Abstract
We report 13C magic angle spinning NMR observation of photochemically induced dynamic nuclear spin polarization (photo- CIDNP) in the reaction center (RC) of photosystem II (PS2). The light-enhanced NMR signals of the natural abundance 13C provide information on the electronic structure of the primary electron donor P680 (chlorophyll a molecules absorbing around 680 nm) and on the pz spin density pattern in its oxidized form, P680⨥. Most centerband signals can be attributed to a single chlorophyll a (Chl a) cofactor that has little interaction with other pigments. The chemical shift anisotropy of the most intense signals is characteristic for aromatic carbon atoms. The data reveal a pronounced asymmetry of the electronic spin density distribution within the P680⨥. PS2 shows only a single broad and intense emissive signal, which is assigned to both the C-10 and C-15 methine carbon atoms. The spin density appears shifted toward ring III. This shift is remarkable, because, for monomeric Chl a radical cations in solution, the region of highest spin density is around ring II. It leads to a first hypothesis as to how the planet can provide itself with the chemical potential to split water and generate an oxygen atmosphere using the Chl a macroaromatic cycle. A local electrostatic field close to ring III can polarize the electronic charge and associated spin density and increase the redox potential of P680 by stabilizing the highest occupied molecular orbital, without a major change of color. This field could be produced, e.g., by protonation of the keto group of ring V. Finally, the radical cation electronic structure in PS2 is different from that in the bacterial RC, which shows at least four emissive centerbands, indicating a symmetric spin density distribution over the entire bacteriochlorophyll macrocycle.
Photosystem II (PS2) is a multisubunit membrane protein complex that consists of 25 different proteins (1). At the heart of this multisubunit complex is the PS2 reaction center (RC), comprising the D1 and D2 polypeptides. These two polypeptides bind the cofactors that are involved in the light-driven primary electron transfer process (2). On illumination, a special Chl a molecule or aggregate of Chl a molecules absorbing around 680 nm (P680) is initially brought into its first electronically excited singlet state and donates the energized electron within a few picoseconds to a pheophytin (Phe) molecule to form the radical pair state P680⨥Phe⨪. Phe⨪ then passes an electron to a bound plastoquinone molecule (QA) within 300 ps. The P680⨥ is the strongest oxidizing agent known in living nature (P680/P680⨥ ≈ 1.2 V). P680⨥ is reduced within nano- to microseconds by a redox active tyrosine (YZ) at position 161 of the D1 protein. YZ⋅ is reduced by a tetra-manganese cluster, which stores the oxidation equivalents needed for oxidizing water to molecular oxygen.
A high resolution structure of PS2 has not yet been published. On the basis of the similarity to the purple bacterial RC, models of the structure of the RC of PS2 have been proposed (3–6). They are in line with recent electron microscopic investigations on RCs of PS2 (7) and take into account the spectroscopic evidence (reviewed in ref. 8) that PS2 does not contain a tightly coupled “special pair” like P in the bacterial RC. The presentation of a structural model is expected soon from crystallographic data (9).
Despite the fact that the RC of PS2 has been investigated in recent years with a variety of spectroscopic tools, no clear picture of the process of the emission of an electron from the electronically excited state of the primary electron donor exists. In addition, a detailed understanding of the molecular mechanism of the inhibition of the back reaction, which is probably due to the high exothermic reaction enthalpy pushing the system into the inverted Marcus region, is missing. Finally, it is not clear how the high redox potential of the P⨥ is established, which suggests an essentially different structure and environment of the primary electron donor. However, these special features of P680 are not yet known. Comparison of the mechanism of electron emission and trapping in different natural photosynthetic electron pumps is of great interest, because the extraction of the general features of the electronic structure of highly optimized light-driven electron pumps would open the path to optimize an artificial biomimetic solar-energy conversion device.
Solid-state NMR is a rapidly growing technique in the study of membrane proteins (10) and has already been applied to isotope-labeled bacterial photosynthetic RCs from Rhodobacter sphaeroides (11–14). Recently, a novel application of magic angle spinning (MAS) NMR in photosynthesis research was recognized when photochemically induced dynamic nuclear polarization (photo-CIDNP) signals were observed in a QA-depleted bacterial RC (15–17). It is highly remarkable to obtain strong NMR signals directly from the active site in bacterial RCs even without any kind of isotopic enrichment (16). Photo-CIDNP has been known for a long time in liquid NMR (18). The NMR intensities of nuclei involved in photo-CIDNP are strongly enhanced without affecting the ground-state frequencies. With 15N solid-state NMR, photo-CIDNP signals have been observed arising from bacteriochlorophyll (BChl) and BPhe molecules, as well as from histidine (15, 17). The 13C photo-CIDNP signals from the bacterial RC without any kind of isotope enrichment correspond globally with the response expected for a single BChl molecule (16, 19). Although the exact mechanism of nuclear spin selection and NMR-intensity enhancement by photo-CIDNP in the solid-state NMR spectrum of bacterial RCs is still under discussion (20–22), the observation of photo-CIDNP NMR is a unique tool to provide a coherent molecular picture of the unpaired electron density distribution in the radical cation with atomic selectivity in a single experiment. This is possible by observing the pattern of correlated direct (positive) and induced (negative) atomic spin densities via the 13C nuclei of the aromatic carbons in the π-skeleton (23). These carbons carry pz spin density in the radical cation state and have 13C nuclei with large hyperfine couplings. In addition, CIDNP is a transient event, where effective nuclear spin polarization is established on a time scale of ≈ 20 ns after photo-excitation. These are important advantages over the electron nuclear double resonance (ENDOR) methods that probe the s spin densities mainly for nuclei outside the ring. Here, we present photo-CIDNP spectra of PS2 and compare them with the photo-CIDNP data of the bacterial RC. We show that the photo-CIDNP effect of PS2 is in line with a model of P680 as a Chl a molecule, weakly coupled with other cofactors and strongly interacting with the apoprotein.
Materials and Methods
PS2 Reaction Center Preparation.
Highly purified PS2 reaction centers were prepared from spinach following the method developed by van Leeuwen et al. (24). First PS2 membrane fragments were isolated according to Berthold et al. (25) and suspended in BTS200 [20 mM [bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane (Bistris), pH 6.5/20 mM MgCl2/5 mM CaCl2/10 mM MgSO4/0.2 M sucrose/0.03% (wt/vol) n-dodecyl β-d-maltoside]. PS2 membranes were solubilized with 1.25% (wt/vol) n-dodecyl β-d-maltoside and loaded on a Q-Sepharose column (Amersham Pharmacia) equilibrated with BTS200. Light harvesting complex II was separated from PS2 and removed by extensive washing of the column with BTS200. After subsequent treatment of the resulting PS2 core particles with Triton X-100 on the column and removal of PS2 core antenna proteins, the purified PS2 reaction center complex (D1-D2-cytb559 with about 6 attached chlorophylls and 2 pheophytins) was eluted from the column by using BTS200 containing 75 mM MgCl2 as described earlier (24). No photoreducible QA was found in the purified PS2 RC. For NMR measurements, PS2 RCs were concentrated with a Biomax-30 30,000 NMWL polysulfone membrane filter unit (Millipore). The Chl content of PS2 RC was determined by the method of Arnon et al. (26). PS2 RCs equivalent to ≈2 mg Chl/ml were used for NMR measurements.
Bacterial RC Preparation.
The RCs from R. sphaeroides R26 were isolated as described by Feher and Okamura (27). QA was removed by incubating the RCs at a concentration of 0.6 μM in 4% N,N-dimethyldodecylamine-N-oxide (LDAO), 10 mM o-phenanthroline, and 10 mM Tris buffer (pH 8.0) for 6 h at 26°C, followed by washing on a DEAE column and removal of the reaction centers from the column with 0.5 M NaCl in 10 mM Tris buffer (pH 8.0) containing 0.025% LDAO and 1 mM EDTA (27). Approximately 15 mg of the RC protein complex embedded in LDAO micelles were used for NMR measurements. Quinone removal was tested by monitoring the absorption change at 860 nm, the absorption band of the special pair bacteriochlorophyll, after flash excitation at room temperature.
MAS NMR Measurements.
The NMR experiments were performed by using an MSL-400 NMR spectrometer (Bruker, Karlsruhe, Germany) equipped with a double-resonance MAS probe working at 396.5 MHz for protons and 99.7 MHz for 13C. The sample was loaded into a 4-mm clear sapphire rotor and inserted into the MAS probe. 13C MAS NMR spectra were obtained at a temperature of 225 K. To distinguish the centerbands from the spinning sidebands, photo-CIDNP MAS NMR spectra were recorded with two different spinning frequencies, 4 and 5 kHz. At the start of the experiments, the sample was frozen slowly with liquid nitrogen-cooled bearing gas, using slow spinning of 600 Hz to ensure a homogeneous sample distribution against the rotor wall (12). To obtain spectra under illumination, the sample was continuously irradiated from the side of the spinning sapphire rotor by using an illumination set-up specially designed for the Bruker MAS probe. Modifications of the Bruker MAS NMR probe and the illumination setup will be described elsewhere. The light and dark spectra were collected with a Hahn echo pulse sequence and two-pulse phase modulation (TPPM) proton decoupling (28). A recycle delay of 15 s was used, and a total number of 24,000 scans per spectrum were collected over a period of 24 h.
Quantum Chemical Calculations.
Electronic spin densities were calculated by using Gaussian 98 (Gaussian, Carnegie, PA) on an IBM SP2 scalable computer. The nuclear coordinates for the ethyl-chlorophyllide a were taken from the x-ray structure and used without further optimization (29). In the single-point calculations the LanL2DZ basis set was used (30–33). Density functional theory was implemented by using the BLYP Becke exchange functional (34) and the correlation functional of Lee, Yang, and Parr (35). In the calculations, the expectation values of the spin operators were S = 1/2, with S(S+1) = 3/4. Semiempirical restricted Hartree-Fock quantum chemical calculations on minimal π-skeleton models for Chl a were performed by using the MOPAC 97 code incorporated in the chemoffice package (Cambridgesoft, Cambridge, MA) and by using the MNDO-d parametrization (36–38).
Results and Discussion
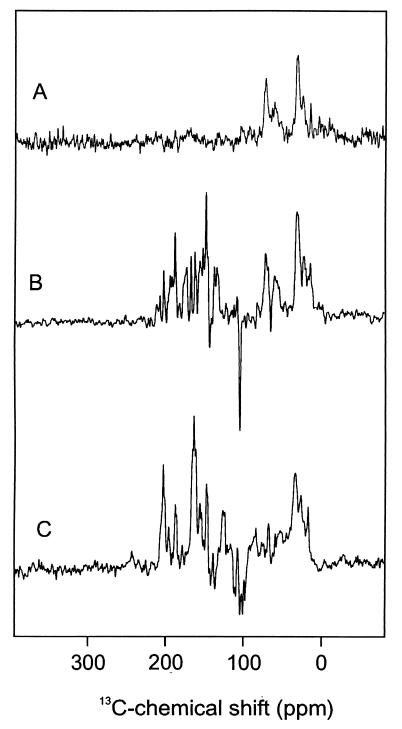
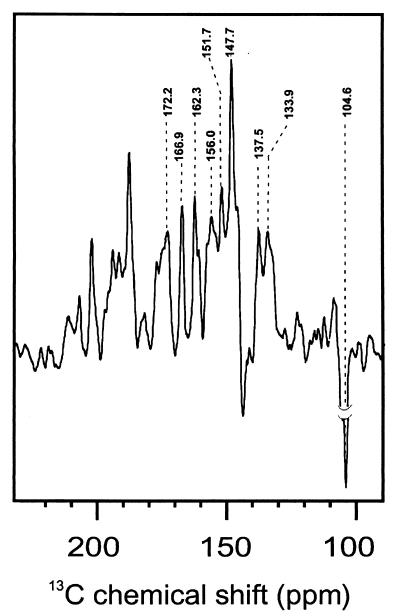
Fig. 1A shows the 13C MAS NMR spectrum of the PS2 sample in the dark. A broad response occurs in the aliphatic region (ca. 0 to 40 ppm). Another broad signal, between 60 and 80 ppm, is due to the C-α of the amino acids. Downfield from 80 ppm, the NMR signal in the dark is insignificant. On illumination of the PS2 with white light, various NMR signals, which were not resolved from the background in the dark spectrum, strongly gain intensity (Fig. 1B). It is indeed remarkable to observe NMR signals of such intensity from the active site of a large membrane enzyme, such as the RC of PS2. A total of nine centerbands was identified (Fig. 2). Except one, all light-induced signals are absorptive (positive). A group of strong absorptive photo-CIDNP signals occurs between 130 and 175 ppm. The single emissive (negative) signal appears at 104.6 ppm.
Figure 1.
13C MAS solid-state NMR spectra. (A) Spectrum of the RC of PS2 in the dark, (B) photo-CIDNP spectrum of the RC of PS2, and (C) photo-CIDNP spectrum of the bacterial RC.
Figure 2.
13C MAS solid-state photo-CIDNP NMR spectra of the RC of PS2.
With moderately fast MAS, spinning sidebands are observed at integral multiples of the rotational frequency, relative to the centerband. These sidebands contain information about the chemical shift anisotropy, i.e., the symmetry of the diamagnetic susceptibility associated with the ground state electron density distribution around the atom. The chemical shift anisotropy for several of the strongest absorptive signals with centerbands between 130 and 175 ppm is estimated as δ ≈ 10 kHz with η ≈ 1, using the method of Herzfeld and Berger (39). These values are characteristic for aromatic carbon atoms (40, 41).
For comparison, we also measured the photo-CIDNP spectrum of a bacterial RC sample (Fig. 1C). The data are in very good agreement with the results reported by Zysmilich and McDermott (16). The centerbands have been tentatively assigned to a single BChl a cofactor (16). As for PS2, the photo-CIDNP intensity gain predominantly involves the aromatic signals between 144 and 165 ppm of the ring carbons that are thought to experience the strongest hyperfine couplings associated with the high pz spin densities during electron transfer. However, there are also significant differences between the data for the two RCs. First, for PS2, only a single strong emissive signal appears, at 104.6 ppm chemical shift. Second, the most downfield centerband signal in the photo-CIDNP spectrum from PS2 is observed at 172.2 ppm, which is outside the range expected for the aromatic ring carbons.
The photo-CIDNP spectrum of Fig. 2 can be compared with the NMR response of Chl a in solution and in the solid state (42, 43). Except for the signal at 172.2 ppm, the centerband signals can be reconciled with a response from a single aromatic cofactor, most likely a Chl a molecule (Table 1). We cannot rule out at this stage that the photo-CIDNP signals originate from more than one Chl a, but they should then be identical in the sense that we do not find a systematic doubling or excess broadening of signals that would indicate the involvement of various Chl a species. The signal observed at 172.2 ppm is anomalous, because it is outside the range expected for aromatic ring carbons. Although an assignment to the carbonyl carbon 13C-13 (3) is in principle possible (Table 1), this should be considered unlikely, because the carbonyl is separated from the ring by a saturated carbon and does not participate in the π-skeleton of the P680⨥. The two strong resolved lines at 169.9 and 162.3 ppm match the ground state response from the aromatic carbons 13C-19 and 13C-14, respectively (Table 1). The broad response at 156.0 ppm may contain up to three lines and can be identified with a response from the 13C-1, 13C-6, and possibly 13C-16, which have very similar chemical shifts. The sharp peak at 151.7 ppm could be assigned to the aromatic 13C-4 or 13C-16 or to both. The most intense positive signal is observed at 147.7 ppm. The chemical shifts of the 13C-9 and 13C-11 match with the observed photo-CIDNP shift. A signal at 137.5 ppm is in line with a response from the 13C-3 and possibly 13C-2. The slight asymmetry of the signal at 133.9 ppm may be caused by an overlap response because of 13C-7 and 13C-12. It is possible that also the carbon 13C-13 adds to the intensity and causes the shoulder at 132.0 ppm. We assign the strong and broad emissive signal at 104.6 ppm to the methine carbons C-15 and C-10. The other methine carbon atoms, C-5 and C-20, which are expected at higher field, are not clearly visible. As it stands, the assignments are tentative and need to be further verified by using isotope-enriched cofactors. The assignment to a Phe a instead of a Chl a cofactor is less convincing, because there are several chemical shifts in the observed spectra that do not match properly with a Phe a (14). The centerbands, e.g., at 172.2, 166.9, 147.7, and 133.9 ppm can be assigned to a Chl a, but not to Phe a. The chemical shift values are also not in line with other cofactors (carotenes or aromatic amino acid side chains). The chemical shifts are at positions expected for neutral and nonradical Chl a molecules. Thus, in the photo-CIDNP spectrum of PS2, the most intense signals come from the aromatic carbon atoms, the most intense absorptive signals are due to the pyrrole α-carbon atoms, and the emissive signal is assigned to methine carbon atoms. These features are analogous to the assignment of the photo-CIDNP data of the bacterial RC to a single BChl a molecule (16).
Table 1.
13C-chemical shifts of the photo-CIDNP signals in PS2 in comparison with published chemical shift data for Chl a
| Chla
|
Photo-CIDNP of
PS2
|
||
|---|---|---|---|
| σliq* | σss† | Atom | σ‡ |
| 189.3 | 190.6 | 131 | |
| 172.7 | 175.3 | 173 | |
| 171.0 | 171.2 | 133 | 172.2 A |
| 167.4 | 170.0 | 19 | 166.9 A |
| 161.4 | 162.0 | 14 | 162.3 A |
| 154.0 | 155.9 | 1 | 156.0 A |
| 155.8 | 154.4 | 6 | |
| 151.4 | 154.0 | 16 | 151.7 A |
| 148.0 | 150.7 | 4 | |
| 147.7 | 147.2 | 11 | 147.7 A |
| 146.1 | 147.2 | 9 | |
| 144.1 | 146.2 | 8 | |
| 139.0 | 137.0 | 3 | 137.5 A |
| 135.5 | 136.1 | 2 | |
| 134.2 | 134.0 | 12 | |
| 134.0 | 133.4 | 7 | 133.9 A |
| 131.5 | 126.2 | 13 | |
| 131.5 | 126.2 | 31 | |
| 118.9 | 113.4 | 32 | |
| 107.1 | 108.2 | 10 | 104.6 E |
| 106.2 | 102.8 | 15 | |
| 100.0 | 98.1 | 5 | |
| 92.8 | 93.3 | 20 | |
The most striking difference between the photo-CIDNP data collected from the two different RCs concerns the number of emissive signals. The spectrum of the bacterial RC shows at least four emissive signals, which are assigned to the response from the methine carbon nuclei 5, 10, 15, and 20 of BChl a (Fig. 1C). The spectrum of PS2 contains only a single but very strong emissive signal at 104.6 ppm. Considering its high intensity and linewidth, as well as the lack of another signal, it is attributed to both the 13C-10 and the 13C-15 methine. The apparent absence of signals from C-5 and C-20, which are expected at significantly higher field, must be due to a lack of induced unpaired spin density at these methine bridges.
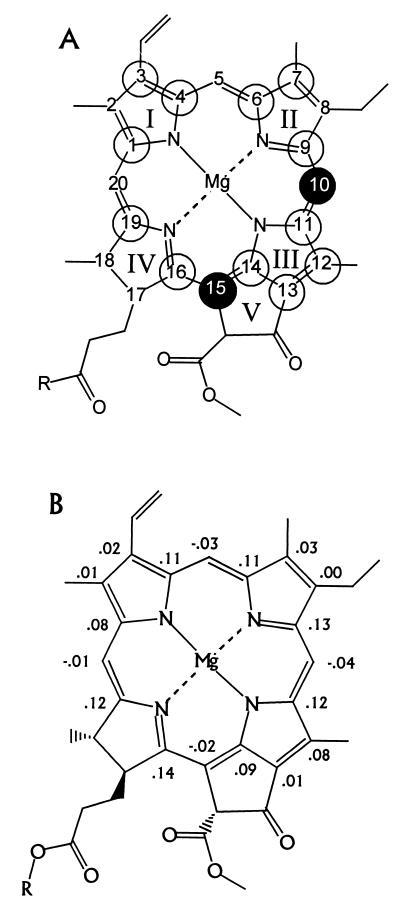
In Fig. 3A open and filled circles indicate the carbons that carry positive and negative spin density according to the comparison in Table 1. The pattern is quite different from the radical cation form of monomeric Chl a in solution and indicates that in the PS2 an asymmetry of the electron spin density distribution over the macrocycle occurs with the maximum spin density shifted toward ring III in the radical cation state. Monomeric Chl a radical cations in solution have been extensively investigated recently, and it was reported that the largest induced spin density is found around the ring II (44). In particular, it was found that the 5-H carries considerable s spin density (45). These results have been reproduced by RHF-INDO/SP calculations (46). Following Petke et al. (47) we have here calculated both the (positive) direct and the (negative) induced pz spin density distribution for a cation radical model by using modern density functional computational methods to include the correlations (Fig. 3B). A good model for Chl a is found in crystalline ethyl-chlorophyllide a (29). In the x-ray structure of this analogue, the Mg is coordinated to the oxygen of a water molecule, and the ring V keto carbonyl is hydrogen bonded to a water molecule. Both water molecules were included in the calculation using their x-ray coordinates. The largest spin densities are calculated for the α-carbons 1, 4, 6, 9, 11, 14, 16, and 19, typically ≈0.1. This result is in line with the pattern presented in Fig. 3A and supports the preliminary assignment of signals given in Table 1. The calculations predict a considerable negative spin density for the C-5, which is not observed with the photo-CIDNP in the PS2. This confirms that the shift of spin density toward the ring III detected with the photo-CIDNP is unusual. It is the major difference between an isolated Chl a, as known from experiment and calculations, and the Chl a in P680 that is resolved on the molecular level.
Figure 3.
Spin density patterns in a chlorophyll a molecule. The numbering is according to the IUPAC nomenclature. (A) The circles indicate the signs of the spin density pattern according to Table 1. Open circles indicate positive signals, and the filled circles at the C-10 and C-15 carbon atoms indicate negative CIDNP intensity enhancement. (B) Spin densities in the π-skeleton for ethyl-chlorophyllide a calculated with density functional theory. The coordinates are taken from the x-ray structure. The Mg is coordinated to H2O, whereas the ring V keto carbonyl is H-bonded to H2O.
It has been shown that protein–cofactor interactions at the keto group of the BChl in the bacterial RC can increase the redox potential of the radical cations by stabilizing the orbital containing the unpaired electron (48). According to the photo-CIDNP, such a type of interaction may also be a predominant factor in generating the unusually high redox potential of 1.2 V of P680⨥. Interaction of the ring V keto group with a static electric field provided by the protein can stabilize spin density in the region of ring III. At the same time, this represents a major strategy to establish the anomalously high redox potential of the P680⨥. In the calculations presented in Fig. 3B, the ring V keto group is hydrogen bonded to a water molecule. Clearly, a regular hydrogen bond is not sufficient to provoke a shift of the spin density to the ring III. This is in line with the work on the bacterial RC (48), where it was found that hydrogen bonding and similarly weak interactions affect the redox potential only moderately by ≈50 mV, an order of magnitude less than the actual value reported for PS2. In this way our calculations imply that, if a protein–cofactor interaction at the ring V keto carbonyl oxygen is responsible for the increased redox potential, it should be very strong.
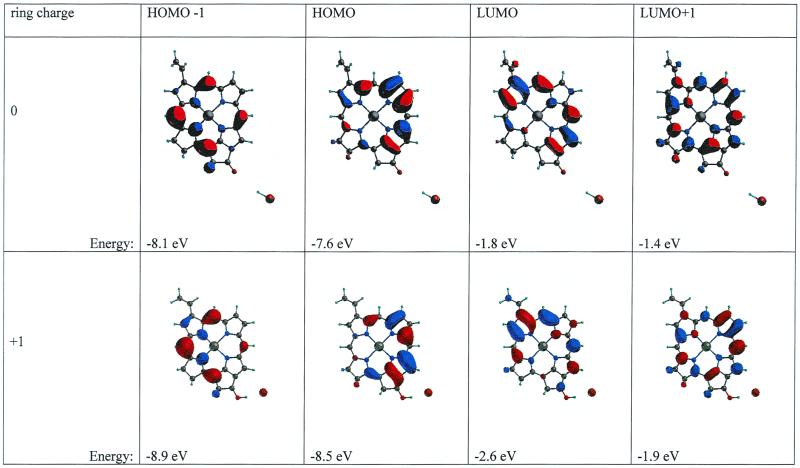
An example of a sufficiently strong interaction mechanism is based on excess charge and illustrated in Fig. 4 (Lower), with semiempirical calculations of molecular orbitals in a minimal model for the π-skeleton in the Chl a. Additional charging of this model can be achieved, e.g., by a very strong hydrogen bond or protonation of the keto group. Although getting reliable energy differences is difficult by such a calculation, it is clear that it produces qualitatively the correct trends. The energy of both the highest occupied molecular orbital (HOMO) and the lowest unoccupied molecular orbital (LUMO) is decreased, which should be equivalent to raising the redox potential while maintaining the same energy gap and color. In line with the CIDNP results, the charge density of the HOMO is rotated toward ring III/V on protonation, whereas the center shifts away from the ring II. This matches the shift of spin density away from ring II toward ring III inferred from the CIDNP. The character of the LUMO−1 and the HOMO+1 appear to change very little.
Figure 4.
Simulation of the effect of charging by protonation by HBr of the ring V keto group in a minimal model for the Chl a π-skeleton (schematic). The upper trace shows the four molecular orbitals (MOs) in the neutral macrocycle, whereas the lower trace shows the same four MOs in the charged macrocycle. The energy of each level in the MNDO-d calculation is listed.
The photo-CIDNP results are difficult to reconcile with an alternative mechanism, because increase of the redox potential is large, ≈0.4 V, and there is no other functional group on the ring that can induce a strong perturbation of the electronic structure of the ring system. We did some additional modeling, such as deprotonation of the C-13 (2) with and without protonation of the keto group (data not shown), and are not able to formulate a better hypothesis within the other constraints of the invariability of the λmax and the shift of the redox potential. In this respect, further calculations indicate that the assignment of the methine carbons will not be affected much by protonation of the ring-V keto group. The model in Fig. 4 indicates that the redox potential of all of the Chl a in PS2 can be shifted by a single straightforward molecular mechanism.§ This is necessary to establish a stable P680 ground state with all of the Chl a in the multimer singly charged having the same redox potential. In this configuration, the P680 radical cation species is formally doubly charged, but it carries only a single additional charge relative to the other singly charged Chl a in the background.
In summary, the photo-CIDNP spectra of PS2 and of the bacterial RC reveal similarities as well as differences. Both spectra show intensive photo-CIDNP enhancement, mainly for the signals from aromatic carbon atoms of a single neutral and nonradical (B)Chl a. The strongest absorptive signals have been assigned in both cases to pyrrole-a carbon atoms. Emissive signals occur only in the region of the methine carbons. A striking difference between the photo-CIDNP spectra of the two RCs is the number of emissive signals: at least four in the bacterial RC, only one in the RC of PS2. The asymmetry in the spin density distribution of P680⨥ is interpreted in terms of a Chl a molecule with weak interaction with other cofactors and a strong interaction with the protein. The unexpected induced spin density localization around ring III suggests the presence of a powerful mechanism for stabilization of the HOMO, leading to the high redox potential of P680⨥, possibly involving the ring V keto oxygen.
Acknowledgments
We thank Marta Germano and Bram Joosten, for the help in the purification of RC, and Silvia Tauw for assistance with computer calculations. J.M. acknowledges a Marie Curie fellowship for Biotechnology of the European Commission (ERB4001 GT97-2589). This work was financially supported by the PIONIER program of the Netherlands Organization for Scientific Research (NWO) and by a Training and Mobility for Researchers (TMR) grant (ERBFMRX CT98-0214).
Abbreviations
- photo-CIDNP
photochemically induced dynamic nuclear spin polarization
- HOMO
highest occupied molecular orbital
- MAS
magic angle spinning
- PS2
photosystem II
- RCs
reaction centers
- P680
chlorophyll a molecules absorbing around 680 nm, the primary electron donor of photosystem II
- BChl
bacteriochlorophyll
- Chl
chlorophyll
- Phe
pheophytin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Article published online before print: Proc. Natl. Acad. Sci. USA, 10.1073/pnas.170138797.
Article and publication date are at www.pnas.org/cgi/doi/10.1073/pnas.170138797
Recently the detection of 16 doubly charged small carotenoid cofactors in a single protein complex, the hexadecameric α-crustacyanin in the carapace of the common lobster, has been reported (49). The charging is thought to be mediated by keto functionalities.
References
- 1.Barber J, Kühlbrandt W. Curr Opin Struct Biol. 1999;9:469–475. doi: 10.1016/S0959-440X(99)80066-9. [DOI] [PubMed] [Google Scholar]
- 2.Diner B A, Babcock G T. In: Advances in Photosynthesis: The Light Reactions. Ort D R, Yocum C F, editors. Vol. 4. Dordrecht, The Netherlands: Kluwer; 1996. pp. 213–247. [Google Scholar]
- 3.Deisenhofer J, Epp O, Miki K, Huber R, Michel H. Nature (London) 1985;318:618–286. doi: 10.1038/318618a0. [DOI] [PubMed] [Google Scholar]
- 4.Svensson B, Etchebest C, Tuffery P, van Kan P, Smith J, Styring S. Biochemistry. 1996;35:14486–14502. doi: 10.1021/bi960764k. [DOI] [PubMed] [Google Scholar]
- 5.Ruffle S V, Donelly D, Blundell T L, Nugent J H A. Photosynth Res. 1992;34:287–294. doi: 10.1007/BF00033446. [DOI] [PubMed] [Google Scholar]
- 6.Xiong J, Subramaniam S, Govindjee Photosynth Res. 1998;56:229–254. [Google Scholar]
- 7.Rhee K H, Morris E P, Barber J, Kühlbrandt W. Nature (London) 1998;396:283–286. doi: 10.1038/24421. [DOI] [PubMed] [Google Scholar]
- 8.van Gorkom H J, Schelvis J P M. Photosynth Res. 1993;38:297–301. doi: 10.1007/BF00046753. [DOI] [PubMed] [Google Scholar]
- 9.Zouni A, Lüneberg C, Fromme P, Schubert W D, Saenger W, Witt H T. In: Photosynthesis: Mechanisms and Effects. Garab G, editor. Dordrecht, The Netherlands: Kluwer; 1998. pp. 925–928. [Google Scholar]
- 10.Marassi F M, Opella S J. Curr Opin Struct Biol. 1999;8:640–648. doi: 10.1016/s0959-440x(98)80157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gebhard R, van der Hoef K, Violette C A, de Groot H J M, Frank H A, Lugtenburg J. Pure Appl Chem. 1991;63:115–122. [Google Scholar]
- 12.Fischer M R, de Groot H J M, Raap J, Winkel C, Hoff A J, Lugtenburg J. Biochemistry. 1992;31:11038–11049. doi: 10.1021/bi00160a013. [DOI] [PubMed] [Google Scholar]
- 13.van Liemt W B S, Boender G J, Gast P, Hoff A J, Lugtenburg J, de Groot H J M. Biochemistry. 1995;34:10229–10236. doi: 10.1021/bi00032a017. [DOI] [PubMed] [Google Scholar]
- 14.Egorova-Zachernyuk T A, van Rossum B, Boender G J, Raap J, Ashurst J, Gast P, Hoff A J, Oschkinat H, de Groot H J M. Biochemistry. 1997;36:7513–7519. doi: 10.1021/bi962770m. [DOI] [PubMed] [Google Scholar]
- 15.Zysmilich M G, McDermott A E. J Am Chem Soc. 1994;116:8362–8363. [Google Scholar]
- 16.Zysmilich M G, McDermott A E. Proc Natl Acad Sci USA. 1996;93:6857–6860. doi: 10.1073/pnas.93.14.6857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zysmilich M G, McDermott A E. J Am Chem Soc. 1996;118:5867–5873. [Google Scholar]
- 18.Roth H D. In: Encyclopedia of Nuclear Magnetic Resonance. Grant D M, Haris R K, editors. Chichester, U.K.: Wiley; 1996. pp. 1337–1350. [Google Scholar]
- 19.Plato M, Lendzian F, Lubitz W, Möbius K. In: The Bacterial Reaction Center II. Breton J, Vermeglio A, editors. New York: Plenum; 1992. pp. 109–117. [Google Scholar]
- 20.McDermott A E, Zysmilich M, Polenova T. Solid State Nuc Mag Res. 1998;11:21–47. doi: 10.1016/s0926-2040(97)00094-5. [DOI] [PubMed] [Google Scholar]
- 21.Jeschke G. J Am Chem Soc. 1998;120:4425–4429. [Google Scholar]
- 22.Polenova T, McDermott A E. J Phys Chem B. 1999;103:535–548. [Google Scholar]
- 23.Grupp A, Höfer P, Käss H, Mehring M, Weizenhofer R, Wegner G. In: Electronic Properties of Conjugated Polymers. Kuzmany H, Mehring M, Roth S, editors. Berlin: Springer; 1987. pp. 156–159. [Google Scholar]
- 24.van Leeuwen P J, Nieveen M C, van de Meent E J, Dekker J P, van Gorkom H J. Photosynth Res. 1991;28:149–153. doi: 10.1007/BF00054128. [DOI] [PubMed] [Google Scholar]
- 25.Berthold D A, Babcock G T, Yocum C F. FEBS Lett. 1981;134:231–234. [Google Scholar]
- 26.Arnon D I. Plant Physiol. 1949;24:1–15. doi: 10.1104/pp.24.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Feher D, Okamura M Y. In: The Photosynthetic Bacteria. Clayton R K, Sistrom W R, editors. New York: Plenum; 1978. , 349–386. [Google Scholar]
- 28.Bennet A E, Rienstra C M, Auger M, Lakshmi K V, Griffin R G. J Chem Phys. 1995;103:6951–6958. [Google Scholar]
- 29.Chow H C, Serlin R, Strouse C E. J Am Chem Soc. 1975;97:7230–7237. [Google Scholar]
- 30.Dunning T H, Hay P J. Modern Theoretical Chemistry. 1976. , ed. H. F. Schaefer, III (Plenum, New York), pp. 1–28. [Google Scholar]
- 31.Hay P J, Wadt W R. J Chem Phys. 1985;82:270–283. [Google Scholar]
- 32.Wadt W R, Hay P J. J Chem Phys. 1985;82:283–298. [Google Scholar]
- 33.Hay P J, Wadt W R. J Chem Phys. 1985;82:299–312. [Google Scholar]
- 34.Becke A D. Phys Rev A. 1988;38:3098–3115. doi: 10.1103/physreva.38.3098. [DOI] [PubMed] [Google Scholar]
- 35.Lee C, Yang W, Parr R G. Phys Rev B. 1988;37:785–789. doi: 10.1103/physrevb.37.785. [DOI] [PubMed] [Google Scholar]
- 36.Stewart J P. J Comput Aided Mol Des. 1990;4:1–67. doi: 10.1007/BF00128336. [DOI] [PubMed] [Google Scholar]
- 37.Dewar M J S, Thiel W. J Am Chem Soc. 1977;99:4899–4905. [Google Scholar]
- 38.Thiel W, Voityuk A A. J Phys Chem. 1996;100:616–624. [Google Scholar]
- 39.Herzfeld J, Berger A E. J Chem Phys. 1980;73:6021–6030. [Google Scholar]
- 40.Veeman W S. Progr Nuclear Magn Reson Spectrosc. 1984;16:193–235. [Google Scholar]
- 41.Metz G, Siebert F, Engelhardt M. Biochemistry. 1992;31:455–462. doi: 10.1021/bi00117a022. [DOI] [PubMed] [Google Scholar]
- 42.Boender G J. Ph. D. thesis. The Netherlands: University of Leiden; 1996. [Google Scholar]
- 43.Abraham R J, Rowan A E. In: Chlorophylls. Scheer H, editor. Boca Raton: CRC; 1991. p. 797. [Google Scholar]
- 44.Käβ H, Bittersmann-Weidlich E, Andreasson L E, Bönigk B, Lubitz W. Chem Phys. 1995;194:419–432. [Google Scholar]
- 45.Käβ H, Lubitz W, Hartwig G, Scheer H, Noy D, Scherz A. Spectrochim Acta. 1998;54A:1141–1156. [Google Scholar]
- 46.Käβ H, Lubitz W. Chem Phys Lett. 1996;251:193–203. [Google Scholar]
- 47.Petke J D, Maggiora G M, Shipman L L, Christoffersen R E. Photochem Photobiol. 1980;31:243–257. [Google Scholar]
- 48.Artz K, Williams J C, Allen J P, Lendzian F, Rautter J, Lubitz W. Proc Natl Acad Sci USA. 1997;94:13582–13587. doi: 10.1073/pnas.94.25.13582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Weesie R J, Jansen F J H M, Merlin J C, Lugtenburg J, Britton G, de Groot H J M. Biochemistry. 1997;36:7288–7296. doi: 10.1021/bi9631982. [DOI] [PubMed] [Google Scholar]