Abstract
16S rRNA gene is one of the preferred targets for resolving species phylogenesis issues in microbiological-related contexts. However, the identification of single-nucleotide variations capable of distinguishing a sequence among a set of homologous ones can be problematic. Here we present ORMA (Oligonucleotide Retrieving for Molecular Applications), a set of scripts for discriminating positions search and for performing the selection of high-quality oligonucleotide probes to be used in molecular applications. Two assays based on Ligase Detection Reaction (LDR) are presented. First, a new set of probe pairs on cyanobacteria 16S rRNA sequences of 18 different species was compared to that of a previous study. Then, a set of LDR probe pairs for the discrimination of 13 pathogens contaminating bovine milk was evaluated. The software determined more than 100 candidate probe pairs per dataset, from more than 300 16S rRNA sequences, in less than 5 min. Results demonstrated how ORMA improved the performance of the LDR assay on cyanobacteria, correctly identifying 12 out of 14 samples, and allowed the perfect discrimination among the 13 milk pathogenic-related species. ORMA represents a significant improvement from other contexts where enzyme-based techniques have been employed on already known mutations of a single base or on entire subsequences.
INTRODUCTION
During the last decades, different nucleic-acid-based detection techniques have been developed in order to employ identification based on single-nucleotide variations in both genotyping and detection experiments on a multiplicity of targets. These techniques allowed distinguishing alleles and correctly assessing the genotype at the single-base level.
In particular, 16S rRNA gene sequences have been used to resolve bacterial phylogeny and taxonomy issues in different contexts. The DNA sequence coding for the small ribosomal subunit has been by far the most common genetic marker employed by the scientific community, because of: (i) its presence in almost all bacteria, often existing as a multi-gene family, or operons; (ii) the function of the 16S rRNA gene has not changed over time, suggesting that random sequence changes are an accurate measure of time (evolution); and (iii) the 16S rRNA gene (more than 1500 bp) is large enough for informatics purposes (1) with large stretches of conserved regions and few different loci.
DNA microarrays represent one of the most popular platforms in molecular technologies, allowing a high-throughput format for the parallel detection of 16S rRNA genes from environmental samples (2). DNA chips have been developed as a preferred device for the identification of different microorganisms based on 16S gene sequences. The multiplicity of species which can be arrayed on a single-DNA chip allows a high multiplexing capability, with the possibility of identifying many different targets at one time (3). Single-base variations by microarray analysis can be detected by differential hybridization techniques using allele-specific oligonucleotide probes (4), or by enzyme-mediated detection methods (5). One of the most critical points of the molecular recognition procedures is the design of the specific probes needed to perform the entire analysis. In genotyping experiments, this is accomplished on the basis of the already-known information about each single-base variation. In detection experiments, on the other hand, in order to explore whether a certain target sequence is present in a DNA sample or not, the main problem is searching for a priori not yet identified specific positions that can discriminate exactly between one target and another.
In hybridization-based techniques, mutations are identified on the basis of the higher thermal stability of the perfectly-matched probes as compared to mismatched probes. Although this has been the most frequently applied technique, it is characterized by many hindrances which make hybridization-based strategy function poorly in high-complexity biological samples. Therefore, for analytical and diagnostic purposes, hybridization is generally combined with some other selection or enrichment procedures. Enzyme-mediated ligation methods, on the other hand, rely on interrogation of a mutation by a couple of oligonucleotides annealing immediately adjacent to each other on a target DNA, with one of the probes having its 3′-end complementary to the point mutation. In this case, the search is for a single base that characterizes a species against all the others in a group of interest. The presence of a point mutation is assessed by the ligation of the two adjacent oligonucleotides, which occurs only when both are correctly base-paired (6). The Ligation Detection Reaction (LDR) (7), for instance, represents a reliable technique for identifying one or more sequences differing by one or more single-base changes, insertions, deletions, or translocations in a plurality of target-nucleotide sequences. This enzymatic in vitro reaction is based on the design of two oligonucleotide probes for each target sequence: a probe specific for the variation (called ‘Discriminating Probe’, or DS), which is 5′-fluorescently labeled, and a 5′-phosphorylated ‘Common Probe’ (or CP), starting one base 3′-downstream of the DS. The previously polymerase chain reaction (PCR)-amplified sample, the oligonucleotide probe pairs and a thermostable DNA ligase are blended to form a mixture: the two probes hybridize consecutively along the template and the DNA ligase joins their ends only in the case of a perfect match. This reaction is cycled to increase product yield. The PCR–LDR approach, usually, is associated to the hybridization onto a Universal Array (UA), where a set of artificial sequences, called Zip-codes are arranged (7). This entire approach was proven to be rapid, flexible and easily adaptable from one target to another, useful, for example, in environmental monitoring (8,9), forensics (10) and the food industry (11,12).
Here we present Oligonucleotide Retrieving for Molecular Applications (ORMA), a series of integrated scripts in Matlab, which performs an accurate search of all the positions able to specifically discriminate one species among homologous ones, based on the 16S rRNA gene sequence. ORMA also performs an accurate selection of high-quality oligonucleotide probes to be used in molecular applications. Automated and computer-based methods can be very useful for performing accurately and rapidly all the requested operations, through the many steps between the original, complete, set of sequences and the final list of application-oriented probes.
The problem of designing specific oligonucleotide probes for the identification of target species has already been addressed by a certain number of software (13–16). At present, there is no preferential reference strategy for designing microarrays for species identification based on 16S rRNA sequences: many authors rely on academic software (17,18), others develop their own scripts (19,20). Among the currently available academic software, ARB (21) and PRIMROSE (22) are very diffused, both being tools implemented specifically on 16S rRNA, structured for interacting with and retrieving sequences from specific databases and operating a probe design on the basis of the phylogenesis of the species under analysis. Also, some commercial software, like Oligo 7 (Molecular Biology Insights, Cascade, CO, USA) (23) or AlleleID (Premier Biosoft, Palo Alto, CA, USA) (24) have been applied for probe design in a pathogen characterization experiment (25). In this article ORMA was used for determining sets of LDR probe pairs in microbiological-related contexts (water safety and food safety applications, respectively). The approach was evaluated and validated using the probe pairs derived from ORMA-determined discriminating positions on a set of cyanobacteria 16S rRNA sequences belonging to 18 different species; the results were compared to those of a previously published study (8). Secondly, a set of LDR probe pairs for the discrimination of 13 mastitis- or intoxication-related pathogens species in bovine milk was designed and experimentally evaluated. The tool, although here applied on 16S rRNA, can be used on any set of highly correlated sequences.
MATERIALS AND METHODS
Algorithm
ORMA scripts were developed under Matlab 6.1 (Mathworks, Natick, MA, USA) environment (Release 12.1). No additional toolboxes are required. All statistical analyses and representations were made by the same software. Probe designs and simulations were run onto a hp Workstation xw4100, with a Dual-core 3.2 GHz. Intel Processor and 2.5 GB RAM. ORMA functions and m-code are available for free upon request.
Overall structure
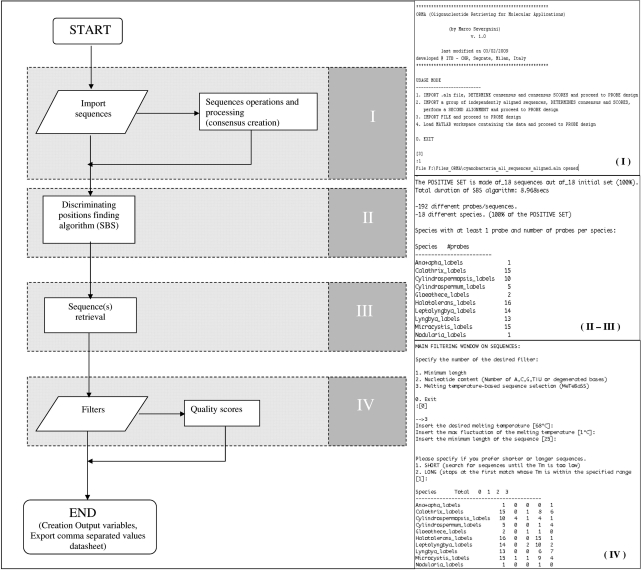
ORMA overall structure is tree-like, with a main function that, sequentially, recalls all the side scripts needed to perform each requested operation. The software also comprises a series of scripts for retrieving oligonucleotide sequences, quality-check them and design probes for different applications, such as Ligase Detection Reaction (LDR) or Minisequencing/Primer Extension probes. The overall procedure is accomplished in four main steps (Figure 1, Supplementary Figure 1): (i) sequence importing and processing; (ii) discriminating positions finding; (iii) designing of the candidate probes, starting from the positions found and (iv) ranking (i.e. assignment of a quality score to each) and exporting of the candidates (in tabular format).
Figure 1.
Block diagram representing the steps through which ORMA works. The four steps described in the main text are highlighted in gray: (I) Sequence importing and consensus creation; (II) Search of the discriminating positions by SBS algorithm; (III) Retrieval of the candidate sequences from the found positions. The actual design depends on the molecular application chosen; (IV) Quality filtering and ranking of the candidate probes. On the right, in boxes, example screenshots (probe pair design on cyanobacteria dataset) are given for each step. Steps (II) and (III) are indistinguishable in ORMA output and have been represented together. Please note that for visualization purposes only a part of the total 18 sequences are represented.
(i) Sequence import and processing
The search for discriminating positions on 16S rRNA starts from the import of a set of already-aligned sequences (which can be optionally used for the creation of consensus sequences, grouping them in homogeneous clusters, before being used for the discriminating position search algorithm). Standard multiple-alignment formats (Clustal-like, Multi Sequence Files, or aligned FASTA format) can be used. A careful check of multiple alignment scores should be made, in order to avoid designs on sequence datasets of distantly related species, which can occur in base misalignments. The scripts also include a procedure for consensus determination from a set of user-defined sequences, according to four different rules: (a) majority rule, in which the consensus base is the most frequently present in the aligned sequences and no degenerated bases are used. In case of equal occurrences, ‘N's are used in the consensus; (b) threshold rule ‘simple’, which assigns a specific base to the corresponding position in the consensus only if its frequency is above a given threshold. Different thresholds can be set for gaps and bases. Degenerated bases are not used and are substituted by ‘N's in the consensus; (c) threshold rule ‘complex’, which comprises also degenerated bases. The algorithm is the same as point (b) option, but requires a threshold for substituting positions with multiple bases above the threshold with the corresponding IUPAC code degenerated base and (d) ARB-like algorithm, with separate thresholds for gaps and bases. All the bases above the given threshold are used to compute eventual degenerated bases.
For each of these four options, consensus score accuracy is calculated, as the percentage of original sequences that carried the same base as the consensus in each position.
(ii–iii) Design of candidate probes
We have implemented a Single Base Seeker (SBS) algorithm, for the determination of positions able to discriminate one sequence among a set of homologous ones. The discriminating position finding procedure can be summarized as follows in four basic steps: (a) Choice of a user-defined subset of sequences of the dataset (indicated as the ‘positive set’). The remaining sequences are used as a group of the discriminating positions must be different from; these are addressed, in the present article, as the ‘negative set’. ‘Positive’ and ‘Negative’ sets differ for the fact that every consensus in the ‘positive set’ group will be subjected to probe design, whereas those of the ‘negative set’ will not; (b) For each sequence, determination of a list of the positions of non-degenerated bases; (c) For each position on point b, calculation of a score as the sum of all the sequences carrying the same base as the considered sequence, in the same position. If the only sequence carrying the base is the tested one, the position is set as discriminating and (d) Re-calculation of the score on point c, substituting to each (eventual) degenerated base its two or three alternatives (an ‘N’ automatically flags the position as non-discriminating).
ORMA, then, retrieves the sequences flanking each of the putative discriminating positions. Actual oligonucleotide design is dependent on the molecular application chosen. The maximum length and the thermodynamic model for calculation of the parameters of the probes can be specified by the user. For the LDR experiments here described, two oligonucleotide probes are designed, one upstream (Discriminating Probe, DS, comprising the discriminating position) and one downstream (Common Probe, CP) of each position.
(iv) Discriminating position related filters and scores
The putative discriminating positions and related candidate probes are subjected to a series of constraints and quality filters. The software keeps track of all the designed candidates, assigning a quality score, depending on how many filters they pass. The current options of the script on the discriminant base are: (a) limiting the range of positions, in order to exclude candidates insisting on positions too close to the 5′- or 3′-end of the sequences, where, usually, the majority of errors in the alignment or characterization of the sequences occur and (b) testing the presence of other species with probes insisting on the same position, thus excluding eventual interactions between a single CP and multiple DS, with subsequent non-specificity. The candidate probes can also be filtered and ranked according to their thermodynamic properties (length, melting temperature, number of degenerated bases, low complexity regions), evidencing the candidates having a certain length, a melting temperature comprised in a user-specified range, having no more than the inputted number of degenerated bases (which can be a real issue for the oligonucleotide specificity), having short homopolymeric regions and not comprising short tandem repeats. Then, ORMA calculates some specific statistics for the qualitative evaluation of the candidates designed on consensus sequences, compared to the original dataset (i.e. the subset of sequences from which every consensus is built): (a) the intra-group score, as the number of initial sequences having the same discriminating base as the consensus and (b) the inter-group score, as the number of sequences other than those used for that consensus having the same discriminating base as the candidate one. This latter score is calculated only when the consensus were created inside ORMA, starting from a single-global alignment. These scores allow the choice of probes that best discriminate between the target and the non-target sequences (i.e. having the highest intra-group and the lowest inter-group score). The software output can be exported as a comma-separated spreadsheet reporting: (a) the list of all the discriminating bases, grouped per species, with absolute (referring to the global alignment) and relative (referring to the specific consensus) positions of the discriminating base, and the base distributions of all the other consensus sequences in the same position; (b) the thermodynamic parameters of the candidate probe pairs, including the Tm, the length of DS and CP probes and the number of degenerated bases in each and (c) the qualitative filtering and the specificity-related scores, including the sequence score, as the average of the consensus scores along all the bases constituting the DS and CP, with penalties for degenerated bases.
Experimental data
Cyanobacteria dataset experiment
The complete cyanobacteria 16S rRNA data set comprised a total of 352 sequences, which were organized by phylogenetical similarity and grouped in a total of 18 clusters, as described in (8). Multiple alignments of all the sequences was performed by ClustalW (26) and the resulting file was imported into ORMA, where 18 consensus, one per cluster, were built. Consensus sequences were determined following the ‘ARB-like’ algorithm (as described in ‘Materials and Methods’ section and in Supplementary Methods), setting 50% as the threshold for gaps and 40% as the threshold for other bases. Melting temperature calculations followed the ‘salt-adjusted’ method, with 50 mM Na+ and 0% formamide. Candidate probe pairs were filtered on the basis of their length (minimum 25 nt, maximum 60 nt per probe), melting temperature (63–68°C) and number of degenerated bases (maximum 4), on both DS and CP. The best probe pairs for all the species were selected, according to their best intra- and inter-group scores. We required that no less than 80% of the sequences constituting each of the 18 clusters carried the same base as the consensus in the candidate discriminating position (intra-group score). When only one candidate was designed or the intra-group score of the best candidate was below 80%, we still picked that candidate for further evaluations. On the other hand, the inter-group score was set to be below a 2% threshold, with the same exceptions as above. The ‘Unicyano’ probe, which allowed the identification of any of the species in the study, was the one proposed by Castiglioni et al., with minor refinements for adjusting its melting temperature. At first, the LDR mix made by all probe pairs (250 fmol/µl each probe) was tested on specific synthetic templates (perfectly complementary to each probe pair) to assess the feasibility of the LDR procedure with the ORMA-designed probe pairs. Then, a total of 14 DNA samples, corresponding to 13 cyanobacteria species (kindly provided by MIDI_CHIP project partners, http://www.cip.ulg.ac.be/midichip/) (Table 1), were tested in duplicate, independent, LDR experiments, with both ORMA and Castiglioni et al. probe pairs.
Table 1.
Cyanobacterial samples and related LDR results for Castiglioni et al. probes and ORMA-designed ones
| Group | Sample ID | Strain/Clone name | Geographic origin | Sequencing classification (score)c | LDR resultsb |
|
|---|---|---|---|---|---|---|
| Castiglioni et al. probes | ORMA probes | |||||
| Calothrix | 1 | Calothrix sp.strain PCC 7714 | Small pool, Aldabra Atoll, India | Specific (2/2) | Specific (2/2) | |
| Cylindrospermopsis | 2 | Cylindrospermopsis 1LT32S01a | Trasimeno Lake, Italy | Specific (2/2) | Specific (2/2) | |
| Cylindrospermum | 3 | Cylindrospermum stagnale PCC 7417 | Soil, greenhouse, Stockholm, Sweden | Aspecific (2/2) | Specific (2/2) | |
| Halotolerans | 4 | Cyanothece sp.strain PCC 7418 | Solar Lake, Israel | Aspecific (1/2), Specific (1/2) | Specific (2/2) | |
| Leptolyngbya | 5 | Leptolyngbya sp.strain 0BB 30S02 | Bubano Basin, Imola, Italy | Specific (2/2) | Specific (2/2) | |
| Microcystis | 6 | Microcystis aeruginosa PCC 9354 | Little Rideau Lake, Ontario, Canada | Specific (1/2) No signal (1/2) | Specific (2/2) | |
| Nodularia | 7 | Nodularia 3SD7S01a | Svalbard Islands, Norway | Aspecific (2/2) | Specific (2/2) | |
| Nostoc | 8 | Nostoc sp.strain PCC 7107 | Shallow pond, Point Reyes, CA, USA | Nostoc (100%) | Aspecific (2/2) | Non-specific (0/2) |
| 9 | Nostoc sp.strain PCC 8114 | Water bloom, Lake Hepet.on, Morris Co, NJ, USA | Cylindrospermum (58%) | Non-specific (0/2) | Non-specific (0/2) | |
| Planktothrix | 10 | Planktothrix sp.strain 2 | Lake Markusbölefjärden, Åland Islands, Finland | Specific (2/2) | Specific (2/2) | |
| Prochlorococcus+ Synechococcus | 11 | Prochlorococcus marinus PCC 9511 | Mediterranean Sea | Specific (2/2) | Specific (2/2) | |
| 12 | Synechococcus sp.strain Hegewald 1974-30 | Lake Kuusjärvi, Saukkolahti, Finland | Aspecific (1/2), Specific (1/2) | Specific (2/2) | ||
| Spirulina | 13 | Spirulina major PCC 6313 | Brackish water, Berkeley, CA, USA | Aspecific (2/2) | Specific (2/2) | |
| Synechocystis | 14 | Synechocystis sp.strain PCC 7008 | Shallow pond, Point Reyes, CA, USA | Non-specific (0/2) | Specific (2/2) | |
Where sequencing has been performed, the result of the classification is also reported. Sample ID refers to the numbers used in Figure 2.
aClonal DNA from environmental sample.
bSpecific indicates that only the probe corresponding to the species was present; non-specific means that no probe was present (except for the universal cyanobacteria probe); aspecific means that the species-specific probe was present, but also other probes showed an IF significantly above background signal. The number of replicates is reported within brackets.
cAccording to RDP II database, release 9.60.
Milk-pathogen dataset experiment
Milk pathogens-related 16S sequences were retrieved from RDP-Ribosomal Database Project II (release 9.51, http://rdp.cme.msu.edu/) (27) for a total of 738 sequences and divided into 13 subgroups, according to their phylogenetic classification. Only sequences of length >1200 bp and flagged as of ‘good’ quality were retrieved. Each subgroup was aligned independently in ClustalW, since the overall number of 16S sequences was >500 (above the maximum limit of the alignment tool) and imported into ORMA. The consensus sequence for each group was calculated with the same parameters specified for the cyanobacteria data set. Then, a new multiple-alignment step was performed before proceeding to actual probe design. One probe pair for each of the main six subspecies of the Streptococcus group (Streptococcus agalactiae, S. bovis, S. equi, S. canis, S. dysgalactiae S. uberis) was designed; moreover, the Staphylococcus aureus probe pair was designed independently from all the remaining coagulase negative Staphylococci (grouped in the ‘Staphylococcus, no aureus’ probe), because of its relationship with outbreaks of mastitis in dairy ruminants (28) and with major health issues, like food-related intoxications (29). In order to have the best homogeneity among the species within each group, the design was actually performed in three rounds: (a) Salmonella spp. was aligned against Escherichia coli and related spp. consensus sequence only; (b) S. canis was aligned against Streptococcus group sequences only; (c) All the remaining positions were selected considering the alignment of all other subspecies. One probe pair per species was designed, except for Campylobacter spp. for which two probe pairs were evaluated in terms of reproducibility and specificity. The thermodynamic parameters were the same described for the cyanobacteria data set, except for the melting temperature, which was required to be in the range 67–69°C. The inter-group score of the candidates was required to be above a threshold of 80%, as in the cyanobacteria dataset. Probe pair specificity was checked by both RDP II database and BLAST (Basic Local Alignment Search Tool, http://www.ncbi.nlm.nih.gov/blast/Blast.cgi) (30) analysis, carefully examining the 3′-region of the discriminating probe, in order to exclude any interaction between probe pairs targeting different species. LDR probe pairs were mixed at a final concentration of 1 pmol/μl and tested on 13 DNAs from ATCC reference strains (LGC Promochem, Middlesex, UK) and bacterial collections (Supplementary Table 1). Genomic DNA was extracted following the protocol described by (31), PCR amplified and analyzed in duplicate, by separated LDR reactions.
PCR and LDR/Universal Array approach
Complete experimental procedures concerning the amplification of 16S rRNA sequences (including primers and thermal cycling), LDR mixes, Universal Arrays preparation and hybridization are reported in Supplementary Data.
Data analysis
All arrays were scanned with ScanArray 5000 scanner (Perkin Elmer Life Sciences, Boston, MA, USA), at 10 μm resolution, with different acquisition parameters on both laser power and photo-multiplier gain, in order to avoid saturation. Intensities of fluorescence (IF) were quantitated by ScanArray Express 3.0 software, using the ‘Adaptive circle’ option, letting diameters vary from 60 to 300 μm. No normalization procedures on the IFs were performed.
To assess whether a probe pair was significantly above the background (i.e. was ‘present’ or not), we performed a one-sided t-test (α = 0.01). At the same time, also the type II error was calculated and 1-β used as the estimate of the power of the statistical test. The null distribution was set as the population of ‘Blank’ spots (e.g. with no oligonucleotide spotted, n = 6) IFs. Two times the standard deviation of pixel intensities of the same spots was added to obtain a conservative estimate. For each Zip-code, we considered the population of the IFs of all the replicates (n = 4) and tested it for being significantly above the null-distribution (H0: μtest = μnull; H1: μtest > μnull).
Signal-to-noise ratios, SNRp and SNRnp were calculated, for each ‘present’ and ‘non-present’ probe pairs, respectively, indicating the ratio between the mean IF of each probe pair and the mean ‘Blank’ IF, divided on the probe-type.
RESULTS AND DISCUSSION
Searching, designing and selecting oligonucleotide probes for molecular applications experiments on sets of highly similar sequences, such as the 16S rRNA, is a non-trivial procedure, which involves many complex and time-consuming steps. In this article, this procedure was accomplished by the use of ORMA, an integrated architecture of Matlab scripts. The 16S rRNA, a gene sequence of more than 1500 bp, is the preferred genomic target for analyses in the microbiological field (17–20). It should be noted that 16S region is commonly used in taxonomical classifications involving in silico alignment and procedures for its two basic properties: (i) 16S presents highly conserved regions which can be used to correctly align all the sequences in the database; (ii) on the other side, 16S presents highly polymorphic regions that can be used in clusterization, phylogenetic tree construction and molecular discrimination of microbiological families even very close one to each other (32). Use of an automated method for discriminating positions determination, probe retrieval and filtering has obvious and evident advantages over the manual design, often used in previously published papers (8,33–35). These advantages become more significant with increasing dimension of the databases and of the sequences length. ORMA can perform all these operations with user-specified parameters in an automated way and calculates a series of qualitative parameters which help in the choice of candidate probes that best discriminate between the sequences of the positive and those of the negative set. The general idea of these scores is to distinguish the sequences/groups which are of interest in a given experiment from those who aren’t and that can potentially have a cross-contamination with the positive set, because they could be amplified by PCR, contributing to the molecular complexity of the sample. In this article, performances of ORMA were evaluated by considering the experimental evidences coming from the design of LDR probe pairs on two different 16S rRNA datasets. First, a new set of cyano-specific probe pairs was designed and compared to the original one (8), generated on the same database of sequences. Then, the tool was used to setup LDR probe pairs for the identification of pathogenic species present in bovine milk.
Cyanobacteria dataset
Species-specific probe pairs were designed in a single round, starting from the whole dataset of 352 ClustalW-aligned cyanobacteria 16S rRNA sequences, imported, converted and grouped into 18 group-specific consensus sequences by ORMA. Calculated consensus sequences were highly similar, (ClustalW score = 87.31 ± 2.13, n = 18), had a high consensus score (average score 89.20 ± 4.16, n = 352) and a very low content of degenerated bases (average < 2%, max = 6%). ORMA identified a total of 192 candidate probe pairs for the 18 species, with an overall duration of the whole procedure of less than 5 min (Table 2). More tests on speed performances of the SBS algorithm on simulated data available as Supplementary Data and Supplementary Figure 2. One probe pair per species was chosen, according to its ranking after ORMA filtering steps. The probe pair for Anabaena + Aphanizomenon group was flagged as inadequate by ORMA, having six degenerated bases in the CP, which could negatively influence its thermodynamical properties. However, this probe pair insisted on the only discriminating position found for that cluster. The mix containing all probe pairs was tested on the corresponding synthetic templates and, as expected, all except Anabaena+Aphanizomenon gave positive results. Duplicate LDR experiments on 18 probe pairs (17 species-specific + 1 universal) were carried out on 14 16S rRNA PCR products. We performed side-by-side tests of the same DNA samples by the two probe pairs datasets, ORMA and the one described in Castiglioni et al., comparing their performances and specificity.
Table 2.
List of probe pairs for the cyanobacteria experiment, associated Zip-codes and major thermodynamic parameters
| Oligo name | Species | Discr Base pos Fulla | Real Posb | Zip code | Discrim oligo | Common probe | Length of DS | Length of CP | Tm DS | Tm CP | Number of Deg bases DS | Number of Deg bases CP | Score | Intra- group Score | Inter- group score | Seq DS Score | Seq CP Score | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Calothrix _z_36 | Calothrix | 1116 | 93 | 36 | GGTGAGTAACGCGTGAGAATCTGT | CTTYAGGTCGGGGACAACAGTT | 24 | 22 | 65.2 | 63.1 | 0 | 1 | 10 | 3 (3) | 100% | 0 (349) | 0% | 100 | 100 |
| Cylindrospermopsis_z_28 | Cylindrospermopsis | 1560 | 543 | 28 | CGTAAAGGGTCTGCAGGTGGA | ACTGAAAGTCTGCTGTTAAAGAGTTTG | 21 | 27 | 63.3 | 63.7 | 0 | 0 | 10 | 3 (3) | 100% | 3 (349) | 1% | 100 | 100 |
| Cylindrospermum_z_29 | Cylindrospermum | 2133 | 1062 | 29 | GTTTTTAGTTGCCAGCACTTCGGG | TGGGCACTCTAGAGAGACTGC | 24 | 21 | 65.2 | 63.3 | 0 | 0 | 10 | 2 (2) | 100% | 0 (350) | 0% | 100 | 100 |
| Halotolerans_z_13B | Halotolerans | 1634 | 584 | 13B | CTGGTGYGCTAGAGGGCGAC | AGGGGTAGAGGGAATTCCCAG | 20 | 21 | 65.6 | 63.3 | 1 | 0 | 10 | 8 (8) | 100% | 0 (344) | 0% | 95.00 | 100 |
| Leptolyngbya_z_37 | Leptolyngbya | 1202 | 185 | 37 | GTGAAATGTTWTWTYGCCTGAGGATGAA | CTCGCGTCTGATTAGCTAGTTGG | 28 | 23 | 65.0 | 64.6 | 3 | 0 | 10 | 5 (5) | 100% | 0 (347) | 0% | 93.57 | 100 |
| Microcystis _z_1B | Microcystis | 1581 | 524 | 1B | GTCAGCCAAGTCTGCYGTCAAAT | CAGGTTGCTTAACGACCTAAAGGC | 23 | 24 | 63.8 | 65.2 | 1 | 0 | 10 | 91 (91) | 100% | 0 (261) | 0% | 99.09 | 99.27 |
| Nodularia_z_23B | Nodularia | 1239 | 211 | 23B | TAGCTAGTAGGTGTGGTAAAAGCG | CACCTAGGCGACGATCAGTAG | 24 | 21 | 63.5 | 63.3 | 0 | 0 | 10 | 28 (30) | 93% | 14 (322) | 4% | 98.19 | 99.52 |
| Nostoc_z_32 | Nostoc | 1886 | 825 | 32 | GGGGAGTACGCCGGCAACG | GTGAAACTCAAAGGAATTGACGGG | 19 | 24 | 66.0 | 63.5 | 0 | 0 | 10 | 5 (6) | 83% | 4 (346) | 1% | 97.37 | 100 |
| Prochl+Synech _z_3B | Prochlorococcus+ Synechococcus | 1475 | 426 | 3B | CTTGAGGAATAAGCCACGGCTAAT | TCCGTGCCAGCAGCCGCG | 24 | 18 | 63.5 | 65.2 | 0 | 0 | 10 | 86 (86) | 100% | 0 (266) | 0% | 98.60 | 99.94 |
| Planktotrix _z_21B | Planktotrix | 1558 | 510 | 21B | GGGCGTAAAGAGTCCGTAGGTA | GTCATCCAAGTCTGCTGTTAAAGAG | 22 | 25 | 64.0 | 64.1 | 0 | 0 | 10 | 11 (11) | 100% | 0 (341) | 0% | 100 | 100 |
| Spirulina_z_11B | Spirulina | 2473 | 1350 | 11B | CACACCATGGAAGCTGGCAACA | TCCGAAGTCGTTACTCCAACYKTT | 22 | 24 | 64.0 | 63.5 | 0 | 2 | 10 | 10 (11) | 91% | 1 (341) | 0% | 63.64 | 64.39 |
| Synechocystis _z_31 | Synechocystis | 1602 | 576 | 31 | GTTAAAGAATGGAGCTTAACTCCATAG | GAGCGGTGGAAACTGCAAGAC | 27 | 21 | 63.7 | 63.3 | 0 | 0 | 10 | 8 (9) | 89% | 2 (343) | 1% | 97.94 | 97.88 |
| UniCyano_z_8 | UniCyano | 1330 | 304 | 8 | CCTACGGGAGGCAGCAGTG | GGGAATTTTCCGCAATGGGCG | 19 | 21 | 63.8 | 63.3 | 0 | 0 | 10 | 18 (18) | 100% | – | – | 100 | 100 |
| Gloeothece_z_35 | Gloeothece | 1857 | 795 | 35 | GCCGAAGCTAACGCGTTAAGTC | TCCCGCCTGGGGAGTACGC | 22 | 19 | 64.0 | 66.0 | 0 | 0 | 10 | 3 (5) | 60% | 0 (347) | 0% | 97.27 | 100 |
| Lyngbya _z_34 | Lyngbya | 1120 | 112 | 34 | AGTAACGCGTGAGAATCTGCCTTA | GGGTCGGGGACAACCACCG | 24 | 19 | 63.5 | 66.0 | 0 | 0 | 10 | 3 (3) | 100% | 1 (349) | 0% | 100 | 100 |
| Phormidium_z_33 | Phormidium | 1440 | 309 | 33 | TGGGAAGAAAGTTGTGAAAGCAGC | CTGACGGTACCAGAGGAATCAG | 24 | 22 | 63.5 | 64.0 | 0 | 0 | 10 | 2 (2) | 100% | 0 (350) | 0% | 100 | 100 |
| Thrichodesmium_z_27 | Thricodesmium | 1139 | 112 | 27 | CCTTCAGGTCTGGGACAACAGAA | GGAAACTTCTGCTAATCCCGGATG | 23 | 24 | 64.6 | 65.2 | 0 | 0 | 10 | 7 (7) | 100% | 0 (345) | 0% | 99.38 | 99.40 |
| Woronichinia_z_5B | Woronichinia | 1299 | 285 | 5B | GCAGCCACACTGGAACTGAGAA | ACRGTCCAGACTCCTACGGG | 22 | 20 | 64.0 | 63.5 | 0 | 1 | 10 | 2 (2) | 100% | 0 (350) | 0% | 100 | 98.75 |
| Ana+Apha_z_38 | Anabaena+ Aphanizomenon | 1989 | 923 | 38 | ACCTTACCAAGGCTTGACATGTCA | CGAATYCYGTWGAAAKATRGRAGTG | 24 | 25 | 63.5 | 63.3 | 0 | 6 | 8.3 | 62 (68) | 91% | 0 (284) | 0% | 99.20 | 95.59 |
‘Len DS’ (or ‘Len CP’) is the probe length; ‘Tm’ is the melting temperature; ‘Deg bases’ is the number of degenerated bases within each probe; ‘Score’ is proportional to the number of quality checks each probe passed (10 means all, 8.3 is five out of six); ‘Inter-group score ‘and ‘Intra-group score’ evaluate the probe pair specificity (full description in the text); ‘Seq score’ is the score of the consensus sequence (as reported in the text). The exact probes sequence from ORMA is reported. For synthesis purpose, any degenerated base was substituted with inosine (I). The first 11 specific +1 universal probes corresponded to probes which were actually tested on cyanobacteria samples. The last six species were tested only on the synthetic templates. Anabaena + Aphanizomenon probe pair did not show any signal, probably due to high number of degenerated bases in the sequence of the common probe.
aThe reported position refers to the absolute position in the multiple alignment.
bThe ‘Real Position’ refers to the position in the single consensus per species.
Probe pairs used in Castiglioni et al. identified correctly (P < 0.005, average beta power of the test: 0.85) 6 out of 14 analyzed DNAs (in both duplicate LDR), whereas other two completely failed. Six other DNAs somehow showed a degree of aspecificity (i.e. the correct probe pair was present, but non-specific probe pairs were also called present) (Table 1, Figure 2). Cyanobacteria universal probe pair was called as statistically over the background in all the experiments. Evaluations on ratio of signal intensities suggested that hybridizations went well and were not responsible for the aspecificity. In fact, excluding non-specific signals, SNRnp had an average value of 1.18 ± 0.61 and SNRp varied between 10 and 680, with an average of about 149 (data not shown). The Anabaena + Aphanizomenon probe pair of Castiglioni et al. study resulted specific on both synthetic and environmental samples (data not shown). This probe pair, however, was designed with its DS insisting on a position which did not discriminate univocally the Anabaena + Aphanizomenon consensus from the consensuses of the other species. Thus, it would never be identified by ORMA as discriminating (because of the way the algorithm is built). Instead, the presence of some internal mismatches (especially the one on the second base before the 3′-end of the DS) is probably the reason for this finding. In fact, the mismatch gives instability to the 3′-end of the DS when annealing on the 16S rRNA sequences of species other than those of Anabaena + Aphanizomenon cluster, impeding the ligase to join the two adjacent end of the DS and CP oligonucleotides.
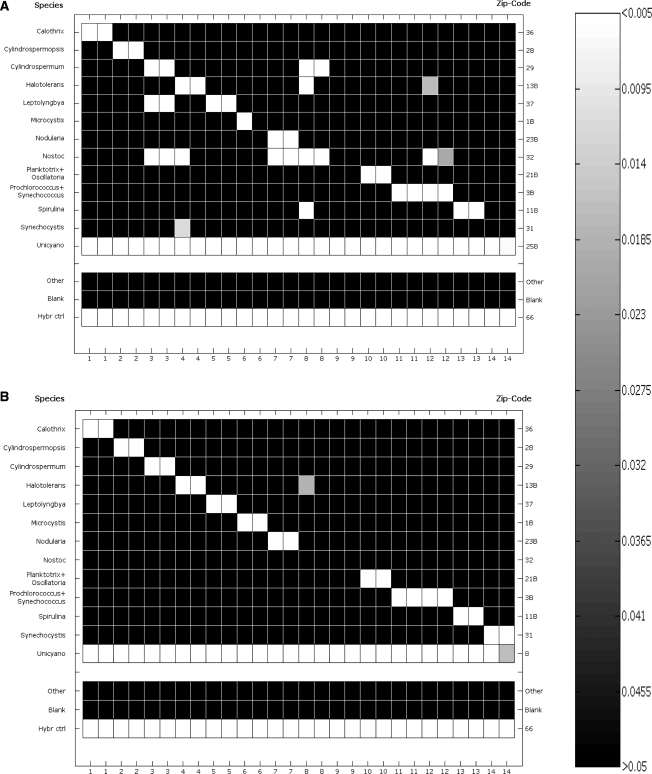
Figure 2.
Heat maps of P-values deriving from the duplicate LDR experiments on cyanobacteria dataset. (A) Castiglioni et al. probes; (B) ORMA-designed probes. The scale varies between non-significance (>0.05) to high-significance (<0.005). On the x-axis, the IDs of the tested samples (see Table 1 for full description) are reported. On the y-axis, the probe pair name is reported. The line ‘Other’ represents the mean of all the remaining Zip-codes in the universal arrays that were not associated to any actual probe. Experiments on Nostoc samples were repeated twice on different DNAs because of the failure of the first test. Halotolerans probe pair in one replicate of sample 8 (classified as Nostoc) has a P-value of 0.02, above the threshold of 0.01 chosen for significance.
ORMA designed probe pairs have been capable of correctly identifying (P < 0.005, average beta power of the test: 0.85) 12 out of 14 analyzed cyanobacteria samples, on both replicates. Also in this experimental set, the cyanobacteria universal probe pair was called as statistically over the background in all the experiments (as expected, since this probe pair and the ones used in Castiglioni et al. coincided). Performances of the LDR procedure, in terms of signal-to-noise ratios were comparable to those obtained with the Castiglioni et al. probe set, having a SNRnp of 1.1 ± 0.26 and a SNRp ranging from 7 to 387 (average ∼131) (data not shown), indicating a certain variability. In this case, we had no signs of aspecificity in the experiments (Figure 2), even in those cases which were critical with Castiglioni et al. probe pairs. In fact, probes were chosen in order to maximize the intra-group similarity (i.e. having the maximum number of sequences in the positive set carrying the discriminating base) and minimize the possibility of an inter-group cross-talk (i.e. having a minimum number of sequences in the negative set carrying the discriminating base) (Figure 3). The average of intra-group scores of the candidates was 95.1% ± 10.1% (n = 17), varying in the range 60–100%. The minimum value was that of the cluster of Gleotheceae, in which we had only five sequences, whereas 13 out of the 17 clusters were characterized by a score of 100%. Inter-group scores, on the other hand, were always very low, with an average of 0.4% ± 1% (n = 17). Thus, where ORMA probe pairs failed, we had a false negative call (with the cyanobacteria universal probe pair called as ‘present’), but not a false positive. Experiments on the two Nostoc DNAs gave no results on the species-specific probe pair; anyway the presence of a cyanobacterial DNA was correctly assessed by the Universal probe pair. Sequencing of the two products revealed that one of them has been correctly classified by microbiological methods, whereas the other DNA was very uncertain and classified as ‘cylindrospermum’ (58% confidence) by RDP ‘Classifier’ tool, release 9.60. [On 22 May 2008, RDP II database for cyanobacteria (release 9.61) underwent a major change in hierarchical classification of the species. The taxonomies here presented refer to older versions, which at present, can be found within genus GpI of family Family I, philum cyanobacteria.] Both sequences found very little similarity with our probe pairs, with internal mismatches and a different base in the discriminating position. Anyway, the probe pair itself was successfully tested on the synthetic template. The failure of ORMA-designed Anabaena + Aphanizomenon probe pair suggests the possibility of making a re-design in the near future, increasing the number of sequences of the database and improving the information content of the dataset. Another strategy could be designing probe pairs on sub-clusters of Anabaena + Aphanizomenon, building new consensuses from more homogeneous groups; in this way, the presence of such two species would be assessed by multiple probe pairs and not only by one.
Figure 3.
Graphical comparison between the Castiglioni et al. and the ORMA-designed probe pair (DS+CP) on Cylindrospermum species, aligned in ClustalW with Cylindrospermum strain sequences (Cy*) and the Leptolyngbya strain sequences (Leptolyngbya* and Lpg*). The part of each probe flanking the discriminating position is highlighted in red (Castiglioni et al. probe pair) or green (ORMA). The bases aligned with the discriminating base are marked by a black box. In Castiglioni et al. probe pair, the discriminating position was found also on some Leptolyngbya strains, whereas in ORMA probe pair, the discriminating position is unique to all Cylindrospermum sequences. Absolute positions of the bases in the alignment are reported on the top ruler.
Milk-pathogens dataset
16S rRNA sequences of pathogens contaminating bovine milk or related to bovine mastitis were used to design LDR oligonucleotide probes by ORMA, providing a further confirmation of its reliability and specificity. In this study, three round of design were actually performed, in order to have the best homogeneity between the species used in each round. A single round would have caused the loss of discriminating positions due to misalignment of some species (e.g. Salmonella) which are somehow different from all others. ORMA found a total of 392 candidate positions (34, 4 and 354 in the design for Salmonella, S. canis and all remaining species, respectively), which were selected according to the quality ranking scores assigned by ORMA. In this experiment, ORMA calculated only the intra-group score, but not the inter-group score, because of the fact that the sequences for each group were imported separately and the software was unable to recall the position corresponding to discriminating ones in all the sequences constituting each of the consensuses. The candidate probes were all characterized by an optimal specificity of the discriminating base, as suggested by the intra-group scores which were above 90% in 11 out of 14 cases. The scores were, in any case, above the fixed threshold of 80%, having an average of 94.0% ± 6.9% (n = 14). Also in this case, the lowest score (i.e. 80%) was that of the cluster (i.e. S. equi) constituted by the lowest number of sequences (n = 5). The final evaluation on the candidate probe pairs was made by RDP and BLAST checks, because of the multiplicity of species, whose 16S rRNA gene was amplified by means of universal primers, potentially present in milk-derived matrixes and the lack of a complete internal negative set in ORMA. The probes were slightly longer than the ones on cyanobacteria dataset, with an average length of about 40 nt, with very homogeneous melting temperatures (mean Tm = 67.6 ± 0.4, n = 28) and a very low number of degenerated bases (only the DS probe for S. equi had 1 degenerate base) (Table 3). The consensus scores for both the DS and CP confirmed the overall quality of the probes (average score of 96.5 ± 4.2, n = 28, with 60% of the probes having a score >99%).
Table 3.
List of probe pairs for the milk-pathogens experiment and major thermodynamic parameters
| Oligo name | Species | Discr Base pos | Real Pos | Zip code | Discrim oligo | Common probe | Length of DS | Length of CP | Tm of DS | Tm CP | #Deg bases DS | #Deg bases CP | Score | Intra- group score | Seq DS score | Seq CP score | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Bacillus_z_10 | Bacillus spp. | 880 | 862 | 10 | GCTAAGTGTTAGAGGGTTTCCGCCCTTT AGTGCTGAAGT | TAACGCATTAAGCACTCCGCCTGGGGAG TACGG | 39 | 33 | 67.6 | 68.1 | 0 | 0 | 10 | 313 (313) | 100% | 99.87 | 99.90 |
| S_equi_z_12 | Strept. equi | 224 | 178 | 12 | CTAATACCGCATAAAAGTGGTTGACCC ATGTTAACNATTTAAAAGGAGCAACA | GCTCCACTATGAGATGGACCTGCGTTGT ATTAGCTAGTTG | 53 | 40 | 67.5 | 67.6 | 1 | 0 | 10 | 4 (5) | 80% | 88 | 99.50 |
| S_agal_z_15 | Strept. agal | 87 | 78 | 15 | CGTGCCTAATACATGCAAGTAGAACGCT GAGGTTTGGTGTTTA | CACTAGACTGATGAGTTGCGAACGGGT GAGTAACGC | 43 | 36 | 67.4 | 67.9 | 0 | 0 | 10 | 17 (18) | 94% | 92.25 | 99.69 |
| S_bovis_z_16 | Strept. bovis | 91 | 81 | 16 | GTGCCTAATACATGCAAGTAGAACGCTG AAGACTTTAGCTTGCTAA | AGTTGGAAGAGTTGCGAACGGGTGAGT AACGCGTAG | 46 | 36 | 67.2 | 67.9 | 0 | 0 | 10 | 19 (22) | 86% | 92.98 | 98.11 |
| S_uberis_z_19 | Strept. uberis | 223 | 192 | 19 | CGCATGACAATAGGGTACACATGTACCC TATTTAAAAGGGGCAAA | TGCTTCACTATGAGATGGACCTGCGTTGT ATTAGCTAGTTGG | 45 | 42 | 67.3 | 67.4 | 0 | 0 | 10 | 5 (5) | 100% | 98.22 | 99.52 |
| Staph_aureus_z_2 | Staph aureus | 222 | 219 | 2 | CCGGATAATATTTTGAACCGCATGGTTCA AAAGTGAAAGACGGTC | TTGCTGTCACTTATAGATGGATCCGCGCT GCATTAGCTAG | 45 | 40 | 67.3 | 67.6 | 0 | 0 | 10 | 61 (62) | 98% | 99.39 | 99.80 |
| Mycoplasma_z_20 | Mycoplasma spp. | 906 | 848 | 20 | CATCGACGCAGCTAACGCATTAAATGAT CCGCCTGAGT | AGTACGTTCGCAAGAATAAAACTTAAAG GAATTGACGGGGATCCG | 38 | 45 | 67.7 | 67.3 | 0 | 0 | 10 | 51 (51) | 100% | 98.71 | 98.47 |
| Staphylococcus_z_21 | Staphylococcus (no aureus) | 208 | 186 | 21 | GAAACCGGAGCTAATACCGGATAATATA TTGAACCGCATGGTTCAAT | AGTGAAAGACGGTTTTGCTGTCACTTATA GATGGATCCGCG | 47 | 41 | 67.2 | 67.5 | 0 | 0 | 10 | 41 (49) | 84% | 96.79 | 97.51 |
| E_coli _z_28 | E_coli and related species | 484 | 469 | 28 | GTTGTAAAGTACTTTCAGCGGGGAGGAA GGGAGTAAAGTTAATAC | CTTTGCTCATTGACGTTACCCGCAGAAGA AGCACCG | 45 | 36 | 67.3 | 67.9 | 0 | 0 | 10 | 10 (11) | 91% | 90.91 | 90.91 |
| S_canis_z_3 | Strept. canis | 474 | 469 | 3 | GATCGTAAAGCTCTGTTGTTAGAGAAGA ACGGTAATGGGAGTGGAAAAC | CCATTATGTGACGGTAACTAACCAGAAA GGGACGGCTAACTAC | 49 | 43 | 68.7 | 68.3 | 0 | 0 | 10 | 6 (6) | 100% | 99.66 | 100 |
| S_dysgal_z_4 | Strept. dysgal | 1061 | 1039 | 4 | GTCTAGAGATAGGCTTTCCCTTCGGGG CAGG | AGTGACAGGTGGTGCATGGTTGTCGTC AGCTCG | 31 | 33 | 67.0 | 68.1 | 0 | 0 | 10 | 55 (55) | 100% | 99.65 | 100 |
| Salmonella_z_5 | Salmonella spp. | 258 | 251 | 5 | CCATCAGATGTGCCCAGATGGGATTAGC TTGTTGGTGA | GGTAACGGCTCACCAAGGCGACGATCCC | 38 | 28 | 67.7 | 67.2 | 0 | 0 | 10 | 41 (41) | 100% | 99.87 | 100 |
| Campylob_1_z_8 | Campylobacter spp. | 179 | 148 | 8 | CCCTACACAAGAGGACAACAGTTGGAAA CGACTGCTAATACT | CTATACTCCTGCTTAACACAAGTTGAGTA GGGAAAGTTTTTCGGTG | 42 | 46 | 67.4 | 67.2 | 0 | 0 | 10 | 71 (78) | 91% | 90.60 | 90.75 |
| Campylob_2_z_9 | Campylobacter spp. | 233 | 192 | 9 | CTCTATACTCCTGCTTAACACAAGTTGA GTAGGGAAAGTTTTTCGG | TGTAGGATGAGACTATATAGTATCAGCT AGTTGGTAAGGTAATGGCTTAC | 46 | 50 | 67.2 | 67.0 | 0 | 0 | 10 | 71 (78) | 91% | 90.75 | 89.59 |
The exact probes sequence from ORMA is reported. For synthesis purpose, any degenerated base was substituted with inosine (I). The description of the reported columns is the same as those in Table 2.
The procedure showed optimal specificity, with excellent signal-to-noise ratios, as shown in detail in the article of Cremonesi and co-workers (36). Results were in complete concordance with sample identification made by ATCC; only probes associated to the supposed species were present (P-values always <0.005), whereas all remaining probes were well below any acceptable P-value for the t-test (Figure 4). In this dataset, SNRp varied from 4.31 to 238.3, with an average of 34.28; at the same time, SNRnp varied between 0.12 and 0.83, with an average of 0.48 ± 0.18. The two probes on Campylobacter species (insisting on two different positions) performed nearly the same in terms of specificity, both giving P-values far below the acceptance threshold of 0.01, whereas performances in terms of signal intensity varied, with one probe having average IFs about 2-fold higher than the other, in both replicates, suggesting a somehow different sensitivity in the two competing probes.
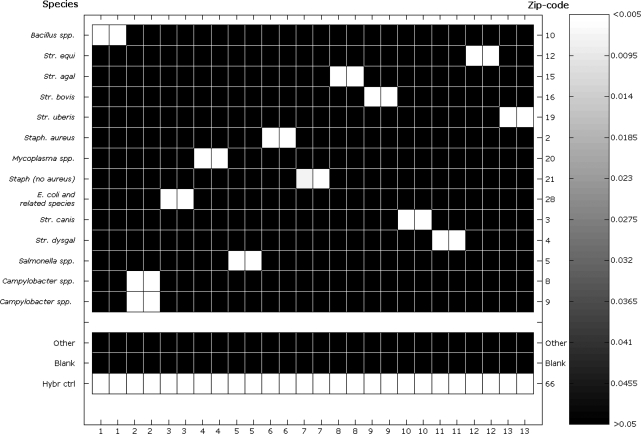
Figure 4.
Heat map of P-values deriving from the duplicate LDR experiments on milk pathogen dataset. The scale varies between non-significance (>0.05) to high-significance (<0.005). The line ‘Other’ represents the mean of all the remaining Zip-codes in the universal arrays that were not associated to any actual probe. Complete association between samples numbers and names is given in Supplementary Table 2.
Thus, ORMA helped in developing a reliable PCR-LDR-UA assay, which allowed the identification of pathogenic species in milk, based only on 16S rRNA gene, whereas other assays (37) needed multiple genes. The molecular procedure permitted the discrimination between the most frequently isolated or emerging pathogens in mastitis (e.g. S. aureus, S. agalactiae, S. uberis), or potentially dangerous for human health (e.g. E. coli and related species, Salmonella, S. aureus and Bacillus spp). Streptococcus spp. was identified at the species level, even in the cases, like the one of S. uberis and S. parauberis, where the molecular identification on the basis of the 16S rRNA gene required PCRs with species-specific primers. Moreover, the ORMA-based LDR technique represents a significant improvement of the existing detection methods for Mycoplasma spp. strains (38), known to be contagious causes of intramammary infection in herds, overcoming the long and laborious standard-detection methods based on microbiological procedures (36). These results confirmed the ability of this tool to determine discriminating positions in complex datasets.
Important remarks
The ability to identify ‘fingerprint’ positions within a set of homologous sequences, like those of 16S rRNA gene, is the main feature of ORMA. To achieve optimal results, the starting set of sequences should be carefully selected, because, if sequences are characterized by many low-similarity regions, the determination of terminally discriminating position could be biased by badly aligned subsequences. In that case, a different algorithm (actually not included, but under development) for the determination of detection probes by means of the hybridization strategy, can be more appropriated. On the other side, using sequences nearly identical one to each other can cause the opposite behavior, where no discriminating positions can be determined. A careful grouping of the sequences in clusters (as we did for both of our examples, building 18 consensus out of 352 sequences in cyanobacteria dataset and 13 consensus out of 752 sequences in milk pathogens dataset) is strongly suggested. In this latter application three rounds of design were applied, in order to compensate the non-perfect homogeneity of some species. Experimental results demonstrated the correctness of this approach and the specificity of the probe pairs obtained with this design strategy. Experimental data on the 16S rRNA cyanobacteria and milk-pathogens dataset demonstrated that ORMA specifically addressed discriminating positions within a set of highly similar sequences. Nonetheless, our tool identified a total of 192 and 392 candidate positions, respectively. The intra- and inter- group scores were demonstrated to be very helpful in determining the best probes for discrimination and avoiding cross-talk between species.
ORMA is a bioinformatic tool for the search and determination of single-discriminating positions among a set of highly homologous sequences and represents a significant improvement from other contexts where enzyme-based techniques have been employed on already known single-nucleotide polymorphisms (SNPs) (39) or on entire subsequences (11). This unique feature makes ORMA completely different from all the other available software for probe design in detection experiments. During the past years, academic software for species detection have been developed. ProDesign (13) is a tool based on a ‘spaced seed algorithm’ for the determination of probes capable of discriminating multiple pathogenic species, at different hierarchical levels. Similarly, YODA (14) performs design tasks on complete genomes against non-target species. TOFI-beta (15) implements a suffix-tree-based algorithm for isolating suitable candidate probes from a target genome and filters the list according to thermodynamical and specificity requirements. These three software are implemented for the design of probes for hybridization-based detection assays. PathogenMIPer (16), instead, is based on a different strategy (i.e. molecular inversion assays), which starts from the selection of unique sequences on a reduced dataset and then does a global comparison to all those potentially matching.
All these software perform smart designs where the probes have to be selected on the whole genomic DNA; this is the typical pipeline in contexts where no pre-selection of the target sequences has been made, which is not the case of ORMA. In fact, in both the presented datasets, the molecular complexity of the genomic material has been reduced by PCR on the 16S rRNA. The probe pairs design, then, was performed only on the basis of a specific subset of the whole 16S dataset, limited for the specific environment in which the target species have to be detected: cyanobacteria DNA was selected and amplified by cyano-specific PCR primers, while milk pathogens 16S rRNA sequences, although amplified by universal primers, were compared only to context-specific species. The double check in RDP and BLAST, performed after the complete probe pair design by ORMA, confirmed that our choice to work with such a reduced dataset was indeed correct, because the detected species accounted for the majority of the biological diversity present in the target matrix (i.e. milk). Moreover, many of the aforementioned software perform the specificity checks by extensive BLAST searches, which is a reasonable choice for designing specific probes starting from the whole genomic DNA; in case of datasets with limited complexity (or in which the complexity has been reduced by means of molecular procedures), this approach results too computationally intensive and unnecessary for the scope. ARB (21) and PRIMROSE (22) are tools widely used for the classification and the phylogenesis of bacterial species, structured for interacting with databases specific for the same molecular target (i.e. 16S rRNA) and operate a probe design on the basis of the phylogenesis of the species under analysis. None of these two software, however, is built specifically for the determination of discriminating positions within a set of very similar sequences and they provide probe design functionality only for hybridization assays or PCR primers. When used for probe design in detection application, the strategies are based on internal mismatches or on unique stretches of nucleotides (40). In this case, the discrimination power resides more in the decreased melting temperature of mismatched duplexes, rather than on a perfectly matched base pair between the probe and the target. Although our tool was applied on the design of probes for a specific technique (LDR) and on a specific target gene (16S rRNA), the software is not limited to this combination. LDR technique approach implied the retrieval of a pair of sequences, one of which (the DS probe) insisted on the discriminating position, whereas the other (the CP probe) is designed to anneal one base 3′-downstream of the discriminating position; the design of probes for minisequencing application would have implied only the determination of one probe with its 3′-end one base before the variation. At the same time, the design of a reporting probe for a TaqMan Real-time PCR assay would have implied the determination of one oligo with the single-base variation in the middle of the sequence. Due to its modular structure and to the straightforwardness of other applications from the already implemented one, probe sets retrieval and filtering methods could be easily added, starting from the discriminating positions found by the SBS algorithm. A further extension to hybridization probes/standard PCR primer design will be evaluated, changing the strategy for determining the positions to be tested. Provided that the initial database of sequences is accurate, updated and complete as much as possible, ORMA can retrieve discriminating positions and design specific probes on every set of sequences. Its implementation, in fact, is not based on an internal database of sequences (which are, instead, retrieved and loaded from external resources) and can be extended to any gene. In any case, the database should be critically built by only context-specific sequences. Standard procedures, like PCR with specific primers, can help in isolating only the subsets of sequences which constitute the actual database from those completely unrelated to the biological context under investigation, avoiding any interference with actual probes, as exemplified by the cyanobacteria dataset experiment. Sequences of off-target or distantly-related species could negatively act in the process of multiple-alignment, leading to poorly aligned datasets and biased designs. Since the databases, cyanobacterias in particular, are constantly and frequently upgraded, ORMA capability of determining discriminating positions can be refined, depending on the completeness of the initial datasets (both positive and negative set). Moreover, the continuous changing of classification and the addition of new sequences make an exhaustive and definitive design of the best cyanobacteria probes absolutely not trivial.
ORMA represents a good alternative solution to the troublesome problem of searching specific positions within a large set of homologous 16S rRNA sequences and provides tools for performing a series of probe-related operations, such as sequence retrieval, filtering and scoring, allowing the user to have a set of candidates on which the actual and definitive selection can be done. The calculation on intra- and inter-group scores allows the selection of highly specific probes for molecular applications, covering the highest number of species of the positive set and having the lowest interaction with the negative set. In silico checks versus public databases (e.g. RDP or BLAST) are necessary only in case of lack of a reference among the sequences imported in ORMA or when the species eventually present in the biological context under study are too many for being comprised into a reasonably small negative set (e.g. all the microorganisms potentially present in bovine milk). Appropriate experimental designs, comprising context-specific PCRs for reducing the molecular complexity of the target can also be helpful. A complete set of major thermodynamic parameters are reported in the output, helping the researcher to carefully select the best probes. Our data assessed and demonstrated the performances of ORMA in designing probes for molecular applications on 16S rRNA gene and their feasibility for experimental use, with improved specificity and sensitivity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
FIRB 2003 [RBLA03ER38_004 (‘NG-LAB’)]; ‘Regione Lombardia’ [Contract n 962, ‘Safe Milk’]. Funding for open access charge: FIRB 2003 [RBLA03ER38_004 (‘NG-LAB’)].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Marco Candela, Annamaria Piano, Alessia Cariani and Giulio Zanaroli at the University of Bologna for helpful hints and discussions for developing the software. Dr Stefano Ventura and the partners of MIDI_CHIP European Project in the Fifth Framework Programme of the European Community are acknowledged for sharing their data and samples. They also thank Maria Vurchio for her help in administrative issues.
REFERENCES
- 1.Patel JB. 16S rRNA gene sequencing for bacterial pathogen identification in the clinical laboratory. Mol. Diagn. 2001;6:313–321. doi: 10.1054/modi.2001.29158. [DOI] [PubMed] [Google Scholar]
- 2.Bodrossy L, Sessitsch A. Oligonucleotide microarrays in microbial diagnostics. Curr. Opin. Microbiol. 2004;7:245–254. doi: 10.1016/j.mib.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 3.Hacia JG. Resequencing and mutational analysis using oligonucleotide microarrays. Nat. Genet. 1999;21(Suppl 1):42–47. doi: 10.1038/4469. [DOI] [PubMed] [Google Scholar]
- 4.Saiki RK, Walsh PS, Levenson CH, Erlich HA. Genetic analysis of amplified DNA with immobilized sequence-specific oligonucleotide probes. Proc. Natl Acad. Sci. USA. 1989;86:6230–6234. doi: 10.1073/pnas.86.16.6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Syvänen AC. Accessing genetic variation: genotyping single nucleotide polymorphisms. Nat. Rev. Genet. 2001;2:930–942. doi: 10.1038/35103535. [DOI] [PubMed] [Google Scholar]
- 6.Grossman PD, Bloch W, Brinson E, Chang CC, Eggerding FA, Fung S, Iovannisci DM, Woo S, Winn-Deen ES. High-density multiplex detection of nucleic acid sequences: oligonucleotide ligation assay and sequence-coded separation. Nucleic Acids Res. 1994;22:4527–4534. doi: 10.1093/nar/22.21.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gerry NP, Witowski NE, Day J, Hammer RP, Barany G, Barany F. Universal DNA microarray method for multiplex detection of low abundance point mutations. J. Mol. Biol. 1999;292:251–262. doi: 10.1006/jmbi.1999.3063. [DOI] [PubMed] [Google Scholar]
- 8.Castiglioni B, Rizzi E, Frosini A, Sivonen K, Rajaniemi P, Rantala A, Mugnai MA, Ventura S, Wilmotte A, Boutte C, et al. Development of a universal microarray based on the ligation detection reaction and 16S rrna gene polymorphism to target diversity of cyanobacteria. Appl. Environ. Microbiol. 2004;70:7161–7172. doi: 10.1128/AEM.70.12.7161-7172.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rantala A, Rizzi E, Castiglioni B, De Bellis G, Sivonen K. Identification of hepatotoxin-producing cyanobacteria by DNA-chip. Environ. Microbiol. 2008;10:653–664. doi: 10.1111/j.1462-2920.2007.01488.x. [DOI] [PubMed] [Google Scholar]
- 10.Belgrader P, Barany F, Lubin M. Development of a multiplex ligase detection reaction DNA typing assay. In Proceedings of the Sixth International Symposium on Human Identification. 1995. [Last accessed date 20th May 2009.]. http://www.promega.com/geneticidproc/ussymp6proc/belgrad.htm.
- 11.Bordoni R, Mezzelani A, Consolandi C, Frosini A, Rizzi E, Castiglioni B, Salati C, Marmiroli N, Marchelli R, Rossi Bernardi L, et al. Detection and quantitation of genetically modified maize (Bt-176 transgenic maize) by applying ligation detection reaction and universal array technology. J. Agric. Food Chem. 2004;52:1049–1054. doi: 10.1021/jf034871e. [DOI] [PubMed] [Google Scholar]
- 12.Chessa S, Chiatti F, Ceriotti G, Caroli A, Consolandi C, Pagnacco G, Castiglioni B. Development of a single nucleotide polymorphism genotyping microarray platform for the identification of bovine milk protein genetic polymorphisms. J. Dairy Sci. 2007;90:451–464. doi: 10.3168/jds.S0022-0302(07)72647-4. [DOI] [PubMed] [Google Scholar]
- 13.Feng S, Tillier ER. A fast and flexible approach to oligonucleotide probe design for genomes and gene families. Bioinformatics. 2007;23:1195–1202. doi: 10.1093/bioinformatics/btm114. [DOI] [PubMed] [Google Scholar]
- 14.Nordberg EK. YODA: selecting signature oligonucleotides. Bioinformatics. 2005;21:1365–1370. doi: 10.1093/bioinformatics/bti182. [DOI] [PubMed] [Google Scholar]
- 15.Vijaya Satya R, Zavaljevski N, Kumar K, Reifman J. A high-throughput pipeline for designing microarray-based pathogen diagnostic assays. BMC Bioinformatics. 2008;9:185. doi: 10.1186/1471-2105-9-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thiyagarajan S, Karhanek M, Akhras M, Davis RW, Pourmand N. PathogenMIPer: a tool for the design of molecular inversion probes to detect multiple pathogens. BMC Bioinformatics. 2006;7:500. doi: 10.1186/1471-2105-7-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Behr T, Koob C, Schedl M, Mehlen A, Meier H, Knopp D, Frahm E, Obst U, Schleifer K, Niessner R, et al. A nested array of rRNA targeted probes for the detection and identification of enterococci by reverse hybridization. Syst. Appl. Microbiol. 2000;23:563–572. doi: 10.1016/s0723-2020(00)80031-4. [DOI] [PubMed] [Google Scholar]
- 18.Maynard C, Berthiaume F, Lemarchand K, Harel J, Payment P, Bayardelle P, Masson L, Brousseau R. Waterborne pathogen detection by use of oligonucleotide-based microarrays. Appl. Environ. Microbiol. 2005;71:8548–8557. doi: 10.1128/AEM.71.12.8548-8557.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kochzius M, Nölte M, Weber H, Silkenbeumer N, Hjörleifsdottir S, Hreggvidsson GO, Marteinsson V, Kappel K, Planes S, Tinti F, et al. DNA microarrays for identifying fishes. Mar. Biotechnol. 2008;10:207–217. doi: 10.1007/s10126-007-9068-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stedtfeld RD, Wick LM, Baushke SW, Tourlousse DM, Herzog AB, Xia Y, Rouillard JM, Klappenbach JA, Cole JR, Gulari E, et al. Influence of dangling ends and surface-proximal tails of targets on probe-target duplex formation in 16S rRNA gene-based diagnostic arrays. Appl. Environ. Microbiol. 2007;73:380–389. doi: 10.1128/AEM.01785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, et al. ARB: a software environment for sequence data. Nucleic Acids Res. 2004;32:1363–1371. doi: 10.1093/nar/gkh293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ashelford KE, Weightman AJ, Fry JC. PRIMROSE: a computer program for generating and estimating the phylogenetic range of 16S rRNA oligonucleotide probes and primers in conjunction with the RDP-II database. Nucleic Acids Res. 2002;30:3481–3489. doi: 10.1093/nar/gkf450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rychlik W. OLIGO 7 primer analysis software. Methods Mol. Biol. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- 24.Apte A, Singh S. AlleleID: a pathogen detection and identification system. Methods Mol. Biol. 2007;402:329–346. doi: 10.1007/978-1-59745-528-2_17. [DOI] [PubMed] [Google Scholar]
- 25.Pingle MR, Granger K, Feinberg P, Shatsky R, Sterling B, Rundell M, Spitzer E, Larone D, Golightly L, Barany F. Multiplexed identification of blood-borne bacterial pathogens by use of a novel 16S rRNA gene PCR-ligase detection reaction-capillary electrophoresis assay. J. Clin. Microbiol. 2007;45:1927–1935. doi: 10.1128/JCM.00226-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cole JR, Chai B, Farris RJ, Wang Q, Kulam-Syed-Mohideen AS, McGarrell DM, Bandela AM, Cardenas E, Garrity GM, Tiedje JM. The ribosomal database project (RDP-II): introducing myRDP space and quality controlled public data. Nucleic Acids Res. 2007;35:D169–D172. doi: 10.1093/nar/gkl889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bergonier D, De Crémoux R, Rupp R, Lagriffoul G, Berthelot. X. Mastitis of dairy small ruminants. Vet. Res. 2003;34:689–716. doi: 10.1051/vetres:2003030. [DOI] [PubMed] [Google Scholar]
- 29.Su YC, Lee Wong A. Current perspectives on detection of Staphylococcal Enterotoxins. J. Food Prot. 1997;60:195–202. doi: 10.4315/0362-028X-60.2.195. [DOI] [PubMed] [Google Scholar]
- 30.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 31.Cremonesi P, Castiglioni B, Malferrari G, Biunno I, Vimercati C, Moroni P, Morandi S, Luzzana M. Technical Note: improved method for rapid DNA extraction of mastitis pathogens directly from milk. J. Dairy Sci. 2006;89:163–169. doi: 10.3168/jds.S0022-0302(06)72080-X. [DOI] [PubMed] [Google Scholar]
- 32.Plays T, Nakamura LK, Cohan FM. Discovery and classification of ecological diversity in the bacterial world: the role of DNA sequence data. Int. J. Syst. Bacteriol. 1997;47:1145–1156. doi: 10.1099/00207713-47-4-1145. [DOI] [PubMed] [Google Scholar]
- 33.Hashimoto M, Hupert ML, Murphy MC, Soper SA, Cheng YW, Barany F. Ligase detection reaction/hybridization assays using three-dimensional microfluidic networks for the detection of low-abundant DNA point mutations. Anal. Chem. 2005;77:3243–3255. doi: 10.1021/ac048184d. [DOI] [PubMed] [Google Scholar]
- 34.Carnevale EP, Kouri D, DaRe JT, McNamara DT, Mueller I, Zimmerman PA. A multiplex ligase detection reaction-fluorescent microsphere assay for simultaneous detection of single nucleotide polymorphisms associated with Plasmodium falciparum drug resistance. J. Clin. Microbiol. 2006;45:752–761. doi: 10.1128/JCM.01683-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Long WH, Xiao HS, Gu X-M, Zhang Q-H, Yang H-J, Zhao G-P, Liu J-H. A universal microarray for detection of SARS coronavirus. J. Virol. Methods. 2004;121:57–63. doi: 10.1016/j.jviromet.2004.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cremonesi P, Pisoni G, Severgnini M, Consolandi C, Moroni P, Raschetti M, Castiglioni B. Pathogens Detection in Milk Samples by LDR-Mediated Universal Array Method. J. Dairy Science. 2009 doi: 10.3168/jds.2008-1773. In press. [DOI] [PubMed] [Google Scholar]
- 37.Wang X-W, Zhang L, Jin LQ, Jin M, Shen ZQ, An S, Chao FH, Li J-W. Development and application of an oligonucleotide microarray for the detection of food-borne bacterial pathogens. Appl. Microbiol. Biotechnol. 2007;76:225–233. doi: 10.1007/s00253-007-0993-x. [DOI] [PubMed] [Google Scholar]
- 38.Cremonesi P, Vimercati C, Pisoni G, Perez G, Ribera AM, Castiglioni B, Luzzana M, Ruffo G, Moroni P. Development of DNA extraction and PCR amplification protocols for detection of Mycoplasma bovis directly from milk samples. Vet. Res. Commun. 2007;1:225–227. doi: 10.1007/s11259-007-0011-x. [DOI] [PubMed] [Google Scholar]
- 39.Consolandi C, Frosini A, Pera C, Ferrara GB, Bordoni R, Castiglioni B, Rizzi E, Mezzelani A, Bernardi LR, De Bellis G, et al. Polymorphism analysis within the HLA-A locus by universal oligonucleotide array. Hum. Mutat. 2004;24:428–434. doi: 10.1002/humu.20098. [DOI] [PubMed] [Google Scholar]
- 40.Bodrossy L, Stralis-Pavese N, Murrell JC, Radajewski S, Weilharter A, Sessitsch A. Development and validation of a diagnostic microbial microarray for methanotrophs. Environ. Microbiol. 2003;5:566–582. doi: 10.1046/j.1462-2920.2003.00450.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.