Abstract
Lsm1 is a component of the Lsm1-7 complex involved in cytoplasmic mRNA degradation. Lsm1 is over-expressed in multiple tumor types, including over 80% of pancreatic tumors, and increased levels of Lsm1 protein have been shown to induce carcinogenic effects. Therefore, understanding the perturbations in cell process due to increased Lsm1 protein may help to identify possible therapeutics targeting tumors over-expressing Lsm1. Herein, we show that LSM1 over-expression in the yeast Saccharomyces cerevisiae inhibits growth primarily due to U6 snRNA depletion, thereby altering pre-mRNA splicing. The decrease in U6 snRNA levels causes yeast strains over-expressing Lsm1 to be hypersensitive to loss of other proteins required for production or function of the U6 snRNA, supporting a model wherein excess Lsm1 reduces the availability of the Lsm2-7 proteins, which also assemble with Lsm8 to form a complex that binds and stabilizes the U6 snRNA. Yeast strains over-expressing Lsm1 also display minor alterations in mRNA decay and demonstrate increased susceptibility to mutations inhibiting cytoplasmic deadenylation, a process required for both 5′-to-3′ and 3′-to-5′ pathways of exonucleolytic decay. These results suggest that inhibition of splicing and/or deadenylation may be effective therapies for Lsm1-over-expressing tumors.
INTRODUCTION
Tumors of the pancreas pose a critical problem in eliminating mortality due to cancer in that incidence and morbidity rates for this disease are nearly equal (NCI SEER database, http://seer.cancer.gov/statfacts/html/pancreas.html). Even with recent technological advances in genomic analysis, the overall relative 5-year survival rate of pancreatic tumors from 1996 to 2004 was 5.1%, and trend analysis of the period from 2003 to 2005 revealed no significant changes in mortality rate (NCI SEER database, http://seer.cancer.gov/statfacts/html/pancreas.html). Given that the current prognosis for patients with these tumors is dismal, it is vital that we search for novel therapeutics targeting this disease.
In 1997, Lsm1 was identified through subtractive hybridization cloning in pancreatic cancer cells (1) and was shown to be over-expressed in 87% of pancreatic cancers. Subsequently, its over-expression has been described in 40% of prostate cancers (2), a subset (15–20%) of breast cancers that are amplified at the 8p11-12 region (3,4) and most recently in lung cancers and mesotheliomas (5). The direct involvement of Lsm1 in carcinogenesis in these tissues has been demonstrated through analyses of Lsm1's effects on growth and anchorage dependence (2,6,7), contact inhibition (2), autocrine activity (7) and tumor establishment and metastases (2,6,8,9). The increase in Lsm1 levels in these tumors is moderate (about 2- to 5-fold) (7), suggesting that subtle changes in the levels of Lsm1 can affect the growth properties of mammalian cells. Thus, it is important to elucidate the processes affected by LSM1 over-expression in order to provide new targets for therapeutic development against pancreatic and other Lsm1-over-expressing cancers.
Lsm1 over-expression could affect cellular metabolism in several manners. For example, Lsm1 over-expression has been suggested to destabilize certain tumor suppressor transcripts, allowing for carcinogenesis (2). This model is based on the fact that Lsm1 in yeast and humans assembles with the Lsm2-Lsm7 proteins to form a heteroheptameric Lsm1-7 complex that binds mRNAs, components of the decapping machinery, and promotes mRNA decapping and degradation (10–14). Alternatively, Lsm1 over-expression might inhibit the function of the related Lsm2-8 complex, wherein the Lsm1 protein is replaced by the Lsm8 protein. The Lsm2-8 complex binds the 3′-end of the U6 snRNA protecting it from degradation and thereby allowing normal rates of pre-mRNA splicing (15–17). Consistent with Lsm1 over-expression affecting the nuclear Lsm2-8 complex, over-expression of LSM1 in budding yeast increased the cytoplasmic localization of Lsm7p (18). Hence, over-expression of LSM1 may actually reduce U6 levels and selectively influence splicing, allowing for carcinogenesis.
To understand how Lsm1 over-expression influences cell processes, we took advantage of the conservation of Lsm1 function in both budding yeast and humans to determine how Lsm1 over-expression affects RNA metabolism in yeast. We found that over-expression of LSM1 in the yeast Saccharomyces cerevisiae leads to defects in pre-mRNA splicing, which is caused by decreased levels of the U6 snRNA. The splicing defect causes yeast strains over-expressing Lsm1 to be hypersensitive to loss of other components required for maintaining levels of U6 snRNA. Moreover, yeast strains over-expressing Lsm1 are more susceptible to mutations inhibiting cytoplasmic deadenylation, which is normally a prerequisite for mRNA decay. These results suggest that inhibition of splicing and/or deadenylation may be effective therapies for LSM1-over-expressing tumors.
MATERIALS AND METHODS
Yeast strains, growth conditions and plasmids
The genotypes of the strains used are listed in Supplementary Table S3. Cells were cultured in either yeast extract/peptone medium or synthetic medium supplemented with appropriate amino acids and 2% sugar (sucrose or galactose) and were grown at 30°C. Yeast strains were transformed as previously described (19) and maintained in the appropriate selective media. Over-expression studies were performed by culturing strains continuously in galactose. Plasmids utilized in this study are found in Supplementary Table S4.
Plasmid construction
The GAL LSM1 2 μ plasmid was constructed by amplifying the LSM1 coding region 64 nt prior to its start through 240 nt following the stop. A BamHI restriction site immediately upstream of the 5′-end and a SalI restriction site at the 3′-end of this product facilitated its ligation to a GAL 2 μ vector (pRP861) and placed the LSM1 gene under transcriptional control of a GAL promoter. The GAL LSM1 CEN plasmid was constructed by amplifying the LSM1 coding region 59 nt prior to its start through 240 nt following the stop. A SacI restriction site immediately upstream of the 5′-end and a SalI restriction site at the 3′-end of this product facilitated its ligation to a GAL CEN vector (pRP23) and placed the LSM1 gene under transcriptional control of a GAL promoter.
RNA analysis
RNA analyses were performed as previously described (20). Total RNA was isolated (21) from midlog cultures grown in appropriate media, and 20 μg of each sample were electrophoresed on 6% acrylamide, 8 M Urea gels. Northern blots were performed using the indicated oligonucleotides radiolabelled with 32P at the 5′-end. Oligonucleotides used in this study are detailed in Supplementary Table S5.
For decay time course experiments, transcriptional shutoff was achieved by resuspending galactose-induced cultures in media containing 4% dextrose (22) and then collecting samples over a brief time course.
Protein analysis
Midlog cultures were collected and harvested for protein analysis. Samples were lysed using 5 M urea, boiled, then vortexed in glass beads for 5 min. A solution of 125 mM Tris–Cl pH 6.8, 2% SDS was added at 2.5× the volume of 5M urea used, and this was vortexed into the mixture, then samples were boiled a second time. Collected lysate was clarified by spinning at 16 000 RCF, and the supernatant was resuspended in protein loading buffer (0.05 M Tris pH 6.5, 1% SDS, 0.01% bromophenol blue, 10% glycerol), boiled, and run on a 12% Tris–SDS acrylamide gel. Gels were transferred to nitrocellulose and probed using standard Western blotting protocols using an antibody to Lsm1 (a generous gift of Allen Sachs and Karsten Weis) and an anti-rabbit secondary coupled to HRP (Pierce). Lsm1 signal was revealed using Pierce SuperSignal West Dura and exposing the blots to film and developing in a film processor (Konica).
Films were scanned into .tif format using an HP Scanjet Pro flatbed scanner, and images were analyzed and quantitated in Adobe Photoshop following the method outlined at http://www.lukemiller.org/journal/2007/08/quantifying-western-blots-without.html. References from Supplementary Tables are Table S3 (23–25) and Table S4 (26–28).
RESULTS
LSM1 over-expression in yeast can affect cell growth
In order to understand the effects of LSM1 over-expression in budding yeast, we first expressed LSM1 from the GAL promoter on a CEN plasmid in a wild-type yeast strain. To determine the degree of over-expression, we utilized antisera against Lsm1 to determine the increase in the Lsm1 protein levels as compared to a vector only control strain. We observed that strains transformed with a GAL-LSM1 centromere plasmid showed ∼3× the levels of wild-type yeast strains (Figure 1A and B), an increase in levels similar to what is seen in various human tumor cell lines. We observed that cells carrying this plasmid exhibited a slight decrease in growth as compared to the same strain carrying the vector backbone, although the strains were still able to grow at some rate (Figure 1C, uppermost panel). Thus, over-expression of LSM1 inhibits, but does not completely prevent, the growth of wild-type yeast strains.
Figure 1.
Moderate levels of Lsm1 affect cell growth. (A) LSM1 over-expression on a GAL CEN plasmid yields 2- to 4-fold protein expression. (B) Quantitative analysis of Lsm1 over-expression. (C) LSM1 over-expression on a GAL CEN plasmid affects growth and yields synthetic growth defects with strains mutant for U6 snRNP function. (A, B, C) OE CEN = Lsm1 expressed on GAL CEN plasmid (pRP1851). V = CEN vector (pRP23). (A) Western blot. Strains were transformed as indicated. ‘Lsm1 protein’ labels the position of Lsm1 band that was quantitated. Asterisk marks a cross-reacting band used for normalization. WT = yRP841, lsm1Δ = yRP1365. (B) Quantitation of protein levels. Relative intensities were compared to yield fold increase in Lsm1 protein levels over wild-type control. Four comparisons were made of OE CEN to v. Error bars represent standard deviation (see ‘Materials and Methods’ section, protein analysis for details on protocols used in A and B). (C) BY4741(WT) and isogenic deletion strains transformed as indicated and were frog ponded by 10-fold dilutions and plated on plates containing galactose and grown at 30°C.
LSM1 over-expression shows genetic interactions with proteins affecting the U6 snRNP and mRNA deadenylases
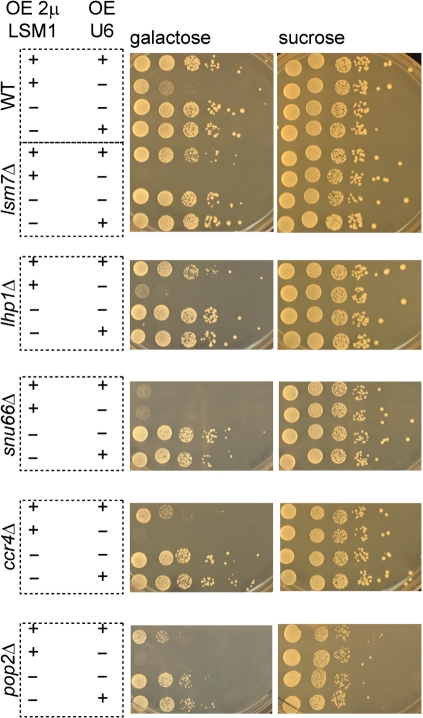
There are two likely possibilities for how LSM1 over-expression might affect cell function. First, since the Lsm1-7 complex functions in the control of mRNA decapping, over-expression of LSM1 might alter the stoichiometry of components of the decapping complex, enhancing or inhibiting mRNA decapping and thereby affecting growth. Alternatively, increased levels of Lsm1p could deplete the nuclear Lsm2-8 complex, thus affecting cell growth through inhibition of splicing. To consider these possibilities, we first examined the effects of Lsm1 over-expression from the GAL CEN plasmid in lsm7Δ and lhp1Δ strains, which are both compromised for U6 levels (15, 29–32). We observed that over-expression of Lsm1 strongly limited the growth of both lsm7Δ and lhp1Δ strains, with the strongest effect seen with lsm7Δ (Figure 1C, lower panels).
In order to extend this genetic analysis, we decided to over-express Lsm1 from the GAL promoter on a multicopy 2 μ plasmid, thereby increasing the probability of us detecting additional genetic interactions. We observed that over-expression from the GAL 2 μ plasmid gave a stronger inhibition of growth than the GAL centromere plasmid (compare Figure 1C uppermost panel and Figure 2A), although the cells do still grow at a reduced rate (Figure 2A). Using this system of expressing LSM1 from the GAL 2 μ plasmid, we then examined Lsm1 over-expression in a variety of yeast strains lacking non-essential proteins that function in pre-mRNA splicing or mRNA degradation. A complete list of these mutants and their phenotypic effects observed with LSM1 over-expression is provided in Supplementary Tables S1 and S2.
Figure 2.
LSM1 2 μ over-expression affects cell growth. (A) LSM1 over-expression on a GAL 2 μ plasmid affects growth of wild-type yeast. (B) LSM1 over-expression on a GAL 2 μ plasmid yields synthetic growth defects with strains mutant for U6 snRNP or (C) deadenylase functions. BY4741(WT) (A) and isogenic deletion strains (B,C) transformed with either the GAL-LSM1 2 μ plasmid construct (pRP1840, OE) or GAL 2 μ plasmid control (pRP861, V) were frog ponded by 10-fold dilutions and plated on plates containing the indicated sugar and grown at 30°C.
Our broader genetic analysis revealed that factors interacting with U6 snRNA were indeed important for growth when Lsm1 was over-expressed. Expressing LSM1 from the GAL 2 μ plasmid still demonstrated strong synthetic growth defects with strains deleted for LSM7 or LHP1 (Figure 2B). Furthermore, snu66Δ, which removes a component of the U4/U6/U5 tri-snRNP (33,34), also showed a strong exacerbation of the growth defect observed when LSM1 was over-expressed from the GAL 2 μ plasmid (Figure 2B). Taken together, these results suggest that LSM1 over-expression may interfere with growth by inhibiting U6 snRNA biogenesis or function.
We also observed that over-expressing LSM1 displayed strong synthetic growth defects with deletions of the CCR4 and POP2 genes, which are key components of the predominant mRNA deadenylase (35). As seen in Figure 2C, expression of GAL-LSM1 on a 2 μ plasmid in strains deleted for CCR4 or POP2 inhibited growth more strongly than over-expression in a wild-type strain (Figure 2A). Significant synthetic growth defects were not demonstrated with deletions in many other decay factors (Supplementary Table S1), including strains deleted for PAT1, DHH1, and EDC3 (Figure 2C), which encode factors that enhance decapping and/or translational repression (13,22,36,37). The enhanced toxicity of Lsm1 over-expression in the ccr4Δ and pop2Δ strains suggests that mRNA decay might be altered when Lsm1 is over-expressed, increasing the relative requirement for deadenylation.
LSM1 over-expression induces defects in splicing
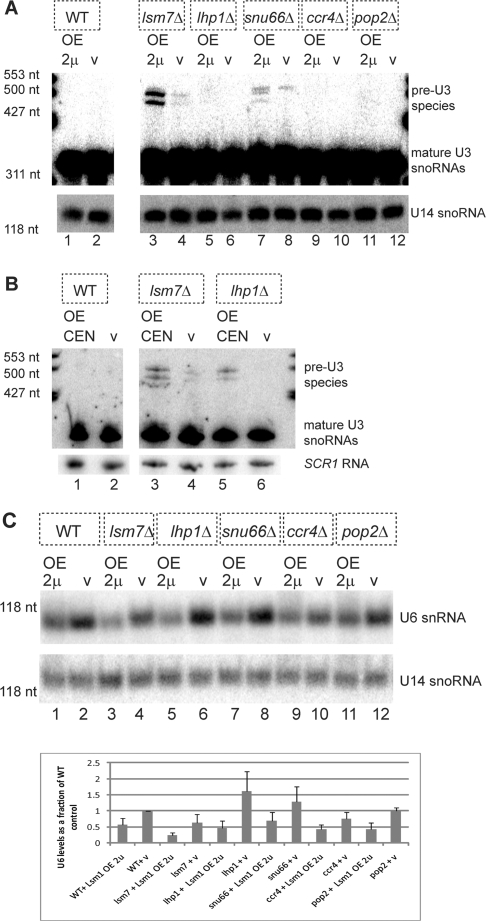
Since strains lacking LSM7, LHP1, and SNU66 are defective for U6 or U4/U6/U5 snRNP function (29,32,34), we hypothesized that LSM1 over-expression might further interfere with splicing in these deletion strains. To assess the effects of Lsm1 over-expression on splicing we examined the accumulation of the intron-containing precursor to the U3 snoRNA, which is a sensitive measure of splicing in budding yeast (29). In this analysis, we also included the ccr4Δ and pop2Δ strains, in case these mutations had some previously unobserved effect on splicing. For this experiment, cells containing the GAL-LSM1 2 μ plasmid, or a vector control, were continuously grown in galactose, cells were harvested at mid-log, and the RNA was analyzed on northern blots for the U3 snoRNA.
We observed that over-expression of Lsm1 in a wild-type strain did not lead to accumulation of the intron-containing precursor to the U3 snoRNA (pre-U3 snoRNA). However, over-expression of Lsm1 in an lsm7Δ strain, increased the accumulation of the pre-U3 snoRNA seen in this strain (Figure 3A, compare lanes 3 and 4). Similarly, a snu66Δ strain accumulated small amounts of the pre-U3 snoRNA, and these levels increased with LSM1 over-expression (Figure 3A, compare lanes 7 and 8). In an lhp1Δ strain, no pre-U3 snoRNA was evident, and small amounts accumulated with Lsm1 over-expression, which were visible on longer exposures (Figure 3A, compare lanes 5 and 6). Finally, ccr4Δ and pop2Δ strains failed to accumulate pre-U3 snoRNA with or without LSM1 over-expression (Figure 3A, examine lanes 9 through 12). Moreover, in a similar analysis, over-expressing LSM1 on the GAL CEN plasmid demonstrated similar synthetic splicing defects in strains deleted for LSM7 or LHP1 as compared to vector only controls (Figure 3B, compare lane 3–4 and lane 5–6). These results indicate that Lsm1 over-expression can lead to defects in splicing, even at moderate levels of over-expression as observed in tumors (7), which are most easily revealed in strains lacking proteins affecting U6 snRNA biogenesis or function.
Figure 3.
LSM1 over-expression inhibits splicing by affecting U6 snRNA. (A) LSM1 over-expression (GAL 2 μ) inhibits splicing of U3 snoRNA in some U6 snRNP defective strains. (B) LSM1 over-expression (GAL CEN) inhibits splicing of U3 snoRNA in some U6 snRNP defective strains. OE 2 μ = GAL-LSM1 2 μ plasmid construct (pRP1840). OE CEN = GAL-LSM1 CEN plasmid (pRP1851). V = appropriate vector control (pRP861 or pRP23). (A, B, C) Total RNA (20 μg) from BY4741(WT) and isogenic deletion strains transformed as indicated were electrophoresed on 8 M Urea, 6% acrylamide gels and blotted by northern using a probe for U3 snoRNA. U14 snoRNA or SCR1 RNA is included as a normalization control. Marker sizes are indicated. (C) Upper panel. LSM1 over-expression depletes levels of U6 snRNA. Separate blot, processed as in (A), but probed for U6 snRNA. Lower panel. Histogram of U6 levels (normalized to U14 levels) expressed as a fraction of levels for wild-type expressing the control plasmid. Results are an average of three experiments with error bars representing the standard deviation.
LSM1 over-expression leads to a decrease in U6 snRNA levels
A simple hypothesis for how Lsm1 over-expression leads to defects in splicing is that Lsm1 over-expression reduces the levels of the Lsm2-8 complex, which binds to and stabilizes the U6 snRNA (17,29). This hypothesis predicts that the levels of U6 snRNA should be reduced by Lsm1 over-expression. To test this prediction, we examined the levels of U1, U2, U4, U5 and U6 snRNAs in both wild-type and various mutant strains over-expressing Lsm1.
We observed that over-expressing LSM1 on a GAL 2 μ plasmid had little effect on U1, U2, U4 and U5 snRNA levels (Figure 3C and data not shown), but U6 levels were decreased in all cells over-expressing LSM1. Specifically, in wild-type cells over-expressing LSM1 on a GAL 2 μ plasmid, U6 snRNA levels decreased by an average of 2-fold (Figure 3C, compare lane 2 to 1 and histogram), and this level of U6 depletion was also observed when moderately over-expressing LSM1 on a GAL CEN plasmid (data not shown). Moreover, and consistent with the genetic interactions, U6 levels were most depleted (4.5-fold less on average, Figure 3C histogram) in an LSM7 deletion background over-expressing LSM1 on a GAL 2 μ plasmid when compared to wild-type expressing a vector control (Figure 3C, examine lanes 3 and 2 and histogram). In these two instances, the results imply that LSM1 over-expression exerts its inhibition of growth through a reduction of U6 levels, yielding an inhibition of splicing. However, the levels of U6 snRNA in lhp1Δ, snu66Δ, ccr4Δ, and pop2Δ strains over-expressing LSM1 on a GAL 2 μ plasmid were not consistently lower than that of a wild-type strain over-expressing LSM1. These results suggest that the consequences of Lsm1 over-expression on growth can be made more significant without further reductions in the U6 level. In the case of the snu66Δ, this effect may be due to the assembly of a defective U4/U6/U5 tri- snRNP, whereas the growth inhibition in the ccr4Δ and pop2Δ strains may be due to defects in mRNA decay (see ‘Discussion’ section).
Depletion of U6 is responsible for synthetic growth and splicing defects in wild-type, lsm7Δ and lhp1Δ strains over-expressing LSM1
The decrease in U6 snRNA levels with Lsm1 over-expression suggests that the inhibition of growth in wild-type and possibly various mutant strains could be due to the decreased U6 snRNA levels. This hypothesis predicts that the growth and splicing defects due to Lsm1 over-expression should be reversed by increasing levels of U6. To test this possibility, we introduced a high-copy plasmid expressing U6 snRNA into each strain and examined its effects on growth and splicing in the presence and absence of Lsm1 over-expression from a GAL 2 μ plasmid.
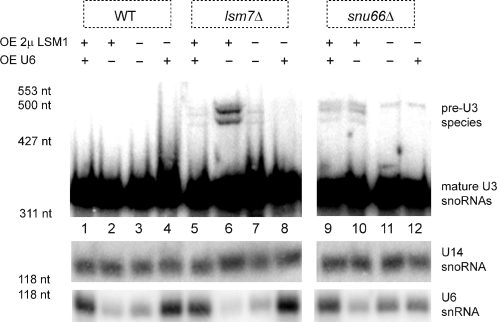
Our results revealed that at least some of the growth defects due to LSM1 over-expression are attributable to low levels of U6 snRNA. First, over-expression of U6 snRNA suppressed the growth defect seen in wild-type cells due to Lsm1 over-expression (Figure 4, uppermost panel). Second, consistent with its severe growth defects and U6 snRNA reduction, lsm7Δ over-expressing LSM1 was nearly completely rescued by the increased U6 expression (Figure 4) and U6 snRNA over-expression suppressed the splicing defect seen with Lsm1 over-expression in this strain (Figure 5, compare lanes 5, 6 and 7). Third, a similar complementation of synthetic growth defects was observed in lhp1Δ over-expressing LSM1 when U6 was over-expressed (Figure 4). These observations indicate that at least some of the growth and splicing defects in LSM1-over-expressing strains arise from depletion in the levels of U6.
Figure 4.
Growth defects due to LSM1 over-expression can be rescued by increasing levels of U6. Increasing U6 levels rescues synthetic growth defects in wild-type, lsm7Δ and lhp1Δ strains. BY4741(WT) and isogenic deletion strains. Transformation of strains as indicated: ‘+’ in a column indicates expression of either GAL-LSM1 2 μ (pRP1840, OE 2 μ LSM1) and/or pSNR6 (OE U6), ‘–’ indicates that the control vector was used. Frog ponds are by 10-fold dilutions onto plates containing the indicated sugar, grown at 30°C.
Figure 5.
Synthetic splicing defects due to LSM1 over-expression can be rescued by increasing levels of U6. Total RNA (20 μg) from BY4741(WT) and isogenic deletion strains transformed as indicated: ‘+’ in a row indicates expression of either GAL-LSM1 2 μ (pRP1840, OE 2 μ LSM1) and/or pSNR6 (OE U6), ‘–’ indicates that the control vector was used. Samples were electrophoresed on 8 M Urea, 6% acrylamide gels and blotted by northern using a probe for U3 snoRNA. U14 snoRNA is included as a normalization control. U6 snRNA probe confirms its over-expression. Marker sizes are noted.
However, the growth defects in the snu66Δ strain were not rescued by over-expression of U6 snRNA (Figure 4). We speculate that the synthetic growth defects in the snu66Δ strain may be due to the assembly of a U6 snRNA that is compromised in function due to loss of both the Snu66p and the Lsm2-8 complex. Consistent with that possibility, over-expression of U6 snRNA did not suppress the splicing defect seen in the snu66Δ strain (Figure 5, compare lanes 9, 10 and 11). This suggests that in the absence of the Snu66 protein, the Lsm2-8 complex may contribute directly to U6 snRNP function (see ‘Discussion’ section).
We observed that the ccr4Δ and pop2Δ strains were partially rescued by over-expression of U6 snRNA (Figure 4). This suggests that the growth defects in these strains is in part due to reduced U6 snRNA levels and in part due to alterations in deadenylation, which might affect some aspect of mRNA decay.
Lsm1 over-expression does not globally alter mRNA decay
The strong synthetic growth defects seen in the pop2Δ and ccr4Δ strain with over-expression of Lsm1 on a GAL 2 μ plasmid led us to hypothesize that Lsm1 over-expression could also alter mRNA decay in some manner. This was also supported by the fact that the strong synthetic growth defects in ccr4Δ and pop2Δ over-expressing LSM1 was only partially rescued by increasing U6 levels. To test if Lsm1 over-expression affects decapping, we examined the effects of LSM1 over-expression on mRNA decay in a wild-type strain using the MFA2pG mRNA, which is a commonly used reporter for mRNA decapping in yeast (38). In order to analyze the decay of MFA2pG mRNA, we expressed it as a low-copy, galactose-inducible promoter fusion along with our GAL-LSM1 2 μ plasmid. Cells were grown in galactose and transcription was repressed by the addition of 4% dextrose (22), with samples taken for RNA analysis over a brief time course to avoid impacting the pool of Lsm1 protein whose half-life has been reported to be 76 min (39). We observed no significant change in the decay rate of the MFA2pG mRNA in a wild-type strain over-expressing Lsm1 as compared to a vector control (t1/2 less than 5′ for both). Similarly, over-expressing Lsm1 at moderate levels on a GAL-LSM1 CEN plasmid did not affect the decay profile of the MFA2pG mRNA (data not shown). This indicates that Lsm1 over-expression does not have a strong effect on the decay of all mRNAs although it remains possible that Lsm1 over-expression affects the decay of a subset of mRNAs.
DISCUSSION
Lsm1 over-expression alters pre-mRNA splicing
In this work, we provide several lines of evidence that Lsm1 over-expression can inhibit cell growth in budding yeast by affecting the biogenesis and/or function of the U6 snRNA. First, over-expression of Lsm1 inhibited growth and decreased the levels of the U6 snRNA (Figures 1, 2 and 3). Second, the toxicity of the Lsm1 over-expression, and in certain strains its impact on splicing, was increased in the lsm7Δ, lhp1Δ and snu66Δ strains, all of which impact in some manner on the function/biogenesis of the U6 snRNP (Figures 1, 2 and 3). Third, over-expression of the U6 snRNA rescued growth and the splicing defect in some strains mutant for U6 snRNP function (Figures 4 and 5). The simplest interpretation of these observations is that Lsm1 over-expression impacts on U6 snRNA by depleting the levels of the Lsm2-8 complex due to competition between Lsm1 and Lsm8 for the Lsm2-7 complex members. This interpretation is also consistent with synthetic growth defects, as well as depletion of U6 snRNA levels, seen when LSM1 was over-expressed in an lsm8-1 mutant and with the observation that LSM1 over-expression increases the concentration of Lsm7p in cytoplasmic foci, presumably reflecting an increased formation of the Lsm1-7p complex (18).
Our results suggest that Lsm1 over-expression can impact on the U6 snRNP in two manners depending on other alterations in the strains. In the lsm7Δ strain, the primary effect is on the levels of the U6 snRNA, and over-expression of the U6 snRNA can suppress the growth defect seen in these cases. This is consistent with other observations that in an otherwise wild-type strain the predominant role of the Lsm2-8 complex is to enhance the stability of the U6 snRNA (29,40). However, in the snu66Δ, over-expression of the U6 snRNA fails to suppress the Lsm1 growth and splicing phenotypes resulting from Lsm1 over-expression (Figures 4 and 5). The simplest interpretation here is that when Snu66, a component of the U4/U6/U5 tri-snRNP, is missing, the Lsm2-8 complex now plays a more important role in the tri-snRNP's function in splicing. Thus, even when U6 snRNA is over-expressed any resulting U4/U6/U5 complex lacking both the Snu66 and Lsm2-8 proteins would be defective for function.
A clear implication of these observations is that changes in Lsm1 levels might impact on splicing in pancreatic tumors. Our studies reveal that when Lsm1 is over-expressed in yeast at moderate levels comparable to that observed in cancer cells (7), splicing is altered due to a depletion in U6 snRNA levels. In this light, it is notable that alterations in the splicing machinery have been previously described in human pancreatic tumors and transgenic mouse models. For instance, a serine/arginine protein kinase involved in splicing, SRPK1, has been shown to be upregulated in pancreatic tumors and its downregulation correlated to decreased proliferation and increased apoptosis in these tumors (41). In addition, an analysis of genomic changes in an Ela-c-myc transgenic mouse model for pancreatic cancer revealed that splicing factors and spliceosome-related genes were part of a major class of genes upregulated in primary tumors and liver metastatic regions as compared to normal pancreas (42). Hence, it is possible that LSM1 over-expression also contributes to changes in splicing patterns by altering the stoichiometry of the spliceosomal machinery, allowing for carcinogenesis to occur in pancreatic cells. An implication of this analysis, and the increased toxicity of Lsm1 over-expression in yeast strains compromised for U6 snRNA function, is that therapies directed at reducing U6 snRNA biogenesis and/or function might be effective therapies for any tumor over-expressing Lsm1.
LSM1 over-expression decreases cell viability when deadenylation is inhibited
We also provide evidence that Lsm1 over-expression leads to a change in the cells’ requirement for different mRNA decay factors. Specifically, we observed that the toxicity of the Lsm1 over-expression was increased in ccr4Δ and pop2Δ strains (Figure 1C), which are compromised for the predominant cytoplasmic deadenylase (35). Moreover, the synthetic growth defects in these deadenylase mutants could only be partially rescued by increasing U6 snRNA levels (Figure 4). Overall, these results indicate that some process of mRNA decay is altered by Lsm1 over-expression, perhaps due to the assembly of mRNA decay complexes that are defective in function. However, since we did not observe an alteration in the MFA2pG mRNA, any alterations in degradation must be limited to subsets of mRNAs.
Prior studies in human cells do suggest that alterations in the decay of subsets of mRNAs may exist in Lsm1 over-expressing cancers. Array analyses of how Lsm1 over-expression affects the transcriptome (2,7) have demonstrated that increased Lsm1 expression alters levels of certain transcripts, some of which encode factors important in carcinogenesis. However, only one example is published providing an mRNA decay analysis. The p21/Cip1 mRNA, encoding a cyclin-dependent kinase inhibitor (43), was shown to be stabilized by targeted reduction of Lsm1 in a cancer cell line, yet this did not correspond to an increase in its protein expression (2). Thus, it is still unclear whether therapeutic effects achieved with Lsm1 targeting in cancer cells are due to altering the decay of specific transcripts. Nevertheless, the synthetic lethality of Lsm1 over-expression in strains defective in mRNA deadenylation implies that therapeutic agents targeting deadenylase activity could be possible mechanisms for the treatment of tumors with over-expression of Lsm1.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health [R37 GM45443 to R.P.]; National Institutes of Health [T32 CA09213 to N.L.]. Funding for open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr Sandra L. Wolin for the generous gift of the pSNR6 plasmid, Dr Pamela A. Silver for providing the GAL 2 μ plasmid used in our studies (pRP861) and Drs Alan B. Sachs and Karsten Weis for providing the Lsm1 antibody. Finally, we thank all members of the Parker lab for their valuable feedback and useful discussions in the preparation of this manuscript.
REFERENCES
- 1.Schweinfest CW, Graber MW, Chapman JM, Papas TS, Baron PL, Watson DK. CaSm: an Sm-like protein that contributes to the transformed state in cancer cells. Cancer Res. 1997;57:2961–2965. [PubMed] [Google Scholar]
- 2.Fraser MM, Watson PM, Fraig MM, Kelley JR, Nelson PS, Boylan AM, Cole DJ, Watson DK. CaSm-mediated cellular transformation is associated with altered gene expression and messenger RNA stability. Cancer Res. 2005;65:6228–6236. doi: 10.1158/0008-5472.CAN-05-0650. [DOI] [PubMed] [Google Scholar]
- 3.Garcia MJ, Pole JC, Chin SF, Teschendorff A, Naderi A, Ozdag H, Vias M, Kranjac T, Subkhankulova T, Paish C, et al. A 1 Mb minimal amplicon at 8p11-12 in breast cancer identifies new candidate oncogenes. Oncogene. 2005;24:5235–5245. doi: 10.1038/sj.onc.1208741. [DOI] [PubMed] [Google Scholar]
- 4.Yang ZQ, Streicher KL, Ray ME, Abrams J, Ethier SP. Multiple interacting oncogenes on the 8p11-p12 amplicon in human breast cancer. Cancer Res. 2006;66:11632–11643. doi: 10.1158/0008-5472.CAN-06-2946. [DOI] [PubMed] [Google Scholar]
- 5.Watson PM, Miller SW, Fraig M, Cole DJ, Watson DK, Boylan AM. CaSm (LSm-1) overexpression in lung cancer and mesothelioma is required for transformed phenotypes. Am. J. Respir. Cell. Mol. Biol. 2008;38:671–678. doi: 10.1165/rcmb.2007-0205OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelley JR, Brown JM, Frasier MM, Baron PL, Schweinfest CW, Vournakis JN, Watson DK, Cole DJ. The cancer-associated Sm-like oncogene: a novel target for the gene therapy of pancreatic cancer. Surgery. 2000;128:353–360. doi: 10.1067/msy.2000.107605. [DOI] [PubMed] [Google Scholar]
- 7.Streicher KL, Yang ZQ, Draghici S, Ethier SP. Transforming function of the LSM1 oncogene in human breast cancers with the 8p11-12 amplicon. Oncogene. 2007;26:2104–2114. doi: 10.1038/sj.onc.1210002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan Y, Rubinchik S, Watson PM, Kelley JR, Fraser MM, Wood AL, Dong JY, Gillanders WE, Boylan AM, Watson DK, et al. Establishing a murine pancreatic cancer CaSm model: up-regulation of CaSm is required for the transformed phenotype of murine pancreatic adenocarcinoma. Mol. Ther. 2005;11:363–372. doi: 10.1016/j.ymthe.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 9.Yan Y, Rubinchik S, Wood AL, Gillanders WE, Dong JY, Watson DK, Cole DJ. Bystander effect contributes to the antitumor efficacy of CaSm antisense gene therapy in a preclinical model of advanced pancreatic cancer. Mol. Ther. 2006;13:357–365. doi: 10.1016/j.ymthe.2005.06.485. [DOI] [PubMed] [Google Scholar]
- 10.Boeck R, Lapeyre B, Brown CE, Sachs AB. Capped mRNA degradation intermediates accumulate in the yeast spb8-2 mutant. Mol. Cell Biol. 1998;18:5062–5072. doi: 10.1128/mcb.18.9.5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bouveret E, Rigaut G, Shevchenko A, Wilm M, Seraphin B. A Sm-like protein complex that participates in mRNA degradation. EMBO J. 2000;19:1661–1671. doi: 10.1093/emboj/19.7.1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingelfinger D, Arndt-Jovin DJ, Luhrmann R, Achsel T. The human LSm1-7 proteins colocalize with the mRNA-degrading enzymes Dcp1/2 and Xrnl in distinct cytoplasmic foci. RNA. 2002;8:1489–1501. [PMC free article] [PubMed] [Google Scholar]
- 13.Tharun S, He W, Mayes AE, Lennertz P, Beggs JD, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 14.Tharun S, Parker R. Targeting an mRNA for decapping: displacement of translation factors and association of the Lsm1p-7p complex on deadenylated yeast mRNAs. Mol. Cell. 2001;8:1075–1083. doi: 10.1016/s1097-2765(01)00395-1. [DOI] [PubMed] [Google Scholar]
- 15.Achsel T, Brahms H, Kastner B, Bachi A, Wilm M, Luhrmann R. A doughnut-shaped heteromer of human Sm-like proteins binds to the 3′-end of U6 snRNA, thereby facilitating U4/U6 duplex formation in vitro. EMBO J. 1999;18:5789–5802. doi: 10.1093/emboj/18.20.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ryan DE, Stevens SW, Abelson J. The 5′ and 3′ domains of yeast U6 snRNA: Lsm proteins facilitate binding of Prp24 protein to the U6 telestem region. RNA. 2002;8:1011–1033. doi: 10.1017/s1355838202026092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vidal VP, Verdone L, Mayes AE, Beggs JD. Characterization of U6 snRNA-protein interactions. RNA. 1999;5:1470–1481. doi: 10.1017/s1355838299991355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spiller MP, Reijns MA, Beggs JD. Requirements for nuclear localization of the Lsm2-8p complex and competition between nuclear and cytoplasmic Lsm complexes. J. Cell Sci. 2007;120:4310–4320. doi: 10.1242/jcs.019943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 20.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 21.Caponigro G, Muhlrad D, Parker R. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell Biol. 1993;13:5141–5148. doi: 10.1128/mcb.13.9.5141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Coller JM, Tucker M, Sheth U, Valencia-Sanchez MA, Parker R. The DEAD box helicase, Dhh1p, functions in mRNA decapping and interacts with both the decapping and deadenylase complexes. RNA. 2001;7:1717–1727. doi: 10.1017/s135583820101994x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brachmann CB, Davies A, Cost GJ, Caputo E, Li J, Hieter P, Boeke JD. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 24.Hatfield L, Beelman CA, Stevens A, Parker R. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell Biol. 1996;16:5830–5838. doi: 10.1128/mcb.16.10.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 26.Fernandez CF, Pannone BK, Chen X, Fuchs G, Wolin SL. An Lsm2-Lsm7 complex in Saccharomyces cerevisiae associates with the small nucleolar RNA snR5. Mol. Biol. Cell. 2004;15:2842–2852. doi: 10.1091/mbc.E04-02-0116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- 28.Lee MS, Henry M, Silver PA. A protein that shuttles between the nucleus and the cytoplasm is an important mediator of RNA export. Genes Dev. 1996;10:1233–1246. doi: 10.1101/gad.10.10.1233. [DOI] [PubMed] [Google Scholar]
- 29.Mayes AE, Verdone L, Legrain P, Beggs JD. Characterization of Sm-like proteins in yeast and their association with U6 snRNA. EMBO J. 1999;18:4321–4331. doi: 10.1093/emboj/18.15.4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spiller MP, Boon KL, Reijns MA, Beggs JD. The Lsm2-8 complex determines nuclear localization of the spliceosomal U6 snRNA. Nucleic Acids Res. 2007;35:923–929. doi: 10.1093/nar/gkl1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verdone L, Galardi S, Page D, Beggs JD. Lsm proteins promote regeneration of pre-mRNA splicing activity. Curr. Biol. 2004;14:1487–1491. doi: 10.1016/j.cub.2004.08.032. [DOI] [PubMed] [Google Scholar]
- 32.Pannone BK, Xue D, Wolin SL. A role for the yeast La protein in U6 snRNP assembly: evidence that the La protein is a molecular chaperone for RNA polymerase III transcripts. EMBO J. 1998;17:7442–7453. doi: 10.1093/emboj/17.24.7442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stevens SW, Abelson J. Purification of the yeast U4/U6.U5 small nuclear ribonucleoprotein particle and identification of its proteins. Proc. Natl Acad. Sci. USA. 1999;96:7226–7231. doi: 10.1073/pnas.96.13.7226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stevens SW, Barta I, Ge HY, Moore RE, Young MK, Lee TD, Abelson J. Biochemical and genetic analyses of the U5, U6, and U4/U6 x U5 small nuclear ribonucleoproteins from Saccharomyces cerevisiae. RNA. 2001;7:1543–1553. [PMC free article] [PubMed] [Google Scholar]
- 35.Tucker M, Valencia-Sanchez MA, Staples RR, Chen J, Denis CL, Parker R. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. doi: 10.1016/s0092-8674(01)00225-2. [DOI] [PubMed] [Google Scholar]
- 36.Coller J, Parker R. General translational repression by activators of mRNA decapping. Cell. 2005;122:875–886. doi: 10.1016/j.cell.2005.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kshirsagar M, Parker R. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics. 2004;166:729–739. doi: 10.1534/genetics.166.2.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 39.Belle A, Tanay A, Bitincka L, Shamir R, O'Shea EK. Quantification of protein half-lives in the budding yeast proteome. Proc. Natl Acad. Sci. USA. 2006;103:13004–13009. doi: 10.1073/pnas.0605420103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pannone BK, Kim SD, Noe DA, Wolin SL. Multiple functional interactions between components of the Lsm2-Lsm8 complex, U6 snRNA, and the yeast La protein. Genetics. 2001;158:187–196. doi: 10.1093/genetics/158.1.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hayes GM, Carrigan PE, Beck AM, Miller LJ. Targeting the RNA splicing machinery as a novel treatment strategy for pancreatic carcinoma. Cancer Res. 2006;66:3819–3827. doi: 10.1158/0008-5472.CAN-05-4065. [DOI] [PubMed] [Google Scholar]
- 42.Thakur A, Bollig A, Wu J, Liao DJ. Gene expression profiles in primary pancreatic tumors and metastatic lesions of Ela-c-myc transgenic mice. Mol. Cancer. 2008;7:11. doi: 10.1186/1476-4598-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Y, Hannon GJ, Zhang H, Casso D, Kobayashi R, Beach D. p21 is a universal inhibitor of cyclin kinases. Nature. 1993;366:701–704. doi: 10.1038/366701a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.