Abstract
It has been proposed that the hepatitis C virus (HCV) internal ribosome entry site (IRES) resides within a locked conformation, owing to annealing of its immediate flanking sequences. In this study, structure probing using Escherichia coli dsRNA-specific RNase III and other classical tools showed that this region switches to an open conformation triggered by the liver-specific microRNA, miR-122. This structural transition, observed in vitro, may be the mechanistic basis for the involvement of downstream IRES structural domain VI in translation, as well as providing a role of liver-specific miR-122 in HCV infection. In addition, the induced RNA switching at the 5′ untranslated region could ultimately represent a new mechanism of action of micro-RNAs.
INTRODUCTION
In a linear genomic map, the hepatitis C virus (HCV) internal ribosome entry site (IRES) (domains II–IV, bases 40–368) (1) is flanked upstream by domain I (bases 1–40), which is recognized by a liver-specific microRNA (miR-122) that acts as an enhancer of replication (2) (Figure 1A). Evidence accumulated over the last 10 years indicates that IRES downstream sequences in the coding region, particularly in domains V and VI (bases 368–510) are important for viral viability (3–6) through an unknown RNA structure-dependent mechanism.
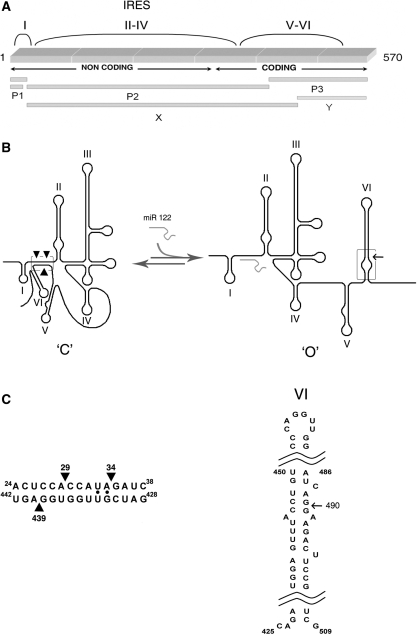
Figure 1.
Schematic representation of 1–570 RNA. (A) Primary structure, where mapping of band X is indicated relative to 1–570 RNA and previously published products of the RNase III reaction with RNA in the closed conformation (‘C’). (B) Proposed secondary structures. Left: closed (‘C’) conformation. Solid arrowheads indicate the RNase III cleavages in the LRA. Right: linear open (‘O’) conformation; new cleavage in stem-loop VI indicated with an arrow; (C) detailed sequences of the double helical elements recognized by RNase III.
In the secondary structure representation, functional (5), structural (7) and phylogenetic (6) studies indicate that bases 428–442 within the coding region interact with the complementary 5′ unstructured sequence, 24–38, forming a long-range annealing (LRA) motif that constrains the IRES in a closed ‘C’ conformation (Figure 1B and C) known to inhibit viral translation (5,6). Alternatively, earlier work suggested a structure within the core-coding domain, called stem-loop VI, in which bases 428–442 are paired with bases 495–508 (8) (Figure 1B and C), yielding an open conformation, which is designated ‘O’ herein in contraposition to the ‘C’ conformation.
We hypothesized that switching between the alternative ‘C’ and ‘O’ structures could regulate the ability of an HCV RNA molecule to participate in various biological processes (7). This idea was favored by subsequent descriptions of interactions between the most abundant liver microRNA, miR-122 and the 22–28 HCV region (2). Because this region overlaps with sequence 24–38, which anneals at long distance with 428–442, we hypothesized that miR-122 would inhibit LRA formation and thus promote the structural transition to stem-loop VI. This study investigates this possibility.
MATERIALS AND METHODS
RNA transcripts
The DNA templates for HCV RNA transcripts were derived from the plasmid vector pN(1–4728) Bluescript, which contains nt 1–4728 of HCV, under the T7 promoter. All RNA transcripts include the first base of the HCV genome and end at positions 402 (Aat II NEB), 466 (PCR product) and 570 (Blp I NEB). The PCR product ending at base 466 was generated by amplification of pN(1–4728) with the upstream primer 5′-CGCGGATCCTAATACGACTCACTATAGGCCGCCCCGATTGGGGGCGA (which serves to introduce the T7 promoter in the PCR product) and the downstream primer 5′-GGGCCCCTGCGCGGCAACAG.
In vitro transcription and internal labeling
To obtain internally labeled substrates for the cleavage assays, 1–2 µg of DNA template was transcribed in vitro for 1 h at 37°C with T7 RNA polymerase (Promega) and [α-32P]GTP (Perkin Elmer) followed by a 10-min treatment with 0.04 U/µl RNase-free RQ1 DNAse I (Promega) at 37°C (9). Cellulose CF11 chromatography was used to eliminate DNA, dsRNA fragments and unincorporated nucleotides (10). Transcripts were then purified by gel electrophoresis under denaturing conditions on 4% polyacrylamide gels containing 7 M urea. Bands were visualized by autoradiography, excised from the gel and eluted in buffer (100 mM Tris–HCl, pH 7.5 and 10 mM EDTA, pH 7.5). The concentration of radioactive transcripts was determined by calculating the amount of incorporated [α-32P]GTP based on quick count measures.
The 21-base transcripts representing the wt miR-122 and mutants were synthethised according to a T7 RNA polymerase transcription protocol in a 50 µl reactions overnight at 37°C (9). Synthetic DNA oligodeoxinucleotides used as transcription templates were obtained from the Instituto de Parasitología y Biomedicina (C.S.I.C) facility. Transcripts were labelled by the inclusion of a [α-32P]GTP labeled in the reaction mixture. Transcription reactions were subsequently subjected to RQ1 DNase I treatment, phenol extraction and ethanol precipitation. The transcripts were purified by electrophoresis in 10% polyacrilamide gel containing 7 M urea, and eluted and measured identically to larger in vitro transcripts.
5′ end labeling
5′ end-labeled RNA transcript was obtained in a standard transcription reaction with two differences, the labeled triphosphate nucleotide was [γ32P]GTP (Perkin Elmer), and the concentration of cold GTP in the nucleotide reaction mix was lowered to a quarter of the standard amount. Subsequent purification was performed as described above.
3′ end labeling
Unlabeled transcripts prepared in standard transcription reactions were subsequently reacted as follows: aliquots containing a molar ratio of 2:1 pmol of 5′ [32P]pCp (Perkin Elmer) with respect to unlabeled RNA were incubated with T4 RNA ligase (Amersham Bioscience). The reaction was carried out in 10 µl of 50 mM Tris–HCl (pH 7.5), 10 mM MgCl2, 10 mM DTT, 1 mM ATP, 0.01% BSA, 0.1% PEG, 20 U RNasin (Promega) and 4 U/µl T4 RNA ligase. The reaction mixture was incubated for 4 days at 4°C. The labeled RNA was purified again using the electrophoretic procedure described above.
RNase III cleavage assay
Escherichia coli RNase III is a nuclease specific for dsRNA (11,12). The salt and buffer conditions used in all our experiments on HCV RNA cleavage by RNase III were the same as those used previously to detect RNase P cleavage of a tRNA-like structure near the AUG start triplet (13); these are known as secondary conditions of cleavage for RNase III (14). HCV RNA substrate was pre-heated at 90°C for 1 min before addition of reaction buffer (10 mM HEPES–KOH [pH 7.5], 10 mM Mg[AcO]2 and 100 mM NH4[AcO]) and then left to cool down to room temperature. Cleavage reactions were performed with 20 U RNasin, and 0.0005 U/µl or 0.001 U/µl (final concentration [FC]) of E. coli RNase III (Ambion) in the presence of 2 µg/µl of yeast tRNA (Ambion), and were carried out in a volume of 10 µl at 37°C for 1 h. These optimal conditions were used in all the experiments and are referred to throughout the text as ‘standard conditions’. However, when the reactions were performed in the presence of rabbit reticulocyte lysate (supplemented with a mixture of 0.01 mM aa minus Cys), RNA was extracted successively with phenol and chloroform/isoamyl alcohol and precipitated. The cleavage products were separated on 4% denaturing polyacrylamide gels and visualized by autoradiography on Kodak BioMax MR or Fuji Super RX film.
For the kinetic studies, samples were treated as above and scaled-up to 100 µl reaction volumes. Aliquots of 10 µl were drawn at 0, 10, 20, 30, 40, 50 and 60 min and analyzed on polyacrylamide gels, as described above. Product bands were quantified using a Storm PhosphorImager (Amersham-Pharmacia Biotech) as follows: percent RNase III cleavage product (A) = (A product)/(starting material + ∑products) × 100. Fold increase of P1X was measured as: P1X product at 60 min in the desired conditions divided by P1X product in the control lane at 60 min, run in the same gel.
RNase H cleavage assay
The salt and buffer conditions used for E. coli RNase H (Ambion) digestion were the same as those used in the RNase III cleavage assay. HCV RNA substrate was pretreated identically. Cleavage reactions were performed in the presence of 20-mer DNA oligodeoxynucleotides (15, 150 and 1500 nM) with 20 U RNasin in the presence of 2 µg/µl of yeast tRNA (Ambion) and 0.5 U/µl of RNase H, and were carried out in a volume of 10 µl at 37°C for 1 h. Cleavage products were separated on 4% denaturing polyacrylamide gels and visualized by autoradiography.
Analysis on non-denaturing gels
HCV RNA was prepared as described above. We compared the buffer and salt conditions commonly used for RNA conformation assays, which contain 20 mM Tris–Ac (pH 7.6), 10 mM MgAcO and 100 mM NaCl (TMN 1X), with our standard conditions. Our conditions provided similar or even better results; hence, they were selected for the subsequent reactions. In all cases, just before addition of the probe, 2 µg of carrier tRNA was added per reaction. Annealing reactions were incubated for 1 h at 37°C and transferred to an ice bucket. The RNA was left on ice for at least 10 min before re-suspending in loading buffer, consisting of 60% glycerol, TMN 1×, 0.4 µg/µl yeast tRNA, 0.4% (w/v) xylene cyanol, and 0.4% (w/v) bromophenol blue, and was then used for gel analysis. Non-denaturing gels were 0.8 mM thick and contained 6% polyacrylamide, 50 mM Tris–Ac, pH 8.3 and 10 mM MgAcO. The gels were run at constant amperage of 12 mA for 24 h at 4°C, and autoradiographed.
Structural mapping with the single- or double-stranded RNases, T1 and V1
Various concentrations were prepared of RNase T1 (Calbiochem) (0.01, 0.001, 0.0001, 0.00001 μg/μl) and RNase V1 (Ambion) (0.0005 and 0.001 U/μl). Analytical amounts of 3′ end-labeled 32pCp 1–570 RNA with and without different probes were digested with these RNase fractions and the digestion products electrophoresed on a denaturing 4% and 10% 7 M urea-containing polyacrylamide gel. The optimal RNase T1 and V1 digestion conditions were found to be 0.0005 μg/μl in a 20-min reaction and 0.001 U/μl in a 30-min reaction at 37°C, respectively. Adequate band separation to detect all differences between the electrophoretic lanes was achieved when bromophenol blue dye reached 80% of the length of a 4% polyacrylamide–urea gel. These conditions were used for the subsequent analyses of 1–570 RNA alone or pre-incubated with the DNA or RNA probes at several concentrations before adding the nucleases T1 or V1. Preincubation was performed for 1 h at 37°C.
To allow identification of the cleavage sites, a parallel run was performed with a 3′ end-labeled 1–570 RNA ladder generated with either limited alkaline hydrolysis or RNase T1 degradation in denaturing conditions. In the alkaline hydrolysis reactions, aliquots containing 104 c.p.m. of 3′ end-labeled RNA were incubated with 2 µg of carrier tRNA in 0.2 M NaHCO3–Na2CO3 (pH 9) and 1 mM EDTA for 5 min at 90°C. The RNase T1 reaction in denaturing conditions was performed with similar amounts of labeled substrate in the presence of 7 M urea at 55°C for 5 min. All reactions were stopped with one volume of loading buffer and maintained in dry ice until loading on 4% polyacrylamide-urea sequencing gel.
RNA sequencing using RACE
Two bands from T1 RNase digestion gel were purified and sequenced by the Rapid Amplification of cDNA Ends (RACE) procedure (ROCHE 5′/3′RACE kit 2nd generation). 5′ RACE reactions were performed according to the manufacturer′s recommendations. The HCV-specific oligonucleotide used for first-strand cDNA synthesis was 5′-542CTTGGGGATAGGCTGTCGCCT522, and the one used for PCR was 5′-521TCCACGGGGTTGCGACCGCT502.
Synthetic RNA oligonucleotides
The complete miR-122 RNA sequence is 5′-UGGAGUGUGACAAUGGUGUUUGU. The underlined portion (5′-UGGAGUGUGA) is a 10-mer oligoribonucleotide containing the 7 nt of the seed sequence. The remaining portion (5′-CAAUGGUGUUUGU) is miR-122 RNA minus the seed sequence. All oligoribonucleotides used were synthesized by IDT Integrated DNA Technologies.
Protein quantification
Protein concentration was determined using the Bio-Rad protein microassay procedure (Bio-Rad Laboratories).
RESULTS
Detection of a new RNase III cleavage event in 1–570 RNA indicates the potential of this region to form an alternative RNA structure
Previous RNase III cleavage experiments involved a 570-base HCV RNA transcript (nt 1–570). Figure 1A is a schematic drawing of the 1–570 RNA fragment showing the final reaction products characterized (P1, P2 and P3), located according to their corresponding positions. The autoradiogram in Figure 2, lane 2, reproduces an RNase III cleavage of a 1–570 RNA substrate resulting in partial digestion products (P2P3 and P1P2) and complete digestion products (P1, P2 and P3). It was demonstrated that these cleavages are only possible after annealing of both IRES flanking sequences (7). In the course of that experiment, we observed an additional minor band that appeared in a time-dependent manner [Figure 7 in reference (7)], but was not examined at that time. The band, which has a mobility between the two large partial product bands, P2P3 and P1P2, becomes a prominent band in the gel when, as in the present case, the reaction is performed at a higher RNase III concentration. The appearance of this band, referred to herein as band ‘X’, and its partner product, band Y, which comprise the total 1–570-base transcript, can only be observed when nearly all the 1–570 substrate has been processed and P2P3 becomes a dominant high molecular weight band. We propose two possible explanations for this: (i) Band X may correspond to a less sensitive site for RNase III in 1–570 RNA fragments, or (ii) Band X could be the result of an additional cleavage of P2P3 by RNase III, which does not occur in the 1–570 RNA substrate. This second option would indicate the presence of alternative structures within 1–570 RNA.
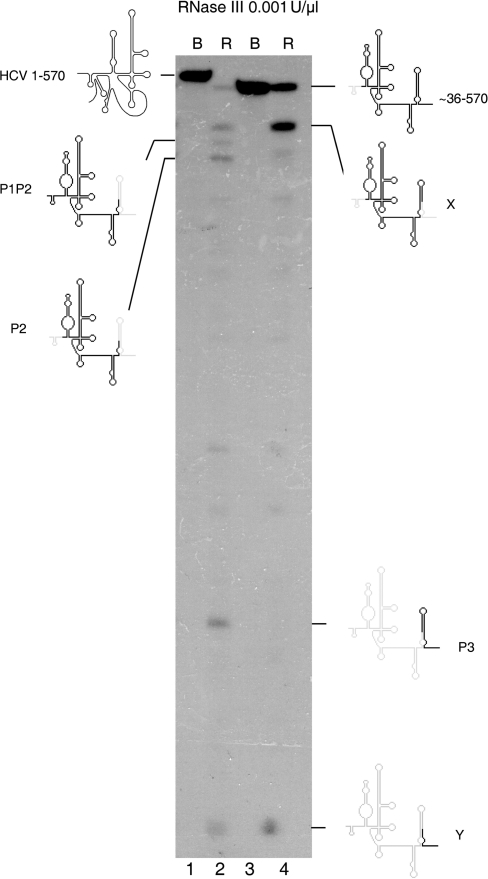
Figure 2.
The new RNase III cleavage band detected in 1–570 RNA transcript (lanes 1–2) is indicated by X, and is shown to be highly promoted in a 5′ truncated fragment ∼36–570 (lanes 3–4). Lanes 1 and 3 are 1–570 and ∼36–570 RNA transcripts incubated in buffer, and lanes 2 and 4 are the same transcripts incubated with 0.001 U/μl of RNase III.
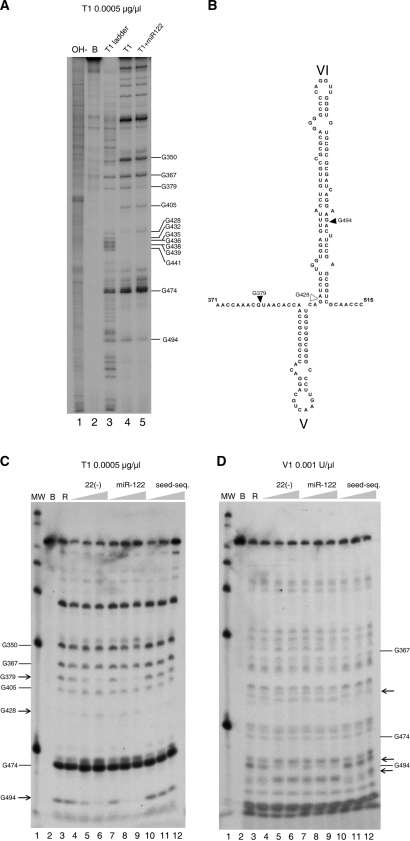
Figure 7.
(A) ‘G’ sequence determination of 3′ end-labeled 1–570 RNA in our standard conditions of cleavage by parallel running of RNA degradation with RNase T1 0.0005 µg/µl in denaturing conditions (lane 3), in standard buffer (lane 4) and in the presence of miR-122 at 15 nM (lane 5). Lanes 1 is an alkali ladder degradation and lane 2, the RNA incubated in standard buffer alone. Gs are identified at the right of the gel. (B) Secondary structure of stem loop VI and its boundary regions summarizing the position with differential reactivity for RNase T1. Increased resistance is indicated by solid triangles and increased sensitivity by blank triangles. Nucleotide numbering is used, as in Figure 1. (C) Evaluation of the effect of increasing concentrations of probes on the T1 nuclease pattern of cleavage. Lane 1 is a MW marker. Control incubation of 1–570 RNA in the buffer (lane 2), or after addition 0.0005 µg/µl of RNase T1 (lane 3). In subsequent lanes, before addition of the T1 RNase, RNAs were pre-incubated for 1 h with increasing concentrations of ODN 22(−) of 15 nM, 150 nM and 1500 nM (lanes 4–6), or miR-122 at final concentrations of 1.5 nM, 15 nM and 150 nM (lanes 7–9) and a 10-mer oligoribonucleotide carrying the 7 nucleotides of the miR-122 seed sequence to a final concentration of 1.5 nM, 15 nM and 150 nM (lanes 10–12). (D) Same as panel B, but the RNase used was double-stranded RNase V1. Arrows indicate the changes in sensitivity to the nuclease.
To investigate potential structural transitions within 1–570 RNA, we set up a parallel enzymatic reaction of 1–570 RNA and a 5′ truncated ∼36–570 HCV RNA transcript (nearly identical in size to P2P3) with RNase III. We found that band X was the main product of the reaction with the truncated transcript, where LRA formation is impeded (Figure 2, lanes 3 and 4). This is the key result that indicates that band X is not a direct product of 1–570 RNA cleavage, but instead, is a product of P2P3 cleavage.
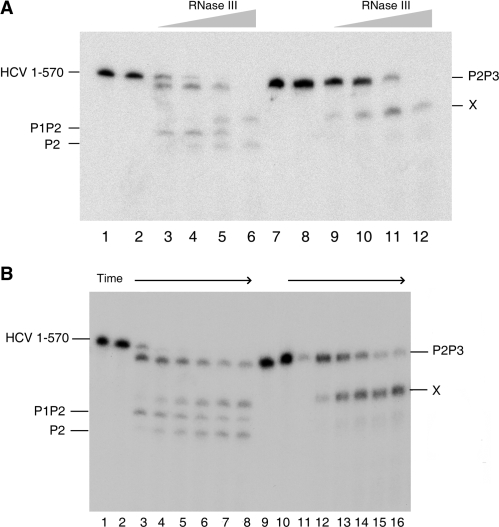
Additional support was provided by titration experiments with RNase III on 1–570 RNA and purified P2P3. These fragments were incubated in the presence of increasing concentrations of RNase III, and the resulting products were analyzed in parallel migration in gel electrophoresis. As is shown in Figure 3A lanes 3–6, RNase III cleaves 1–570 RNA to produce P2P3 at an enzyme concentration 10-fold lower than the concentration needed to release band X. In contrast, both gel-purified and endogenously produced P2P3 incubated with RNase III releases the X band at an enzyme concentration similar to that producing P2P3 from 1–570 RNA (Figure 3A). In addition, a correlation was found between P2P3 decrease and band X formation curves. This demonstrates that both the LRA motif and its alternative conformation form stable helices, equally sensitive to RNase III, and strongly indicates that band X derives from P2P3. It also further supports our previous data that the LRA within 1–570 RNA is the major structural conformation.
Figure 3.
(A) A new RNase III cleavage detected in 1–570 RNA (lanes 1–6) is highly promoted in its larger cleavage product, P2P3 (lanes 7–12). 1–570 RNA and P2P3 were titrated for cleavage by E. coli RNase III at four concentrations: 0.0001 U/µl (lanes 3 and 9), 0.0005 U/µl (lanes 4 and 10), 0.001 U/µl (lanes 5 and 11) and 0.005 U/µl (lanes 6 and 12). Lanes 1 and 7 are RNA alone, whereas 2 and 8 are RNA incubated with reaction buffer alone. The new cleavage band is indicated by X. (B) Parallel kinetic analysis of E. coli RNase III cleavage of 1–570 RNA (lanes 1–8) and P2P3 RNA (lanes 9–16). Lanes 1 and 9, respectively, show 1–570 RNA and P2P3 transcripts alone, incubated on ice; Lanes 2–8 represent sequentially 10, 20, 30, 40, 50 and 60 min of incubation at 0.0005 U/µl of RNase III; Lanes 10–16 represent the same incubation times as two to eight at the same enzyme concentration.
The new RNase III cleavage event is specific
Since HCV RNA is not a canonical large dsRNA substrate for RNase III, it must be proven that band X is the specific result of RNase III activity and not of ssRNase-specific contaminant activities or otherwise non-canonical actions of the commercial RNase III preparation. Therefore, for band X, we identified the characteristic RNase III chemistry of cleavage, which leaves the 5′ P and 3′ OH end groups in the newly generated termini, using three different assays that included phosphatase, kinase and ligase treatment, as described in the Supplementary Figure 1A, B and C.
Sequence determination of band X
The exact 5′ and 3′ termini of band X were determined by cyclization of band X RNA, followed by reverse transcription and sequencing across the ligated RNA junction. Cleavage positions were 5′ A29/A34 and 3′ G490, which concurred with the results of direct RNA fingerprinting analysis of band X (Supplementary Figure 2B). Fingerprinting analysis (Supplementary Figure 2A) of the rest of the larger RNase III cleavage products demonstrated that their sequence corresponded to the already characterized bands (P2 P3, P1P2, p2 and P3), and that there is any new band product.
The A29/A34 position corresponds to a dsRNA region formed by the described LRA that produces RNase III cleavage at a low enzyme concentration. The new cleavage event between G489 and G490 maps approximately across from the 428–442 sequence in the secondary structure of stem-loop VI (Figure 1C). This suggests that stem-loop VI is an alternative conformation to the LRA when LRA formation is impaired.
Factors involved in switching between alternative structures
The results described in the preceding paragraphs suggest that the 428–442 sequence can switch between two mutually exclusive base pairing conformations, either interacting with a ssRNA sequence in domain I of the LRA motif (allowing RNase III to recognize the LRA) or forming the basal part of stem-loop VI (allowing X cleavage). Because destabilization of the ‘C’ conformation and stabilization of the new alternative structure have to be achieved mechanistically to allow switching, we subsequently assessed the role of cis-acting and trans-acting sequences on switching between the ‘C’ and ‘O’ conformations (Figure 1B).
Cis-acting elements
To delineate the sequence elements participating in the switch, two sets of RNase III and RNase H structure-dependent cleavage reactions were performed using an oligodeoxynucleotide ‘blocking’ assay and truncated fragments, as described below.
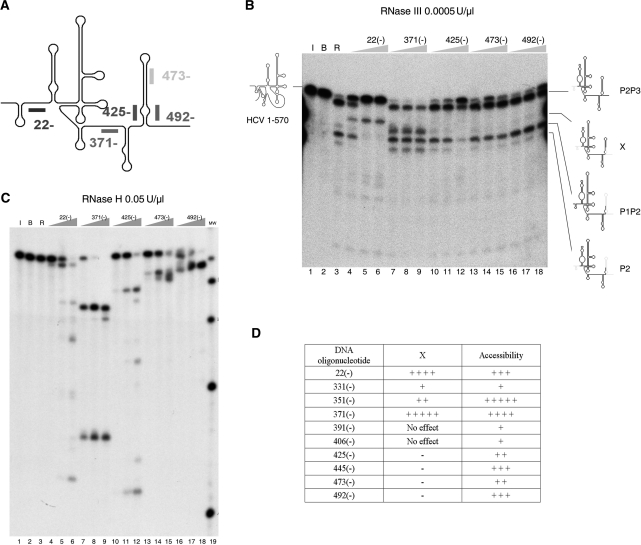
We analyzed the RNase III cleavage pattern of 1–570 RNA in the presence of oligonucleotide (ODN) 22(−) and a consecutive set of 20-mer DNA oligonucleotides complementary to 1–570 RNA from domain IV to domain VI. The results are described in Figure 4B. ODN 22(−) inhibited LRA and promoted cleavage at the X site, producing a band containing fragments P1X that migrated more slowly than band X alone (characterization is provided in the ‘Trans-acting element: miR-122’ section). ODNs 473(−) and 492(−) inhibited the small percentage of cleavage at the X site produced at 0.005 U/µl RNase III concentration and had no other effect on the cleavage pattern. ODNs 473(−) and 492(−) were not expected to cause any alteration in the ‘C’ conformation. When the experiment was repeated with a higher concentration of RNase III (0.001 U/µl), so that the alternative ‘O’ structure is formed more quickly and in a higher percentage, the mix of ODN 473(−) and 492 (−) was then able to block band X formation (Supplementary Figure 3). The key result of this section is provided by ODN 425(−). The 428–442 sequence is theoretically involved in the formation of both structures, the LRA motif and stem-loop VI, and ODN 425(−) effectively inhibited production of all RNase III-dependent products. Unexpectedly, ODN 371(−) stimulates cleavage at both the LRA and X site (Figure 4B, lanes 7–9). The RNA folding application of the Mfold program (http://mfold.bioinfo.rpi.edu/) indicates possible folding of 1–570 RNA with an interaction between part of the sequence blocked by 371(−) and the key ‘two partner’ sequence, 428–442, which can hybridize with either 20–40 or the right arm of stem-loop VI. Specifically, the 372ACCAAAACG379 sequence, which spans interdomains IV and V and is commonly drawn as an ssRNA region, might base pair with the 431CGUUGGU437 sequence, which constitutes the basal part of the left strand of stem-loop VI in the ‘O’ form. If this interaction were to occur, it would disturb stem-loop VI and inhibit band X formation, or sequester the distal sequence for LRA formation, impeding cleavage at the LRA helix of the ‘O’ conformation. Thus, by blocking the 371–390 fragment, adequate formation of both the ‘C’ and ‘O’ structures would occur, favoring RNase III cleavage at both helices. Hence, the 372A-G379 sequence may be an additional element participating in the switch. Additional information consistent with this hypothesis was obtained in the T1 analysis (see ‘Results’ section, ‘Partial T1 and V1 digestions’).
Figure 4.
(A) Schematic drawing of 1–570 RNA, indicating a subset of 20-base ODNs, each complementary to an HCV IRES region beginning with the residue number shown; the (−) sign indicates complementary to the viral sequence. (B) Cleavage reaction of 1–570 RNA with E. coli RNase III in the presence of a set of ODNs complementary to the viral sequences: 22(−), 371(−), 425(−), 473(−) and 492(−). Cleavage reactions were performed at 0.0005 U/µl of RNase III. Lanes 1 and 2: 1–570 RNA alone incubated on ice and incubated with buffer, respectively. Lane 3: control cleavage reaction without oligonucleotide. Lanes 4–6: cleavage reaction in the presence of increasing concentrations of ODN 22(−): (lane 4: 15 nM; lane 5: 150 nM; lane 6: 1500 nM). The same for lanes 7–9, 10–12, 13–15 and 16–18 for ODN 371(−), 425(−), 473(−) and 492(−), respectively. RNA molecular weight markers of 1–502 and 1–466 nt in length are indicated by a line on the left of the gel fragments. (C) Analysis of E. coli RNase H digestion products of 1–570 RNA annealed with complementary ODNs. Lane 1 is RNA incubated on ice, lane 2 incubated in buffer and lane 3, buffer with 0.5 units of E. coli RNase H. Lanes 4 to 18: the annealing reaction was performed with a set of 20-mer ODNs, 22(−), 371(−), 425(−), 473(−) and 492(−), at increasing concentrations of 15, 150 and 1500 nM, as indicated at the top of the gel, and treated with 0.5 U/µl of RNase H. Lane 19 is a commercial radiolabeled ladder of RNA fragments of 100, 200, 300, 400, 500, 750 and 1000 bases in length, which will be referred to as ‘MW marker’ in the remaining figure legends. Only fragments 100–500 bases in size appear in this gel image. (D) Table summarizing the changes in reactivity of 1–570 RNA to RNase III in the presence of complementary ODNs. The ‘oligonucleotide’ column indicates the positions where DNA oligonucleotides hybridize to RNA. The ‘X’ column shows the relative activation of RNase III cleavage at the X site (including fragments X and P1X) in 1–570 RNA by the presence of ODNs. The ‘accessibility’ column indicates the relative sensitivities of 1–570 RNA to DNA-mediated RNase H cleavage.
Oligonucleotides complementary to stem-loop IV and V had no effect on the LRA or switching (Figure 4D). A parallel RNase H analysis, performed for all the oligonucleotides used, indicated that (at least, at the highest concentration) the oligonucleotides were able to anneal to 1–570 RNA (Figure 4C) to saturation or, in the case of ODNs 473(−) (Figure 4C, lane 15), 331(−) and 391(−) (data not shown), to 80% of the starting material. This supports the conclusion that an absence of an effect of an oligonucleotide on RNase III activity is not due to an inability to hybridize to HCV RNA.
Parallel RNase III and RNase H analysis was performed on 1–570 RNA and transcripts consecutively shortened at their 3′ ends (HCV RNA 1–466 and HCV RNA 1–402) in the presence of ODN 22(−) (Supplementary Figure 4). The results indicate that, in the absence of the right strand of stem-loop VI, RNase III cleavage in the LRA is faster and competition between ODN 22(−) and the LRA is more difficult, whereas the absence of both strands of stem-loop VI greatly favors ODN 22(−) annealing at its complementary site (Supplementary Figure 4B). Taken together, these experiments indicate a competitive interplay between sequence 20–40 and the right strand of stem-loop VI to anneal with sequence 425–445.
Trans-acting element: miR-122
As described above, two mutually exclusive and readily distinguishable conformations of 1–570 RNA are evident from the RNase III products. Because the proximal region of the LRA (bases 24–28) is involved in the interaction with specific liver miR-122 (2), this molecule is a good candidate to favor the alternative conformation by simple competition with the LRA. Examination of the RNase III products obtained after induction of the ‘O’ conformation with ODN 22(−) or miR-122 revealed a band pattern that indicated that cleavage positions were at similar positions as that obtained in the control reaction (Figure 5, lanes 4–6), except for the appearance of a band that migrated more slowly than band X and was not present in the reactions without probes. This band could occur as follows: ODN 22(−) and miR-122 both carry complementary sequences in the 20–40 region of 1–570 RNA, and by annealing to it, the alternative ‘O’ conformation and cleavage at the X site are induced. However, concomitantly, the annealed DNA forms a hybrid DNA/RNA, and the miR-122 forms a short duplex miR-122/HCV RNA, which are no longer substrates for RNase III, and also partially abolish RNase III cleavage within the 20–40 sequence (i.e. between P1 and P2). Thus, in the presence of the probes, a new fragment starting at position 1 instead of at 27/34, and yielding a band length equivalent to P1X (1–490) is expected. The results of Figure 5 demonstrate that the new RNase III cleavage product band observed in the internally labeled transcript (lanes 5 and 6), contains the 5′ triphosphate group (lanes 10 and 11), as is visualized in the γ-32P-labeled transcript. It also shows that the 3′ end position of the new band coincides with the previously characterized bands. In the lanes in which RNA was internally labeled or 3′ end-labeled, no differentiating product band appeared. In addition, attending to the 3′ end-labeled reactions, the band that joins band X to give the complete 1–570-base transcript (which we call band Y) increased in intensity in the ODN 22(−) and miR-122-treated lanes relative to the untreated sample (Figure 5, lanes 14–16). Taken together, these findings indicate that the new band contains nt 1 of HCV 1–570 RNA and the 3′ end of band X; therefore, the fragment is P1X.
Figure 5.
Characterization of an induced RNase III cleavage product of 1–570 RNA by ODN 22(−) and miR-122. Autoradiogram showing a parallel run of RNase III cleavage reaction and control reactions for differentially radiolabeled transcripts of 1–570 RNA: internally (lanes 2–6), in the 5′ end (lanes 7–11), or in the 3′ end (lanes 12–16). RNAs were either kept on ice (lanes 2, 7 and 12), incubated in standard conditions without the enzyme (lanes 3, 8 and 13), treated with RNase III alone (lanes 4, 9 and 14), or treated with RNase III plus either ODN 22(−) at 150 nM final concentration (lanes 5, 10 and 15) or miR-122 at 15 nM final concentration (lanes 6, 11 and 16). Previously identified RNA fragments are indicated on the right. Lanes 1 and 17 are MW markers.
Kinetic analysis of E. coli RNase III cleavage of 1–570 HCV RNA in the presence or absence of miR-122
We previously demonstrated that products P2P3 and P2 represent the ‘C’ conformation, and products X and P1X are characteristic of the ‘O’ conformation. Because several RNase III cleavage events occurred in the same molecule, we examined the efficiency of cleavage and order of events in the miR-122-induced transition from ‘C’ to ‘O’, using kinetic analysis.
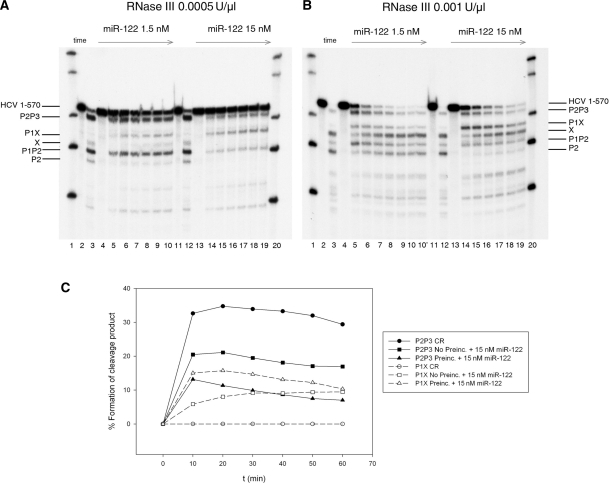
Two concentrations of enzyme (0.0005 and 0.001 U/µl) were employed and three types of 1–570 HCV RNA substrate preparations: (i) alone; (ii) pre-incubated with different concentrations of miR-122 (1.5, 15 and 150 nM) during 60 min, with subsequent addition of RNase III; and (iii) without pre-incubation. The RNase III kinetic pattern of cleavage for a subset of experiments is shown in gels (Figure 6A and B). The results of these gels are summarized as the percentage formation of cleavage products: P2P3, P1X, P and X as a function of time (Figure 6C).
Figure 6.
(A) Cleavage Kinetics of 1–570 RNA by E. coli RNase III at 0.0005 U/µl in the presence of 1.5 nM miR-122 (lanes 4–10) and 15 nM miR-122 (lanes 13–19). In this set of reactions, miR-122 was added to standard reaction, incubated for 1 hour at 37°C and then digested by the enzyme. In each individual reaction profile (lanes 4–10 and lanes 13–19) from left to right, each line represents time 0, 10, 20, 30, 40, 50 and 60 min incubation with RNase III. Lanes 2 and 11 RNA incubated on ice. Lanes 3 and 12 are control reactions without miR-122. Lanes 1 and 20 are MW century markers. (B) Same as panel A but reactions were performed at 0.001 U/µl E. coli RNase III. Bands are indicated by lines at the right of the figure. (C) Graphic representation of the time course processing of P2P3 and P1X by E. coli RNase III (0.001 U/µl) at different conditions: control reactions (CR), non-pre-incubated miR-122 reactions, or reactions pre-incubated with 15 nM miR-122.
The fold increase on stimulating P1X band production in the presence of miR-122 was measured at the last time point of the reaction, as described in Material and methods section. 1–570 RNA pre-incubated with miR-122 in condition (ii) was cleaved by RNase III to produce P1X at a higher rate than 1–570 RNA alone (Figure 6). In reactions without preincubation, the effect of miR-122 on activating the P1X band was less pronounced at 0.001 U/µl RNase III and undetectable at 0.005 U/µl (Figure 6).
The major RNase III single cleavage product of the ‘C’ conformation (P2P3) and the newly identified cleavage product induced in the presence of miR-122 (P1X) in the 0.001 U/µl RNase III and miR-122 15 nM reaction were compared in a kinetics analysis (Figure 6C). In the absence of miR-122, there was rapid production of P2P3 in the first 10 min. miR-122 significantly decreased the rate of P2P3 formation, suggesting that miR-122 competes with formation of the ‘C’ conformation. P1X can be only observed in the presence of miR-122. The initial kinetics of this effect was lower in miR-122 reactions without preincubation. These results are highly indicative that miR-122, after annealing with HCV 1–570 RNA induces the ‘O’ conformation.
Confirmation and characterization of the miR-122-induced RNA switch sequence by different methods
Because a secondary structure switch is at the basis of the 1–570 RNA conformational change, and there is evidence that the miR-122 sequence alone can induce switching, we evaluated the effect of miR-122 on the 1–570 RNA structure with three different methods that specifically respond to modifications of stable RNA:RNA duplexes. These include partial digestion with the single- and double-stranded nucleases T1 and V1, electrophoretic mobility in non-denaturing gels and the effect of mutated miR-122 on the RNase III cleavage pattern.
Partial RNase T1 and V1 digestion
Mapping the regions involved in the switch
Single- and double-stranded RNA-specific digestions were carried out under conditions leading to partial cleavage in order to evaluate whether conformational changes deduced from previous analyses could be visualized by this classical methodology and mapped to specific bases. We focused on the hypothetical key ‘two-partner’ sequence G428-U442, which should theoretically be displaced by competition from long range annealing with sequence A24-C38 by the ODN 22(−) or miR-122 sequences.
Comparative analysis of RNase T1 partial digestion of 1–570 RNA alone or pre-incubated with miR-122
RNase T1 partial digestions of the 3′ radiolabeled 1–570 RNA alone or pre-incubated with a miR-122 sequence at 150 nM were run in parallel with a sequence ladder of 1–570 RNA treated with alkali and heat or with RNase T1 in denaturing conditions (Figure 7A). In the analysis of the changes, base G428 showed a newly acquired sensitivity after miR-122 incubation. These results confirm the hypothesis that ODN 22(−) and miR-122 can induce changes in the ‘secondary status’ of 1–570 RNA at a long distance, which is consistent with the proposed RNA conformational switch.
While change G428 resides in the left strand of stem loop VI, there is another change at base G494, which maps across the helix in the complementary strand of the newly formed stem loop VI, indicated in Figure 7B. Decrease in cleavage at base G494 suggest an increase in stem loop VI formation as expected. Of particular interest is the increase in resistance of base G379 to RNase T1 attack after switch induction. This base forms part of the 372A-G379 sequence, which appeared in the RNase III/RNase H analysis as a third element participating in the switch. Changes in reactivity at other positions indicate that other RNA fragments reorganize because of the switching. Further study is needed to determine the relationship between the overall conformational switch we propose and the specific secondary structure variation.
Effect of different concentrations of ODN 22(−), miR-122 and the miR-122 seed sequence on the pattern of RNases T1 and V1
Figure 7C and D show a comparative analysis pattern of RNase T1 and V1 reactivity on 1–570 RNA alone or pre-incubated with several probes. ODN 22(−) and miR-122 are observed to have nearly identical qualitative stimulatory effects on switching, although quantitatively, miR-122 was 10-fold more effective than ODN 22(−); both exerted a dose-dependent effect. Regarding the effects on the RNase V1 pattern, ODN 22(−) as well as miR-122 promote similar changes. In agreement with the RNase T1 results, the changes are concentrated in the region around G494 in the left and right strand of stem loop VI. The seed sequence alone was found to have a far less pronounced ability to promote changes in the T1 pattern. The effects were limited to slight V1 nuclease resistance around position 494 only when the highest concentration of seed sequence was used.
Non-denaturing gels
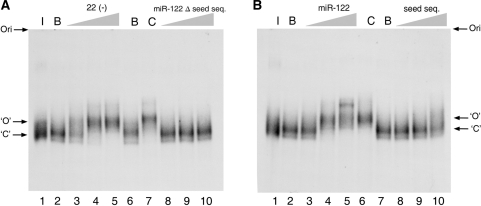
Native gels were used to investigate the conformational change from ‘C’ to ‘O’ of 1–570 RNA in the presence of specific probes. Three potential oligonucleotide inducers, ODN 22(−), miR-122 and miR-122 seed sequence and a theoretically unreactive control probe, truncated miR-122 (lacking the seed sequence) were added to 0.6-nM samples of internally labeled 1–570 RNA, incubated and analyzed in non-denaturing acrylamide gels, as is described in Material and Methods section. As is seen in Figure 8A and B, addition of only buffer to the starting 1–570 RNA concentrates the radioactive label from a fuzzy smear to a more delineated band, consistent with a report demonstrating that Mg++ is required for the 5′ region of HCV RNA to achieve proper folding (15). Incubation of HCV RNA with increasing concentrations of ODN 22(−) at 15, 150 and 1500 nM (Figure 8A, lanes 3–5) induced a band that migrated more slowly than the untreated samples, showing a sharp transition between 15 and 150 nM. Incubations with miR-122 at 1.5, 15 and 150 nM (Figure 8B, lanes 3–5) showed a similar position for the retarded band migration (Figure 8, lane 6), except that a very slow migrating band appeared at the highest miR-122 concentration used. Complete band mobility retardation was attained repeatedly at a final concentration of 15 nM. The miR-122 seed sequence alone had a faint effect. At the most concentrated condition, the band slowed to a slightly retarded smear (Figure 8B, lanes 8–10). The miR-122 sequence lacking the seed sequence had no effect at any concentration (Figure 8A, lanes 8–10).
Figure 8.
Analysis of induced conformational changes of 1–570 RNA on non-denaturing gel electrophoresis. (A) 1–570 RNA was resuspended at 0.6 nM in each of the following conditions: in water and kept on ice (lane 1), in standard buffer reaction alone (lanes 2 and 6), together with ODN 22(−) at a final concentration of 15 nM, 150 nM and 1500 nM (lanes 3–5), or with miR-122 minus seed sequence at increasing final concentrations of 1.5 nM, 15 nM or 150 nM (lanes 8–10). Lane 7, control ODN 22(−) at 1500 nM. (B) Ice-incubated control RNA is in lane 1, and buffer-incubated RNA controls are in lanes 2 and 7. RNA was incubated together with miR-122 at a final concentration of 1.5 nM, 15 nM and 150 nM (lanes 3–5). Lane 6, control 22(−) or together with seed sequence (5′ U GGAGUGU GA 3′) at concentrations of 1.5 nM, 15 nM and 150 nM (lanes 8–10). ‘ORI’ marks the origin of the electrophoresis. Arrows marked with ‘O’ and ‘C’ indicate open and close conformations respectively.
In another experiment, we changed the annealing conditions between the probes and 1–570 RNA. We found that if 1–570 RNA were preheated to 90°C in water and slowly cooled with a mixture of our standard buffer plus miR-122 or seed sequence, miR-122 at 1.5 nM was able to promote mobility retardation of all of the 1–570 RNA counts (data not shown). Because we had previously shown that annealing of ODN 22(−) with HCV RNA was favored in these conditions (16), this result, together with the previous findings, confirmed that the limiting factor in promoting the RNA switch is the ability of the probe to anneal with the viral RNA target. However, the effect of the seed sequence alone on retardation was only slightly more evident than in the experiments that did not include annealing during the preheating step (data not shown).
A control was performed to rule out that 1–570 RNA retardation in the experiments above was due to the increase in weight contributed by the annealed probe, even though it represents only about 3.5% in mass and charge. A 20-mer DNA, ODN 188(−), which hybridizes to completion with 1–570 RNA in apical stem-loop III at positions 188–207, promoted no mobility changes (Supplementary Figure 5A).
Effect of miR-122 seed sequence mutations on 1–570 RNA cleavage by RNase III
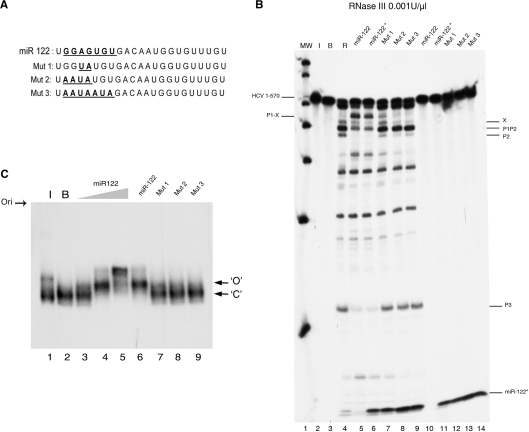
We determined the effect of the presence of mutations within the miR-122 seed sequence on the RNase III reaction and native gel electrophoresis of 1–570 RNA. We used 15 nM of all the probes whose sequences are described in panel A of Figure 9. We found that 2 mutations in the middle of the seed sequence (mutant 1) sufficed to drastically reduce the switch induction effect of miR-122 (Figure 9B, lane 7). This effect was totally abolished in the variants carrying 4 or 7 mutations (mutants 2 and 3, Figure 9, lanes 8 and 9, respectively). It was also observed that, unlike miR-122, these probes were unable to stabilize the retarded band in the native gel electrophoresis (Figure 9C, lanes 7–9). These results indicate that switch induction is a highly specific result of miR-122 interaction with 1–570 RNA through its seed sequence.
Figure 9.
Effect of miR-122 seed sequence mutations on 1–570 RNA cleavage by RNase III (A) The miR-122 sequence and mutants 1, 2 and 3. Seed sequence mutations are indicated in bold and underlined. (B) Autoradiogram shows a 0.001 U/µl RNase III cleavage reaction of 1–570 RNA fragment alone (lane 4), or in the presence of 15 nM of an RNA sequence corresponding to the following: miR-122 (chemically synthesized and unlabeled in lane 5 or in vitro-transcribed from a DNA template and internally labeled in lane 6, referred to in the figures as miR-122*); or mutants of miR-122 transcribed from a DNA template and internally labeled: mut 1 (lane 7), mut 2 (lane 8), mut 3 (lane 9). Lane 1 is a MW marker, lane 2 is 1–570 RNA alone maintained on ice, and lane 3 is RNA incubated in reaction buffer. Lanes 10–14 are equivalent to lanes 6–8, but incubated in the absence of the enzyme. The miR-122-induced cleavage products are indicated at the left of the gel. The migration position of miR-122 and mutants are identified at the bottom of the gel. (C) Non-denaturing gel autoradiography of 1–570 RNA alone incubated on ice (Lane 1) or in buffer (Lane 2); or incubated for 1 h at 37°C with increasing final concentrations of miR-122: 1.5 (lane 3), 15 nM (lane 4) and 150 nM (lane 5); or with 15 nM final concentrations of mut 1 (lane 6), mut 2 (lane 7), mut 3 (lane 8) or mut 4 (lane 9).
DISCUSSION
In this study, the transition between two double-helical elements is characterized in the context of a conformational change of 1–570 HCV RNA and its dynamic in vitro modulation by the liver-specific microRNA, miR-122. These conformational changes involve the HCV IRES, which switches from a closed ‘circular’ conformation ‘C’ (5,7), formed by LRA (bases 24–38 in inter-domains I-II with bases 428–442 in the basal part of domain VI) to an open ‘O’ form (where bases 428–442 interact with bases 494–508; i.e. those forming the basal part of stem-loop VI). The in vitro switch is governed by the interaction of miR-122, which disfavors the LRA. Liver miR-122 has been demonstrated to modulate viral abundance (2) and recently, to stimulate translation (17) when viral RNA fragments containing stem-loop VI are used. This led us to propose that the 1–570 RNA conformational change induced by miR-122 is an integral regulatory component of the viral genome.
The conformational switch we propose solves the apparent conflict between studies supporting the closed conformation (5–7) and the report describing stem-loop VI (8). Kinetic analysis in our previous study (7) showed that at least, 80% of the molecules were in the closed conformation. In apparent contrast, a structural study of RNA in the core coding region indicated that the G428-U442 sequence forms the basal part of the large stem-loop VI, whose presence is incompatible with the closed conformation (8). The conflict disappears if we consider that the RNA transcript fragment used in the study did not include the most proximal region of HCV RNA, which is needed for the closed conformation, and propose that 1–570 RNA is a conformationally dynamic molecule, able to switch between two conformations. A similar conflict was identically resolved with the discovery of two alternating structures in HIV leader RNA (18).
Upon miR-122 binding, changes in the accessibility of 1–570 RNA to RNase H, T1 and V1 are mostly located downstream of the IRES and are consistent with transition from the ‘C’ to ‘O’ conformation. Therefore, we conclude that the mobility changes seen on gel shift analysis are related to changes in the overall conformation. Thus, translation activation after miR-122 induction (17) might be related to increased accessibility to the small ribosomal subunit or to other initiation factors (i.e. eiFIII) in the ‘O’ conformation.
miR-122 is a viral inducer that interacts in tandem with the proximal 23–40 nt of the viral sequence (19), coinciding with the region involved in the LRA. Regarding the influence of miR-122 on the population of the two conformomers in vitro, several factors seem of interest: (i) the low ratio of miR-122 to 1–570 RNA (3:1) needed to induce the switch. This is consistent with the differing representation of these two molecules in infected hepatocytes [>103 for miR-122 (20) to <100 for positive-stranded HCV RNA (21)]; (ii) the seed sequence, which we proved to be essential for miR-122 activity, is unable to promote the switch alone (unless it is used at a high concentration); (iii) the miR-122-induced switch is reversible, whereas the switch induced by the complementary ODN 22(−) is not. These three points indicate that the switch is not promoted by simple Watson-Crick pairing competition. More complicated interaction of miR-122 by 1–570 RNA is suggested, through a process that may be more similar to bacterial riboswitches.
The results of the RNase III reaction in the presence of miR-122 and RRL suggest that the conformational switch is maintained in a specific biological context (Supplementary Figure 6). This prompted us to propose that the 1–570 RNA switch in the presence of miR-122 may represent the structural correlate of the recently described positive effect that miR-122 exerts on HCV translation in RRL and cell cultures (17). If this is the case, the data provide a role for RNA in the proximal part of the CORE region, whose unknown structure-dependent function has been described as essential for the virus in a chimpanzee model (3).
An intriguing question is whether the interaction of miR-122 with 1–570 RNA suffices for this biological effect (i.e. with miR-122 acting as a ligand in a manner similar to small molecule-induced riboswitches in bacteria) or protein factors are involved. In any case, the activity of miR-122 as a conformational switch-inducer departs from the concept of microRNA regulation of mRNA translation in eukaryotes. While that reported effect is inhibitory and occurs through direct binding to the 3′ UTR of mRNA, in the present case it involves activation at the 5′ UTR.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Ministerio de Ciencia e Innovación BIO2007-60106, BIO2004-06114 Proyecto Excelencia Comunidad Autónoma de Andalucía CVI-03050; FISS (CIBERehd); FIPSE 36549/06 for work in Granada; National Institutes of Health grant NIH/NIDA K01 DAO18262 for work in Doylestown, Pennsylvania. Funding for open access charge: BIO-2007, 60106 (IPB-LN) CSIC. MCyT.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Drs Encarnación Martínez-Salas, Josep Vilardell and Esteban Domingo for their helpful comments on the manuscript. We are grateful to Drs Cristina Romero and Francisco Muñoz for their help in the experimental procedures. Finally, the authors acknowledge the initial contribution to this study of Tchering Shorden and Dr Nerea Beguiristain, presented in Figure 2 of the Supplementary Data, which was done at Dr Hugh D Robertson's laboratory in Cornell University, New York.
REFERENCES
- 1.Reynolds JE, Kaminski A, Carroll AR, Clarke BE, Rowlands DJ, Jackson RJ. Internal initiation of translation of hepatitis C virus RNA: the ribosome entry site is at the authentic initiation codon. RNA. 1996;2:867–878. [PMC free article] [PubMed] [Google Scholar]
- 2.Jopling CL, Yi M, Lancaster AM, Lemon SM, Sarnow P. Modulation of hepatitis C virus RNA abundance by a liver-specific MicroRNA. Science. 2005;309:1577–1581. doi: 10.1126/science.1113329. [DOI] [PubMed] [Google Scholar]
- 3.McMullan LK, Grakoui A, Evans MJ, Mihalik K, Puig M, Branch AD, Feinstone SM, Rice CM. Evidence for a functional RNA element in the hepatitis C virus core gene. Proc. Natl Acad. Sci. USA. 2007;104:2879–2884. doi: 10.1073/pnas.0611267104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vassilaki N, Friebe P, Meuleman P, Kallis S, Kaul A, Paranhos-Baccala G, Leroux-Roels G, Mavromara P, Bartenschlager R. Role of the hepatitis C virus core+1 open reading frame and core cis-acting RNA elements in viral RNA translation and replication. J. Virol. 2008;82:11503–11515. doi: 10.1128/JVI.01640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim YK, Lee SH, Kim CS, Seol SK, Jang SK. Long-range RNA-RNA interaction between the 5′ nontranslated region and the core-coding sequences of hepatitis C virus modulates the IRES-dependent translation. RNA. 2003;9:599–606. doi: 10.1261/rna.2185603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda M, Rijnbrand R, Abell G, Kim D, Lemon SM. Natural variation in translational activities of the 5′ nontranslated RNAs of hepatitis C virus genotypes 1a and 1b: evidence for a long-range RNA-RNA interaction outside of the internal ribosomal entry site. J. Virol. 1999;73:4941–4951. doi: 10.1128/jvi.73.6.4941-4951.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beguiristain N, Robertson HD, Gomez J. RNase III cleavage demonstrates a long range RNA: RNA duplex element flanking the hepatitis C virus internal ribosome entry site. Nucleic Acids Res. 2005;33:5250–5261. doi: 10.1093/nar/gki822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tuplin A, Evans DJ, Simmonds P. Detailed mapping of RNA secondary structures in core and NS5B-encoding region sequences of hepatitis C virus by RNase cleavage and novel bioinformatic prediction methods. J. Gen. Virol. 2004;85:3037–3047. doi: 10.1099/vir.0.80141-0. [DOI] [PubMed] [Google Scholar]
- 9.Milligan JF, Uhlenbeck OC. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- 10.Franklin RM. Purification and properties of the replicative intermediate of the RNA bacteriophage R17. Proc. Natl Acad. Sci. USA. 1966;55:1504–1511. doi: 10.1073/pnas.55.6.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robertson HD. A Nuclease Specific for Double-Stranded RNA. Virology. 1967;12:718–719. doi: 10.1016/0042-6822(67)90048-7. [DOI] [PubMed] [Google Scholar]
- 12.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol. Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 13.Nadal A, Martell M, Lytle JR, Lyons AJ, Robertson HD, Cabot B, Esteban JI, Esteban R, Guardia J, Gomez J. Specific cleavage of hepatitis C virus RNA genome by human RNase P. J. Biol. Chem. 2002;277:30606–30613. doi: 10.1074/jbc.M203595200. [DOI] [PubMed] [Google Scholar]
- 14.Dunn JJ. RNase III cleavage of single-stranded RNA. Effect of ionic strength on the fideltiy of cleavage. J. Biol. Chem. 1976;251:3807–3814. [PubMed] [Google Scholar]
- 15.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 16.Lyons AJ, Lytle JR, Gomez J, Robertson HD. Hepatitis C virus internal ribosome entry site RNA contains a tertiary structural element in a functional domain of stem-loop II. Nucleic Acids Res. 2001;29:2535–2541. doi: 10.1093/nar/29.12.2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henke JI, Goergen D, Zheng J, Song Y, Schuttler CG, Fehr C, Junemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300–3310. doi: 10.1038/emboj.2008.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jopling CL, Schutz S, Sarnow P. Position-dependent function for a tandem microRNA miR-122-binding site located in the hepatitis C virus RNA genome. Cell Host Microbe. 2008;4:77–85. doi: 10.1016/j.chom.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sarasin-Filipowicz M, Krol J, Markiewicz I, Heim MH, Filipowicz W. Decreased levels of microRNA miR-122 in individuals with hepatitis C responding poorly to interferon therapy. Nat. Med. 2009;15:31–33. doi: 10.1038/nm.1902. [DOI] [PubMed] [Google Scholar]
- 21.Agnello V, Abel G, Knight GB, Muchmore E. Detection of widespread hepatocyte infection in chronic hepatitis C. Hepatology. 1998;28:573–584. doi: 10.1002/hep.510280240. [DOI] [PubMed] [Google Scholar]