Abstract
Recombinant inbred lines (RILs) derived from Caenorhabditis elegans wild-type N2 and CB4856 are increasingly being used for mapping genes underlying complex traits. To speed up mapping and gene discovery, introgression lines (ILs) offer a powerful tool for more efficient QTL identification. We constructed a library of 90 ILs, each carrying a single homozygous CB4856 genomic segment introgressed into the genetic background of N2. The ILs were genotyped by 123 single-nucleotide polymorphism (SNP) markers. The proportion of the CB4856 segments in most lines does not exceed 3%, and together the introgressions cover 96% of the CB4856 genome. The value of the IL library was demonstrated by identifying novel loci underlying natural variation in two ageing-related traits, i.e. lifespan and pharyngeal pumping rate. Bin mapping of lifespan resulted in six QTLs, which all have a lifespan-shortening effect on the CB4856 allele. We found five QTLs for the decrease in pumping rate, of which four colocated with QTLs found for average lifespan. This suggests pleiotropic or closely linked QTL associated with lifespan and pumping rate. Overall, the presented IL library provides a versatile resource toward easier and efficient fine mapping and functional analyses of loci and genes underlying complex traits in C. elegans.
INTRODUCTION
Over the last decades, Caenorhabditis elegans has become a versatile model organism for unravelling genetic mechanisms underlying many diverse phenotypes such as stress response, ageing, host–pathogen interaction, behaviour and even learning and memory. Gene discovery in C. elegans is based on the wealth of advanced genetic methods and is facilitated by the ease of forward and reverse genetics. Although standard forward and reverse genetic analyses have led to extensive knowledge about individual genes and their specific molecular functions in pathways, we still know little about their effects on natural phenotypic variation and the overall genetic architecture of complex traits including the total number of genes, typical effect sizes for alleles or the interactions between them (1,2).
Mapping natural genetic variation in C. elegans has recently received increased attention using approaches based on quantitative trait loci (QTL) analysis using recombinant inbred lines (RIL). In QTL mapping, phenotypic traits are compared with the molecular marker genotypes of a segregating population to search for genomic regions (also called QTL) associated with the trait variation (3). For instance in C. elegans, not only QTLs have been mapped to study genetic determinants of natural variation in sensitivity to volatile anesthetics (4) and oxygen levels (5), but variations in other complex traits have also been mapped such as longevity, reproduction, body growth rate and gene expression (6–10). A QTL approach was used also to investigate phenotypic plasticity, pleiotropy, genotype–environment interactions and epistasis in C. elegans life histories (6–9,11–12).
However, mapping analyses in RILs have their own drawbacks because of the masking effects of major QTL and epistatic interactions of multiple QTL (13). In addition to RILs, introgression lines (ILs) are a more powerful resource facilitating high-resolution QTL mapping (14,15). In principle, the genome of an IL (which can also be referred to as a congenic strain or near isogenic line) is composed of a recipient genome contributed by one of the parental strains and a short, homozygous segment of the donor genome contributed by another, genetically distinct, parental strain. In this way, the difference in the phenotype of the recipient (background) strain and the IL can be precisely attributed to the introgressed locus. Without the confounding effects of other segregating QTLs that occur in RIL populations, the effect of the locus of interest can be determined with high accuracy. The additional value of IL libraries is that the selected ILs can be further backcrossed to the recipient parent and the resulting set of lines with shorter introgressions (sub-ILs) can be used to provide high-resolution information on the QTL architecture. ILs were successfully applied in plants to dissect QTLs of large effects into multiple, tightly linked loci (16,17) and facilitate subsequent positional cloning (18,19) providing clues for the molecular basis of gene functions.
Here, we report the first genome-wide library of C. elegans ILs developed from N2 Bristol (recipient) and CB4856 (donor) strains. The N2 Bristol strain is the standard strain for genetic and developmental studies. Its genome was fully sequenced in 1998 and provided the first genomic platform to study metazoan gene function (20). We chose the Hawaiian CB4856 wild type as a donor strain because of its highest genetic divergence from N2 compared to other wild isolates and a rapidly growing interest in mapping natural genetic polymorphisms underlying many diverse phenotypes. CB4856 and N2 differ by one single-nucleotide polymorphism (SNP) per 840 bp (21,22) and show a high overall difference in gene content as indicated by the comparative genomic hybridization (23). Numerous phenotypic differences between these two strains include clumping and bordering behaviour (24), ethanol responses (25), RNAi sensitivity (26), pathogen susceptibility (27) and some life-history traits (7).
Genetic and mutant analyses in N2 led to extensive knowledge about many complex human disease pathways. On the other hand, we have become more aware of the profound role of genetic and environmental interactions behind complex disease phenotypes (28). It is, therefore, of fundamental importance to understand the exact mechanisms underlying background modifiers. In mutation mapping studies using SNPs of the CB4856 strain, it has been frequently observed that the penetrance of mutations can be greatly modified by the CB4856 background indicating a high potential of the N2/CB4856 IL library for the mechanistic studies of the modifier genes. Several strategies for such studies have already been proposed and can be readily implemented (1,29).
Overall, the genome-wide C. elegans IL library is a unique and powerful resource facilitating the development of the new direction of complex trait mapping in C. elegans. Here, we genetically characterize the constructed CB4856/N2 IL library and complement this description with phenotypic analyses of lifespan (LSP) and pharyngeal pumping rate and subsequent mapping of these traits. Both traits are correlated in N2 in the sense that pharyngeal pumping rate declines cause a decline in survival probability suggesting a shared regulatory system (30).
MATERIALS AND METHODS
Construction of ILs
Three hermaphrodites of each selected (parental) RIL (WN17, WN19, WN28, WN29, WN57, WN64, WN77, WN80; Supplementary Figure 1) were backcrossed to N2 males (six males per hermaphrodite). Depending on the proportion of the CB4856 segments in the parental RIL, three to five rounds of backcrossing were performed and followed by 10 generations of selfing to obtain homozygous strains. This procedure resulted in the creation of ∼500 lines (85 derived from each RIL) with a strongly reduced, but unknown contribution of donor segments. All developed lines were initially subjected to a rough genotype screening with 7–12 SNP markers. The markers were distributed evenly over all CB4856 segments previously identified in their parental RIL. Only the lines with CB4856 alleles detected on no more than one chromosome were used for detailed genotyping. These lines (∼15 lines per parental RIL) were fully genotyped with 123 evenly spaced SNPs.
This procedure resulted in the identification of ∼20 ILs homozygous for a single CB4856 segment. Some of these ILs and several other lines with few homozygous donor segments were further backcrossed to N2 and subsequently selfed (the protocol as mentioned above) to construct ILs with either shorter or uniquely located CB4856 segments. The lines with more than one introgression are not included in the presented IL library. All lines are stored as frozen stocks at −80°C (31).
SNP markers
The SNP markers and the genotyping method were the same as previously applied for creating the RIL library (10). Briefly, all markers were selected on the C. elegans SNP data website (http://www.genome.wustl.edu/genome/celegans/celegans_snp.cgi). For chromosomes I, II and III, we selected 20 evenly spaced markers, for chromosomes IV, V and X we selected 22, 21 and 20 markers, respectively. We selected easily detectable (i.e. with a common restriction enzyme) SNP markers with high PSNP values (PSNP ≥ 0.7). Suspected SNP mistypings were checked for a second time.
Nematode culturing
During the IL construction, the nematodes were grown, backcrossed and selfed at 20°C in six-well plates filled with the nematode growth medium, NGM, containing 2% agar and seeded with Escherichia coli OP50 (31). The derived lines were subsequently cultured at 15°C on 6 cm petri dishes with NGM and E. coli.
Prior to the LSP and pumping rate assays, the ILs, N2 and CB4856 strains were grown on 9-cm Petri dishes at 20°C for 2 weeks and synchronized by bleaching (32). Synchronization was performed during 1 day in three batches with the lines randomly assigned to each batch. After ∼40 h, 12 hermaphrodites in fourth larval stage (L4) per IL were randomly picked and placed in a 6 cm Petri dish for phenotyping. All experiments were carried out in Elbanton climate chambers (Elbanton, NL, USA). Temperature was monitored with a Tinytag Transit temperature logger (Gemini Data Loggers, UK). All measurements were conducted using a dissecting microscope.
Pumping rate
Pumping rate was defined as the number of contractions in the terminal bulb in a 15 s period and recalculated to the number of contractions per minute. This was repeated three times for each nematode, and 12 nematodes from each plate per IL were measured by two observers. Pumping rate was measured on Days 4, 6, 8 and 10 after hatching.
Lifespan
The LSP measurement started with 12 nematodes per IL (in the fourth larval stage). In the first week, the number of surviving individuals was counted every day and after that, every second day. The nematodes were assumed dead if no reaction was observed after repeated touching with a hair. The nematodes were transferred daily to a new plate until the reproduction was finished. If they were observed to have crawled up the wall of a plate and have died there, they were removed from the measurement. The same was done when a nematode was observed to have died due to unnatural causes, such as developing a ‘bagging’ phenotype, where eggs hatch inside the adult.
Bin mapping
Per marker, the ILs containing a CB4856 allele were grouped into bins. Also, the ILs containing a CB4856 marker +1 or −1 of the marker under study were selected. The trait values of the grouped ILs were than tested against N2, by a two-sided t-test (assuming unequal variance) for average LSP and by linear regression for the decrease in pumping rate in time. The pumping rate coefficient (see Supplementary Table 1) of this regression equation was used for QTL mapping. A further decrease in QTL size was done by obtaining the locus that contained the most individual ILs significantly different from N2.
RESULTS
Construction of a C. elegans IL population
We constructed a population of 90 ILs, each containing a single CB4856 introgression in the N2 background. Together these ILs contain almost the whole CB4856 genome. The library was created by backcrossing a limited set of RILs of the CB4856 x N2 population (10) with N2 (Supplementary Figure 1). These RILs were selected based on a low proportion of CB4856 segments (∼30%). After backcrossing and selfing, the genotypes of the obtained ILs were determined by the 123 markers used to genotype the CB4856 x N2 RILs (Supplementary Table 1).
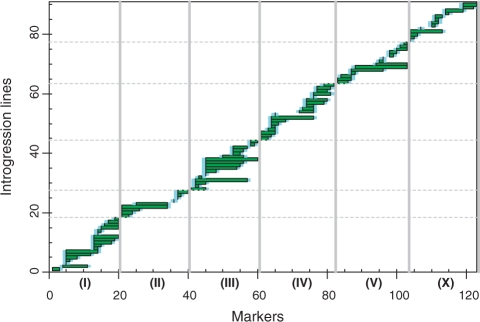
Genetic constitution of the IL library is presented in Figure 1. The CB4856 segments in the different ILs predominantly overlapped; however, a minor fraction of adjacent introgressions did not overlap. We estimated that a maximum of ∼3.8 Mb (megabase) was not covered by our set of ILs giving the set >96% coverage of the CB4856 genome. Furthermore, we obtained 32 ILs with two separate CB4856 segments (Supplementary Table 1).
Figure 1.
Genotypes of the N2 x CB4856 introgression line (IL) population showing genome-wide coverage (Supplementary Table 1). The CB4856 introgressions depicted in green and light blue indicate the position of the crossover. Chromosomes are indicated next to the axes.
The median segment length, defined as the distance between the two markers having N2 alleles flanking the CB4856 introgression, was ∼3.5 Mb (average ∼4.5 Mb) with a minimum of ∼1.45 Mb and a maximum of ∼16.3 Mb. Collectively, these 90 ILs have 79 distinguishable loci with a median length of ∼1.7 Mb (average ∼2.0 Mb; min.: ∼0.6 Mb; max.: ∼7.0 Mb).
Loci affecting LSP and pumping rate
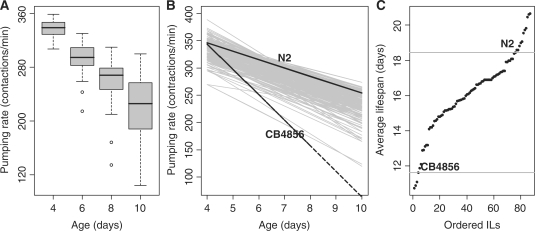
To benchmark the use of our population of ILs in identifying QTLs, we measured LSP and pharyngeal pumping (PP) of 86 ILs giving a genome-wide coverage (Supplementary Table 1). A relation was found between LSP and PP, where pumping decreased in aging worms across the ILs (Figure 2A and B). This is in agreement with similar studies carried out in N2 by Huang et al. (30). The average PP at each time point was also comparable to the values reported by Huang et al. (30). The average PP across the IL population decreased and the variation increased as the worms got older (Figure 2A). We found variation among ILs in absolute PP (Figure 2B) as well as in the rate of decrease in time. CB4856 showed a sharper decrease in pumping than N2. Most ILs had a PP decrease rate in between the parental strains, mostly similar to N2 (Figure 2B). We also found a large difference for LSP across the ILs, varying from 10 days to 21 days, extremes larger and smaller than the parental strains (Figure 2C).
Figure 2.
Decrease in pumping rate over time and lifespan. (A) Distribution of the pumping rate averages per IL on Days 4, 6, 8 and 10. (B) Regression lines of pumping rate over time of the individual ILs (grey lines), CB4856 and N2 (black lines). (C) ILs ordered by average lifespan and the average lifespan of CB4856 and N2 (grey horizontal lines).
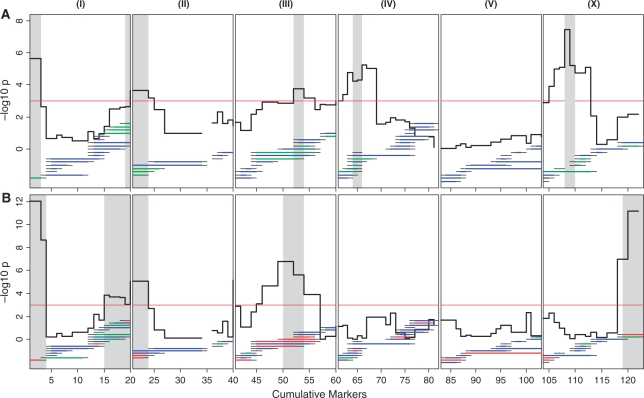
Obtaining the QTLs for both traits can give an estimate for the level of genetic intertwinement by colocation of QTLs. While mutant studies show a large decrease in LSP when a gene affecting PP is functionally compromised, such as reported by (30), not much is known about natural occurring alleles affecting this relation. Bin mapping of average LSP resulted in six QTLs, which all have a LSP-shortening effect on the CB4856 allele (Figure 3). Although CB4856 has a shorter average LSP than N2, the difference between CB4856 and N2 is less than the sum of effects of the loci detected with the ILs (6.8 days versus 26.3 days, respectively). This indicates that the effects of some LSP affecting loci are caused by the interaction between the CB4856 allele and the N2 background. The detection of QTL for LSP on chromosomes II, IV and X confirms the position of similar QTL detected by (6,9,11) although these studies were based on a mapping population derived from different parental strains. The QTLs on chromosomes I and III have not been previously reported and might harbour novel loci and candidate genes affecting LSP. We found five QTLs for the decrease in PP, of which four colocated with QTLs found for average LSP (Figure 3). This suggests pleiotropic or closely linked QTL affecting LSP and PP.
Figure 3.
Bin mapping of QTLs for average lifespan (A) and pumping rate (B) in the N2 x CB4856 IL population. Significance of a bin locus is indicated by the black line. QTLs determined by the bin with the ILs significantly different from N2 are indicated by the grey areas. Threshold (P = 0.001) is indicated by the red horizontal line. Individual ILs are shown by the green, blue or red horizontal lines that indicate their CB4856 introgression position. The thin black lines at the end of each introgression are the regions between the CB4856 and N2 marker and harbour the crossover. Green lines indicate the ILs with the trait values significantly lower than N2 (P < 0.01) and red lines show which ILs showed higher trait values than N2 (P < 0.01).
DISCUSSION
We report the first construction and properties of a genome-wide mapping library consisting of 90 ILs in C. elegans, a powerful resource to study complex traits in this model species. The choice of the parental strains of our IL library is one of the major factors ensuring a high power of genetic dissection. While the overall level of polymorphism among wild isolates of C. elegans is relatively low, the genetic distance between the N2 and CB4856 is high. In fact, CB4856 is reported to show the highest genetic distance from N2 compared to other wild isolates (21). CB4856 exhibits genetic isolation from all other global C. elegans populations that were demonstrated to intercross and recombine (33). This divergence is also reflected by the profound differences in the traits like formation of the copulatory plug (34,35) and many life-history traits (7,8). The use of the N2 strain as the recipient for CB4856 introgressions is additionally justified by the common use of N2 in C. elegans research and the wealth of available genetic and genomic tools.
In both RILs and ILs, the mapping resolution depends on the number of lines used and the frequency of recombination. In the case of RILs, the recombination frequency in the lines is fixed and can be increased by either adding more RILs or by intercrossing lines before fixation by inbreeding. In ILs, the resolution can be increased by minimizing the introgressed segment of each IL. Consequently, to maintain genome-wide coverage more lines are required. Because RILs and ILs have different genetic makeups, they both require different mapping tools. Overall, the recombination frequency in RILs is higher compared to an equal number of ILs. Therefore, fewer lines are required for analysis to obtain the same resolution. Each RIL has more than one introgressed fragment, and, on average, each genomic region is represented by the same number of parental genotypes. For these reasons, replication of individual RILs is often not necessary. Moreover, the multiple introgressions per RIL allow the detection of genetic interactions between loci (epistasis). Additionally, QTL of large effect might hide the detection of QTL with small additive effects. In contrast, ILs increase the power to detect small-effect QTL, but the presence of a single introgressed fragment does not allow testing for genetic interactions.
The introgressed loci within the ILs are mostly much smaller than the QTL intervals obtained with the C. elegans RIL population (10) increasing the resolution, while reducing the number of candidate genes and effort of fine mapping. The current IL library provides a relatively high mapping resolution, as compared to other genome wide IL populations. For example, there are many more donor segments compared to recently developed ILs of mice. These are called consomic strains because they are chromosome substitution strains and represent the most recent addition to the mouse genetic resources aiming to genetically analyse QTL (36,37). So far there are only a few other genome-wide IL libraries available in barley (38) and Arabidopsis (13). In Arabidopsis, 40 lines derived from Ler and Cvi parental accessions contained a single introgression, while 52 lines carried several Cvi fragments in a Ler background. The genetic length of the introgression fragments ranged from 31.7 to 5.2 cM. Lines with multiple Cvi fragments carried a main large introgression and several much smaller Cvi fragments. They selected a core set of 25 lines covering >90% of the genome.
In principle, there are two main approaches of QTL mapping using IL libraries. In the first one, a number of ILs are selected based on the location of QTLs previously identified with the use of another mapping population (e.g. RILs) and analysed to confirm and fine-map the putative QTLs. The second approach, applied in this paper, involves direct library-wide screens or bin mapping. The advantage of the first approach is that it might result in the initial detection of QTLs interacting with another locus, which might become undetectable when introgressed to a common background. These QTLs can be further analysed with the lines carrying combined donor segments constructed from multiple ILs. The second approach, in turn, allows the detection of the QTLs of small effects due to the reduced background noise and does not require primary QTL analysis with other mapping populations (39).
While the examination of 123 SNP markers indicated no undesired CB4856 segments in the presented ILs (except for a single marker in ewIR28 and ewIR75), the presence of additional short donor segments within a region flanked by two adjacent markers (or the regions between the ends of the chromosomes and the closest markers) cannot be entirely excluded. Genotyping using high-density marker would allow the detection of the additional CB4856 segments and taking them into account in QTL analysis. Nevertheless, the current average distance between markers is 2.38 cM and the probability of crossovers within such a small genomic region is low. Consequently, undetected CB4856 segments are expected to be rare.
Currently, one of the greatest challenges of QTL research is the development of strategies and resources that allow for the efficient and accurate dissection of QTLs of small effects with the aim to detect causal genes. The presented genomic library of ILs is the first step to achieve these goals and significantly improve the QTL detection process in C. elegans.
Although beyond the scope of this article, the LSP QTL on chromosome X may guide us toward gene identification underlying LSP. This QTL harbours npr-1. This gene is known to be polymorphic between CB4856 and N2 (24). The two alleles cause a difference in pathogen susceptibility (27) and social feeding (24). The difference in pathogen susceptibility was measured by a shorter LSP of lines carrying the CB4856 allele of npr-1. It is likely that LSP of C. elegans grown on E. coli (OP50) also depends on the allele type of npr-1. This could explain the reduced LSP of the IL ewIR81 carrying the CB4856 allele of npr-1. Moreover, this line shows significant more clumping behaviour than N2 confirming the functional difference between the N2 and CB4856 npr-1 alleles. This IL library might therefore be useful to further focus on this candidate gene, and, for instance to search for epistatic interactions with other loci using crosses between different ILs.
The present IL library was used to search for novel loci associated with LSP and pumping rate to illustrate the potential application of this resource. We believe that the library will accelerate the discovery of natural polymorphisms underlying complex traits and will lead to a better understanding of the mechanisms behind the observed phenotypic variation in C. elegans. The ILs are freely available on request by sending an email to the corresponding author J.K.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU FP7 PANACEA 222936 and NWO (the Netherlands Organisation for Scientific Research) for providing the resources for this research, as project 835.80.008. Funding for open access charge: Wageningen University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Bas van de Waterbeemd, Ana Quilez-Moraga and Pieter Cornelis for technical support and the Caenorhabditis Genetics Center for providing the nematode strains N2 and CB4856.
REFERENCES
- 1.Kammenga JE, Phillips PC, De Bono M, Doroszuk A. Beyond induced mutants: using worms to study natural variation in genetic pathways. Trends Genet. 2008;24:178–185. doi: 10.1016/j.tig.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 2.Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O'Brien W, Courtland H-W, Jepsen KJ, Kirby A, Kulbokas EJ, et al. Genetic architecture of complex traits: large phenotypic effects and pervasive epistasis. Proc. Natl Acad. Sci. USA. 2008;105:19910–19914. doi: 10.1073/pnas.0810388105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Slate J. Quantitative trait locus mapping in natural populations: progress, caveats and future directions. Mol. Ecol. 2005;14:363–379. doi: 10.1111/j.1365-294X.2004.02378.x. [DOI] [PubMed] [Google Scholar]
- 4.Van Swinderen B, Shook DR, Ebert RH, Cherkasova VA, Johnson TE, Reis RJ, Crowder CM. Quantitative trait loci controlling halothane sensitivity in Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1997;94:8232–8237. doi: 10.1073/pnas.94.15.8232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Persson A, Gross E, Laurent P, Busch KE, Bretes H, De Bono M. Natural variation in a neural globin tunes oxygen sensing in wild Caenorhabditis elegans. Nature. 2009;458:1030–1033. doi: 10.1038/nature07820. [DOI] [PubMed] [Google Scholar]
- 6.Ayyadevara S, Ayyadevara R, Hou S, Thaden JJ, Reis RJHS. Genetic mapping of quantitative trait loci governing longevity of Caenorhabditis elegans in recombinant-inbred progeny of a Bergerac-BO x RC301 interstrain cross. Genetics. 2001;157:655–666. doi: 10.1093/genetics/157.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gutteling EW, Riksen JAG, Bakker J, Kammenga JE. Mapping phenotypic plasticity and genotype-environment interactions affecting life-history traits in Caenorhabditis elegans. Heredity. 2007;98:28–37. doi: 10.1038/sj.hdy.6800894. [DOI] [PubMed] [Google Scholar]
- 8.Gutteling EW, Doroszuk A, Riksen JAG, Prokop Z, Reszka J, Kammenga JE. Environmental influence on the genetic correlations between life-history traits in Caenorhabditis elegans. Heredity. 2007;98:206–213. doi: 10.1038/sj.hdy.6800929. [DOI] [PubMed] [Google Scholar]
- 9.Shook DR, Johnson TE. Quantitative trait loci affecting survival and fertility-related traits in Caenorhabditis elegans show genotype-environment interactions, pleiotropy and epistasis. Genetics. 1999;153:1233–1243. doi: 10.1093/genetics/153.3.1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Álvarez OA, Gutteling EW, Tijsterman M, Fu J, Riksen JAG, Hazendonk E, Prins P, Plasterk RHA, Jansen RC, et al. Mapping determinants of gene expression plasticity by genetical genomics in C. elegans. PLOS Genet. 2006;2:e222. doi: 10.1371/journal.pgen.0020222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ayyadevara S, Ayyadevara R, Vertino A, Galecki A, Thaden JJ, Reis RJS. Genetic loci modulating fitness and life span in Caenorhabditis elegans: categorical trait interval mapping in CL2a X Bergerac-BO recombinant-inbred worms. Genetics. 2003;163:557–570. doi: 10.1093/genetics/163.2.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shook DR, Brooks A, Johnson TE. Mapping quantitative trait loci affecting life history traits in the nematode Caenorhabditis elegans. Genetics. 1996;142:801–817. doi: 10.1093/genetics/142.3.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keurentjes JJB, Bentsink L, Alonso-Blanco C, Hanhart CJ, Vries HB-D, Effgen S, Vreugdenhil D, Koornneef M. Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics. 2007;175:891–905. doi: 10.1534/genetics.106.066423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paran I, Zamir D. Quantitative traits in plants: beyond the QTL. Trends Genet. 2003;19:303–306. doi: 10.1016/S0168-9525(03)00117-3. [DOI] [PubMed] [Google Scholar]
- 15.Flint J, Valdar W, Shifman S, Mott R. Strategies for mapping and cloning quantitative trait genes in rodents. Nat. Rev. Genet. 2005;6:271–286. doi: 10.1038/nrg1576. [DOI] [PubMed] [Google Scholar]
- 16.Fridman E, Liu YS, Carmel-Goren L, Gur A, Shoresh M, Pleban T, Eshed Y, Zamir D. Two tightly linked QTLs modify tomato sugar content via different physiological pathways. Mol. Genet. Genom. 2002;266:821–826. doi: 10.1007/s00438-001-0599-4. [DOI] [PubMed] [Google Scholar]
- 17.Thomson MJ, Edwards JD, Septiningsih EM, Harrington SE, McCoucH SR. Substitution mapping of dth1.1, a flowering-time quantitative trait locus (QTL) associated with transgressive variation in rice, reveals multiple sub-QTL. Genetics. 2006;172:2501–2514. doi: 10.1534/genetics.105.050500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clark RM, Wagler TN, Quijada P, Doebley J. A distant upstream enhancer at the maize domestication gene tb1 has pleiotropic effects on plant and inflorescent architecture. Nat. Genet. 2006;38:594–597. doi: 10.1038/ng1784. [DOI] [PubMed] [Google Scholar]
- 19.Clee SM, Yandell BS, Schueler KM, Rabaglia ME, Richards OC, Raines SM, Kabara EA, Klass DM, Mui ET-K, Stapleton DS. Positional cloning of Sorcs1, a type 2 diabetes quantitative trait locus. Nat. Genet. 2006;38:688–693. doi: 10.1038/ng1796. [DOI] [PubMed] [Google Scholar]
- 20.C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 21.Koch R, van Luenen H, van der Horst M, Thijssen KL, Plasterk RHA. Single nucleotide polymorphisms in wild isolates of Caenorhabditis elegans. Genome Res. 2000;10:1690–1696. doi: 10.1101/gr.gr-1471r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swan KA, Curtis DE, McKusick KB, Voinov AV, Mapa FA, Cancilla MR. High-throughput gene mapping in Caenorhabditis elegans. Genome Res. 2002;12:1100–1105. doi: 10.1101/gr.208902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maydan JS, Flibotte S, Edgley ML, Lau J, Selzer RR, Richmond TA, Pofahl NJ, Thomas JH, Moerman DG. Efficient high-resolution deletion discovery in Caenorhabditis elegans by array comparative genomic hybridization. Genome Res. 2007;17:337–347. doi: 10.1101/gr.5690307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Bono M, Bargmann CI. Natural variation in a neuropeptide Y receptor homolog modifies social behavior and food response in C. elegans. Cell. 1998;94:679–689. doi: 10.1016/s0092-8674(00)81609-8. [DOI] [PubMed] [Google Scholar]
- 25.Davies AG, Bettinger JC, Thiele TR, Judy ME, McIntire SL. Natural variation in the npr-1 gene modifies ethanol responses of wild strains of C. elegans. Neuron. 2004;42:731–743. doi: 10.1016/j.neuron.2004.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Tijsterman M, Okihara KL, Thijssen K, Plasterk RHA. PPW-1, a PAZ/PIWI protein required for efficient germline RNAi, is defective in a natural isolate of C. elegans. Curr. Biol. 2002;12:1535–1540. doi: 10.1016/s0960-9822(02)01110-7. [DOI] [PubMed] [Google Scholar]
- 27.Reddy KC, Andersen EC, Kruglyak L, Kim DH. A polymorphism in npr-1 is a behavioral determinant of pathogen susceptibility in C. elegans. Science. 2009;323:382–384. doi: 10.1126/science.1166527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kristensen VN, Edvardsen H, Tsalenko A, Nordgard SH, Sørlie T, Sharan R, Vailaya A, Ben-Dor A, Lønning PE, Lien S, et al. Genetic variation in putative regulatory loci controlling gene expression in breast cancer. Proc. Natl Acad. Sci. USA. 2006;103:7735–7740. doi: 10.1073/pnas.0601893103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Milloz J, Duveau F, Nuez I, Félix M-A. Intraspecific evolution of the intercellular signaling network underlying a robust developmental system. Genes Dev. 2008;22:3064–3075. doi: 10.1101/gad.495308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Huang C, Xiong C, Kornfeld K. Measurements of age-related changes of physiological processes that predict lifespan of Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 2004;101:8084–8089. doi: 10.1073/pnas.0400848101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lewis JA, Fleming JT. Basic culture methods. In: Epstein HF, Shakes DC, editors. Caenorhabditis elegens: Modern Biological Analysis of an Organism. Inc., San Diego, CA: Academic Press; 1995. pp. 3–29. [Google Scholar]
- 32.Emmons SW, Klass MR, Hirsh D. Analysis of the constancy of DNA sequences during development and evolution of the nematode Caenorhabditis elegans. Proc. Natl Acad. Sci. USA. 1979;76:1333–1337. doi: 10.1073/pnas.76.3.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rockman MV, Kruglyak L. Recombinational landscape and population genomics of Caenorhabditis elegans. PLOS Genet. 2009;5 doi: 10.1371/journal.pgen.1000419. 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodgkin J, Doniach T. Natural variation and copulatory plug formation in Caenorhabditis elegans. Genetics. 1997;146:149–164. doi: 10.1093/genetics/146.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palopoli MF, Rockman MV, TinMaung A, Ramsay C, Curwen S, Aduna A, Laurita J, Kruglyak L. Molecular basis of the copulatory plug polymorphism in Caenorhabditis elegans. Nature. 2008;454:1019–1022. doi: 10.1038/nature07171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takada T, Mita A, Maeno A, Sakai T, Shitara H, Kikkawa Y, Moriwaki K, Yonekawa H, Shiroishi T. Mouse inter-subspecific consomic strains for genetic dissection of quantitative complex traits. Genome Res. 2008;18:500–508. doi: 10.1101/gr.7175308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gregorová S, Divina P, Storchova R, Trachtulec Z, Fotopulosova V, Svenson KL, Donahue LR, Paigen B, Forejt J. Mouse consomic strains: exploiting genetic divergence between Mus m. musculus and Mus m. domesticus subspecies. Genome Res. 2008;18:509–515. doi: 10.1101/gr.7160508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schmalenbach I, Körber N, Pillen K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor. Appl. Gen. 2008;117:1093–1106. doi: 10.1007/s00122-008-0847-7. [DOI] [PubMed] [Google Scholar]
- 39.Iakoubova OA, Olsson CL, Dains KM, Ross DA, Andalibi A, Lau K, Choi J, Kalcheva I, Cunanan M, Louie J, et al. Genome-tagged mice (GTM): two sets of genome-wide congenic strains. Genomics. 2001;74:89–104. doi: 10.1006/geno.2000.6497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.