolog, and the SNF2H-containing complex hACF reposition nucleosomes and leave their structure intact. The data also support previous observations indicating that BRG1 and the BRG1-containing complex hSWI/SNF produce an elevated and spread-out DNA accessibility on nucleosomes. However, our detailed single-molecule analysis shows that this overall accessibility is a consequence of heterogeneity in translational nucleosome positions along with a significant number of nucleosomes showing a reduced (sub-nucleosomal-size) DNA protection that extends to the end of the DNA fragment. These data rule out the necessity for creating stable loops as a mechanism for allowing access to internal sites and support the hypothesis that SWI/SNF family complexes are especially potent when moving nucleosome position.
MATERIALS AND METHODS
Nucleosome substrate preparation
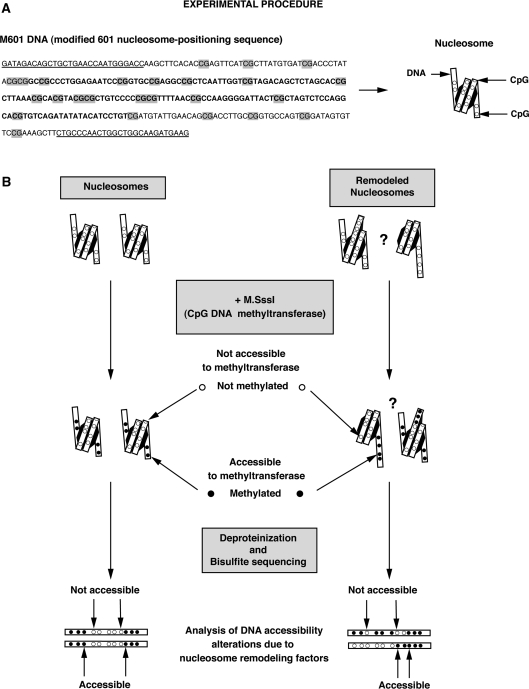
The modified 601 (M601) DNA sequence was generated using 242-bp of the 282-bp ‘601’ strong nucleosome-positioning sequence (38). Mutations were introduced such that five new CpG sites were created and the construct was flanked with HindIII sites and GRP78 sequence primers were added using these sites (see sequence below).
DNA sequence
GATAGACAGCTGCTGAACCAATGGGACCAAGCTT CACACCGAGTTCATCGCTTATGTGATCGACCCTATACGCGGCCG CCCTGGAGAATCCCGGTGCCGAGGCCGCTCAATTGGTCGT AGACAGCTCTAGCACCGCTTAAACGCACGTACGCGCTGTCCCCCG CGTTTTAACCGCCAAGGGGATTACTCGCTAGTCTCCAGGCACGTGTCAG ATATATACATCCTGTCGATGTATTGAACAGCGACCTTGCCGGTGCC AGTCGGATAGTGTTCCGAAAGCTTCTGCCCAACTGGCTG GCAAGATGAAG.
The DNA fragment was amplified by PCR (primers underlined above) using PCR Master Mix (promega) supplemented with cloned Pfu polymerase (Stratagene) followed by a phenol/chloroform step prior to DNA precipitation. The DNA was assembled into mononucleosomes by standard salt dialysis using histones purified from HeLa cells. Low histone:DNA ratios were chosen to retrieve a more homogeneous population of centrally-positioned nucleosomes. Since these conditions favored the appearance of a second position, the nucleosome substrate was purified by subjecting the assembly to a 10%–30% glycerol gradient for 18 h at 35 000 rpm using a Beckman SW55Ti rotor. Pure fractions were pooled and dialyzed against nucleosome remodeling buffer (see below).
Proteins purifications
The hSWI/SNF complex was essentially purified as described previously using a cell line expressing FLAG-tagged Ini1 (39). Recombinant FLAG-tagged BRG1, FLAG-tagged SNF2H and hACF (FLAG-tagged SNF2H + untagged hACF1) were expressed in exponentially growing Sf9 cells using the baculovirus system. Cells extracts were prepared in BC buffer [10% Glycerol, 20 mM HEPES, pH 7.9, 0.4 mM EDTA, supplemented with protease inhibitors (complete tablets, Roche)] containing 150 (BC 150) or 200 mM NaCl (BC 200). Cells were lysed by freeze-thaw cycles in liquid nitrogen and lysates were cleared by centrifugation prior to adding M2-affinity gel (Sigma). Extracts were incubated for about 4 h at 4°C and the affinity gel was recovered by centrifugation and transferred to Econo columns (Bio-Rad), washed twice with at least 10 resin-volumes of: BC 200, BC 500, BC 1000 and once with about 10 resin-volumes of BC 500, BC 200 and finally with about of 10 resin-volumes of BC 100 prior to elution with BC 100 containing 0.25 mg/ml of FLAG-peptide (Sigma). The hACF complex was further purified over a 5 ml HiTrap Q HP column (Amersham Biosciences) to separate non-complexed SNF2H away from the complex. Proteins were then concentrated using Amicon Ultra centrifugal filter devices (Millipore), aliquoted and stored at −80°C.
Restriction enzyme-accessibility assays
REA reactions were initiated by the addition of 50 nM of 32P-labeled M601 nucleosomes to a mix containing a remodeling enzyme (SNF2H: 530 nM; hACF: 160 nM; BRG1: 690 nM and hSWI/SNF 68 nM) and 25 U of MfeI in addition to 0, 2.5 or 5 U of M.SssI at 30°C (as specified in Figure 2) and 2 mM ATP. Aliquots were removed at various times and quenched in 1.5 volumes of stop mix (10% glycerol, 70 mM EDTA, 20 mM Tris, pH 7.9, 2% SDS and xylene cyanol). Cut DNA was separated from uncut DNA on a 4.5% native polyacrylamide gel (0.5× TBE) after deproteinization at 37°C for at least 1 h with 20 μg proteinase K (Novagen). Data were quantified using the ImageQuantTL software (GE Healthcare Life Sciences).
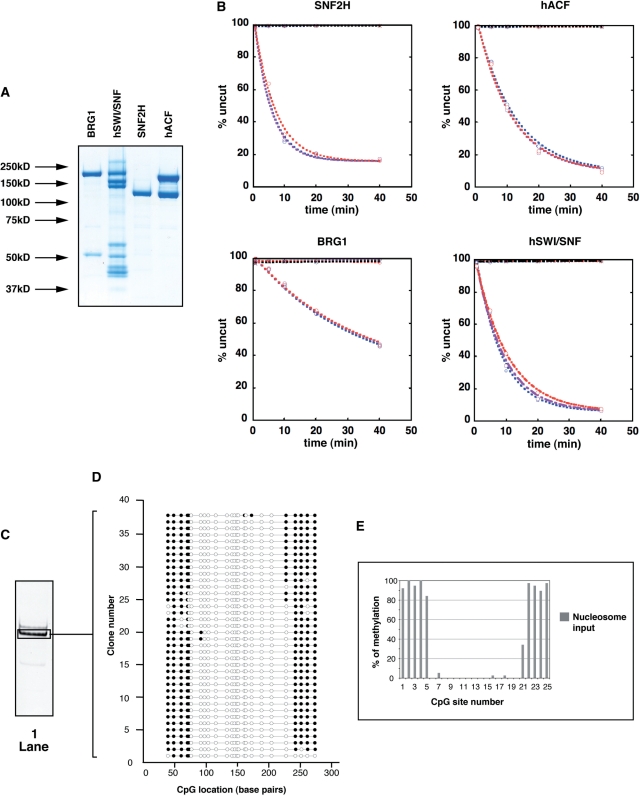
Figure 2.
(A) Proteins used in the nucleosome remodeling assays were separated by SDS-PAGE and visualized by Coomassie Blue staining. Molecular masses are indicated on the left and enzymes indicated on top. (B) Analysis of remodeling activities in the presence of M.SssI. The restriction enzyme-accessibility (REA) assays measured the ability of the remodeling factors to expose an MfeI restriction site at bp position 108 (31-bp away from the first protected site) in the absence or presence of M.SssI. M601 nucleosomes (50 nM) were incubated with MfeI (25 U) in the absence (blue curves) or the presence (2.5 U, purple curves; 5 U, red curves) of M.SssI. Reactions in the absence or presence of remodeling enzyme are specified by diamond or circle plots, respectively (as indicated on top of each panel). Reactions in the presence of remodeler but absence of ATP are plotted in black. Enzymes were used at the following concentrations: SNF2H: 530 nM; hACF: 160 nM; BRG1: 690 nM and hSWI/SNF 68 nM. Curve fits of the data (obtained from averaging at least two independent experiments) were achieved using first-order exponential decay using an apparent endpoint of the reactions with the KaleidaGraph software. kobs for SNF2H = 0.18 ± 0.01 min−1 without M.SssI, 0.19 ± 0.01 min−1 + 2.5 U M.SssI and 0.15 ± 0.01 min−1 + 5 U M.SssI. kobs for hACF= 0.08 ± 0.01 min−1 without M.SssI, 0.09 ± 0.01 min−1 + 2.5 U M.SssI and 0.09 ± 0.01 min−1 + 5 U M.SssI. kobs for BRG1 = 0.02 ± 0.001 min−1 without M.SssI, 0.019 ± 0.001 min−1 + 2.5 U M.SssI and 0.02 ± 0.001 min−1 + 5 U M.SssI. kobs for hSWI/SNF = 0.13 ± 0.01 min−1 without M.SssI, 0.11 ± 0.01 min−1 + 2.5 U M.SssI and 0.1 ± 0.005 min−1 + 5 U M.SssI. (C) Nucleosomes (50 nM) were incubated as in Figure 3, but addition of remodeler was omitted. Nucleosomes were then methylated with of M.SssI (5 U), separated by native gel electrophoresis and visualized by ethidium bromide staining. The gel area excised and used for analysis is delimited by a black frame. (D) Schematic representation of individual DNA molecules. Bisulfite-converted DNA from the excised gel slice (black frame, lane 1) was amplified by PCR, cloned and sequenced. Each line represents the sequence of individual DNA clones and the circles represent CpG dinucleotides. Methylated and unmethylated CpGs are indicated by filled (black) and open circles, respectively. (E) The frequency of methylation was determined at given CpG sites by averaging methylation for all the DNA molecules showed in panel D and expressed as a percentage. The position of the CpGs relative to the DNA sequence is indicated on the X-axis and clone numbers are indicated on the Y-axis.
DNA methyltransferase-accessibility assay
Nucleosome remodeling reactions were performed in nucleosome remodeling buffer (10 mM HEPES, pH 7.9, 50 mM NaCl, 3 mM MgCl2, 3% Glycerol, 1 mM Dithiothreitol). Approximately 100 nM of nucleosomes were incubated with increasing concentrations of remodeling factor (SNF2H: 6, 19, 57 nM; hACF: 8, 24, 72 nM; BRG1: 55, 170, 500 nM; hSWI/SNF: 6, 19, 56 nM) after addition of 2 mM MgCl2 or 1 mM ATP/3 mM MgCl2 for 1 h at 30°C in a final volume of 20 μl. Reaction were stopped by addition of 10 mM ADP and incubation on ice for 10 min. Methylation of the remodeled products was performed by incubation at 37°C for 15 min after addition of 5 U M.SssI and S-adenosylmethionine to a final concentration of 160 μM (New England BioLabs). After addition of competitor plasmid DNA, samples were further incubated on ice for 10 min and separated by native gel electrophoresis for about 2 h and stained with ethidium bromide for 10 min. Major bands were excised and eluted from the gel by overnight incubation in 400 μl of 10 mM Tris pH 8.0, 1 mM EDTA at 55°C. The DNA was then deproteinized with phenol/chloroform, precipitated and subjected to bisulfite treatment.
Bisulfite treatment procedure
Ten to fifty nanograms of DNA was bisulfite converted according to (40) with a few minor changes. Briefly, DNA was resuspended in 25 μL of nuclease free water and denatured by incubation at 95°C for 20 min. Then 0.6 M NaOH was added and the solution was incubated at 45°C for 20 min. Meanwhile, 2.08 g NaHSO3 (Wako), 0.67 g (NH4)2SO3•H20 (Wako) and 5.0 mL of 50% (NH4)HS03 (Wako) were mixed and heated at 90°C to obtain a solution of pH 5.2–5.3 (when measured at ambient temperature). Two hundred and eighty two microliters of the 10 M bisulfite solution was added to the alkali-denatured DNA. The mixture was then incubated at 90°C for 10 min. Samples were then purified using the Wizard Miniprep Kit (Promega) and DNA was eluted in 50 μL of 80°C nuclease free water. Five microliters of 3 M NaOH was then added to the purified samples and the solution was incubated at 40°C for 20 min. Samples were then ethanol precipitated.
Cloning of bisulfite-treated DNA
Bisulfite converted M601 was PCR amplified using the following primers: forward, 5′-GATAGATAGTTGTTGAATTAATGGGATT-3′ and reverse, 5′-CTTCATCTTACCAACCAATTAAACAA-3′. PCR conditions were as follows: 95°C for 3 min, followed by 20 cycles of 95°C for 1 min, 60°C for 1 min and 72°C for 1 min. A final incubation was done at 72°C for 10 min. M601 was then cloned into the pCR2.1-TOPO vector using the TOPO TA Cloning Kit (Invitrogen) according to manufacturer's protocol. After sequencing of the individual molecules, the data were analyzed using BiQ Analyzer software (41) and figures were generated using the CpG Bubble Chart Generator. Sequences showing no methylation were discarded.
RESULTS
A novel in vitro assay for analyzing ATP-dependent nucleosome remodeling
To characterize how ATP-dependent remodeling occurs on individual nucleosomes, we methylated remodeled templates at exposed CpG sites with the M.SssI DNA methyltransferase, gel-purified and sequenced individual examples. This approach was based upon studies by Kladde and Simpson who initially developed a method using M.SssI to monitor the average DNA accessibility in yeast (42,43). Binding of proteins to DNA with high affinity protects the DNA from being methylated. Hence, M.SssI can be used to probe where there are accessible CpG base pairs (bp) in a nucleosome or a remodeled nucleosome. To achieve single molecule resolution, this method was extended to analyze the methylation pattern by using conventional bisulfite treatment, PCR, cloning and sequencing of individual DNA fragments (44,45).We and others have applied this technology to map nucleosome position and binding of transcription factors in vivo (45–48). Here, we have developed an in vitro system to study the stable changes that occurred in reconstituted mononucleosomes after remodeling.
Formation of a canonical nucleosome renders the histone-associated DNA inaccessible to methylation at CpG sites by M.SssI (42,43). Remodeling alters the position of nucleosomes and potentially the conformation of DNA on the surface of the nucleosome. By studying the accessibility of CpG sites to methylation on individual remodeled products, we were able to compare how these products differ from the starting material and how distinct remodeling enzymes generate different types of products.
To increase the resolution of our assay, we modified the ‘601’ nucleosome positioning sequence (38) so that it contained more CpG dinucleotides and used this as a template to assemble nucleosomes (Figure 1). We refer to this altered template as M601 (modified 601). M601 DNA was assembled into mononucleosomes by salt dialysis using histones purified from HeLa cells. The major nucleosome species that formed under our assembly conditions was isolated on a glycerol gradient and found to have the same nucleosome position as previously observed for the 601 sequence as judged from its methylation pattern (Figure 2C and D, see ‘Materials and Methods’ section).
Figure 1.
(A) M601 DNA sequence used in the remodeling assays. GRP78-gene primer sequences flanking the DNA construct are underlined and CpG dinucleotides are highlighted in grey. The residues in bold denote the approximate position of the protection caused by the histone octamer on the nucleosome substrate. (B) Schematic representation of the procedure used to perform single molecule analyses of the remodeled products. Nucleosomes were assembled using the M601 DNA. After incubation with (or without) nucleosome remodeling factor, remodeled nucleosomes were methylated with the M.SssI CpG methyltransferase to create a footprint of DNA accessibility (the DNA methyltransferase only methylates cytosine residues that are not bound to the histones). After native electrophoresis, excision and elution from the gel, nucleosomes were deproteinized and the nucleosomal DNA molecules subjected to conventional bisulfite treatment to reveal their methylation pattern. Changes in these patterns were compared to the input nucleosome to assess alterations in histone–DNA contacts resulting from the action of the remodeling factors.
Isolation and characterization of remodeling proteins
We chose to compare the nucleosome remodeling activities of SNF2H and BRG1 along with complexes containing each of these proteins. These two families of complexes are abundant in mammalian cells and previous studies have shown that their products behave differently from one another. We expressed and purified FLAG-tagged BRG1 and SNF2H ATPases as well as the SNF2H-containing human ACF (hACF) complex using Sf9 cells. The BRG1-containing hSWI/SNF complex was purified from a HeLa cell line expressing a FLAG-tagged version of the Ini-1 subunit (Figure 2A).
We tested the activity of each remodeling enzyme in the presence or absence of M.SssI using the restriction enzyme-accessibility (REA) assay. This was done to compare relative activities of the studied proteins and to determine whether the amount of M.SssI used in these experiments altered remodeling activity. While it is known that M.SssI does not alter the position of canonical nucleosomes, we wished to control for the possibility that M.SssI might have an interaction with remodeled nucleosomes that might alter the properties of these nucleosomes. We reasoned that if the DNA methyltransferase influenced the distribution of remodeled nucleosomes this enzyme would consequently affect the rates of the reactions catalyzed by the remodeling factors. As shown Figure 2B, M.SssI does not affect the reaction rates of any of the tested remodelers. Remodeling reactions in the presence of limited as well as higher M.SssI concentrations necessary to efficiently methylate DNA were indistinguishable from reactions without M.SssI (compare curves +2.5 U and +5 U to 0 U, respectively). Therefore, we conclude that the DNA methyltransferase can be used as probe to analyze remodeled nucleosomes.
Comparison of SNF2H remodeled products and the canonical nucleosome
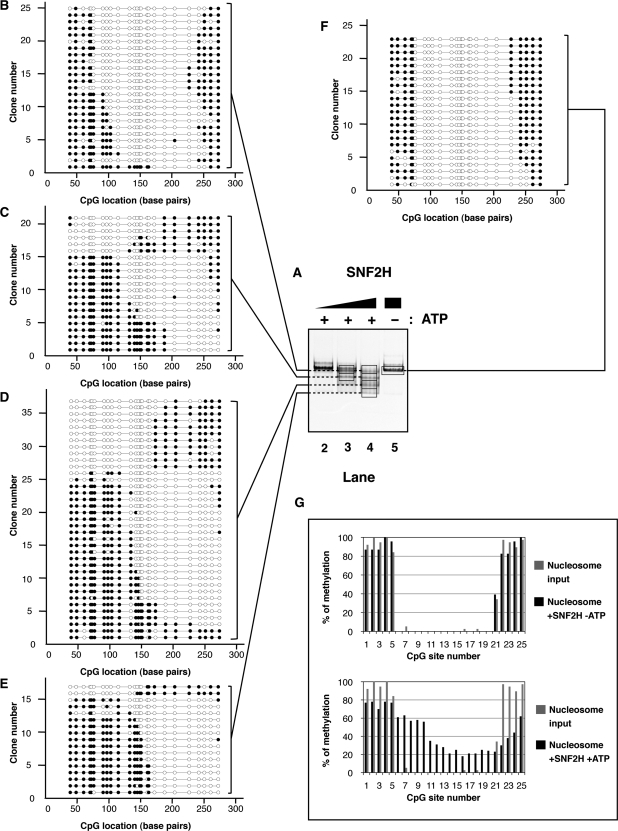
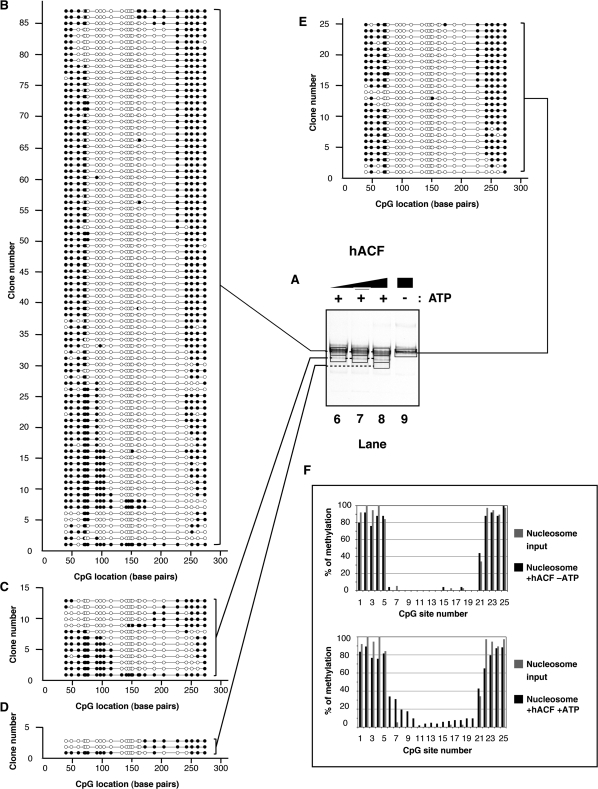
ISWI-family factors such as SNF2H have been shown to maintain the canonical structure of nucleosomes after remodeling. These enzymes alter DNA access primarily by sliding the nucleosome (49–51). To determine the spectrum of products generated by SNF2H, we characterized 100 individual nucleosomes resulting from the remodeling reactions. Mononucleosomes were incubated in the presence or absence of SNF2H and the reactions were stopped with ADP. The remodeled products were then methylated with M.SssI. After addition of competitor DNA, the nucleosomes were separated using native electrophoresis (Figures 2C, lane 1 and 3A, lanes 2–5). Major bands were excised, their contents eluted from the gel and the nucleosomal DNA was deproteinized prior to performing a standard bisulfite treatment to deaminate the non-methylated cytosine residues. Molecules were amplified using PCR, cloned and sequenced to reveal which cytosine residues were methylated. DNA accessibility on single molecules was inferred accordingly. The results are shown indicating accessible residues by filled circles (Figure 2D). To reflect the proportion of the species formed after the remodeling reactions, we have adjusted the number of sequences displayed for each species relative to the intensity of that species on the native gel (Figure 3B–F). Since similar numbers of clones were initially analyzed for each of these species, we randomly excluded clones for underrepresented species; we present these excluded clones in supplemental information (Figures S1 and S2).
Figure 3.
(A) Native electrophoresis of SNF2H remodeled products. Nucleosomes (∼100 nM) were incubated with increasing concentrations of SNF2H (lane 2: 6 nM; lane 3: 19 nM; lanes 4 and 5: 57 nM) in the presence (lanes 2–4) or absence (lane 5) of ATP (1 mM) as indicated on top, for 1 h at 30°C. Reactions were stopped by addition of ADP (10 mM) and incubation on ice for 10 min. Methylation of the remodeled products were performed as follow. The reactions were incubated for 15 min at 37°C after addition of M.SssI (5 U) and S-adenosylmethionine (160 μM). The remodeling reactions were separated by native gel electrophoresis after addition of competitor plasmid DNA and visualized by ethidium bromide staining. Gel areas excised and used for analysis are delimited by a black frame. Back frames are connected by dashed lines when gel slices were combined. (B–F) Schematic representation of individual DNA molecules remodeled by SNF2H. Bisulfite-converted DNAs from gel slices (black frames, lanes 3–5) were amplified by PCR, cloned and sequenced. Individual DNA clones are represented as described in Figure 2D. The number of remodeled molecules shown is proportional to the average intensity of the bands generated after remodeling at enzyme concentration allowing maximal remodeling in three independent experiments. (G) Frequency of methylation at a given CpG site. Upper panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panel F (reaction without ATP). Lower panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panels B–E (reactions with ATP). In both the upper and lower panels, frequencies obtained from the nucleosome substrate in the absence of remodeler (Figure 2E) are shown in grey for comparison.
Addition of recombinant SNF2H to the reaction in the absence of ATP led to a modest increase (about 12%) in methylation at the CpG #5 compared to the nucleosome alone (Figure 3F and G, upper panel). Otherwise, both the nucleosome in the absence of remodeler and the reaction without ATP showed protection expected from a nucleosome positioned over the M601 sequence. Addition of both SNF2H and ATP resulted in changes in DNA accessibility on most of the molecules (Figure 3A, lanes 2–4 and panels B–E). The protection caused by the histone octamer was translated towards either end of the DNA. Interestingly, SNF2H moved the nucleosomal protection with a noticeable sequence bias (Figure 3, panels B–E, note the increased movement to the ‘right’ side of the fragment in this depiction). Averaging the changes in methylation at individual CpG sites confirms that overall SNF2H preferentially positioned the histone octamer between CpG #11 (bp: 134) and CpG #23 (bp: 252) exposing the DNA between CpG #6 and #10 (between bp 77 and 116, respectively; Figure 3C–E and G lower panel). Quantification of methylation site accessibility on all analyzed molecules indicates that overall SNF2H remodeling increased DNA accessibility by about 14%.
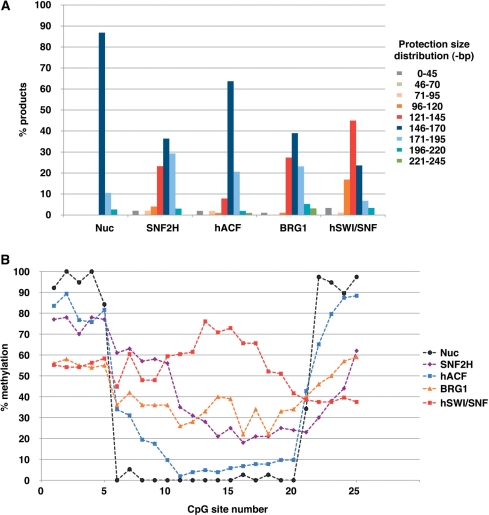
These results are in general agreement with previous studies showing that nucleosome mobilization by ISWI-type remodelers does not produce nucleosomes with a non-canonical structure (49–51). Some individual molecules, however, displayed patterns not easily reconcilable with a canonical structure. Taking into account the irregularity of CpG spacing throughout the fragment, we calculated an upper limit for how much DNA was protected on each molecule by noting the limits of where methylation occurred. While almost 90% of the input nucleosomes showed a protection pattern with a maximal size ranging between 146 and 170 bp, <40% of the nucleosomes remained in this size distribution after remodeling by SNF2H. Surprisingly, more than 20% of the remodeled nucleosomes displayed a smaller protection than expected for a canonical nucleosome (between 121 and 145-bp), while others showed protection >171-bp (see Summary in Figure 7A).
Figure 7.
(A) Size distribution of the protections observed after incubation without (Nuc) or with (as indicated on the X-axis) remodeling factor. Bars indicate the percentage of sequences showing a protection in the following size ranges: between 0–45, 46–70, 71–95, 96–120, 121–145, 146–170, 171–195, 196–220 and 221–245-bp, as indicated by the color code on the right. (B) Direct comparison of average DNA accessibility generated by nucleosome remodeling factors (as indicated by the color code on the right). Methylation averages at given CpGs obtained for nucleosome (Figure 2E), SNF2H (Figure 3G, lower panel), hACF (Figure 4F, lower panel), BRG1 (Figure 5G, lower panel) and hSWI/SNF (Figure 6F, lower panel) are displayed as curves for clarity.
Analysis of remodeled products formed by hACF
The remodeling properties of ISWI motor proteins change upon association with non-catalytic subunits (10,49,50,52–56). Therefore we examined the impact of SNF2H association with the hACF1 protein on the outcome of nucleosome remodeling. Incubation of this complex with nucleosomes without ATP resulted in very small changes in accessibility, under these conditions (Figure 4E and F, upper panel). This complex is active as measured by the REA assay (Figure 2B), however, this complex had a limited (yet noticeable) impact on nucleosome electrophoretic mobility when ATP was added to the reaction (Figure 4A, compare lane 9 to lanes 6–8). This was expected as this complex has been shown to favor a steady state in which most remodeled mononucleosomes appear centrally located (57) similar to the starting substrate in this experiment.
Figure 4.
(A) Native electrophoresis of hACF remodeled products. Nucleosomes (∼100 nM) were incubated with increasing concentrations of hACF complex (lane 6: 8 nM; lane 7: 24 nM; lanes 8 and 9: 72 nM) in the presence (lanes 6–8) or absence (lane 9) of ATP (1 mM) as indicated on top. Reactions were handled identically and in parallel to samples in Figure 3. (B–E) Schematic representation of individual DNA molecules remodeled by hACF. Bisulfite-converted DNAs from gel slices (black frames, lanes 6–9) were amplified by PCR, cloned and sequenced. Individual DNA clones are represented as described in Figure 3. (F) Frequency of methylation at a given CpG site. Upper panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panel E (reaction without ATP). Lower panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panels B–D (reactions with ATP). In both the upper and lower panels, frequencies obtained from the nucleosome substrate in the absence of remodeler (Figure 2E) are shown in grey for comparison.
Nucleosomes isolated from the most prominent species formed after remodeling showed regions of methylation protection mostly at central positions, but these positions were more heterogeneous than the starting material (compare Figure 2D to 4B–D; Figure 4F, lower panel). Nucleosomes isolated from the minor species formed after the remodeling reaction showed significant translational movement away from the central position. These data demonstrate that individual products from the hACF reaction are very different in position than products of the reaction catalyzed by isolated SNF2H, confirming previous conclusions based upon analysis of the entire reaction. We also noticed a slight increase (8%) in the number of nucleosomes with a protection between 121–145-bp, although less of these nucleosomes smaller than a canonical size were seen with hACF than with SNF2H (23%). Additionally, there was a bias toward moving the nucleosomes toward the ‘right’ side of the fragment, suggesting a sequence bias for this complex similar to that seen with isolated SNF2H.
BRG1 and hSWI/SNF create broadly distributed protection patterns that can be subnucleosomes in size
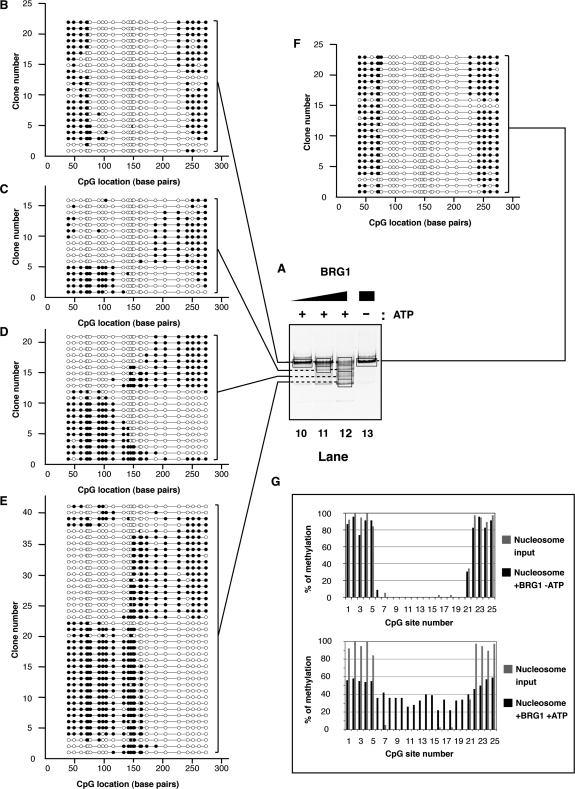
Several comparative studies suggest that the ISWI- and SWI/SNF-subfamilies perform different functions when remodeling nucleosomes (34,58–60). Specifically, the SWI/SNF family complexes can efficiently create access to sites internal to the starting nucleosome. To determine whether this is caused by creating products with DNA that has been ‘looped’ away from the histone octamer or products that are dramatically repositioned on fragment to open the central region, we examined approximately 100 individual species that had been remodeled by BRG1 or by hSWI/SNF.
Incubation of centrally-positioned M601 nucleosomes with the BRG1 enzyme without ATP did not lead to detectable changes in nucleosome mobility or nucleosomal DNA accessibility, under these conditions (Figure 5A, lane 13, F and G, upper panel). When ATP was present most nucleosomes were remodeled and the nucleosomal protection was translocated from the center to either end of the DNA fragment (Figure 5A, compare lane 13 to 10–12 and compare panel F to B–E). Interestingly, unlike observed with SNF2H, BRG1 did not show an obvious directional bias in relocating the nucleosomal protection (compare Figure 3C–E to Figure 5C–E). As a result, BRG1 generated relatively uniform overall accessibility as demonstrated by quantification of methylation across the entire population of nucleosomes (Figure 5G, lower panel). This observation is in agreement with our previous studies showing that BRG1 generates multiple remodeled products with different sequences of DNA exposed as measured by nuclease digestion (33). However, the use of the methyltransferase revealed that this overall DNA accessibility results from generating a broad spectrum of octamer positions compared to remodelers such as SNF2H, with no evidence of significant loop formation.
Figure 5.
(A) Native electrophoresis of BRG1 remodeled products. Nucleosomes (∼100 nM) were incubated with increasing concentrations of BRG1 (lane 10: 55 nM; lane 11: 170 nM; lanes 12 and 13: 500 nM) in the presence (lanes 10–12) or absence (lane 13) of ATP (1 mM) as indicated on top. Reactions were handled identically and in parallel to samples in Figure 3. (B–F) Schematic representation of individual DNA molecules remodeled by BRG1. Bisulfite-converted DNAs from gel slices (black frames, lanes 10–13) were amplified by PCR, cloned and sequenced. Individual DNA clones are represented as described in Figure 3. (G) Frequency of methylation at a given CpG site. Upper panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panel F (reaction without ATP). Lower panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panels B–E (reactions with ATP). In both the upper and lower panels, frequencies obtained from the nucleosome substrate in the absence of remodeler (Figure 2E) are shown in grey for comparison.
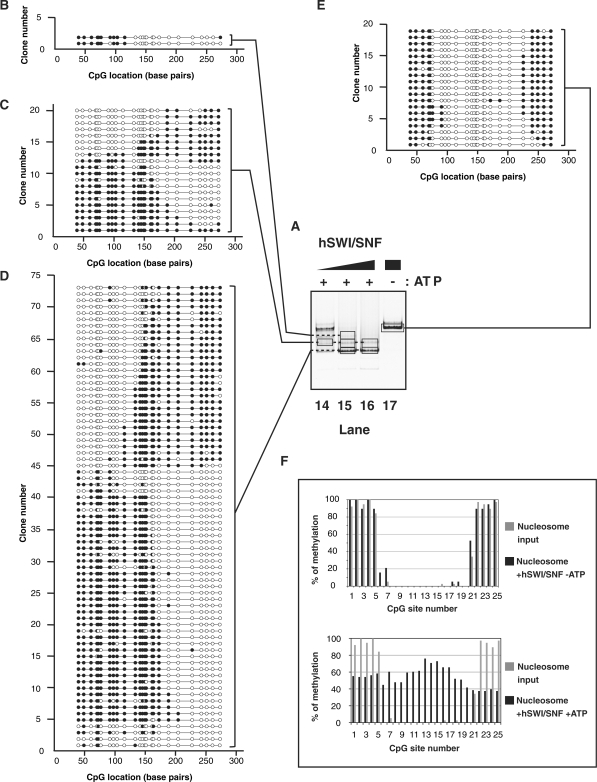
While BRG1 alone is able to remodel nucleosomes, the efficiency of remodeling is significantly greater with the entire hSWI/SNF complex (61). We therefore were interested in determining the spectrum of remodeled species created by hSWI/SNF and determining how it differed from the products created by BRG1. Addition of hSWI/SNF to the nucleosome in the absence of ATP noticeably increased methylation of the CpG sites #6 and #7 (by 16% and 21%, respectively) compared to unreacted nucleosomes. A similar increase occurred at the other entry/exit point of the nucleosome, at CpG #21 (from 34% to 53%; Figure 6A, lane 17, 6E and F, upper panel). Interestingly, this increase in accessibility was observed either on one or both sides of the same nucleosome (Figure 6E).
Figure 6.
(A) Native electrophoresis of hSWI/SNF remodeled products. Nucleosomes (∼100 nM) were incubated with increasing concentrations of hSWI/SNF complex (lane 14: 6 nM; lane 15: 19 nM; lanes 16 and 17: 56 nM) in the presence (lanes 14–16) or absence (lane 17) of ATP (1 mM) as indicated on top. Reactions were handled identically and in parallel to samples in Figure 3. (B–E) Schematic representation of individual DNA molecules remodeled by hSWI/SNF. Bisulfite-converted DNAs from gel slices (black frames, lanes 14–17) were amplified by PCR, cloned and sequenced. Individual DNA clones are represented as described in Figure 3. (F) Frequency of methylation at a given CpG site. Upper panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panel E (reaction without ATP). Lower panel: the frequency of methylation was determined by averaging methylation for all the DNA molecules showed in panels B–D (reactions with ATP). In both the upper and lower panels, frequencies obtained from the nucleosome substrate in the absence of remodeler (Figure 2E) are shown in grey for comparison.
Upon addition of ATP, the hSWI/SNF complex created a broad redistribution of the DNA protection (Figure 6A, lanes 14–16 and panels B–D). Notably, the size distribution of the protections contrasted with the distribution seen with SNF2H and hACF remodeled products. More than 60% of the molecules showed a protection smaller than 146-bp (17% between 96 and 120 bp, and 45% between 121 and 145-bp, Figure 7A). It was evident that hSWI/SNF is particularly efficient at exposing the sites which were around the pseudo-dyad axis of the starting positioned nucleosome (Figures 6F, lower panel and 7B). This resulted from the propensity of hSWI/SNF to move the nucleosome toward an end of the DNA fragment and to create a protected region smaller than observed with a canonical nucleosome. Interestingly, we noticed a qualitative difference between the remodeling activities of BRG1 and hSWI/SNF, as the ATPase alone was not able to create as many nucleosomes in the smallest size category as the complex. In fact, only about 30% of the BRG1 products harbored a protection smaller than 146-bp while hSWI/SNF generated twice as much of these products (compare Figures 5E and 6D, and see summary in Figure 7A). In addition to creating a greater spectrum of products, hSWI/SNF caused an approximate 3–4% increase in free DNA (Figure 6A, lane 16.) In conclusion, by reducing the size of the protection produced by the histones and by causing its widespread distribution (Figure 6C and D), the hSWI/SNF complex drastically increased the overall DNA accessibility along the entire nucleosomal DNA fragment when accessibility was determined on the average population (Figures 6F, lower panel and 7B). Similar results were obtained when whole reactions were analyzed prior to electrophoresis (data not shown), indicating that we had not missed a significant remodeled product by focusing our analysis on the major products seen on the native gels. Altogether, these data show that broad nucleosomal DNA accessibility can be achieved without creating stable loops on the histone octamer surface.
DISCUSSION
The ability to examine multiple single products generated by ATP-dependent chromatin remodeling proteins is useful because these complexes generate heterogeneous remodeled products. This heterogeneity has prevented a clear elucidation of the nature of the products of remodeling reactions. Previous characterization of the bulk products of remodeling reactions has led to several important conclusions: many remodelers can change the translational position of nucleosomes; ISWI-family remodeling complexes tend to create a product with characteristics of standard nucleosomes; SWI/SNF family remodeling complexes can remove nucleosomes from DNA and can create nucleosomal structures with altered characteristics and topology. By characterizing several hundred individual products of the remodeling reactions of two predominant families of remodeling factors, we were able to retrieve a wide range of information concerning directionality and sequence bias of nucleosome repositioning, size of the protection caused by the remodeled nucleosomes, and DNA accessibility on single molecules and in the population. Individual products of the SWI/SNF family were found to have an unusually short region of DNA protected by the histones. While ISWI complexes primarily produce species with the characteristics of a repositioned canonical nucleosome, they were also able to create species with shorter stretches of protected DNA than seen with canonical nucleosomes.
The characteristics of the remodeled products observed here confirm previous observations that complexes in the ISWI and SWI/SNF families produce different outcomes when remodeling nucleosomes, and expand upon the nature of those differences. Both families of complexes generated DNA protections that were more heterogeneous in size than the starting nucleosome. The greatest product heterogeneity was generated by the intact hSWI/SNF complex, while less heterogeneity was observed with hACF. In addition, the ISWI family reactions showed a distinct bias in the direction in which the nucleosome was moved, in contrast to the hSWI/SNF remodeling reactions. ISWI-related enzymes are known to show a directional bias that depends upon the length of DNA on either side of the nucleosome. However, the starting substrate was flanked by 77 and 80-bp while SNF2H and hACF are only able to discriminate DNA lengths that are shorter than 40 and 60-bp, respectively (57). Therefore, as observed previously (62), the ISWI enzymes likely show a preference for a particular DNA feature, as can hSWI/SNF on certain templates (63). Altogether, these results add further evidence that these two families of remodeling proteins harness the energy of ATP hydrolysis differently.
Our data validate, at the level of the individual products, previous studies showing that ISWI-family factors slide nucleosomes while leaving their structure intact (49–51). We found, however, that a proportion of individual SNF2H-remodeled nucleosomes exhibited smaller protections than expected for a canonical nucleosome. One potential explanation for this observation is that SNF2H might disrupt the interaction between DNA and the H3-αN′- and possibly the H2A C-terminal tails and affect the angles of DNA entry/exit of the nucleosome. As a result, the protection caused by the histones might be reduced. Previous studies suggest that canonical nucleosomes are partially unwrapped 2–10% of the time (64), an observation that might relate to our findings. However we did not observe significant shortening of the DNA protection in the absence of remodeling factor under our assay conditions. It is also possible that remodeling altered the phasing of the CpG dinucleotides such that they become more exposed to the DNA methyltransferase. In some instances, remodeling might have been efficient enough to move the histone octamer off of the end of the DNA, a model that has been proposed previously for SWI/SNF function.
hSWI/SNF remodeling mainly created molecules where the DNA protection was confined to either end of the DNA and most molecules harbored a protection size smaller than that of a canonical nucleosome. Canonical nucleosome core particles are defined by about 146-bp of DNA wrapped around a histone octamer in a left-handed superhelix (65). This DNA superhelix interacts with the histone octamer at 14 sites, referred to as superhelix locations (SHLs) −0.5 to −6.5 on one side and 0.5 to 6.5 on the other side of the nucleosome. In our experiments, the majority of starting mononucleosomes showed a protection between 146 and 170-bp while more than 60% of the SWI/SNF remodeled products only had between 96 and 145-bp protected (Figure 7A). Thus, our data demonstrates that hSWI/SNF can create products with significantly fewer histone–DNA contacts than the canonical structure.
This non-canonical structure explains how SWI/SNF can create broad access to DNA throughout the nucleosome. Previous studies had indicated that smaller MNase resistant fragments are produced following BRG1 remodeling and it had been proposed that this might have resulted from stable loops of DNA on the surface of the histone octamer (33,34). If such loops were common, then many octamers would be expected to harbor protected regions of DNA in two or more stretches. Instead, visualization of the individual species shows that a majority of these remodeled species have little protected DNA and indicates no evidence of loop formation, as we did not observed additional patches of protection. Importantly, we note that averaging the methylation patterns we obtained recapitulates the previously observed overall DNA accessibility on the remodeled products. This provides support to the notion that our procedure does not lead to missing accessible structures. Furthermore, quantification of DNA accessibility on the individual hSWI/SNF remodeled mononucleosomes explains why broad accessibility was observed in these preceding studies using nucleases after remodeling. Importantly, the data rules out the necessity for looping to allow access to internal sites (Figure 7B).
There are two main categories of changes that could explain why less DNA is protected in the hSWI/SNF remodeled products compared with canonical nucleosomes. First, the conformation or the integrity (or both) of the histone octamer might be altered, resulting in a different path of the DNA superhelix around the histones. Second, the octamer might remain unchanged with the DNA path changing or moving to create lowered protection. Regarding the first possibility, very little is known as to whether the histone octamer can adopt alternative (non-canonical) conformations when bound to DNA. A recent study proposed that upon torsional stress, nucleosomes can switch from a left-handed to a right-handed conformation [these particles were termed ‘reversomes’ (66)]. While it is known that SWI/SNF remodeling causes significant topological changes in chromatin, it is not known whether this complex could catalyze such a chiral transition. In previous studies, Allfrey and colleagues hypothesized that nucleosomes could be converted into a U-shaped structure called a ‘lexosome’ in order to facilitate transcription. This model was based on observations that the two histone H3 cysteine residues (at position 110 which are normally inaccessible in a nucleosome) were found to be reactive to thiol-reagents in transcribed rDNA genes. However, this structure was later shown to entail a histone loss (67,68). Further investigations are needed to address the significance of such altered structures and their relevance to remodeling.
Changes to the histone octamer that would decrease histone contacts include dissociation of one or two (H2A–H2B) dimers to produce hexamers or tetramers. Earlier studies imply that octamer disruption is not required for SWI/SNF-family enzymes to alter nucleosomal DNA accessibility and dissociation of the histone octamer structure is not a necessary consequence of remodeling (23,28,69). Yet, (H2A–H2B) dimer loss can occur upon remodeling (23,70) although it may be DNA sequence-dependent or require the presence of a histone chaperone (71–73). Histone dimer loss has not been observed using the 601 DNA sequence from which we derived the construct used in our study (37). The remodeled products that are generated in our study appear to maintain an intact octamer (as judged by staining of the histones, data not shown) at the low ionic strength (50 mM NaCl) used.
If the histone octamer remains intact and in its normal configuration in the remodeled products, previous studies imply that the DNA must maintain contact with each of the core histones to maintain this stability; otherwise the (H2A–H2B) dimers are not expected to be stably associated to the (H3–H4)2 tetramer under these ionic conditions (74). Several models for an altered DNA path have been proposed. As we mainly observed smaller regions of protection located at the end of the DNA fragment, our data are consistent with models in which a canonical histone octamer has been ‘pushed’ off of the end of the DNA (23,24,27,75). These models imply that charged surfaces on the histone octamer become exposed after remodeling. These could bind to stretches of free DNA either in cis (intramolecularly) or in trans (intermolecularly). If DNA was bound to these exposed surfaces in cis, the DNA would loop back, whereas if DNA was bound in trans, the binding would promote the formation of nucleosome dimers (17,18,21,23,75). In the latter case, if this DNA were already associated with histones, the observed protection would not be altered on the individual molecules examined here. Otherwise, looping back would be expected to create a protection at the DNA end opposite to where the nucleosome is located. Our data suggest that this is either rare, transient or that less than 40-bp (the closest methylation site from a DNA end) of the looped-back DNA is bound to the histones. Since 94-bp (but not 90-bp) of DNA are sufficient to assemble stable sub-nucleosomal particles (76), it is tempting to speculate that about 47-bp of DNA on one side of the nucleosome would be sufficient to preserve the octamer structure if the other half of the octamer were bound with DNA. This latter consideration would indicate that the octamer could be ‘pushed off’ of the end of the DNA up to the SHL −4.5 or +4.5 without histone dimer loss.
To address these issues further it would be useful to use methylation to study DNA accessibility on arrays of nucleosomes. This would allow exploring the possibility that SWI/SNF has the potential to force nucleosomes to invade each other (77) or lead to the formation of novel species containing more than one octamer (22). We have investigated the feasibility of this approach using defined trinucleosomal templates. Preliminary results obtained after ATP-dependent remodeling of a defined trinucleosome demonstrate that changes in patterns of accessibility can be detected on these longer individual molecules (data not shown). Significant work will be needed to interpret these patterns. Nevertheless, the technology described here can be used on mononucleosomes or extended to the use of longer nucleosomal arrays. This approach should prove useful in further mechanistic studies of all classes of complexes that impact nucleosome structure, not just those involved in remodeling.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
American Cancer Society postdoctoral fellowship (to T.B.M.); National Institutes of Health grant CA82422 (to P.A.J.); National Institutes of Health grant GM48045 (to R.E.K.). Funding for open access charge: GM48045.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Joe Garlick for excellent technical assistance with protein expression, Jerome Déjardin, Matthew D. Simon, Jonathan H. Dennis and Caroline J. Woo for critical reading of the manuscript, Lance Davidow and John Morris for helping with initial bioinformatics analyzes. We are grateful to Mark A. Miranda for creating the CpG Bubble Chart Generator Program.
REFERENCES
- 1.Taylor IC, Workman JL, Schuetz TJ, Kingston RE. Facilitated binding of GAL4 and heat shock factor to nucleosomal templates: differential function of DNA-binding domains. Genes Dev. 1991;5:1285–1298. doi: 10.1101/gad.5.7.1285. [DOI] [PubMed] [Google Scholar]
- 2.Hayes JJ, Wolffe AP. The interaction of transcription factors with nucleosomal DNA. Bioessays. 1992;14:597–603. doi: 10.1002/bies.950140905. [DOI] [PubMed] [Google Scholar]
- 3.Gronbaek K, Hother C, Jones PA. Epigenetic changes in cancer. APMIS. 2007;115:1039–1059. doi: 10.1111/j.1600-0463.2007.apm_636.xml.x. [DOI] [PubMed] [Google Scholar]
- 4.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 5.van Vugt JJ, Ranes M, Campsteijn C, Logie C. The ins and outs of ATP-dependent chromatin remodeling in budding yeast: biophysical and proteomic perspectives. Biochim. Biophys. Acta. 2007;1769:153–171. doi: 10.1016/j.bbaexp.2007.01.013. [DOI] [PubMed] [Google Scholar]
- 6.Racki LR, Narlikar GJ. ATP-dependent chromatin remodeling enzymes: two heads are not better, just different. Curr. Opin. Genet. Dev. 2008;18:137–144. doi: 10.1016/j.gde.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eisen JA, Sweder KS, Hanawalt PC. Evolution of the SNF2 family of proteins: subfamilies with distinct sequences and functions. Nucleic Acids Res. 1995;23:2715–2723. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Identification of multiple distinct Snf2 subfamilies with conserved structural motifs. Nucleic Acids Res. 2006;34:2887–2905. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boyer LA, Logie C, Bonte E, Becker PB, Wade PA, Wolffe AP, Wu C, Imbalzano AN, Peterson CL. Functional delineation of three groups of the ATP-dependent family of chromatin remodeling enzymes. J. Biol. Chem. 2000;275:18864–18870. doi: 10.1074/jbc.M002810200. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Bulger M, Pazin MJ, Kobayashi R, Kadonaga JT. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell. 1997;90:145–155. doi: 10.1016/s0092-8674(00)80321-9. [DOI] [PubMed] [Google Scholar]
- 11.Varga-Weisz PD, Wilm M, Bonte E, Dumas K, Mann M, Becker PB. Chromatin-remodelling factor CHRAC contains the ATPases ISWI and topoisomerase II. Nature. 1997;388:598–602. doi: 10.1038/41587. [DOI] [PubMed] [Google Scholar]
- 12.Cote J, Quinn J, Workman JL, Peterson CL. Stimulation of GAL4 derivative binding to nucleosomal DNA by the yeast SWI/SNF complex. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 13.Kwon H, Imbalzano AN, Khavari PA, Kingston RE, Green MR. Nucleosome disruption and enhancement of activator binding by a human SW1/SNF complex. Nature. 1994;370:477–481. doi: 10.1038/370477a0. [DOI] [PubMed] [Google Scholar]
- 14.Imbalzano AN, Kwon H, Green MR, Kingston RE. Facilitated binding of TATA-binding protein to nucleosomal DNA. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 15.Logie C, Peterson CL. Catalytic activity of the yeast SWI/SNF complex on reconstituted nucleosome arrays. EMBO J. 1997;16:6772–6782. doi: 10.1093/emboj/16.22.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Imbalzano AN, Schnitzler GR, Kingston RE. Nucleosome disruption by human SWI/SNF is maintained in the absence of continued ATP hydrolysis. J. Biol. Chem. 1996;271:20726–20733. doi: 10.1074/jbc.271.34.20726. [DOI] [PubMed] [Google Scholar]
- 17.Schnitzler G, Sif S, Kingston RE. Human SWI/SNF interconverts a nucleosome between its base state and a stable remodeled state. Cell. 1998;94:17–27. doi: 10.1016/s0092-8674(00)81217-9. [DOI] [PubMed] [Google Scholar]
- 18.Lorch Y, Cairns BR, Zhang M, Kornberg RD. Activated RSC-nucleosome complex and persistently altered form of the nucleosome. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 19.Phelan ML, Schnitzler GR, Kingston RE. Octamer transfer and creation of stably remodeled nucleosomes by human SWI-SNF and its isolated ATPases. Mol. Cell Biol. 2000;20:6380–6389. doi: 10.1128/mcb.20.17.6380-6389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lorch Y, Zhang M, Kornberg RD. Histone octamer transfer by a chromatin-remodeling complex. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 21.Ulyanova NP, Schnitzler GR. Inverted factor access and slow reversion characterize SWI/SNF-altered nucleosome dimers. J. Biol. Chem. 2007;282:1018–1028. doi: 10.1074/jbc.M609473200. [DOI] [PubMed] [Google Scholar]
- 22.Ulyanova NP, Schnitzler GR. Human SWI/SNF generates abundant, structurally altered dinucleosomes on polynucleosomal templates. Mol. Cell Biol. 2005;25:11156–11170. doi: 10.1128/MCB.25.24.11156-11170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kassabov SR, Zhang B, Persinger J, Bartholomew B. SWI/SNF unwraps, slides, and rewraps the nucleosome. Mol. Cell. 2003;11:391–403. doi: 10.1016/s1097-2765(03)00039-x. [DOI] [PubMed] [Google Scholar]
- 24.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling through directional DNA translocation from an internal nucleosomal site. Nat. Struct. Mol. Biol. 2005;12:747–755. doi: 10.1038/nsmb973. [DOI] [PubMed] [Google Scholar]
- 25.Jaskelioff M, Gavin IM, Peterson CL, Logie C. SWI-SNF-mediated nucleosome remodeling: role of histone octamer mobility in the persistence of the remodeled state. Mol. Cell Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schnitzler GR, Cheung CL, Hafner JH, Saurin AJ, Kingston RE, Lieber CM. Direct imaging of human SWI/SNF-remodeled mono- and polynucleosomes by atomic force microscopy employing carbon nanotube tips. Mol. Cell Biol. 2001;21:8504–8511. doi: 10.1128/MCB.21.24.8504-8511.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flaus A, Owen-Hughes T. Dynamic properties of nucleosomes during thermal and ATP-driven mobilization. Mol. Cell Biol. 2003;23:7767–7779. doi: 10.1128/MCB.23.21.7767-7779.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bazett-Jones DP, Cote J, Landel CC, Peterson CL, Workman JL. The SWI/SNF complex creates loop domains in DNA and polynucleosome arrays and can disrupt DNA-histone contacts within these domains. Mol. Cell Biol. 1999;19:1470–1478. doi: 10.1128/mcb.19.2.1470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang Y, Smith CL, Saha A, Grill SW, Mihardja S, Smith SB, Cairns BR, Peterson CL, Bustamante C. DNA translocation and loop formation mechanism of chromatin remodeling by SWI/SNF and RSC. Mol. Cell. 2006;24:559–568. doi: 10.1016/j.molcel.2006.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lia G, Praly E, Ferreira H, Stockdale C, Tse-Dinh YC, Dunlap D, Croquette V, Bensimon D, Owen-Hughes T. Direct observation of DNA distortion by the RSC complex. Mol. Cell. 2006;21:417–425. doi: 10.1016/j.molcel.2005.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lorch Y, Zhang M, Kornberg RD. RSC unravels the nucleosome. Mol. Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 32.Guyon JR, Narlikar GJ, Sif S, Kingston RE. Stable remodeling of tailless nucleosomes by the human SWI–SNF complex. Mol. Cell Biol. 1999;19:2088–2097. doi: 10.1128/mcb.19.3.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narlikar GJ, Phelan ML, Kingston RE. Generation and interconversion of multiple distinct nucleosomal states as a mechanism for catalyzing chromatin fluidity. Mol. Cell. 2001;8:1219–1230. doi: 10.1016/s1097-2765(01)00412-9. [DOI] [PubMed] [Google Scholar]
- 34.Fan HY, He X, Kingston RE, Narlikar GJ. Distinct strategies to make nucleosomal DNA accessible. Mol. Cell. 2003;11:1311–1322. doi: 10.1016/s1097-2765(03)00192-8. [DOI] [PubMed] [Google Scholar]
- 35.Flaus A, Owen-Hughes T. Mechanisms for ATP-dependent chromatin remodelling: farewell to the tuna-can octamer? Curr. Opin. Genet. Dev. 2004;14:165–173. doi: 10.1016/j.gde.2004.01.007. [DOI] [PubMed] [Google Scholar]
- 36.Cairns BR. Chromatin remodeling: insights and intrigue from single-molecule studies. Nat. Struct. Mol. Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shundrovsky A, Smith CL, Lis JT, Peterson CL, Wang MD. Probing SWI/SNF remodeling of the nucleosome by unzipping single DNA molecules. Nat. Struct. Mol. Biol. 2006;13:549–554. doi: 10.1038/nsmb1102. [DOI] [PubMed] [Google Scholar]
- 38.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 39.Sif S, Stukenberg PT, Kirschner MW, Kingston RE. Mitotic inactivation of a human SWI/SNF chromatin remodeling complex. Genes Dev. 1998;12:2842–2851. doi: 10.1101/gad.12.18.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shiraishi M, Hayatsu H. High-speed conversion of cytosine to uracil in bisulfite genomic sequencing analysis of DNA methylation. DNA Res. 2004;11:409–415. doi: 10.1093/dnares/11.6.409. [DOI] [PubMed] [Google Scholar]
- 41.Bock C, Reither S, Mikeska T, Paulsen M, Walter J, Lengauer T. BiQ Analyzer: visualization and quality control for DNA methylation data from bisulfite sequencing. Bioinformatics. 2005;21:4067–4068. doi: 10.1093/bioinformatics/bti652. [DOI] [PubMed] [Google Scholar]
- 42.Kladde MP, Simpson RT. Positioned nucleosomes inhibit Dam methylation in vivo. Proc. Natl Acad. Sci. USA. 1994;91:1361–1365. doi: 10.1073/pnas.91.4.1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kladde MP, Simpson RT. Chromatin structure mapping in vivo using methyltransferases. Methods Enzymol. 1996;274:214–233. doi: 10.1016/s0076-6879(96)74019-3. [DOI] [PubMed] [Google Scholar]
- 44.Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL. A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc. Natl Acad. Sci. USA. 1992;89:1827–1831. doi: 10.1073/pnas.89.5.1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fatemi M, Pao MM, Jeong S, Gal-Yam EN, Egger G, Weisenberger DJ, Jones PA. Footprinting of mammalian promoters: use of a CpG DNA methyltransferase revealing nucleosome positions at a single molecule level. Nucleic Acids Res. 2005;33:e176. doi: 10.1093/nar/gni180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gal-Yam EN, Jeong S, Tanay A, Egger G, Lee AS, Jones PA. Constitutive nucleosome depletion and ordered factor assembly at the GRP78 promoter revealed by single molecule footprinting. PLoS Genet. 2006;2:e160. doi: 10.1371/journal.pgen.0020160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin JC, Jeong S, Liang G, Takai D, Fatemi M, Tsai YC, Egger G, Gal-Yam EN, Jones PA. Role of nucleosomal occupancy in the epigenetic silencing of the MLH1 CpG island. Cancer Cell. 2007;12:432–444. doi: 10.1016/j.ccr.2007.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Appanah R, Dickerson DR, Goyal P, Groudine M, Lorincz MC. An unmethylated 3′ promoter-proximal region is required for efficient transcription initiation. PLoS Genet. 2007;3:e27. doi: 10.1371/journal.pgen.0030027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamiche A, Sandaltzopoulos R, Gdula DA, Wu C. ATP-dependent histone octamer sliding mediated by the chromatin remodeling complex NURF. Cell. 1999;97:833–842. doi: 10.1016/s0092-8674(00)80796-5. [DOI] [PubMed] [Google Scholar]
- 50.Langst G, Bonte EJ, Corona DF, Becker PB. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell. 1999;97:843–852. doi: 10.1016/s0092-8674(00)80797-7. [DOI] [PubMed] [Google Scholar]
- 51.Kassabov SR, Henry NM, Zofall M, Tsukiyama T, Bartholomew B. High-resolution mapping of changes in histone-DNA contacts of nucleosomes remodeled by ISW2. Mol. Cell Biol. 2002;22:7524–7534. doi: 10.1128/MCB.22.21.7524-7534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tsukiyama T, Palmer J, Landel CC, Shiloach J, Wu C. Characterization of the imitation switch subfamily of ATP-dependent chromatin-remodeling factors in Saccharomyces cerevisiae. Genes Dev. 1999;13:686–697. doi: 10.1101/gad.13.6.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Loyola A, Huang JY, LeRoy G, Hu S, Wang YH, Donnelly RJ, Lane WS, Lee SC, Reinberg D. Functional analysis of the subunits of the chromatin assembly factor RSF. Mol. Cell Biol. 2003;23:6759–6768. doi: 10.1128/MCB.23.19.6759-6768.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vary JC, Jr, Gangaraju VK, Qin J, Landel CC, Kooperberg C, Bartholomew B, Tsukiyama T. Yeast Isw1p forms two separable complexes in vivo. Mol. Cell Biol. 2003;23:80–91. doi: 10.1128/MCB.23.1.80-91.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He X, Fan HY, Narlikar GJ, Kingston RE. Human ACF1 alters the remodeling strategy of SNF2H. J. Biol. Chem. 2006;281:28636–28647. doi: 10.1074/jbc.M603008200. [DOI] [PubMed] [Google Scholar]
- 56.He X, Fan HY, Garlick JD, Kingston RE. Diverse regulation of SNF2H chromatin remodeling by noncatalytic subunits. Biochemistry. 2008;47:7025–7033. doi: 10.1021/bi702304p. [DOI] [PubMed] [Google Scholar]
- 57.Yang JG, Madrid TS, Sevastopoulos E, Narlikar GJ. The chromatin-remodeling enzyme ACF is an ATP-dependent DNA length sensor that regulates nucleosome spacing. Nat. Struct. Mol. Biol. 2006;13:1078–1083. doi: 10.1038/nsmb1170. [DOI] [PubMed] [Google Scholar]
- 58.Aalfs JD, Narlikar GJ, Kingston RE. Functional differences between the human ATP-dependent nucleosome remodeling proteins BRG1 and SNF2H. J. Biol. Chem. 2001;276:34270–34278. doi: 10.1074/jbc.M104163200. [DOI] [PubMed] [Google Scholar]
- 59.Fan HY, Trotter KW, Archer TK, Kingston RE. Swapping function of two chromatin remodeling complexes. Mol. Cell. 2005;17:805–815. doi: 10.1016/j.molcel.2005.02.024. [DOI] [PubMed] [Google Scholar]
- 60.Zofall M, Persinger J, Kassabov SR, Bartholomew B. Chromatin remodeling by ISW2 and SWI/SNF requires DNA translocation inside the nucleosome. Nat. Struct. Mol. Biol. 2006;13:339–346. doi: 10.1038/nsmb1071. [DOI] [PubMed] [Google Scholar]
- 61.Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- 62.Rippe K, Schrader A, Riede P, Strohner R, Lehmann E, Langst G. DNA sequence- and conformation-directed positioning of nucleosomes by chromatin-remodeling complexes. Proc. Natl Acad. Sci. USA. 2007;104:15635–15640. doi: 10.1073/pnas.0702430104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Sims HI, Lane JM, Ulyanova NP, Schnitzler GR. Human SWI/SNF drives sequence-directed repositioning of nucleosomes on C-myc promoter DNA minicircles. Biochemistry. 2007;46:11377–11388. doi: 10.1021/bi7008823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li G, Widom J. Nucleosomes facilitate their own invasion. Nat. Struct. Mol. Biol. 2004;11:763–769. doi: 10.1038/nsmb801. [DOI] [PubMed] [Google Scholar]
- 65.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 66.Bancaud A, Wagner G, Conde ESN, Lavelle C, Wong H, Mozziconacci J, Barbi M, Sivolob A, Le Cam E, Mouawad L, et al. Nucleosome chiral transition under positive torsional stress in single chromatin fibers. Mol. Cell. 2007;27:135–147. doi: 10.1016/j.molcel.2007.05.037. [DOI] [PubMed] [Google Scholar]
- 67.Lavelle C, Prunell A. Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle. 2007;6:2113–2119. doi: 10.4161/cc.6.17.4631. [DOI] [PubMed] [Google Scholar]
- 68.Locklear L, Jr, Ridsdale JA, Bazett-Jones DP, Davie JR. Ultrastructure of transcriptionally competent chromatin. Nucleic Acids Res. 1990;18:7015–7024. doi: 10.1093/nar/18.23.7015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Boyer LA, Shao X, Ebright RH, Peterson CL. Roles of the histone H2A–H2B dimers and the (H3-H4)(2) tetramer in nucleosome remodeling by the SWI–SNF complex. J. Biol. Chem. 2000;275:11545–11552. doi: 10.1074/jbc.275.16.11545. [DOI] [PubMed] [Google Scholar]
- 70.Bruno M, Flaus A, Stockdale C, Rencurel C, Ferreira H, Owen-Hughes T. Histone H2A/H2B dimer exchange by ATP-dependent chromatin remodeling activities. Mol. Cell. 2003;12:1599–1606. doi: 10.1016/s1097-2765(03)00499-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Vicent GP, Nacht AS, Smith CL, Peterson CL, Dimitrov S, Beato M. DNA instructed displacement of histones H2A and H2B at an inducible promoter. Mol. Cell. 2004;16:439–452. doi: 10.1016/j.molcel.2004.10.025. [DOI] [PubMed] [Google Scholar]
- 72.Yang X, Zaurin R, Beato M, Peterson CL. Swi3p controls SWI/SNF assembly and ATP-dependent H2A–H2B displacement. Nat. Struct. Mol. Biol. 2007;14:540–547. doi: 10.1038/nsmb1238. [DOI] [PubMed] [Google Scholar]
- 73.Lorch Y, Maier-Davis B, Kornberg RD. Chromatin remodeling by nucleosome disassembly in vitro. Proc. Natl Acad. Sci. USA. 2006;103:3090–3093. doi: 10.1073/pnas.0511050103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Eickbush TH, Moudrianakis EN. The histone core complex: an octamer assembled by two sets of protein–protein interactions. Biochemistry. 1978;17:4955–4964. doi: 10.1021/bi00616a016. [DOI] [PubMed] [Google Scholar]
- 75.Ramachandran A, Omar M, Cheslock P, Schnitzler GR. Linker histone H1 modulates nucleosome remodeling by human SWI/SNF. J. Biol. Chem. 2003;278:48590–48601. doi: 10.1074/jbc.M309033200. [DOI] [PubMed] [Google Scholar]
- 76.Tatchell K, Van Holde KE. Nucleosome reconstitution: effect of DNA length on nuclesome structure. Biochemistry. 1979;18:2871–2880. doi: 10.1021/bi00580a031. [DOI] [PubMed] [Google Scholar]
- 77.Engeholm M, de Jager M, Flaus A, Brenk R, van Noort J, Owen-Hughes T. Nucleosomes can invade DNA territories occupied by their neighbors. Nat. Struct. Mol. Biol. 2009;16:151–158. doi: 10.1038/nsmb.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.