Abstract
Functional telomeres are required to maintain the replicative ability of cancer cells and represent putative targets for G-quadruplex (G4) ligands. Here, we show that the pentacyclic acridinium salt RHPS4, one of the most effective and selective G4 ligands, triggers damages in cells traversing S phase by interfering with telomere replication. Indeed, we found that RHPS4 markedly reduced BrdU incorporation at telomeres and altered the dynamic association of the telomeric proteins TRF1, TRF2 and POT1, leading to chromosome aberrations such as telomere fusions and telomere doublets. Analysis of the molecular damage pathway revealed that RHPS4 induced an ATR-dependent ATM signaling that plays a functional role in the cellular response to RHPS4 treatment. We propose that RHPS4, by stabilizing G4 DNA at telomeres, impairs fork progression and/or telomere processing resulting in telomere dysfunction and activation of a replication stress response pathway. The detailed understanding of the molecular mode of action of this class of compounds makes them attractive tools to understand telomere biology and provides the basis for a rational use of G4 ligands for the therapy of cancer.
INTRODUCTION
Telomeres are the structures at the end of eukaryotic linear chromosomes. Human telomeres consist of tandem repeats of the hexanucleotide sequence TTAGGG in double strand, except for a terminal 3′ G-rich overhang (1,2). Mammalian telomeres are associated with shelterin, a protein complex that functions to protect DNA ends from being recognized and repaired as double strand breaks and from triggering DNA damage responses (3). In addition, telomeres are transcribed, giving rise to chromatin-associated G-rich transcripts, named TERRA, whose function is still elusive (4,5).
Telomeres can fold into t-loops that may result from invasion of the 3′-overhang into duplex DNA (6) or into G-quadruplex (G4) DNA, an unusual DNA conformation based on guanine quartets (7). These structures might be a source of difficulty for the passage of the replication fork and would need to be resolved to allow DNA replication and telomerase elongation (8). Telomeres have evolved as a puzzling diversity of mechanisms to accommodate the replication problems and to reconstitute capped telomeres. Telomere replication relies on a strong synergy among the conventional replication machinery, telomere protection systems, DNA damage response pathway and chromosomal organization (9). Moreover, experiments performed both in yeast and mammalian cells in vitro indicate that telomere-binding proteins could play an essential role in coordinating telomere replication by preventing stalled forks, which may result in telomere attrition or increased recombination (10,11). Indeed, telomeric complex is dynamic during cell cycle and the binding of telomeric proteins to TTAGGG repeats changes as the cycle progresses (12). Recent studies suggest that replication fork naturally pauses or stalls at the telomeres triggering a transient ATM and Rad3-related (ATR)/ataxia-telangiectasia mutated (ATM) DNA damage response, which is not sufficient to stop cell proliferation but which is likely to be required for a proper telomere processing (13). The transient DNA damage response is essential for telomere processing and recruitment of modifying enzymes. Increasing evidences indicate that RecQ-like helicases Werner's syndrome protein (WRN), Bloom's syndrome protein (BLM) and other factors are capable of removing or remodeling the telomeric structures that can impair fork progression. WRN and BLM helicases are proposed to remove the secondary structure, i.e. a G4 that can be formed in the G-rich strand, while topoisomerase (TOPO) I can rapidly relax the accumulation of positive supercoiling that is transiently generated when replication approaches a t-loop (8,14,15).
The burgeoning knowledge about the structure of telomeres and the roles of various factors involved in telomere maintenance provides several possible targets for pharmacological intervention (16). To date, the area that has received major drug discovery attention is the target of the telomeric G4 structure. G4-interacting agents are small molecules that are able to bind to, and stabilize, the telomeric DNA in a quadruplex conformation, thereby inhibiting telomere extension by telomerase (17). Interestingly, TERRA molecules can fold into G4 structures, suggesting that this telomeric RNA could be a target of G4 ligands (18). In fact, a wealth of data indicates that G4-interacting compounds might disrupt telomere architecture, both in telomerase- and alternative lengthening of telomeres (ALT)-positive tumors, causing immediate and profound effects on cell proliferation (19). The pentacyclic acridinium salt RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is one of the most effective and selective G4 ligands (20). Our group recently demonstrated that in addition to its telomerase inhibitory properties, this drug exerts an anticancer effect by telomeric chromatin alteration (21). Here, we report that stabilization of telomeric DNA quadruplex by RHPS4 can interfere with telomere replication, triggering an ATR-dependent damage response pathway.
MATERIALS AND METHODS
Cells and culture conditions
BJ fibroblasts expressing hTERT and SV40 early region (BJ-HELT) were maintained as previously described (22). The wild-type and ChK2-, p53- or p21-deficient HCT116 cells were obtained by Dr Vogelstein, Johns Hopkins University. The GM847/ATR-wt and GM847/ATR-ki were a generous gift from Dr Rosselli, Institut Gustave Roussy, Villejuif, France. The expression of ATR isoforms was induced by the addition of 1 μg/ml of doxycycline to the medium. All the lines were grown in Dulbecco modified eagle medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal calf serum.
Transfection
For RNA interference experiments, cells were transfected with 100 nM of siATM or siATR or siBLM smart pool (Dharmacon, Chicago, IL, USA) in a 35-mm Petri dishes using Lipofectamine 2000 reagent (Lipofectamine 2000, Invitrogen). siGFP was used as negative control (Dharmacon).
Antibodies
The following antibodies were used: mAb and pAb anti-γH2AX (Upstate, Lake Placid, NY, USA); mAb anti-PCNA and mAb anti-β-actin (Sigma Chemicals, Milano, Italy); mAb anti-TRF2 (Imgenex, San Diego, CA, USA); pAb anti-POT1, pAb anti-S1981 ATM, mAb anti-ATR, pAb anti-BLM, pAb and mAb anti-TRF1 (for immunofluorescence application) (Abcam, Cambridge, UK); pAbs anti-TRF1 (for ChIP application), anti-p21, mAb anti-p53 and pAb anti-ATRIP (Santa Cruz Biotechnology, Santa Cruz, CA, USA); pAb anti-53BP1 (Novus Biologicals, Littleton, CO, USA); mAb anti-BrdU (Becton Dickinson, Heidelberg, Germany); pAb anti-Ser15 p53, pAb anti-Chk1, pAb anti-Ser345 Chk1, pAb anti-Chk2 and pAb anti-Thr68 Chk2 (Cell Signaling); pAb anti-WRN (Bethyl Laboratories, Montgomery, TX, USA); mAb anti-ATM (a generous gift from Dr D. Delia, Istituto Nazionale Tumori, Milano, Italy) and mAb anti-HSP 72/73 (Calbiochem, Cambridge, MA, USA).
In vitro treatments
Treatments were performed 24 h after plating and were used at the following doses: 0.5 μM RHPS4 for different time of exposure, 2 mM hydroxyurea (HU; Sigma) for 3 h, ultraviolet (UV) 10 J/m2, ionizing radiation (IR) 5 Gy. Cell counts and viability (trypan blue dye exclusion) were determined in each experiment. For synchronization, BJ-HELT cells were grown to 50% confluence in 10-cm plates and incubated overnight with 0.5 mM HU. The cells were released by replacing the medium in presence or absence of 0.5 μM RHPS4.
Cell viability
Cell viability was evaluated by using the 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide MTT assay (Sigma) according to manufacturer's instructions.
Western blotting
Western blot and detection were performed as previously reported (23). To check the amount of proteins transferred to nitrocellulose membrane, HSP 72/73 or β-actin was used as control. The relative amounts of the transferred proteins were quantified by scanning the autoradiographic films with a gel densitometer scanner (Bio-Rad, Milano, Italy) and normalized to the related HSP 72/73 or β-actin amounts.
Immunofluorescence
Cells were fixed in 2% formaldehyde and permeabilized in 0.25% Triton X-100 in Phosphate buffered saline (PBS) for 5 min at room temperature. For immunolabeling experiments, cells were incubated with primary antibody, washed in PBS and incubated with the following secondary antibodies: TRITC conjugated Goat anti-Rabbit, FITC conjugated Goat anti-Mouse (Jackson Lab). Nuclei were visualized using DAPI or Hoechst (Sigma). For metaphase chromosomes preparation, cells were treated with 0.1 μg/ml demecolcine (Sigma) of for 4 h and harvested and washed in 75 mM KCl for 5 min at 37°C. After centrifugation, cells were fixed in MetOH and acetic acid in the ratio 3:1 overnight and then spread on slides. Hybridization with Rodamine-coupled Peptide Nucleic Acid (PNA) was performed as described by Lenain et al. (24). Images of metaphases were captured with a 100× objective. Fluorescence signals were recorded by using a Leica DMIRE2 microscope equipped with a Leica DFC 350FX camera and elaborated by a Leica FW4000 deconvolution software (Leica, Solms, Germany) or by confocal analysis obtained with a Zeiss LSM 510 META Laser Scanning Microscope (Zeiss, Oberkochen, Germany).
Chromatin immunoprecipitation (ChIP)
Cells were synchronized at the G1/S boundary as described above. After releasing from the HU block, cells were harvested every 2 h by scraping and were fixed with 1% formaldehyde for 10 min at room temperature. Chromatin immunoprecipitation (ChIP) assay was performed as previously described (21).
For chromatin BrdU incorporation, before harvesting each time point, the cells were incubated with 20 mM BrdU (Sigma) for 1 h. After dot blotting, BrdU incorporation into telomeric DNA was evaluated by western blot analysis.
Flow cytometric analysis
Cell cycle analysis was performed by flow cytometry (Becton-Dickinson) as previously described (25). For the separation of G0/G1 from S–G2/M cells, single-cell suspension was incubated with Hoechst for staining DNA and sorted using FACS Vantage DiVa (BD Biosciences, San Jose, CA, USA). Cell cycle phases were sorted and isolated using electronic gating based on fluorescence emission of Hoechst after excitation with UV laser emitting 350–360 nm UV light.
Progression of cells through the cell cycle phases was analyzed by flow cytometry using BrdU (Becton-Dickinson) incorporation, as previously described (26). Briefly, at different times of RHPS4 treatment, cells were pulsed with BrdU at a final concentration of 10 μM for 15 min, and after the appropriate intervals in BrdU-free medium (from 4 to 16 h) the DNA was denatured. Cells were then incubated with 2 μg/ml of mouse anti-BrdU (clone BMC 9318, Roche Diagnostics Corp., Indianapolis, IN, USA) for 30 min at room temperature, and the BrdU-positive cells were revealed with FITC-conjugated anti-mouse mAb (1:20, Dako, SA, Glostrup, Denmark).
Statistical analysis
The experiments have been repeated from three to five times and the results obtained are presented as means ± SD. Significant changes were assessed by using Student's t-test for unpaired data, and P-values < 0.05 were considered significant.
RESULTS
RHPS4 activates DNA damage response during replication
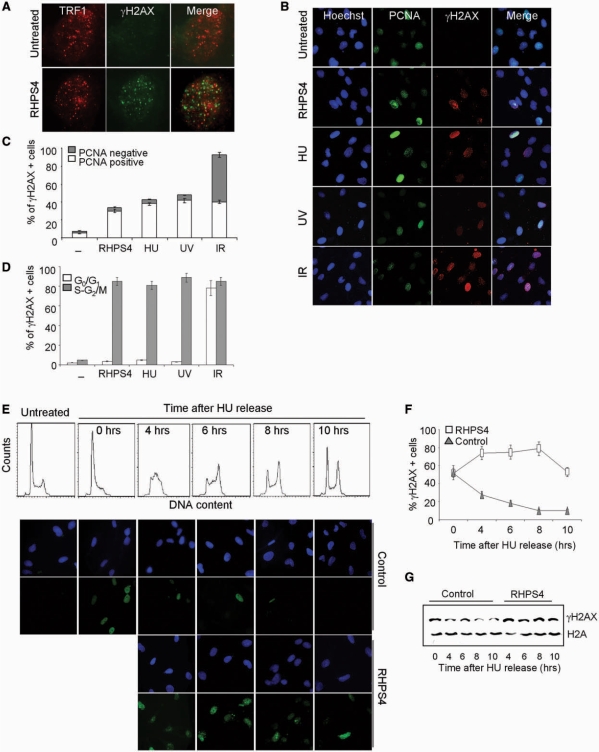
Previously, we showed that exposure of transformed fibroblasts (BJ-HELT cells) to the G4-interactive molecule RHPS4 caused a rapid and specific DNA damage at the telomeres with the formation of several telomeric foci containing damage response factors like phosphorylated H2AX (γH2AX) (21, Figure 1A). Of note, drug-induced phosphorylation of H2AX was restricted to a fraction of RHPS4-treated cells (Figure 1B), suggesting that, similar to treatment with HU and low-UV dose, but in contrast to IRs, RHPS4 can induce a replication-dependent DNA damage. To verify this hypothesis and determine which fraction of the cells formed γH2AX foci, we performed co-immunostaining to γH2AX and the proliferating cell nuclear antigen PCNA, which accumulates in the nucleus during S phase. In the case of RHPS4, as well as of HU and UV, γH2AX foci formation was restricted to PCNA-positive cells, whereas in the case of IR, γH2AX foci formed both in PCNA-positive and -negative cells (Figure 1B and C).
Figure 1.
Replication-dependent induction of damage by RHPS4. (A) BJ-EHLT fibroblasts treated with RHPS4 for 16 h were fixed and processed for IF by using antibodies against γH2AX and TRF1. Representative deconvolution images are reported. (B) BJ-HELT fibroblasts were exposed to the following treatment: 0.5 μM RHPS4 for 16 h, 2 mM HU for 3 h, UV 10 J/m2, IR 5 Gy. Representative images of IF against γH2AX and PCNA were acquired with a Leica Deconvolution microscope (magnification: ×40). (C) Percentage of γH2AX+/PCNA- or γH2AX+/PCNA+ nuclei in the indicated samples. The mean of three independent experiments with comparable results is shown. (D) HeLa cells untreated or exposed to the indicated treatment were sorted by FACS according to the DNA content. The fractions corresponding to cells in G0/G1 and S–G2/M cell cycle phases were cytocentrifuged on cover slips and stained for IF against γH2AX. Histogram represents the percentage of γH2AX-positive cells under different stimulations. The mean of three independent experiments with comparable results is shown. (E) BJ-HELT fibroblasts were treated with a low dose of HU (0.5 mM for 16 h) to block cells at the G1–S boundary. Then, the medium was replaced to release cells and in the treated samples 0.5 μM RHPS4 was added. Cells were fixed at the indicated times for cell cycle analysis (upper panel) and IF against γH2AX. Representative images of IF are reported in the lower panel (magnification: ×63). Percentage of γH2AX-positive nuclei (F) and western blotting analysis of γH2AX (G) in control and RHPS4-treated samples at different times after HU release.
The relationship between γH2AX foci formation and DNA replication was also examined by using cell sorting. For RHPS4, HU and UV, cells in the proliferative compartment (S–G2/M) had the highest γH2AX expression, whereas γH2AX was lowest in the cells in the G0/G1 phase of cell cycle (Figure 1D). In contrast, for IR, γH2AX was equally distributed in all of the cell cycle phases (Figure 1D).
To further confirm that the phosphorylation of H2AX upon RHPS4 treatment is related to replication, BJ-HELT fibroblasts were synchronized by treatment with sub-lethal dose of HU, an agent that causes replication fork arrest due to nucleotide depletion, blocking the cells at the G1/S boundary. Synchronization with HU-induced phosphorylation of H2AX rapidly recovered by release in drug-free medium (Figure 1E and F). Importantly, cells released from the HU block in the presence of RHPS4 showed phosphorylation of H2AX when analyzed during S and G2/M phases (from 4 to 8 h after HU removal) (Figure 1E and F). These results, confirmed by western blotting (Figure 1G), demonstrate that a significative phosphorylation of H2AX is a very early response to RHPS4 when the cells enter into S phase.
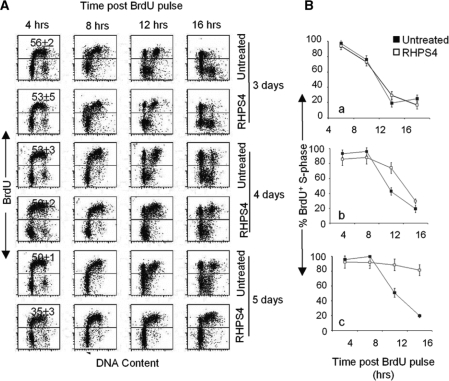
As a consequence of the damage in S phase, cells chronically exposed to RHPS4 showed a delay in the S–G2 transition at Day 4 of drug exposure and at Day 5 most of the cells were arrested (Figure 2A and B). The very early induction of damage (Figure 1E) and the subsequent detrimental effects on cell cycle (Figure 2) suggest that cells have to be chronically exposed to the drug to accumulate enough damages. Indeed, while a short time of RHPS4 exposure is sufficient to phosphorylate H2AX but the cells recover from the damage, long-term treatment creates chronic damages ultimately leading to cell death (Supplementary Figure 1).
Figure 2.
RHPS4 blocks the cells in S–G2 phase of cell cycle. (A) Progression of cells through the cell cycle phases was analyzed by flow cytometry using BrdU. Untreated and RHPS4-treated HeLa cells after 3, 4 and 5 days of drug exposure were pulsed with BrdU for 15 min, and after the appropriate intervals in BrdU-free medium (from 4 to 16 h) the DNA was denatured, incubated with anti-BrdU antibody and the BrdU-positive cells were revealed with FITC-conjugated antibody. The percentage of BrdU-positive cells was reported inside the histogram. (B) The graphs represent the percentage of BrdU-positive S phase cells evaluated at Day 3 (a), 4 (b) and 5 (c) of RHPS4 treatment. The mean of three independent experiments, with SD is shown.
RHPS4 interferes with replication of the telomeres
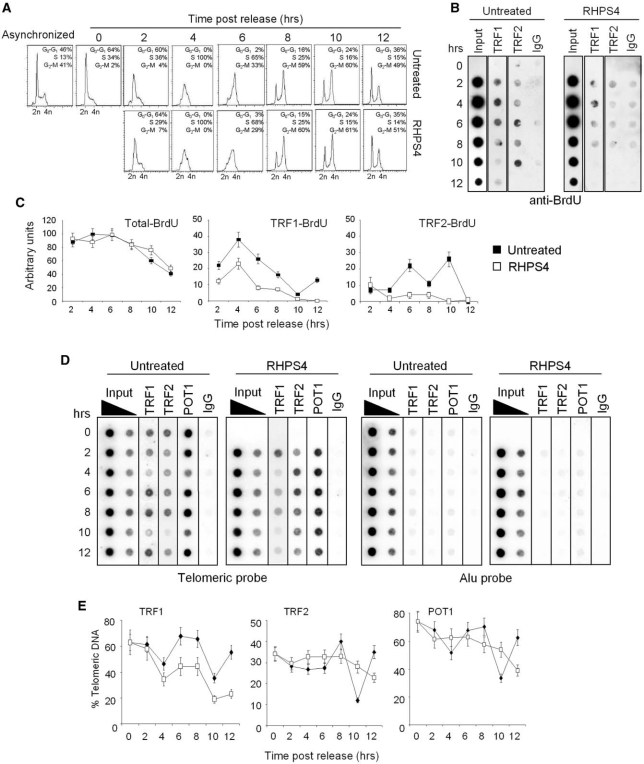
Altogether, the facts that RHPS4 specifically triggers telomere damage and the signaling of RHPS4-induced damages occurs in S phase (Figure 1A) suggest that RHPS4 can interfere with telomere replication. To address this question, we used a combination of BrdU incorporation (to mark replicating DNA) with ChIP assay to measure the amount of replicating DNA associated to telomeric proteins (12). BJ-HELT fibroblasts were synchronized with HU, and released in drug-free or RHPS4-containing medium. Prior to harvest of each individual time point, the cells were incubated with BrdU for 1 h. Cell cycle progression was analyzed by FACS, and DNA content was used as reference for cell cycle phase (Figure 3A). Analysis of untreated samples resulting from immunoprecipitations with antibodies against telomeric-repeat binding factor 1 (TRF1) and TRF2 demonstrated that BrdU was incorporated during S and G2 phases of cell cycle consistent with an early and late telomere replication or telomere processing (12, Figure 3B). The BrdU incorporation pattern was accompanied by transient dissociation of the telomeric proteins: TRF1 showed a decrease at the telomeres in S and G2 phases, while TRF2 and protection of telomeres 1 (POT1) exhibited a dip only during G2 followed a rapid recovery of binding (10, Figures 3D–E). Treatment with RHPS4 did not modify cell cycle progression at the indicated time points (Figure 3A) and cells incorporated BrdU to the same extent of untreated ones (Figure 3B, input lines, and C, left panel). However, RHPS4 markedly decreased the amount of BrdU in the chromatin fragments of immunoprecipitated with TRF1 or TRF2 antibodies (Figure 3B and C). Moreover, the pattern of the three telomeric proteins was remarkably different in RHPS4-treated samples (Figure 3D and E). Indeed, compared with untreated ones there is less telomere-bound TRF1 in RHPS4-treated cells progressing through S and G2 (Figure 3D and E) phases. On the contrary, TRF2 and POT1 remained more stably associated with the telomeric DNA in RHPS4-treated compared with untreated samples. Moreover, the typical dip of two telomeric proteins during the G2 phase followed by the rapid recovery of their binding was not observed in RHPS4-treated cells. Western blotting showed that none of the changes observed both in RHPS4-untreated and treated cells were due to altered levels of protein expression (Supplementary Figure 2). Altogether, our results suggest that RHPS4, by stabilizing G4 DNA at telomeres, can impair fork progression and/or telomere processing.
Figure 3.
RHPS4 impairs telomere replication and alters the dynamics of telomeric proteins. BJ-HELT fibroblasts were synchronized at the G1/S boundary with HU, treated with RHPS4 or released in drug-free medium and harvested every 2 h. Prior to harvest of each individual time point, the cells were incubated with BrdU for 1 h. (A) FACS analysis of untreated and RHPS4-treated BJ-HELT fibroblasts released from G1/S block at the indicated times. (B) ChIP experiments on synchronized BJ-HELT fibroblasts incubated with BrdU. Precipitations were performed with antibodies against TRF1 and TRF2. The precipitated DNA was analyzed by western blotting with anti-BrdU antibody. The total DNA (input) represents 10% of genomic DNA (C) The graphs show the densitometric evaluation of three independent experiments, with error bars indicating the SD. (D) Protein extracts from cells at the indicated times were subjected to ChIP experiments using antibodies against TRF1, TRF2 and POT1. IgG antibody was used as negative control. The total DNA (input) represents 10 and 1% of genomic DNA. Southern blot analysis was performed by using telomeric or ALU repeat-specific probes. (E) The signals obtained were quantified by densitometry, and the percentage of precipitated DNA was calculated as a ratio of input signals and plotted. Four independent experiments were evaluated and error bars indicate the SD.
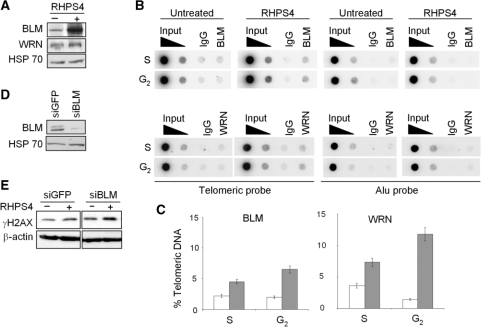
Since WRN and BLM have been proposed to dissociate unusual DNA structures that could block telomere replication and to solve aberrant replication intermediates, we evaluated the effect of RHPS4 on the expression of these two proteins. The results reported in Figure 4A demonstrate that WRN and even more the BLM helicases were increased upon drug treatment. Moreover, both WRN and BLM bound the telomeric sequences more efficiently in RHPS4-treated compared with untreated samples (Figure 4B and C). Functionally, the knock down of BLM by RNA interference strategy (Figure 4D) markedly increased RHPS4-induced damage (Figure 4E), showing that BLM can be required to cop out the replication defects caused by RHPS4.
Figure 4.
BLM and WRN helicases are increased and recruited to the telomeres upon RHPS4 treatment. (A) Western blot analysis of BLM and WRN helicases in BJ-HELT cells untreated or treated with RHPS4 for 96 h. (B) ChIP experiments on BJ-HELT fibroblasts untreated and treated with RHPS4. Protein extracts from cells during S and G2 phases of cell cycle were subjected to ChIP experiments using antibodies against BLM and WRN. IgG antibody was used as negative control. The total DNA (input) represents 10 and 1% of genomic DNA. Southern blot analysis was performed by using telomeric or ALU repeat-specific probes. (C) The signals obtained were quantified by densitometry, and the percentage of precipitated DNA was calculated as a ratio of input signals and plotted. Three independent experiments were evaluated and error bars indicate the SD. (D) Western blot analysis of BLM in siGFP and siBLM-transfected cells. (E) Western blot analysis of γH2AX in siGFP and siBLM transfected cells untreated or treated with RHPS4.
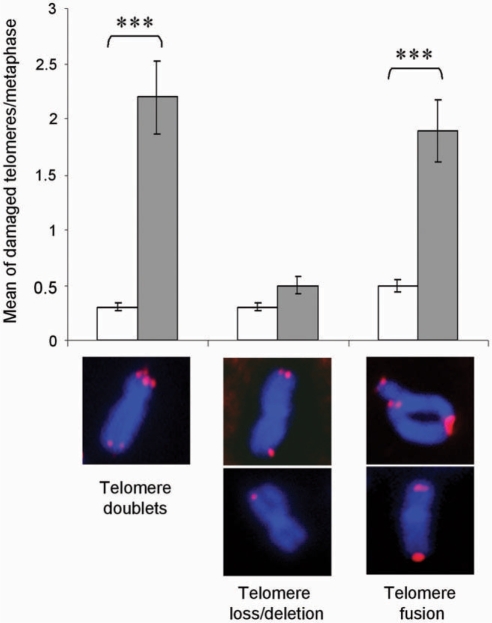
We finally evaluated the consequence of telomere replication defects caused by RHPS4 by analyzing chromosome aberrations after 4 days of treatment, just before induction of cell death. Fluorescence in situ hybridization by using a telomeric PNA probe on metaphase spreads showed that RHPS4 induced telomere instability as revealed by a significant increase of telomere aberration with a mean of 4 ± 0.9 damaged telomeres in RHPS4-treated metaphases versus 1 ± 0.1 in untreated controls. Specifically, RHPS4 caused well-described telomere aberrations: telomere doublets at single chromatid ends and both sister chromatid fusions and/or telomere fusions between two distinct chromosomes (Figure 5). On the contrary, RHPS4 did not induce telomere losses or deletions revealed as a decreased intensity or a lack of telomere signal on one or both sister chromatids (Figure 5).
Figure 5.
RHPS4 induces telomere aberrations. BJ-HELT fibroblasts were treated with 0.5 μM RHPS4 for 96 h and hybridized successively with a telomeric PNA probe (in red) and then counterstained with Hoechst (blue). Histograms show the mean of damaged telomeres per cell in metaphase spreads. ***t-test P-value <0.0001. Representative images of the different telomere aberrations are presented at the bottom.
RHPS4 activates an ATR-dependent checkpoint pathway
We next analyzed the molecular pathway activated by telomere replication defects caused by RHPS4 treatment. Previously we showed that RHPS4-induced phosphorylation of H2AX at the telomeres involves the ATR kinase (21, Supplementary Figure 3). This is in agreement with several data indicating that ATR controls the downstream response to replicational stress-inducing agents (27, Supplementary Figure 4).
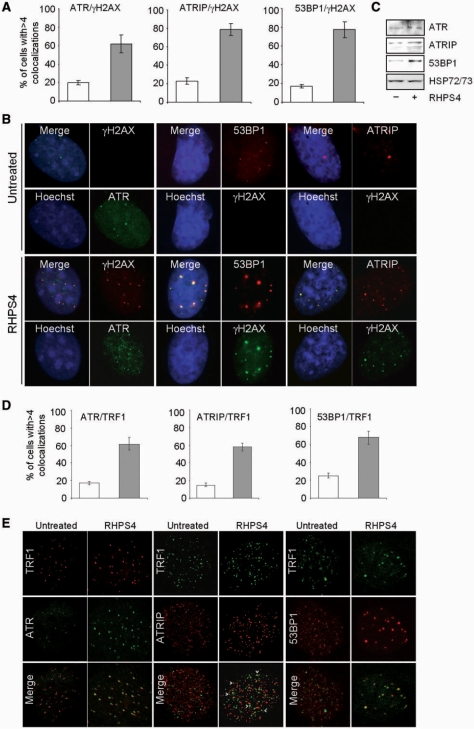
By using biochemical and microscopy techniques, we further demonstrated here that, upon RHPS4 treatment in BJ-HELT cells, ATR, its cofactor ATRIP and the DNA damage response protein 53BP1 were activated and co-localized with γH2AX (Figure 6A–C). Moreover, confocal microscopy experiments revealed that all the ATR and 53BP1 foci are spatially close to TRF1 spots and the signals in part overlap (Figure 6D and E), while to a lesser extent partial co-localizations were observed between ATRIP and TRF1 (see arrow heads). These results suggest that not all the telomeres can have a stalled fork or telomeres/parts of telomeres have not replicated yet.
Figure 6.
RHPS4 activates ATR, its co-factors ATRIP and 53BP1. BJ-HELT fibroblasts were synchronized, treated with 0.5 μM RHPS4 or released in drug-free medium for 6 h and co-immunostained with anti-ATR, ATRIP, 53BP1 antibodies and anti-γH2AX or anti-TRF1. (A) Percentage of cells with more than four co-localizations in control and RHPS4-treated samples. (B) Representative images of IF acquired with a Leica Deconvolution microscope (magnification: ×63). (C) Western blot analysis of ATR, ATRIP and 53BP1. (D) Percentage of cells with more than four co-localizations. (E) Representative images of IF acquired by confocal microscope (magnification: ×63). Control (white bars); RHPS4 (gray bars). The means of three independent experiments with comparable results are shown.
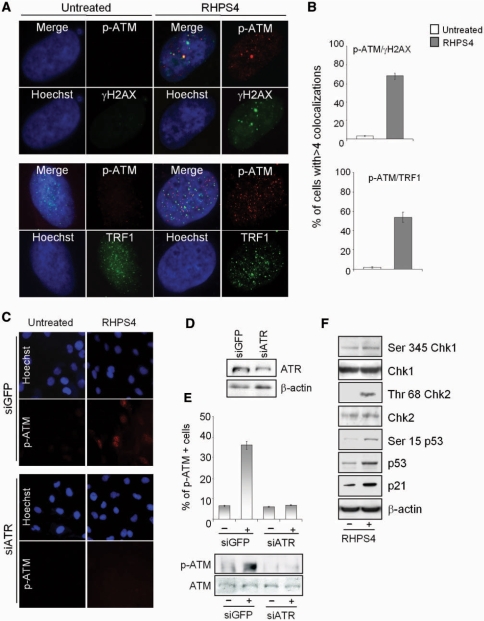
Although ATM is not required for the DNA damage response triggered by RHPS4 (21, Supplementary Figure 3A–D), it is phosphorylated on Ser1981 following the drug treatment and co-localized with damaged telomeres, as revealed by co-immunostaining against γH2AX and TRF1 (Figure 7A and B). This activation is completely abrogated in ATR knock-down cells (Figure 7C–E), placing ATR upstream of ATM and establishing a cross-talk between the two kinases in response to RHPS4. Of note, ATR-dependent ATM phosphorylation is a peculiar characteristic of replicational stress-inducing agents, while ATM works upstream to ATR during IR (27, Supplementary Figure 5).
Figure 7.
RHPS4 triggers an ATR-dependent ATM damage signaling pathway. BJ-HELT fibroblasts were treated with 0.5 μM RHPS4 for 16 h, fixed and co-immunostained with pospho-ATM on Ser1981 (p-ATM) and anti-γH2AX or TRF1 antibodies. Hoechst staining was used to mark nuclei. (A) Representative images acquired by a deconvolution microscopy (magnification: ×100) of p-ATM/γH2AX and p-ATM/TRF1. (B) Percentage of cells with more than four p-ATM/γH2AX and p-ATM/TRF1 co-localizations. The mean of three independent experiments with comparable results is shown. BJ-HELT fibroblasts were transfected with 100 mM of siGFP and siATR. After 48 h, cells were treated with 0.5 μM RHPS4 for 16 h and processed for IF against p-ATM. (C) Representative images of IF acquired with a Leica Deconvolution microscope (magnification: ×40). (D) Western blot analysis of ATR in siGFP and siATR transfected BJ-HELT cells. (E) Percentage of p-ATM-positive cells and western blotting of p-ATM in the indicated samples. (F) Western blot analysis of the phosphorylated form of ChK1, ChK2 and p53 in HeLa cells untreated or treated with RHPS4 for 4 days. The nonphosphorylated forms of the same proteins as well as the expression of p21 are also reported.
Analysis of transducer kinases in HeLa cells (containing active ATM/ATR pathways; 28) revealed that Chk2 was phosphorylated by RHPS4 and p53/p21 molecular death targets were activated (Figure 7F). All these findings strongly demonstrate that RHPS4 fully activates a DNA damage response pathway leading to cell death.
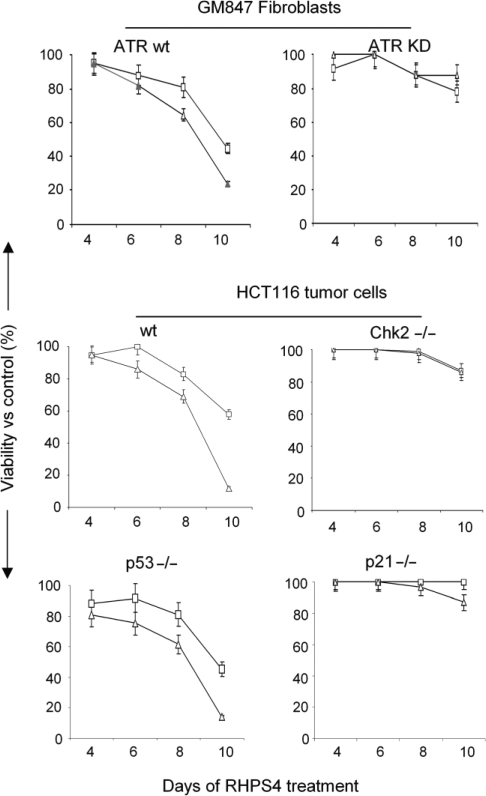
We finally assessed the biological relevance of upstream and downstream kinases by using cell lines with characterized deficiencies. The results demonstrated that RHPS4 induced a dose-dependent reduction of cell viability in transformed fibroblasts expressing inducible wild-type ATR, but not in those expressing kinase inactive ATR (which results in an ATR-dominant negative status). Also HCT116, Chk2 and p21 defective cell lines were resistant to RHPS4 treatment in terms of cell viability (Figure 8). On the contrary, compared with the wild-type cells, the p53 null cell viability was reduced to the same extent (Figure 8). These results demonstrate that while p53 does not influence the response of cells to RHPS4 treatment, the lack of ATR, ChK2 and p21 makes cells resistant to the compound.
Figure 8.
ATR-, ChK2- and p21-defective cells are resistant to RHPS4 treatment. The GM847 and GM847/ATR-KD cells expressing the wild-type and inactive ATR kinase, the wild-type, ChK2- and p21-deficient HCT116 cells and the cells expressing the mutant p53 protein were treated with RHPS4. The graphs represent the viability of RHPS4-treated cells at 0.5 (open square) and 1 μM (open triangle) drug concentration evaluated by MTT assay at the indicated time of treatment. Data are expressed as percentage of cell viability compared with untreated cells. The figures show representative experiments performed in quintuplicate with SDs.
DISCUSSION
RHPS4 is a G4-interactive molecule possessing antitumoral activity because of its ability to rapidly induce telomere damage and cell death independently of telomerase inhibition (21). We reveal here that RHPS4 damages telomeric chromatin during replication triggering an ATR-dependent signaling pathway. Therefore, RHPS4 efficiently impairs the two main pathways of telomere maintenance in dividing cells: elongation by telomerase (29) and telomere replication (this study). This makes this compound particularly suitable to target proliferating cancer cells.
RHPS4 triggers damages in cells during S–G2 phase of cell cycle by interfering with telomere replication. Indeed, we found that RHPS4 specifically reduces the BrdU incorporation in the telomeric chromatin immunoprecipitated with TRF1 and TRF2 antibodies without altering the incorporation of BrdU into total DNA. In addition, RHPS4 triggers a marked alteration of the typical dynamic association of the telomeric proteins TRF1, TRF2 and POT1 occurring during cell cycle (12). Specifically, TRF1 dissociation, which occurs transiently during S–G2 phase is significantly increased by RHPS4 treatment, while, in the meantime, the amount of telomere-bound TRF2–POT1 complex is increased. An explanation of these results is that RHPS4 acts at a point after TRF1 dissociation, i.e. during or just after fork passage, preventing its re-association to the newly formed telomere. Alternatively, RHPS4 might displace TRF1 prior to the passage of the replisome, leading to fork stalling. In these cases, the enhanced binding of TRF2 might simply be the consequence of a lower abundance of TRF1. Another possibility is that the replication damage induced by RHPS4 leads to a specific recruitment of TRF2 and subsequently to TRF1 release. Anyhow, the fact that the timing of dissociation and re-association is different between TRF1 and TRF2–POT1 in response to RHPS4 and is in agreement with previous work showing that the shelterin components can be found in cells as separate subcomplexes (30,31). As revealed by RHPS4 treatment, these subcomplexes might have specific roles during telomere replication.
These results indicate that RHPS4 can impair or delay the timing of telomere replication. As presented in Supplementary Figure 6, one can imagine that RHPS4 interferes with telomere replication because it stabilizes the various types of G4 structures that could be formed at telomeres. In particular, RHPS4 could bind to G4 formed during lagging-strand synthesis or at the 3′-overhang or at the displaced strand of the D-loop, which was proposed to form the basis of t-loop, or in G-loop during TERRA transcription or at TERRA itself. This hypothesis is consistent with our results showing that RHPS4 increases the expression and the telomeric association of BLM and WRN that are principally involved in unwinding G4 DNA as well as other secondary structures that can block fork progression, such as D-loop. The functional role of the BLM helicase has been directly assessed by showing that a reduced expression of BLM markedly increases drug-induced damage. Therefore, it appears that an overexpression of the BLM helicase may provide a means for RHPS4-treated cells to respond to telomere replication defects. In agreement with this view, BLM is often upregulated in cancer cells, probably to accommodate the alterations on replication due to oncogene activation (32).
The G4 structures ‘locked’ by RHPS4 and located in front of the replication fork could create topological barriers (Supplementary Figure 6) leading to fork pausing or stalling, an accumulation of superhelical stress and an alteration of protein dynamics at telomeres. In agreement with this scenario, RHPS4 shows a strong synergistic interaction with camptothecin, a topoisomerase I inhibitor (33), reduces BrdU incorporation at telomeres and alters the binding of telomeric protein during S/G2 (this study).
Telomeric replication defects induced by RHPS4 treatment can result in a recruitment of homologous recombination factors to the stalling forks leading to chromosome rearrangement. Indeed, cells treated with RHPS4 showed an increased frequency of chromosome aberrations, previously described in other contexts in which telomeres were destabilized: telomere doublets are characterized by two telomere arrays (one in terminal position) separated by various lengths of nontelomeric DNA, they could result from an improper T-loop formation, which could recombine in cis with internal sequences of DNA giving rise to double stained telomeres (34), thus in agreement with the hypothesis of RHPS4 interference with terminal processing. Homologous recombination factors are usually recruited at stalling forks. A pause or an arrest of replication fork at telomeres could be resolved by fusion between sister chromatids (8). Moreover, incorrectly capped telomeres are known to give rise to telomeric fusions between different chromosomes (35).
Of note, a short time exposure of tumor cells with low-dose RHPS4 is sufficient to trigger replication defect but not to stop cell proliferation, strongly supporting the conclusion that the S–G2 phase-arrested cells observed upon RHPS4 treatment are a consequence of telomere replication defects, which represent the early event of RHPS4-induced damages. However, long-term exposure of RHPS4 creates chronic damages ultimately leading to cell death.
Analysis of the molecular pathway activated by RHPS4 strongly supports the conclusion that this drug induces a replication stress. Indeed, in our situation, similarly to that triggered by relication stress-inducing agents, such as HU and UV (27), ATR is the main kinase involved in RHPS4 damage, while ATM functions downstream to ATR leading to ChK2/p53/p21 activation. As a result, blockade of ATR or genetic deficiency of ChK2 as well as of p21 inhibits cell death, suggesting an important role for ATR/ChK2/p21 molecules during RHPS4-induced telomere damage. Interestingly, the status of p53 protein does not influence the response of cells to RHPS4 treatment. This is in agreement with our previous results showing that RHPS4 is equally effective in cells expressing both wild-type and mutant p53 (20,21), including the BJ-HELT fibroblasts reported in this article in which p53 and Rb pathways are functionally inactivated by SV40.
In conclusion, this work describes a new mechanism through which G4 ligands can induce telomere damage, raising the intriguing question of the role of G4 DNA in the control of telomere replication. More interestingly, the better characterization of the mode of action of RHPS4 might have implications for the selection of tumors that are more likely responsive to the drug treatment leading to the development of rational and more effective drug combinations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Italian Association for Cancer Research (AIRC); Ministero della Salute, Institut National du Cancer (INCa); Association de Recherche contre le Cancer (ARC). Funding for open access charge: Italian Association for Cancer Research (AIRC).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We are very grateful to Drs Vogelstein, Rosselli, Murnane and Sabatier for cell lines and Mrs Adele Petricca for her helpful assistance in typing the manuscript.
Footnotes
The authors wish it to be known that, in their opinion, the first two authors should be regarded as joint First Authors.
REFERENCES
- 1.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal human chromosomes have long G-rich telomeric overhangs at one end. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Makarov VL, Hirose Y, Langmore JP. Long G tails at both ends of human chromosomes suggest a C strand degradation mechanism for telomere shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 3.de Lange T. Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 4.Azzalin CM, Reichenbach P, Khoriauli L, Giulotto E, Lingner J. Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science. 2007;318:798–801. doi: 10.1126/science.1147182. [DOI] [PubMed] [Google Scholar]
- 5.Schoeftner S, Blasco MA. Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 2008;10:228–236. doi: 10.1038/ncb1685. [DOI] [PubMed] [Google Scholar]
- 6.Griffith JD, Comeau L, Rosenfield S, Stansel RM, Bianchi A, Moss H, de Lange T. Mammalian telomeres end in a large duplex loop. Cell. 1999;97:503–514. doi: 10.1016/s0092-8674(00)80760-6. [DOI] [PubMed] [Google Scholar]
- 7.Oganesian L, Bryan TM. Physiological relevance of telomeric G-quadruplex formation: a potential drug target. Bioessays. 2007;29:155–165. doi: 10.1002/bies.20523. [DOI] [PubMed] [Google Scholar]
- 8.Gilson E, Géli V. How telomeres are replicated. Nat. Rev. Mol. Cell Biol. 2007;8:825–838. doi: 10.1038/nrm2259. [DOI] [PubMed] [Google Scholar]
- 9.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 10.Miller KM, Rog O, Cooper JP. Semi-conservative DNA replication through telomeres requires Taz1. Nature. 2006;440:824–828. doi: 10.1038/nature04638. [DOI] [PubMed] [Google Scholar]
- 11.Ohiki R, Ishikawa F. Telomere-bound TRF1 and TRF2 stall at the replication fork at telomeric repeats. Nucleic Acids Res. 2004;32:1627–1637. doi: 10.1093/nar/gkh309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Verdun RE, Crabbe L, Haggblom C, Karlseder J. Functional human telomeres are recognized as DNA damage in G2 of the cell cycle. Mol. Cell. 2005;20:551–561. doi: 10.1016/j.molcel.2005.09.024. [DOI] [PubMed] [Google Scholar]
- 13.Verdun RE, Karlseder J. The DNA damage machinery and homologous recombination pathway act consecutively to protect human telomeres. Cell. 2006;127:709–720. doi: 10.1016/j.cell.2006.09.034. [DOI] [PubMed] [Google Scholar]
- 14.Shen J, Loeb LA. Unwinding the molecular basis of the Werner syndrome. Mech. Ageing Dev. 2001;122:921–944. doi: 10.1016/s0047-6374(01)00248-2. [DOI] [PubMed] [Google Scholar]
- 15.Crabbe L, Verdun RE, Haggblom CI, Karlseder J. Defective telomere lagging strand synthesis in cells lacking WRN helicase activity. Science. 2004;306:1951–1953. doi: 10.1126/science.1103619. [DOI] [PubMed] [Google Scholar]
- 16.Kelland L. Targeting the limitless replicative potential of cancer: the telomerase/telomere pathway. Clin. Cancer Res. 2007;13:4960–4963. doi: 10.1158/1078-0432.CCR-07-0422. [DOI] [PubMed] [Google Scholar]
- 17.Mergny JL, Riou JF, Mailliet P, Teulade-Fichou MP, Gilson E. Natural and pharmacological regulation of telomerase. Nucleic Acids Res. 2002;30:839–865. doi: 10.1093/nar/30.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Cian A, Gros J, Guédin A, Haddi M, Lyonnais S, Guittat L, Riou JF, Trentesaux C, Saccà B, Lacroix L, et al. DNA and RNA quadruplex ligands. Nucleic Acids Symp. Ser. 2008;52:7–8. doi: 10.1093/nass/nrn004. [DOI] [PubMed] [Google Scholar]
- 19.Biroccio A, Leonetti C. Telomerase as a new target for the treatment of hormone-refractory prostate cancer. Endocr. Relat. Cancer. 2004;11:407–421. doi: 10.1677/erc.1.00764. [DOI] [PubMed] [Google Scholar]
- 20.Leonetti C, Amodei S, D’Angelo C, Rizzo A, Benassi B, Antonelli A, Elli R, Stevens MF, D’Incalci M, Zupi G, et al. Biological activity of the G-quadruplex ligand RHPS4 (3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium methosulfate) is associated with telomere capping alteration. Mol. Pharmacol. 2004;66:1138–1146. doi: 10.1124/mol.104.001537. [DOI] [PubMed] [Google Scholar]
- 21.Salvati E, Leonetti C, Rizzo A, Scarsella M, Mottolese M, Galati R, Sperduti I, Stevens MF, D’Incalci M, Blasco M, et al. Telomere damage induced by the G-quadruplex ligand RHPS4 has an antitumor effect. J. Clin. Invest. 2007;117:3236–3247. doi: 10.1172/JCI32461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brunori M, Mathieu N, Ricoul M, Bauwens S, Koering CE, Roborel de Climens A, Belleville A, Wang Q, Puisieux I, Décimo D, et al. TRF2 inhibition promotes anchorage-independent growth of telomerase-positive human fibroblasts. Oncogene. 2006;25:990–997. doi: 10.1038/sj.onc.1209135. [DOI] [PubMed] [Google Scholar]
- 23.Biroccio A, Amodei S, Benassi B, Scarsella M, Cianciulli A, Mottolese M, Del Bufalo D, Legnetti C, Zupi G. Reconstitution of hTERT restores tumorigenicity in melanoma-derived c-Myc low-expressing clones. Oncogene. 2002;21:3011–3019. doi: 10.1038/sj.onc.1205415. [DOI] [PubMed] [Google Scholar]
- 24.Lenain C, Bauwens S, Amiard S, Brunori M, Giraud-Panis MJ, Gilson E. The Apollo 5′ exonuclease functions together with TRF2 to protect telomeres from DNA repair. Curr. Biol. 2006;16:1303–1310. doi: 10.1016/j.cub.2006.05.021. [DOI] [PubMed] [Google Scholar]
- 25.Citro G, D’Agnano I, Leonetti C, Perini R, Bucci B, Zon G, Calabretta B, Zupi G. c-myc antisense oligodeoxynucleotides enhance the efficacy of cisplatin in melanoma chemotherapy in vitro and in nude mice. Cancer Res. 1998;58:283–289. [PubMed] [Google Scholar]
- 26.Biroccio A, Benassi B, Amodei S, Gabellini C, Del Bufalo D, Zupi G. c-Myc down-regulation increases susceptibility to cisplatin through reactive oxygen species-mediated apoptosis in M14 human melanoma cells. Mol. Pharmacol. 2001;60:174–182. doi: 10.1124/mol.60.1.174. [DOI] [PubMed] [Google Scholar]
- 27.Stiff T, Walker SA, Cerosaletti K, Goodarzi AA, Petermann E, Concannon P, O’Driscoll M, Jeggo PA. ATR-dependent phosphorylation and activation of ATM in response to UV treatment or replication fork stalling. EMBO J. 2006;25:5775–5782. doi: 10.1038/sj.emboj.7601446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukasand J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 29.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent inhibition of telomerase by small-molecule pentacyclic acridines capable of interacting with G-quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 30.Mattern KA, Swiggers SJJ, Nigg AL, Löwenberg B, Houstmuller AB, Zijlmans JMJM. Dynamics of protein binding to telomeres in living cells: implications for telomere structure and function. Mol. Cell Biol. 2004;24:5587–5594. doi: 10.1128/MCB.24.12.5587-5594.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen LY, Liu D, Songyang Z. Telomere maintenance through spatial control of telomeric proteins. Mol. Cell Biol. 2007;27:5898–5909. doi: 10.1128/MCB.00603-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kawabe T, Tsuyama N, Kitao S, Nishikawa K, Shimamoto A, Shiratori M, Matsumoto T, Anno K, Sato T, Mitsui Y. Differential regulation of human RecQ family helicases in cell transformation and cell cycle. Oncogene. 2000;19:4764–4772. doi: 10.1038/sj.onc.1203841. [DOI] [PubMed] [Google Scholar]
- 33.Legnetti C, Scarsella M, Raggio G, Rizzo A, Salvati E, D’Incalci M, Staszewsky L, Frapolli R, Stevens MF, Stoppacciaro A, et al. G-quadruplex ligand RHPS4 potentiates the antitumor activity of camptothecins in preclinical models of solid tumors. Clin. Cancer Res. 2008;14:7284–7291. doi: 10.1158/1078-0432.CCR-08-0941. [DOI] [PubMed] [Google Scholar]
- 34.Philippe C, Coullin P, Bernheim A. Double telomeric signals on single chromatids revealed by FISH and PRINS. Ann. Genet. 1999;42:202–209. [PubMed] [Google Scholar]
- 35.van Steensel B, Smogorzewska A, de Lange T. TRF2 protects human telomeres from end-to-end fusions. Cell. 1998;92:401–13. doi: 10.1016/s0092-8674(00)80932-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.