Abstract
Many proteins that interact with DNA perform or enhance their specific functions by binding simultaneously to multiple target sites, thereby inducing a loop in the DNA. The dynamics and energies involved in this loop formation influence the reaction mechanism. Tethered particle motion has proven a powerful technique to study in real time protein-induced DNA looping dynamics while minimally perturbing the DNA–protein interactions. In addition, it permits many single-molecule experiments to be performed in parallel. Using as a model system the tetrameric Type II restriction enzyme SfiI, that binds two copies of its recognition site, we show here that we can determine the DNA–protein association and dissociation steps as well as the actual process of protein-induced loop capture and release on a single DNA molecule. The result of these experiments is a quantitative reaction scheme for DNA looping by SfiI that is rigorously compared to detailed biochemical studies of SfiI looping dynamics. We also present novel methods for data analysis and compare and discuss these with existing methods. The general applicability of the introduced techniques will further enhance tethered particle motion as a tool to follow DNA–protein dynamics in real time.
INTRODUCTION
Many proteins interact with multiple target sites on DNA to help perform or enhance their specific function. Such simultaneous interactions often result in the formation of DNA loops. Examples are found in DNA replication (1,2), homologous recombination (3,4), transcription regulation (5–7) and the cleaving of double-stranded DNA by many restriction enzymes (RE) (8–11). Restriction endonucleases come in many variants and are categorized by their subunit composition, reaction mechanism and co-factor requirements into four different types, of which Type II is perhaps the best known (12).
In vivo, REs are a major defence mechanism of bacteria against viral infections. Type II REs recognize and bind specific DNA sequences, usually between 4 and 8 bp long. They then hydrolyse the DNA backbone at or near the recognition site, generally using Mg2+ as a cofactor; they do not require ATP to function (13). With Ca2+ as an alternative divalent metal ion cofactor, most of these enzymes can still bind to their recognition sites, often with enhanced affinities, but show no catalytic activity (14,15). Many REs associate with two recognition sites and consequently form DNA loops. DNA-loop formation by REs has been studied extensively by traditional biochemistry methods (11,16–21). These studies have provided detailed information about the roles of DNA looping in enzyme reactions. However, they have yielded only indirect evidence for DNA looping, and thus provide limited insight into the looping dynamics. More recently, several single-molecule biophysical techniques have been developed and applied to directly visualize RE–DNA interactions, elucidating the underlying dynamics (22–26).
The tetrameric Type II RE SfiI has been studied extensively by biochemical methods and it can be considered a model for its Type. The accompanying paper (27) in this issue gives a detailed introduction to, and an extensive analysis of the binding kinetics for this enzyme. In short, by binding to a DNA with two recognition sites first at one site and subsequently at the second site, a single SfiI tetramer traps a loop in the DNA. After loop formation, SfiI hydrolyses the DNA backbone and cleave both DNA strands at both copies of the recognition site (15,17,27).
A RE that binds two sites forms stable loops only in the absence of catalytic activity, which generally entails either the use of Ca2+ (in place of Mg2+) or a protein inactivated by mutation. In the presence of Ca2+, wild-type (WT) SfiI binds almost irreversibly to its recognition site (27) and traps indefinitely stable loops (20), thus precluding any measurement of looping dynamics. Consequently, catalytically inactive mutants of SfiI were constructed that could bind to – but not cleave – DNA in the presence of Mg2+ and one such mutant, D79A, showed rapid association/dissociation kinetics with the recognition sequence (27). It is therefore possible to measure looping dynamics with D79A and Mg2+ in a manner that would not have been possible with WT SfiI and Ca2+.
Here we apply tethered particle motion (TPM), to follow in real time the interaction between SfiI and single DNA molecules (28,29). Using TPM we show that we directly observe and characterize the looping dynamics of SfiI enzymes on individual DNA molecules, while exerting minimal forces on the tether and proteins (30). In this study, we utilize the SfiI D79A mutant to assess the looping kinetics of SfiI. Since visualizing the tethers is done by transmission light microscopy, we observe and track tens of DNA molecules simultaneously in the same field of view. This provides a simple method of increasing statistics not generally available to other single-molecule methods. We compare existing and newly developed data analysis methods (31,32), as well as discuss our results in relation to the detailed biochemical analysis of the reaction rates of SfiI looping by Bellamy et al. (27).
MATERIALS AND METHODS
Proteins and DNA
WT SfiI, the D79A mutant and the DNA plasmids were all purified as described in the accompanying paper (27). Before being added to the reactions, the proteins were first diluted to the requisite concentration in 20 mM Tris–HCl (pH 7.5), 10 mM βME (β-mercaptoethanol), 1 mM spermine, 0.1 mM EDTA, 10% glycerol and 0.2% Triton. All reactions were carried out in Mg2+ buffer [10 mM Tris–HCl (pH 7.5), 25 mM NaCl, 5 mM βME, 100 µg/ml BSA, 100 µg/ml α-casein, 10 mM MgCl2]: for solution assays, the α-casein was omitted; for binding studies with WT SfiI, the MgCl2 was replaced with 2 mM CaCl2.
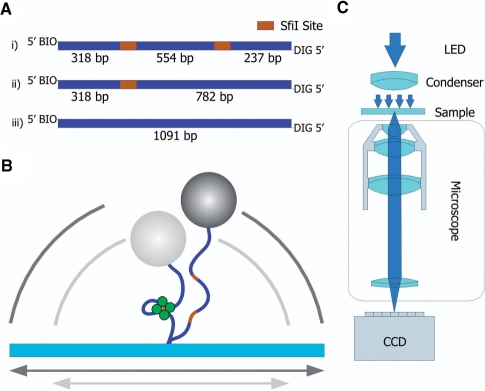
The plasmid pGB1Δ466 is a derivative of pGB1, a 7.65-kb plasmid with two SfiI sites separated by 1023 bp (33): the plasmid was digested with SpeI and MluI (pGB1 carries unique sites for these enzymes in the section between its SfiI sites) followed by blunt-end ligation, to leave a 7.2-kb plasmid with two SfiI sites separated by 554 bp. A section of pGB1Δ466 that spanned the SfiI sites was amplified by PCR, using forward and reverse primers tagged at their 5′ ends with, respectively, biotin and DIG (digitoxin) to give an 1110 bp fragment with two SfiI sites 554 bp apart with approximately similar sized spacers linking to the bead and the surface. The molecule carries a biotin label at the 5′ end of one strand and a DIG label at the 5′ end of the complementary strand (Figure 1A). For control experiments, equivalent PCR products were generated from pGB1/S1 and pSB1 (34), which have, respectively, one or no recognition site for SfiI, to give 1- or 0-site constructs (Figure 1A).
Figure 1.
Schematics TPM experiments. (A) DNA substrates for TPM experiments. Three different DNA substrates are produced to perform the TPM experiments. Each substrate contains a biotin on one 5′ end and a DIG label on the other 5′ end. (i) The two-site substrate, total length 1109 bp, contains two SfiI sites (orange) separated by 554 bp, and is generated by PCR from the pGB1Δ466 plasmid. (ii) The one-site substrate with a total length of 1100 bp was amplified from the pGB1/S1 plasmid. (iii) A substrate containing no SfiI sites was generated by amplifying 1091 bp of the pSB1 plasmid. (B) Schematic drawing of the tethered particle assay. A single DNA molecule is tethered to a glass surface, its remaining free end is labelled with a micro-sized bead. The Brownian motion of the bead is restricted by the length of the DNA molecule. When a SfiI tetramer (green) induces a loop in the DNA, effectively shortening the total DNA tether, the explored volume changes accordingly (from the dark to the grey circle). (C) Schematic drawing of the used instrument. The TPM experiments are performed on an inverted microscope (Nikon TE-2000) using a 100× immersion-oil objective to collect the light. Images are recorded by a CCD camera running at 25 Hz. The acquired images are analysed in real time by a self written image analysis toolkit, to collect the positions of up to 50 beads.
Bulk assay
The kinetics of the DNA cleavage reactions of SfiI in bulk solution were evaluated using final reaction mixtures that contained 5 nM 3H-labelled pGB1Δ466 and 3 nM WT SfiI in 200 µl Mg2+ buffer at 21.5°C. To measure the steady-state phase of these reactions, 10 µl SfiI enzyme [in SfiI dilution buffer: (33)] was added to 190 µl DNA in Mg2+ buffer. Before adding the enzyme, a 15 µl aliquot was removed to serve as the zero time-point. After the addition, further aliquots were removed from the reactions at various times (between 1 and 90 min). The aliquots were vortexed immediately with 10 µl EDTA stop mix to quench the reaction (35). The resulting samples were analysed by electrophoresis through agarose under conditions that separated the supercoiled (SC) substrate from the cleaved DNA products. The concentration of SC DNA left at each time point was determined by scintillation counting (35). Values given here are the means from three independent experiments, with error bars to denote standard errors. The steady-state rate was evaluated by using GRAFIT (Erithacus Software) to fit to a linear slope the zero-order phase of substrate utilization after the initial burst.
To measure the pre-steady-state phase of these reactions, a RQF-63 Quench Flow device (TgK Scientific) was used to mix equal volumes of two solutions to give reactions containing 5 nM pGB1Δ466 and 3 nM wt SfiI in Mg2+ buffer at 21.5°C. Some were conducted by mixing one solution containing twice the requisite concentrations of enzyme and DNA, in EDTA buffer (Mg2+ buffer with 0.2 mM EDTA instead of 10 mM MgCl2), with a second solution containing 20 mM MgCl2. For others, one solution containing 6 nM enzyme and 20 mM MgCl2 was mixed with a second containing 10 nM DNA in EDTA buffer. In both cases, the reactions were quenched after the requisite time delay (250 ms to 40 s) by mixing with a half volume of 0.1 M EDTA (pH 8.0). Note that in some previous studies of this type, the plasmid was cleaved more rapidly in reactions started by adding MgCl2 to the mix of enzyme and DNA, compared to those started with enzyme and DNA in separate solutions and, in the latter, DNA cleavage was sometimes preceded by a lag phase. The quenched samples were analysed and the decline in the concentrations of supercoiled substrate quantified as above. The decline was fitted to a single exponential.
Single molecule assay
The DNA cleavage and looping dynamics of SfiI was studied by TPM. In these experiments DNA molecules were immobilized by attaching the DIG-labelled DNA-end to a glass surface coated with DIG antibody. The other end of the DNA was attached to a 440 nm streptavidin-coated polystyrene bead (Kisker Biotechnology) with a biotin linker (Figure 1B). To prevent the DNA, beads and proteins from sticking to the glass during the experiments, the cell was coated with a layer of casein. The incubation times used and washing buffers were as described in (28).
The TPM assay was conducted in a micro-fluidic flow cell consisting of a microscope slide and a perpendicularly-placed cover slip. Two strips of parafilm were placed between the glasses and melted to create a small flow channel (between 10 and 100 µl depending on the spacing between the strips). We also performed experiments with flow cells assembled using double-sided tape. However, data obtained in these flow cells displayed significant focal drift, most likely caused by slowly detaching tape (gradually increasing the size of the flow cell). The parafilm proved drift resistant and permitted experiments lasting up to 3 h without focal position feedback.
TPM data acquisition
After the sample chamber was prepared, it was mounted a conventional transmission inverted microscope (Nikon eclipse TE2000-U) (Figure 1C). The sample was illuminated using a blue LED (480 nm) and a condenser to achieve maximum contrast. The light was collected by a 100× Nikon immersion-oil objective and imaged on a CCD camera running at video rate.
The position of individual beads within the field of view was measured using a self-written image analysis kit, programmed in LabView (National Instruments). For each frame, the selected tethers were reduced to separate images of 16 by 16 pixels according to their defined region of interest (ROI). Each reduced image was mirrored in both X and Y and correlated with the non-mirrored image to yield the position of the centre of the bead. By using this ‘correlation’ algorithm, we determined the centre of a bead with subpixel accuracy of 2 nm (standard deviation of the spread in positions of a surface-immobilized bead). Moreover, it permitted rapid position detection which enabled us to do all the measurements in parallel and in real time, eliminating the need for fast storage of large data files and post-processing of the video images. This method thus allowed for simultaneous measurements on 50 tethers at 25 Hz, with the resulting data of each measurement being directly accessible for analysis.
Before initiating experiments, we first analysed the spread of the in-plane position distribution of each tethered bead. Beads that were attached by a single DNA tether (i.e. beads having a circular position distribution of the correct size) were selected for tracking SfiI–DNA interactions (32). The position distribution of the correctly tethered bead is a direct measure of several physical properties of the DNA tether, most importantly the effective DNA contour length (36). When a protein induced a loop in the DNA, it reduces the length of the tether and confines the movement of the bead to a smaller region (29,37) (Figure 1B). Thus by tracking the bead movement we measured in real time the looping dynamics induced by proteins. Typically the motion of the beads was followed for 1 h.
Data pre-processing
We used two independent methods to remove in-plane drift of the apparatus. The first method used the information that we gained by tracking multiple tethers in the same field of view. The movement of the centre of all the selected beads (centre of mass) is a measure of in-plane drift. The displacement of the centre of mass was calculated for each frame and subtracted from the individual Brownian motion for each tether.
The second method entailed subtraction of the averaged motion of a single tether over multiple frames from its own trace. The anchor point of each tether was calculated using a running average over 100 frames and was subtracted for each frame. Since we measured a single tether for up to an hour, the centre of its distribution over this time is a measure for the drift in the machine (typically less than 12 nm/min). The second method was used in combination with the first.
TPM data analysis: threshold method
We used a threshold analysis (31) to verify the sizes of the protein-induced DNA loops and obtain an initial measure of the dwell times of the looped and unlooped states. In our method, we converted the position data of each tether to a root mean square motion (RMS) by taking: √[(x − xm)2 + (y − ym)2], with xm and ym the mean values of the two in-plane axis averaged over 100 frames. To average the data, we smoothed the observed RMS values using a Gaussian filter with a standard deviation of σ = 0.5 s. A histogram from the RMS data was created. Such a distribution consisted of two peaks (the unlooped and looped tether lengths) which were fitted to a double normal distribution. The minimum of this function, between the two peaks, is the threshold value for the RMS data. Using this threshold, each data point was assigned to either the looped or unlooped state, yielding the dwell times in the both states. With this method, we distinguished a looped or unlooped state within 17 frames resulting in a time resolution of 0.68 s.
Next, the data was checked for false positives and beads that were (on occasion) stuck to the surface. Beads hardly ever get stuck to the surface because of the surface blocking treatment. Nevertheless, if such events occured, they were distinguishable by very small RMS values (<30 nm) and we then discarded the entire time trace. False positives were caused by transient sticking of the DNA tethers to the glass and are undistinguishable from real loop events by analysing the RMS values. However, they were revealed by a sudden shift in the anchor point of the raw in-plane distribution; these traces were also discarded.
DNA cleavage by SfiI was also analysed with the threshold method. When a tether was cleaved and released by SfiI, the free bead diffused out of its defined region of interest. With the bead gone, the image analysis algorithm no longer was able to determine a bead centre and returned a random position in the ROI. This effect resulted in a sharp peak in the RMS motion, marking the moment that SfiI releases the tethered DNA.
TPM data analysis: hidden Markov method
Dwell times obtained with the threshold method are influenced by the chosen Gaussian filter time, making a comparison with biochemical rates more complicated. Another approach to analyse the data is to look at the underlying statistics between the raw data points, using a hidden Markov model (38). The hidden Markov method, originally developed as a speech recognition algorithm, is well established as a successful tool in the (single-molecule) biophysics field (39,40).
However, if the standard hidden Markov analysis were to be used for TPM analysis, the protocol would assume that the loop formation and loop breakage process is independent of the position of the bead (32). This assumption is invalid; for example, when the tether is fully stretched, the loop formation rate is by definition zero. Recently, Beausang et al. (32) designed a diffusive hidden Markov method (dHMM) that is applicable to analyse correctly TPM experiments. The dHMM analyses the raw data and does not require any averaging of the data in the time domain. Consequently, short-lived looping events are detected and the evaluated dwell times are independent of sampling frequency.
The dHMM algorithm requires two training sets, data from a free and from a permanent looped tether, to develop the underlying statistics. We obtained such training sets from measurements in absence of SfiI enzyme and in the presence of WT SfiI and Ca2+, respectively, followed by position drift corrections as described above. The Ca2+ ions permanently fix the DNA loops induced by WT SfiI [(27): also see below], thus enabling long measurements of the looped state. The algorithm also needs (like any other fitting routine) an initial guess of the input parameters. These input parameters were obtained from the threshold method. Finally, to iterate towards the correct kinetics rates, a simplex solver (by J.F. Beausang, private communication) was used.
RESULTS
DNA cleavage
To test the catalytic activity of WT SfiI in our TPM assay, two types of experiments were performed using buffer containing either magnesium or calcium ions (see ‘Materials and Methods’ section). A sample containing tens of beads attached by single DNA-tethers was prepared and their position distribution was measured in the absence of the protein. Next, the WT enzyme was flown into the chamber while we continuously recorded the state of the DNA-tethers. In the buffer containing Mg2+, the WT enzyme is expected to bind, loop, cleave and eventually detach from the DNA. In the Ca2+ buffer the enzymes are expected to bind and to form a loop in the DNA but not cleave nor dissociate from the DNA, since the catalytic activity of the protein should be fully suppressed in Ca2+ (41,42).
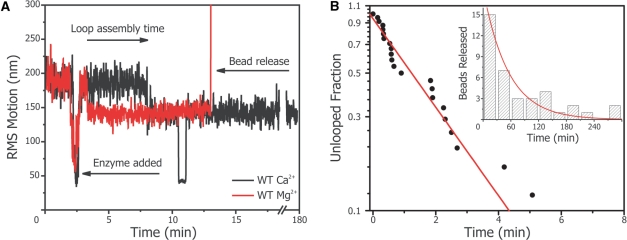
Figure 2A shows a data trace of WT protein in both buffers. It reveals two distinct RMS levels, which are assigned to the free DNA and DNA with a protein-induced loop. In general, we observed within minutes after flowing 0.01 nM WT SfiI into the micro-fluidic chamber that most of the DNA-tethers were looped by the enzyme (loop assembly). In the Ca2+ buffer, SfiI formed a single loop which stays fixed for hours; neither DNA cleavage nor looping dynamics were observed.
Figure 2.
SfiI DNA cleavage analysed with TPM. (A) Time trace of the root mean square (RMS) motion of a single two-site tether. At time t = 2.5 min, 0.01 nM SfiI WT was flown into the chamber, causing the tethers to extend in the flow. The loop assembly time is defined from the addition of the SfiI untill the DNA loop is induced by the protein. If Mg2+is present in the buffer (red trace), the DNA is hydrolysed and bead released is observed by a steep increase in the RMS motion (around t = 14 min). In the buffer containing Ca2+ (black trace), the DNA remained looped for hours (observed for N = 28 tethers). Note the short sticking event of the tether to the glass (at t = 10 min). (B) Cumulative distribution function (CDF) plot of the loop assembly times obtained in the cleavage experiments with the two-site substrate. A concentration of 0.01 nM SfiI WT enzyme in Mg2+ buffer was used to obtain the loop assembly times of DNA loop formation (see main panel). The data is fitted to an exponential (red line). The dwell times of the looped state prior to bead release is also recorded and plotted as a histogram (see inset). The bead release rate is also an exponential distribution (red line in inset).
In Mg2+ buffer most beads were released from their tethers, as deduced from the sharp peak in the RMS value, in the course of an hour. The measured loop assembly times and the bead release times were exponentially distributed indicating, that both are dominated by a single reaction step with rates of, respectively, kloop assembly = 9 ± 1 × 108 M−1 s−1 and kbead release = 3 ± 1 × 10−4 s−1 (Figure 2B) In this case, the loop assembly rate is an upper limit (see ‘Discussion’ section), and indeed is somewhat high for the maximally expected 3D diffusion rate (26,43). On the other hand, the extremely stable complexes of SfiI contrasts with many other RE, which usually cleave and detach within minutes or seconds (43). The slow kbead release has two possible explanations, either SfiI has a slow catalytic rate, i.e. it takes a long time before it hydrolyses the DNA backbone, or the dissociation rate of SfiI from either one of the cleaved sites is the rate limiting step.
To distinguish between the two options, we compared these results with kinetics experiments in bulk solution. Previous solution kinetics performed with WT SfiI and pGB1, a plasmid with two SfiI sites separated by 1 kb, had demonstrated that phosphodiester hydrolysis at 25°C is rapid (0.5 s−1) but the steady-state rate at this temperature is extremely slow (1.7 × 10−4 s−1) compared to that at 50°C [2.5 × 10−2 s−1; (44)]. These results support the fast cleavage but slow dissociation model for SfiI. However, to compare directly with the TPM experiments, the kinetics of DNA cleavage by SfiI were examined in the same reaction buffer and at the same temperature (21.5°C) as the TPM experiments. The DNA used for the kinetic experiments was pGB1Δ466, the template for the TPM substrate: it has two SfiI sites 544 bp apart.
The reactions of WT SfiI were studied under conditions where the presence of a burst phase is readily detected (Figure 3). This was achieved with an enzyme concentration (3 nM) approaching that of the DNA (5 nM) so that enzyme-bound product constituted a significant fraction of total product and at a reaction temperature where SfiI has a very slow turnover rate (44), thus allowing the reactions at elevated enzyme concentrations to be monitored. Under these conditions, WT SfiI cleaved a fraction of the two-site substrate rapidly before entering a slower phase during which the concentration of substrate declined linearly with time (9 × 10−5 mol. DNA per mol. enzyme per sec). The amount of substrate consumed in the initial burst phase was about 70% of the enzyme concentration rather than 100%. This difference was expected, as the addition of SfiI to a two-site substrate leads not only to DNA with one SfiI tetramer bridging the two sites in cis but also to DNA carrying a tetramer at each site, which resists cleavage (20). The presence of the slow phase in this kinetic assay demonstrates that product release is indeed the rate-limiting step of the reaction.
Figure 3.
Solution kinetics of DNA cleavage by SfiI. The reactions contained 5 nM [3H] pGB1Δ466 and 3 nM WT SfiI in Mg2+ buffer at 21.5°C. Displayed is the remaining SC DNA left as a function of time. In the main panel, the linear regime between t = 5 and t = 90 min is fitted to a straight line. The inset shows the pre-steady state kinetics obtained with the rapid quench-flow device: either by pre-mixing WT SfiI with the DNA before initiating the reaction with MgCl2 (black circles); or by adding the DNA to a solution containing the protein and the MgCl2 (open squares). The results are exponentially distributed with decay constants of, respectively, 2.5 × 10−1 s−1 and 1.2 × 10−1 s−1 (black lines).
In order to also obtain the rate of the hydrolytic step, the kinetics of the burst phase of the reaction were analysed by using a rapid quench-flow device (Figure 3). These experiments revealed directly the state of the enzyme-bound DNA throughout the course of the reaction. Here we obtained a hydrolysis rate constant of 0.25 s−1 for the pre-mixed enzyme with DNA and a slower rate of 0.12 s−1 for the reaction starting with the enzyme and DNA in separate solutions. The reduced rate is the result of the binding step that needs to take place before hydrolysis. Hence, the loop assembly rate estimated from these experiments is ∼1 × 108 M−1 s−1 (for additional corrections see ‘Discussion’ section).
DNA looping
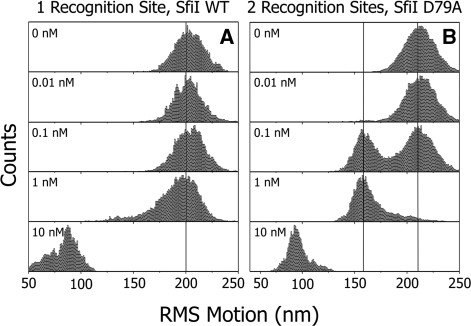
To verify that the observed cleavage by the WT enzyme had indeed occurred via specific loop formation (i.e. a loop formed by a single tetramer bridging two recognition sites), we replaced the original DNA template that had two SfiI sites with a construct that contained one specific binding site (1-site construct: Figure 1A). After addition of ≤1 nM WT enzyme, no looping was observed and all tethers remained intact as expected (Figure 4A). The average RMS value of the tethers was 203 ± 3 nm, identical to the DNA length in the absence of protein indicating that there are no interactions between the bound proteins and the beads. Raising the concentration to 10 nM resulted, however, in small RMS values with a large spread ranging from 50 to 120 nm for individual tethers which is an indication of non-specific DNA condensation (Figure 4A, bottom panel). This experiment was also repeated with a DNA template containing no recognition sites; it showed the same behaviour. Experiments with WT SfiI in Ca2+ buffer using 0-, and 1-site constructs again showed this behaviour; DNA condensation was in all cases observed at [SfiI] >5 nM (data not shown).
Figure 4.
Specific looping of SfiI confirmed with the TPM assay. Histograms of the RMS motion with increasing protein concentration reveal specific looping. Both panels show the data of a single tether and each histogram contains at least 15 × 106 counts. The left panel shows the behaviour of the WT SfiI enzyme on the single site substrate in a buffer containing Ca2+. The tether remains unlooped with increasing WT concentrations up to 1 nM. At 10 nM enzyme, the non-specific interactions of multiple SfiI molecules condense the tether. The right panel shows the equilibrium between looped and unlooped state with increasing D79A concentration in Mg2+ buffer. At 10 nM, D79A also condenses the DNA, similar to the WT enzyme, indicating that the non-specific interactions are not altered by the mutation.
In order to study the looping dynamics of SfiI we needed to prevent the enzyme from cleaving the DNA while permitting recognition of the specific sites. Using Ca2+ as a cofactor, the catalytic activity of a RE can be disabled while specific recognition is maintained (41,42). However, for WT SfiI, Ca2+ induces irreversible binding to the DNA (Figure 2A) (20,27). Instead, we used another approach made possible by the development of the non-cleaving SfiI mutant D79A, as characterized and described in Bellamy et al. in this issue (27). The mutant proved to be catalytic inactive in Mg2+ buffer but it associated with and dissociated from specific DNA on a time scale of seconds. This property was confirmed with the TPM as shown in Figure 4B, which displays the typical double peaked RMS distribution expected for a 2-site construct. At [D79A] < 10 nM, two discrete RMS levels are observed: 211 ± 1 nm and 159 ± 1 nm (average values for a single tether over all concentrations). By comparing these values to the levels observed during the cleavage experiments with the WT enzyme, it is concluded that the first level corresponds to the free tether length (unlooped state) and the lower level corresponds to a specific looped state. As expected, high concentration the D79A mutant displayed non-specific binding identical to the WT protein.
Looping dynamics
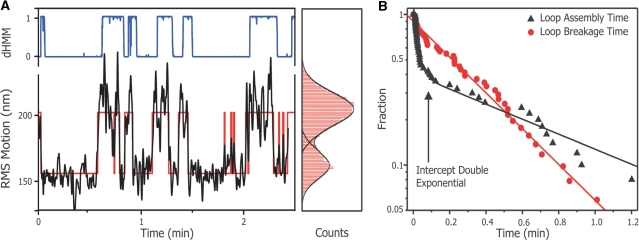
Figure 5A displays a typical trace (black) of the SfiI mutants D79A interacting with a two-site tether in Mg2+ buffer. D79A shows clear switching dynamics from one defined state into the other and vice versa. Also shown (Figure 5A, right panel) is the position distribution histogram fitted with a double Gaussian in order to define the threshold value (see ‘Materials and Methods’ section). This value was used to construct a binary trace (Figure 5A, red line) from which the dwell times were extracted.
Figure 5.
Obtaining looping kinetics with thresholding analysis. (A) Shown are a RMS time trace (left panel, black line) and its corresponding histogram (right panel, red bars) of a single two-site DNA tether in the presence of 0.1 nM D79A in Mg2+ buffer. The histogram is fitted with a double Gaussian (right panel, black lines) and the intersection of the double Gauss is used as a thresholding value. The thresholding results in a binary trace (left panel, red line), from which the dwell times are extracted. Also shown is the probability for each data point to belong to the unlooped distribution (blue line between) as obtained by the diffusive hidden Markov analysis. (B) A CDF plot of the obtained dwell times. The dwell times displayed are from a single tether in the presence of 0.1 nM D79A in Mg2+ buffer, and contain at least 50 data points. The red circles show the distribution of the loop lifetime and are fitted with a single exponential (red line) to obtain the loop breakage rate. The loop assembly times are shown by black triangles and fitted with a bi-exponential decay. The intersection of the two exponentials (arrow) from the bi-exponential fit can be used to estimate the protein dissociation rate of the D79A protein from the DNA.
The dwell time distribution of the looped state, loop break time, fits well to a single exponential indicating that SfiI follows one loop release pathway (Figure 5B). The formation time (i.e. the dwell time between subsequent looping events), on the other hand, only fits a double exponential yielding two different formation speeds.
To explain these two distinct formation speeds we hypothesize that the slow process, defined as loop assembly, consist of the time it takes before a naked tether is occupied by a diffusing SfiI tetramer (protein association), plus the time needed to bring the two sites together to form a loop (loop formation). On the other hand, the fast component is assigned to a tether already having one of its sites occupied by a SfiI molecule and only having to undergo the actual loop formation. This situation would occur when the protein releases one of the DNA sites but stays linked to the other recognition site. Finally, we also obtained the protein dissociation rate from the relative amplitude of the loop assembly events (slow) versus loop formation events (fast). This ratio is directly determined from the location of the intercept of the two exponentials fitted to the formation times (Figure 5B, arrow).
The reaction rates obtained by the threshold method (Supplementary Data) serve as initial guesses for our dHMM analysis. The rates obtained by dHMM are independent of filtering and include short lived looped states. Figure 5A (top) shows a typical dHMM trace corresponding to the raw data. We tested our assumed reaction scheme by investigating all reaction rates as a function of the concentration of the D79A form of SfiI.
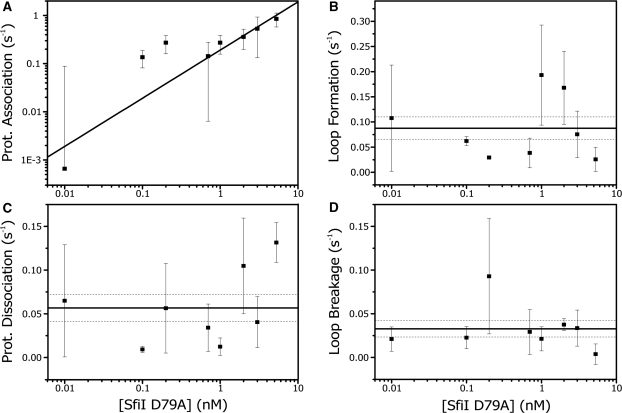
Figure 6A shows that the presumed association is indeed linearly dependent on the protein concentration. Thus the rate of SfiI diffusion to one of the two target sites is determined from the linear fit to the data, resulting in a kprotein association of (2 ± 0.5) × 108 M−1 s−1. Figure 6B and d display the loop formation rate and the loop breakage rate as a function of concentration. As predicted the loop formation is independent of protein concentration (as it is governed by the search time of the DNA only), and has an average value of kformation = 0.1 ± 0.02 s−1. The loop breakage rate is also independent of concentration and has a mean value of kbreakage = 0.03 ± 0.01 s−1. Finally, the average protein dissociation rate, kdissociation, was found to be 0.06 ± 0.02 s−1 (Figure 6C). Combing these rates with those obtained from our biochemical study, we obtain a complete quantitative description of the reaction pathway of SfiI-induced DNA looping and cleavage (Figure 7).
Figure 6.
Looping kinetics obtained by diffusive hidden Markov analysis. Shown are the measured reaction rates as a function of D79A concentration (in Mg2+ buffer): each data point is an average value of at least eight tethers along with the standard error. The rates are obtained using the dHMM analysis with three hidden states. (A) D79A tetramer association with a single recognition site. The data is fitted with a linear function with zero offset (black line), resulting in association rate of 2 ± 0.5 × 108 M−1 s−1 (reduced χ2: 1.4). (B) shows the loop formation rate, which is independent on D79A concentration, with an average value of 0.1 ± 0.02 s−1 (black line, reduced χ2: 2.8). The dotted black lines are the confidences bands as give by the standard error of the average. (C) shows the rate of protein dissociation from one recognition site and its average value: 0.06 ± 0.02 s−1 (black line, reduced χ2: 4.8). Note that, even though the error in this fit is somewhat larger, the residues of the fit are randomly distributed; hence the dissociation rate and the protein concentration are indeed not correlated. (D) gives the loop breakage rate as a function of concentration. The average value is determined at 0.03 ± 0.01 s−1 (black line, reduced χ2: 1.1).
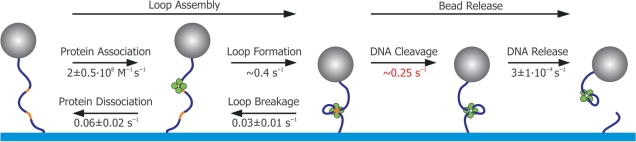
Figure 7.
Reaction pathway of SfiI revealed by combining TPM and solution kinetics. The reaction pathway for DNA cleavage by SfiI can be separated in two stages. The first stage, loop assembly, involves protein association and loop formation: both of these steps are reversible as the protein can let go of one site (to release the loop) or dissociate completely from the DNA. The second stage is irreversible: the DNA is cleaved by the protein and eventually SfiI releases the cleaved DNA. With TPM we obtain all the rates of the first stage in a single measurement (black numbers show corrected values, see ‘Discussion’ section). From the solution kinetics experiments, we find the DNA cleavage rate (red number). Finally, from the TPM cleavage experiments, we acquire the DNA release rate (black number, see ‘Discussion’ section) and complete the whole kinetic scheme. The values obtained by TPM for protein association and dissociation, and for the release of the cleaved DNA, all concur with values acquired from bulk solution kinetics.
DISCUSSION
SfiI reaction pathways
With TPM we were able to directly obtain protein (dis)association-, loop formation-, loop breakage- and DNA (bead) release-rates using both the threshold and the dHMM methods. How do these rates compare to the reaction rates found by bulk biochemical methods? (27). Some of those rates, such as loop formation and breakage cannot be easily obtained by bulk methods while others, DNA hydrolysis for example, is not measurable with the TPM method. Here, we will discuss all the different steps in the reaction pathway and compare the bulk and single-molecule results whenever possible.
To consider the complete DNA cleavage reaction, we first have to take into account that the release of the cleaved DNA product is orders of magnitude slower than DNA cleavage. Therefore, the bead release we measure with TPM ought to be nearly identical with the DNA release after hydrolysis. We confirmed this by determining that the turnover rate of SfiI in free solution on the TPM substrate at 21.5°C is 1 × 10−4 s−1 (Figure 3), and the bead release rate measured with the TPM equals 3 × 10−4 s−1. Note, that the tetramer binds each site with two subunits and the DNA release of one site can occur either non-cooperative or cooperative. The times obtained with the TPM method reflect the release of DNA from the outside subunit of one of the DNA sites (non-cooperative case) or the release of two DNA ends from one of the DNA sites (cooperative case). The solution experiments indicate the release of all four DNA ends (cooperative or not). In either case, the bead release happens with a ratio of 2:1 faster than the solution experiments. Thus, to correctly compare the cleavage rate (TPM) with the turnover rate (solution kinetics), we need to calculate the time that both DNA sites are released by the enzyme. By recognizing that 1/k2-site = 1/k1-site + 2/k1-site (where k2-site is the release from both sites and k1-site denote the release from either of the two bound sites), it follows that k2-site = k1-site/3. So the 3-fold difference observed between the bulk biochemistry rate and the TPM value indicates that the two different methods give an exact match in the rate constant for the release of the cleaved DNA.
Next, the loop assembly pathway consists of two parts, protein association and loop formation, which we will discuss separately. Protein association rates are related to the values found Bellamy et al. in the accompanying paper (27), while the loop formation rate is uniquely obtained from our TPM experiments. The measured loop formation rate, however, needs to be corrected for the volume exclusion effect which is dependent on the size of the bead and the interactions with surface. This exclusion effect causes an effective stretching-force, <Feff>, on the DNA molecule of about 30 fN in these experiments (30). This force changes the statistical properties of the molecule and thus only influences the loop formation rate. In our experiment, <Feff> lowers the loop formation rate by a factor 4 (30), resulting in a corrected loop formation rate of ∼0.4 s−1. The value is similar to those reported for TPM experiments on NarI and NaeI trapping loops of similar size), and is in line with the expected rate for loop formation if this depends entirely on the dynamics of the DNA molecule (28,45,46).
Knowing the loop formation rate permits us to correct the estimated protein association rate from the single-turnover reactions with WT SfiI, to yield a value of ∼2 × 108 M−1 s−1. The value obtained from our single-molecule cleavage experiments needs a different adjustment: we need to take into account the relative long time it takes to flow the enzyme into the sample (about a minute) which makes detecting short events impossible. To correct for this, we assume that the Cumulative Probability Distribution (Figure 2B) only displays events after the first 60 s: hence, we shift the time axis by a similar amount and we obtain an association rate of ∼6 × 108 s−1 M−1. However, these corrections include large errors, which limits the direct comparison of the two rates. Better values (and statistics in the TPM case) be obtained from measurements on the SfiI mutant D79A. Bellamy et al. (27) determined the D79A association rate to be 2 × 108 M−1 s−1: our TPM experiments yielded an identical rate, 2 ± 0.5 × 108 M−1 s−1. Besides the perfect match of the two methods, the association rate determined for D79A is also close to the WT rates. Apparently, the D79A mutation in the binding pocket of the enzyme has no impact on the searching and binding efficiency of the enzyme.
Since SfiI binds as a single tetramer to one site prior to DNA looping, increasing protein concentrations can lead to both recognition sites becoming occupied, which then blocks loop formation (20). By extrapolating the concentration at which the binding rate of the second tetramer is equal to the rate of loop formation, we can estimate when this saturation effect becomes significant. This effect should start at a protein concentration of ∼10 nM. However, at this concentration, the DNA starts to get strongly compacted and no longer displays specific looping. Hence, repression of loop formation by tetramers binding to both specific sites is not observable in our experiments. It is interesting to note that non-specific protein-induced DNA condensation starts around the protein concentration at which the saturation effect is expected to occur (Figure 4). Possibly in the absence of another unoccupied specific site, the two specifically bound proteins form non-specific loops in the DNA that act as nucleation points for further DNA condensation. Previous work of TPM on RE by van den Broek et al. (28) also showed that high RE concentrations result in non-specific DNA compaction.
Bellamy et al. determined the bulk rates of protein dissociation of D79A in Mg2+ and Ca2+ as 0.17 s−1 and 5 × 10−4 s−1, respectively, while no dissociation was observed for the WT enzyme in Ca2+ buffer (27). This lack of dissociation for the WT enzyme in Ca2+ is matched by our TPM experiments in which not a single loop breakage event was observed over a 3-h period (Figure 2A). In Mg2+ buffer, D79A showed rapid dynamics in the TPM and we found a dissociation rate of ∼0.1 s−1. This result is similar to the rate found by Bellamy et al. For D79A in Ca2+ we observed in our TPM traces some dynamics which are of the same order of magnitude as the biochemical study (∼30 min, data not shown). Hence, under all the tested conditions, the rates of TPM and bulk biochemistry methods are again in close agreement.
One could argue that protein dissociation and loop breaking is essentially the same process only governed by a slightly different stochastic probability (29), a factor or 2. Accordingly, the loop breakage rate should be twice as fast as the protein dissociation rate. We can examine this specifically since loop disruption is observed directly in the TPM, making rate determination uniquely accessible. With both rates determined independently, we find that the ratio of the loop breakage rate (0.03 ± 0.01 s−1) to the dissociation rate (0.06 ± 0.02 s−1) is not the 2-fold reduction expected but is instead a 2-fold enhancement. Hence, in accordance with previous biochemical studies (15,35), there must be an energy difference between the protein bound to one recognition sites and that bound to two sites. The biochemical studies had shown that the SfiI tetramer binds two DNA duplexes in a highly cooperative manner: the binding of the first duplex to one of the two DNA-binding sites in the tetramer induces a conformational change that extends to the second DNA-binding site and which causes it to bind its recognition site with a much higher affinity. The enhanced affinity is due primarily to the DNA dissociating slowly from the ES2 complex with two duplexes but rapidly from the ES1 complex with one duplex (35). The dissociation rate noted above is related to the faster of these two rates and the loop breakage rate to the slower.
Improved TPM methods
Using the correlation based bead tracking algorithm we can collect simultaneously data on 50 beads at 25 Hz. This real time data collection rate represents a 50-fold increase compared to our previous TPM study and other recent studies (28,47,48). In addition, tracking multiple beads simultaneously in real time enables us to correct for any drift or slight sample movement. Finally, the parallel data collection and computerized data analysis allows us to fully characterize within one hour all the kinetic rates for a DNA-looping protein in one buffer condition, using one sample chamber.
The introduction of the three-state model used for our dHMM-fit to the TPM data showed that we could obtain parameters not accessible by traditional biochemistry methods. We are able to obtain directly the protein association and dissociation rates from fitting the measured loop lifetimes to a double exponential even though both steps cannot directly be observed in TPM experiments. Finzi et al. also used a bi-exponential fit for the dwell times, but did this to differentiate between the two chemical species of their protein (29).
Although the newly developed dHMM tools (32) have successfully been applied to obtain the kinetic rates from the SfiI TPM traces, there are some remarks to be made. First, the dHMM algorithm requires a set of training data to construct the lookup tables for the fitting. This training data consists of two-time traces: an unlooped DNA tether and a tether fixed for at least 30 min in the looped state. Most RE, however, show fast looping dynamics, and adding Ca2+ does not necessarily stabilize bound complexes for sufficiently long periods. Thus a training set of the looped state can be difficult to obtain or construct. SfiI, fortunately, is an exception to this rule since the WT enzyme forms very stable DNA–protein complexes in the presence of Ca2+, making it a model system for dHMM analysis. Second, like other fit routines, dHMM analysis needs an initial set of guesses to start fitting the data. We demonstrated that our threshold method provides these initial fit coefficients. With these guesses, we could quickly process and fit the raw/unfiltered time traces with the dHMM method.
To conclude, TPM allows us to directly visualize and quantify the complete loop formation process by a single protein binding to two DNA sites on an individual DNA molecule. We have demonstrated that we can measure the two-step process involved in loop assembly, protein association and loop formation by the DNA-bound protein even though the first step does not generate a directly observable signal; i.e. shortening of the DNA tether. Moreover, all the kinetics rates involved in protein-induced loop formation and disruption can be obtained in a single measurement that requires no intervention by the user during the process. Finally, we demonstrate that the reaction rates obtained by TPM all match the rates obtained by biochemical assays in bulk solution, thus providing solid support of TPM as a powerful complementary technique to investigate protein–DNA interactions and their kinetics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fundamenteel Onderzoek der Materie research program ‘Physics of the Genome’; ‘Nederlandse Organisatie voor Wetenschappelijk Onderzoek’ (to N.L. and G.J.L.W.); VICI grant of ‘Nederlandse Organisatie voor Wetenschappelijk Onderzoek’ (to G.J.L.W.); Wellcome Trust [grant 078794] (to S.R.W.B., Y.S.K. and S.E.H.). Funding for open access charge: Wellcome Trust award to the University of Bristol.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Bram van der Broek for his input on the TPM experiments, and John Beausang for his help with the diffusive hidden Markov analysis.
REFERENCES
- 1.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. EMBO J. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dodson M, Roberts J, McMacken R, Echols H. Specialized nucleoprotein structures at the origin of replication of bacteriophage lambda: complexes with lambda O protein and with lambda O, lambda P, and Escherichia coli DnaB proteins. Proc. Natl Acad. Sci. USA. 1985;82:4678–4682. doi: 10.1073/pnas.82.14.4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gellert M, Nash H. Communication between segments of DNA during site-specific recombination. Nature. 1987;325:401–404. doi: 10.1038/325401a0. [DOI] [PubMed] [Google Scholar]
- 4.Moitoso de Vargas L, Pargellis CA, Hasan NM, Bushman EW, Landy A. Autonomous DNA binding domains of lambda integrase recognize two different sequence families. Cell. 1988;54:923–929. doi: 10.1016/0092-8674(88)90107-9. [DOI] [PubMed] [Google Scholar]
- 5.Lewis M, Chang G, Horton NC, Kercher MA, Pace HC, Schumacher MA, Brennan RG, Lu P. Crystal structure of the lactose operon repressor and its complexes with DNA and inducer. Science. 1996;271:1247–1254. doi: 10.1126/science.271.5253.1247. [DOI] [PubMed] [Google Scholar]
- 6.Lloyd G, Landini P, Busby S. Activation and repression of transcription initiation in bacteria. Essays Biochem. 2001;37:17–31. doi: 10.1042/bse0370017. [DOI] [PubMed] [Google Scholar]
- 7.Mastrangelo IA, Courey AJ, Wall JS, Jackson SP, Hough PV. DNA looping and Sp1 multimer links: a mechanism for transcriptional synergism and enhancement. Proc. Natl Acad. Sci. USA. 1991;88:5670–5674. doi: 10.1073/pnas.88.13.5670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Halford SE, Bilcock DT, Stanford NP, Williams SA, Milsom SE, Gormley NA, Watson MA, Bath AJ, Embleton ML, Gowers DM, et al. Restriction endonuclease reactions requiring two recognition sites. Biochem. Soc. Trans. 1999;27:696–699. doi: 10.1042/bst0270696. [DOI] [PubMed] [Google Scholar]
- 9.Deibert M, Grazulis S, Sasnauskas G, Siksnys V, Huber R. Structure of the tetrameric restriction endonuclease NgoMIV in complex with cleaved DNA. Nat. Struct. Biol. 2000;7:792–799. doi: 10.1038/79032. [DOI] [PubMed] [Google Scholar]
- 10.Bath AJ, Milsom SE, Gormley NA, Halford SE. Many type IIs restriction endonucleases interact with two recognition sites before cleaving DNA. J. Biol. Chem. 2002;277:4024–4033. doi: 10.1074/jbc.M108441200. [DOI] [PubMed] [Google Scholar]
- 11.Gowers DM, Bellamy SRW, Halford SE. One recognition sequence, seven restriction enzymes, five reaction mechanisms. Nucleic Acids Res. 2004;32:3469–3479. doi: 10.1093/nar/gkh685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roberts RJ, Belfort M, Bestor T, Bhagwat AS, Bickle TA, Bitinaite J, Blumenthal RM, Degtyarev SK, Dryden DTF, Dybvig K, et al. A nomenclature for restriction enzymes, DNA methyltransferases, homing endonucleases and their genes. Nucleic Acids Res. 2003;31:1805–1812. doi: 10.1093/nar/gkg274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Perona JJ. Type II restriction endonucleases. Methods. 2002;28:353–364. doi: 10.1016/s1046-2023(02)00242-6. [DOI] [PubMed] [Google Scholar]
- 14.Vipond IB, Halford SE. Specific DNA recognition by Ecorv restriction-endonuclease induced by calcium-ions. Biochemistry. 1995;34:1113–1119. doi: 10.1021/bi00004a002. [DOI] [PubMed] [Google Scholar]
- 15.Embleton ML, Williams SA, Watson MA, Halford SE. Specificity from the synapsis of DNA elements by the SfiI endonuclease. J. Mol. Biol. 1999;289:785–797. doi: 10.1006/jmbi.1999.2822. [DOI] [PubMed] [Google Scholar]
- 16.Topal MD, Thresher RJ, Conrad M, Griffith J. NaeI endonuclease binding to pBR322 DNA induces looping. Biochemistry. 1991;30:2006–2010. doi: 10.1021/bi00221a038. [DOI] [PubMed] [Google Scholar]
- 17.Wentzell LM, Halford SE. DNA looping by the Sfi I restriction endonuclease. J. Mol. Biol. 1998;281:433–444. doi: 10.1006/jmbi.1998.1967. [DOI] [PubMed] [Google Scholar]
- 18.Siksnys V, Skirgaila R, Sasnauskas G, Urbanke C, Cherny D, Grazulis S, Huber R. The Cfr10I restriction enzyme is functional as a tetramer. J. Mol. Biol. 1999;291:1105–1118. doi: 10.1006/jmbi.1999.2977. [DOI] [PubMed] [Google Scholar]
- 19.Embleton ML, Siksnys V, Halford SE. DNA cleavage reactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol. 2001;311:503–514. doi: 10.1006/jmbi.2001.4892. [DOI] [PubMed] [Google Scholar]
- 20.Milsom SE, Halford SE, Embleton ML, Szczelkun MD. Analysis of DNA looping interactions by type II restriction enzymes that require two copies of their recognition sites. J. Mol. Biol. 2001;311:515–527. doi: 10.1006/jmbi.2001.4893. [DOI] [PubMed] [Google Scholar]
- 21.Katiliene Z, Katilius E, Woodbury NW. Single molecule detection of DNA looping by NgoMIV restriction endonuclease. Biophys. J. 2003;84:4053–4061. doi: 10.1016/S0006-3495(03)75131-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Seidel R, van Noort J, van der Scheer C, Bloom JG, Dekker NH, Dutta CF, Blundell A, Robinson T, Firman K, Dekker C. Real-time observation of DNA translocation by the type I restriction modification enzyme EcoR124I. Nat. Struct. Mol. Biol. 2004;11:838–843. doi: 10.1038/nsmb816. [DOI] [PubMed] [Google Scholar]
- 23.Yan J, Skoko D, Marko JF. Near-field-magnetic-tweezer manipulation of single DNA molecules. Phys. Rev. E. Stat. Nonlin. Soft Matter Phys. 2004;70:011905. doi: 10.1103/PhysRevE.70.011905. [DOI] [PubMed] [Google Scholar]
- 24.van den Broek B, Noom MC, Wuite GJ. DNA-tension dependence of restriction enzyme activity reveals mechanochemical properties of the reaction pathway. Nucleic Acids Res. 2005;33:2676–2684. doi: 10.1093/nar/gki565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Noom MC, van den Broek B, van Mameren J, Wuite GJL. Visualizing single DNA-bound proteins using DNA as a scanning probe. Nature Methods. 2007;4:1031–1036. doi: 10.1038/nmeth1126. [DOI] [PubMed] [Google Scholar]
- 26.van den Broek B, Lomholt MA, Kalisch SMJ, Metzler R, Wuite GJL. How DNA coiling enhances target localization by proteins. Proc. Natl Acad. Sci. USA. 2008;105:15738–15742. doi: 10.1073/pnas.0804248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bellamy SRW, Kovacheva YS, Zulkipli IH, Halford SE. Differences between Ca2+ and Mg2+ in DNA binding and release by the SfiI restriction endonuclease: implicationas for DNA looping. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp569. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van den Broek B, Vanzi F, Normanno D, Pavone FS, Wuite GJL. Real-time observation of DNA looping dynamics of type IIE restriction enzymes NaeI and NarI. Nucleic Acids Res. 2006;34:167–174. doi: 10.1093/nar/gkj432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Finzi L, Gelles J. Measurement of lactose repressor-mediated loop formation and breakdown in single DNA molecules. Science. 1995;267:378–380. doi: 10.1126/science.7824935. [DOI] [PubMed] [Google Scholar]
- 30.Segall DE, Nelson PC, Phillips R. Volume-exclusion effects in tethered-particle experiments: Bead size matters. Phys. Rev. Lett. 2006;96 doi: 10.1103/PhysRevLett.96.088306. 088306-1/4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Colquhoun D, Sigworth FJ. In: Single Channel Recording. Sakmann B, Neher E, editors. New York and London: Plenum Press; 1983. pp. 191–263. [Google Scholar]
- 32.Beausang JF, Zurla C, Manzo C, Dunlap D, Finzi L, Nelson PC. DNA looping kinetics analyzed using diffusive hidden Markov model. Biophys. J. 2007;92:L64–L66. doi: 10.1529/biophysj.107.104828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wentzell LM, Nobbs TJ, Halford SE. The Sfii restriction-endonuclease makes a 4-strand DNA break at 2 copies of its recognition sequence. J. Mol. Biol. 1995;248:581–595. doi: 10.1006/jmbi.1995.0244. [DOI] [PubMed] [Google Scholar]
- 34.Bellamy SR, Mina P, Retter SE, Halford SE. Fidelity of DNA sequence recognition by the SfiI restriction endonuclease is determined by communications between its two DNA-binding sites. J. Mol. Biol. 2008;384:557–563. doi: 10.1016/j.jmb.2008.09.057. [DOI] [PubMed] [Google Scholar]
- 35.Bellamy SRW, Milsom SE, Kovacheva YS, Sessions RB, Halford SE. A switch in the mechanism of communication between the two DNA-binding sites in the sfil restriction endonuclease. J. Mol. Biol. 2007;373:1169–1183. doi: 10.1016/j.jmb.2007.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nelson PC, Zurla C, Brogioli D, Beausang JF, Finzi L, Dunlap D. Tethered particle motion as a diagnostic of DNA tether length. J. Phys. Chem. 2006;110:17260–17267. doi: 10.1021/jp0630673. [DOI] [PubMed] [Google Scholar]
- 37.Yin H, Landick R, Gelles J. Tethered particle motion method for studying transcript elongation by a single RNA polymerase molecule. Biophys. J. 1994;67:2468–2478. doi: 10.1016/S0006-3495(94)80735-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rabiner LR. A Tutorial on Hidden Markov-Models and selected applications in speech recognition. Proc. IEEE. 1989;77:257–286. [Google Scholar]
- 39.Qin F, Auerbach A, Sachs F. Hidden Markov modeling for single channel kinetics with filtering and correlated noise. Biophys. J. 2000;79:1928–1944. doi: 10.1016/S0006-3495(00)76442-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKinney SA, Joo C, Ha T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 2006;91:1941–1951. doi: 10.1529/biophysj.106.082487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lagunavicius A, Grazulis S, Balciunaite E, Vainius D, Siksnys V. DNA binding specificity of MunI restriction endonuclease is controlled by pH and calcium ions: involvement of active site carboxylate residues. Biochemistry. 1997;36:11093–11099. doi: 10.1021/bi963126a. [DOI] [PubMed] [Google Scholar]
- 42.Engler LE, Sapienza P, Dorner LF, Kucera R, Schildkraut I, Jen-Jacobson L. The energetics of the interaction of BamHI endonuclease with its recognition site GGATCC. J. Mol. Biol. 2001;307:619–636. doi: 10.1006/jmbi.2000.4428. [DOI] [PubMed] [Google Scholar]
- 43.Halford SE. Hopping, jumping and looping by restriction enzymes. Biochem. Soc. Transactions. 2001;29:363–373. doi: 10.1042/bst0290363. [DOI] [PubMed] [Google Scholar]
- 44.Nobbs TJ, Szczelkun MD, Wentzell LM, Halford SE. DNA excision by the SfiI restriction endonuclease. J. Mol. Biol. 1998;281:419–432. doi: 10.1006/jmbi.1998.1966. [DOI] [PubMed] [Google Scholar]
- 45.Vologodskii A, Cozzarelli NR. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J. 1996;70:2548–2556. doi: 10.1016/S0006-3495(96)79826-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jian HM, Schlick T, Vologodskii A. Internal motion of supercoiled DNA: Brownian dynamics simulations of site juxtaposition. J. Mol. Biol. 1998;284:287–296. doi: 10.1006/jmbi.1998.2170. [DOI] [PubMed] [Google Scholar]
- 47.Normanno D, Vanzi F, Pavone FS. Single-molecule manipulation reveals supercoiling-dependent modulation of lac repressor-mediated DNA looping. Nucleic Acids Res. 2008;36:2505–2513. doi: 10.1093/nar/gkn071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wong OK, Guthold M, Erie DA, Gelles J. Interconvertible lac repressor-DNA loops revealed by single-molecule experiments. PLos Biol. 2008;6:2028–2042. doi: 10.1371/journal.pbio.0060232. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.