Abstract
Fibroblast growth factor 1 (FGF1) is involved in muscle development and regeneration. The FGF1 gene contains four tissue-specific promoters allowing synthesis of four transcripts with distinct leader regions. Two of these transcripts contain internal ribosome entry sites (IRESs), which are RNA elements allowing mRNA translation to occur in conditions of blockade of the classical cap-dependent mechanism. Here, we investigated the function and the regulation of FGF1 during muscle differentiation and regeneration. Our data show that FGF1 protein expression is induced in differentiating myoblasts and regenerating mouse muscle, whereas siRNA knock-down demonstrated FGF1 requirement for myoblast differentiation. FGF1 induction occurred at both transcriptional and translational levels, involving specific activation of both promoter A and IRES A, whereas global cap-dependent translation was inhibited. Furthermore, we identified, in the FGF1 promoter A distal region, a cis-acting element able to activate the IRES A-driven translation. These data revealed a mechanism of molecular coupling of mRNA transcription and translation, involving a unique process of IRES activation by a promoter element. The crucial role of FGF1 in myoblast differentiation provides physiological relevance to this novel mechanism. This finding also provides a new insight into the molecular mechanisms linking different levels of gene expression regulation.
INTRODUCTION
The fibroblast growth factor 1 (FGF1 or aFGF) is a member of the heparin-binding growth factor family of proteins with mitogenic, angiogenic, mesoderm-inducing and neurectoderm-modulating activities. FGF1 belongs to the FGF family currently composed of 22 members which have important roles in embryogenesis and differentiation in organisms as diverse as Xenopus laevi and mammals (1,2). FGFs (especially FGF1 and FGF2) are classically considered as mitogens behaving as inhibitors of myoblast differentiation (3). However, intracellular FGF1 has been described as a myoblast differentiation activator (4). Furthermore, although it has been proposed as a negative regulator of muscle development, elevated levels of FGF1 have been observed in regenerating muscle cells of dystrophin-deficient mice (mdx) (5) and in Facioscapulohumoral muscular dystrophy patients (6). Thus, FGF1 is clearly implicated in myogenesis and muscle regeneration, but its role in muscle development is complex and involves non-elucidated mechanisms.
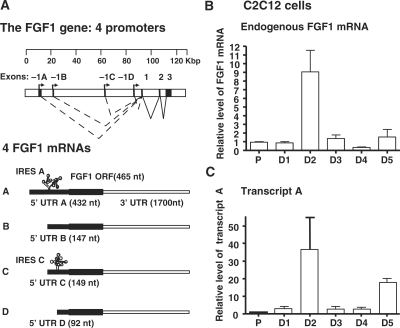
The fgf1 gene, expressing a single protein isoform, has four alternative tissue-specific promoters designed A to D and is subjected to a process of alternative splicing conserved among mammals (7,8). Transcription results in mRNAs differing by their 5′ untranslated region (5′UTR) (Figure 2A). Thus, each promoter leads to synthesis of an mRNA containing a distinct 5′ untranslated exon, suggesting specific translational regulation of FGF1 expression by such 5′UTRs as a consequence of the promoter usage.
Figure 2.
Transcriptional regulation of endogenous FGF1 expression during myoblast differentiation. (A) Schema of the FGF1 gene structure. The gene structure is presented with a scale (kbp). Exons –1A, –1B, –1C and –1D are the alternative exons generated by the use of promoters A, B; C and D, respectively. The four corresponding mRNAs, with different 5′ UTRs A, B, C and D, respectively, are represented below. Transcripts A and C contain IRESs (IRES A and IRES C, respectively). (B) and (C) RT qPCR quantification of endogenous FGF-1 mRNA (B) and of the transcript A (C) standardized to mRL8 RNA in proliferating and differentiating C2C12 myoblasts. Results are expressed relative to the level of proliferating cells and represent mean ± SEM of at least three independent experiments.
Translation in mammalian cells is mainly regulated at the initiation step through the rate-limiting recruitment of ribosomes to mRNA (9). Translation initiation can occur by a cap-dependent or cap-independent mechanism. The former is mediated by the mRNA 5′cap structure and represents the standard mode of translation used by most cellular mRNAs. It is predominantly controlled by the availability of the eukaryotic initiation factor 4F (eIF4F), comprised of the 5′cap binding protein eIF4E, the scaffold protein eIF4G and an ATP-dependent helicase eIF4A (10). eIF4E availability for eIF4F formation is modulated by sequestration by eIF4E-binding proteins (4E-BPs) (11). The most abundant, 4E-BP1, is inactive when hyperphosphorylated by the kinase mTOR and activated when mTOR activity is reduced (12,13).
Cap-independent translation is mostly mediated by mRNA structural elements called IRESs (Internal Ribosomal Entry Sites) (14). IRESs are able to recruit ribosomes either by themselves or with the help of cellular proteins called ITAFs (IRES trans-acting factors) (15). IRESs have been identified in several mammalian mRNAs, mainly in control genes such as growth factors or transcription factors (16). IRESs allow translation of such mRNAs when cap-dependent translation is blocked in conditions of stress or during mitosis (12,17). However they also allow a subtle regulation of mRNA translation in pathological and physiological situations such as hyperglycemia, hormone stimulation, ischemia or brain development (18–21). We have identified IRESs in the FGF1 5′UTRs A and C (Figure 2A) (22).
Fgf1 gene expression is strictly regulated during development and in adulthood (23). Surprisingly, little is known about the molecular mechanisms regulating its expression. While poorly expressed in adult tissues, it can become overexpressed in some pathophysiological situations such as during muscle regeneration (5). Here, we demonstrate that the FGF1, required for myoblast differentiation, is induced during this process as well as in regenerating muscle by a novel mechanism of coupled transcription and translation involving FGF1 promoter A and IRES A.
MATERIALS AND METHODS
Plasmids
Plasmids (P1A-luc, P1B-luc, P1C-luc and P1D-luc) used to measure promoter activities were kindly provided by Dr I.M. Chiu. Plasmids with EMCV and FGF1 IRESs were pCREL, pCRF1AL, pCRF1BL, pCRF1CL and pCRF1DL (22,24). CMV promoter was replaced in pCREL and pCRF1AL FGF1 promoter A amplified from plasmid P1A-luc. For P1A, P1A Δ1–391 and P1A Δ1–682 containing bicistronic constructs, -globin intron, LucR and FGF1 IRES A were inserted in pGL4.12 (Promega, France) downstream from full length or deleted FGF1 promoter A.
Plasmid construction details are available upon request.
Cell culture
C2C12 myoblasts were maintained in Dulbecco's modified Eagle's medium (DMEM) with 20% fetal calf serum in 100-mm diameter dishes at 37°C with 5% CO2. For differentiation, cells were changed into fusion medium (DMEM with 5% horse serum).
Transient transfections were performed in 12-well dishes using 0.5 µg of plasmid with FuGene-6 (Roche Molecular Biochemicals, Mannheim, Germany) and Optimem (Gibco-BRL, Invitrogen, Paisley, United Kingdom).
Small interference RNAs were from Dharmacon siGENOME® SMARTpool® FGF1 siRNA, siGENOME® SMARTpool® eIF4E siRNA and siGENOME® non-targeting siRNA. C2C12 cells were transfected with 20 nM siRNA with Hyperfect transfection reagent (Qiagen).
Mouse muscle regeneration model
Twenty-five microliters of cardiotoxin (Latoxan) at 10 μM, were injected in Tibialis anterior (TA) muscle of 4-week-old female C57BL/6J mice to induce degeneration and regeneration (25). Non-injected TA muscles were used as control.
Two days after cardiotoxin treatment, TA muscles were injected with 50 μg of plasmid (25 μl final volume). Three days later, TA muscles were removed and stored at –80°C.
Ethics statement
All procedures were performed in conformity with the guidelines of the company's local ethics committee and in accordance with the recommendations of the European Accreditation of Laboratory Animal Care Institute. The animal facility (directed by Yara Barreira) has obtained the animal experimentation agreement No B31-555-7.
RNA extraction and real time RT–qPCR
Total RNA was isolated from C2C12 cells using GenElute Mammalian Total RNA kit (Sigma-Aldrich Chimie). After DNase I treatment (DNase I Amplification Grade, Invitrogen), reverse transcription was performed with the High capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). As an internal control, ribosomal L8 (mRL8) RNA was used.
Quantitative PCR was performed on an ABI 7900 sequence detection system (Applied Biosystems) using Sybr Green PCR Master Mix (Applied Biosystems) for detection of FGF1 (all transcripts), FGF1 A and RL8 transcripts.
Primer sequences are available upon request.
Reporter assays
Muscles and C2C12 cells were ground in Passive Lysis Buffer (Promega) using an Ultra-Turrax T25 (Janke and Kunkel, IKA Labortechnik). Quantification of bioluminescence was realised with a luminometer (Centro LB960, Berthold) using the Dual-Luciferase® Reporter Assay (Promega, France).
In vivo imaging and quantification of bioluminescence data
In vivo imaging was performed with a cooled CCD camera (1394 ORCA II, Hamamatsu Photonics) using mice transduced with a bicistronic AAV vector expressing the Firefly luciferase under control of FGF1 IRES A, as previously described (26). Eighteen days after transduction, muscles were cardiotoxin-treated and imaged 3 and 7 days later. Prior to imaging, mice were anesthetized by intraperitoneal injection of Ketamine (12.5 μg/μl) and Xylazine (1%) solutions (10 μl/g body weight). D-luciferin was injected intraperitoneally (50 μg/g BW, Luciferin-EFtm, Promega France), 5 min before measurement of photon emission. Grey scale images and bioluminescence color images were superimposed using the Simple PCI® software (Hamamatsu Photonics). Light output was quantified as the mean grey level per second of time exposure and per region of interest (ROI).
Western blotting
Muscles were homogenized and western blots performed as previously (19).
Primary antibodies were rabbit polyclonal antibodies against FGF1 (F5521, Sigma, 1/500), eIF4E (Cell Signaling, 1/10 000), 4E-BP (Cell Signaling, 1/1500) and mouse monoclonal antibodies against myogenin (F5D, Santa Cruz Biotechnologies, 1/400), Glyceraldehyde 3-phosphate dehydrogenase (GAPDH, Santa Cruz Biotechnologies, 1/200), β-tubulin (Sigma, 1/4000). Secondary antibodies were peroxidase-conjugated- AffiniPure Donkey Anti-Rabbit IgG and -AffiniPure Donkey Anti-Mouse IgG (Jackson ImmunoResearch, Baltimore, 1/20 000).
Protein detection was carried out using Supersignal West Femto Maximum Sensitivity Substrate, SuperSignal West Pico Chemiluminescent Substrate and ECL western blotting substrate (ThermoScientific, USA).
Analysis of 4E-BP1-eIF4E interaction
Cells were harvested in lysis buffer [25 mM Tris–HCl pH 7.4, 50 mM KCl, 5% glycerol, 0.5% Nonidet P-40 and 1 mM DTT supplemented with protease inhibitor mixtures (Roche)] and clarified by centrifugation at 12 000 rpm for 10 min at 4°C. Proteins from the supernatant were quantified using the Bradford method (Bio-Rad). After preclearing cell lysates with 10 μl of Protein G-Agarose conjugate beads for 1 h at 4°C, 50 μg of protein were incubated with 10 μl of eIF4E antibody agarose conjugate (Santa Cruz Biotechnology) overnight at 4°C. Beads were washed five times with lysis buffer and immunoprecipitated proteins eluted with Laemmli sample buffer and subjected to immunoblot analysis.
Statistical analysis
Statistical analysis was performed with Prism® (GraphPad Software Inc.). Unpaired t-tests (two-tailed) were realized to detect significant differences between group means. The level of significance was set at P < 0.05.
RESULTS
FGF1 is induced during myogenesis and required for myoblast differentiation
Treatment of skeletal muscle with the snake venom cardiotoxin (CTX) has been described to induce severe myonecrosis and subsequent muscle regeneration while leaving the innervating nerve intact (25,27). We used this approach to study FGF1 expression in regenerating muscles. We observed that the FGF1 protein level started to increase 3 days after cardiotoxin injection and continued to rise after 7 days. The early induction of FGF1 at Day 3 was concomitant with the expression of myogenin, a specific marker of myotubes (Figure 1A).
Figure 1.
FGF1 expression and knock-down during myogenesis. (A) FGF1 accumulation in mouse regenerating muscles. Western blot analysis was performed using anti-FGF1, anti-myogenin and anti-GAPDH antibodies, on untreated (NT) or treated Tibialis anterior muscles, 3, 5 and 7 days (D3, D5 and D7, respectively) after cardiotoxin (CTX) injection. Muscle No1 and 2 correspond to two different individuals. Myogenin was used as a marker of myoblast differentiation. GAPDH was used as a normalization control. These data correspond to a representative experiment (repeated at least three times). (B) FGF1 accumulation during C2C12 myoblast differentiation. Western blot analysis was performed as in A on proliferating (P) and differentiating myoblasts from Day 1 (D1) to Day 5 (D5) after serum-starvation treatment. These data correspond to a representative experiment (repeated five times). (C) Effect of FGF1 knock-down on myoblast differentiation. C2C12 cells were transfected with siRNA against FGF1 (siFGF1) or siRNA control (siControl). Western blot was performed as above on proliferating (P) and differentiating myoblasts (D2–D5). These data correspond to a representative experiment (repeated at least three times). (D) Effect of FGF1 knock-down on myotube formation. C2C12 cells were transfected with siRNA against FGF1 (siFGF1) or siRNA control (siControl) and myotube formation was followed by contrast phase microscope analysis. These data correspond to a representative experiment (repeated three times).
In order to determine whether FGF1 induction is connected to myoblast differentiation, we used the murine myoblast C2C12 cell line which can be induced to differentiate into myotubes by serum-deprivation (28). We observed that FGF1 starts to accumulate in C2C12 cells after 2 days of serum-deprivation, when myoblasts begin to differentiate into myotubes, as visualized by myogenin expression (Figure 1B).
To evaluate the importance of intracellular FGF1 during myoblast differentiation, we knocked down its expression in C2C12 cells by siRNA transfection experiments. FGF1 knock-down in the early-differentiation stages prevented the strong myogenin induction observed at Days 2 and 3 with the control siRNA (Figure 1C). A smaller induction of myogenin was observed later, at Day 4, concomitantly with an increase of FGF1 expression indicating that FGF1 knock-down was temporary (Figure 1C, right panel).
Myotube formation was observed from Day 2 to Day 5 (Figure 1D). Strikingly, the C2C12 cell aspect was already different at Day 3, since the FGF1 knock-down resulted in anarchical cell layout. Furthermore, this phenotype was aggravated at Days 4 and 5 when formation of myotubes occurred. Indeed, these myotubes appeared irregular and abnormal.
These results showed that FGF1 is induced during muscle differentiation/regeneration and required for correct myoblast differentiation into myotubes. FGF1 knock-down did not prevent myotube formation, but resulted in an anarchical cell layout starting from Day 3 of myoblast differentiation.
The FGF1 promoter A is strongly activated during myogenesis
To investigate the mechanism of FGF1 induction during myoblast differentiation, quantitative real-time RT–PCR (RT–qPCR) analysis was performed to detect the endogenous FGF1 mRNA. Data showed that the total level of FGF1 mRNAs increased 9-fold at Day 2 of differentiation (Figure 2B). The level of transcript A followed the same pattern of induction with a 35-fold increase, suggesting a main contribution of promoter A to FGF1 mRNA induction (Figure 2A and C).
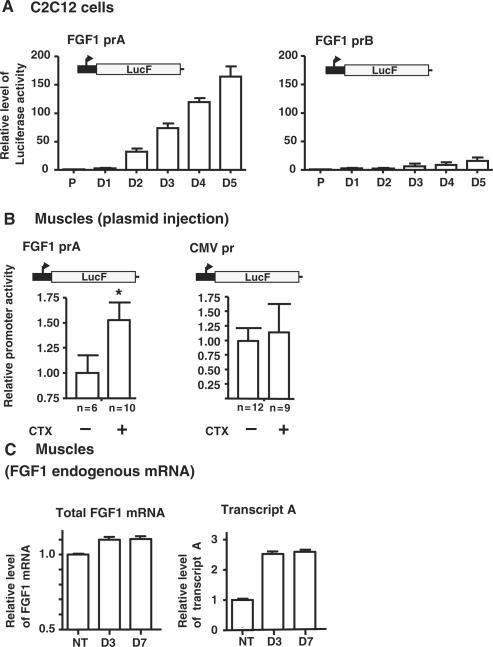
To confirm this hypothesis, activities of the four FGF1 promoters were evaluated using gene reporter assay (Figure 2A and Figure 3). The enzymatic activity of Firefly luciferase (LucF) expressed under the control of each FGF1 promoter was measured in proliferating and differentiating C2C12 cells. Data showed a 41-fold increase of promoter A activity at Day 2, whereas promoter B showed a 3-fold increase (Figure 3A and Supplementary Table I). Regarding promoter C, its activity diminished, while promoter D was always very weak (Supplementary Table I). In contrast to the endogenous transcript A which showed a peak of accumulation at D2, the reporter assay revealed a continuous activation of the promoter A, reaching a 160-fold activation when differentiation into myotubes was completed at Day 5 (Figure 3A and Supplementary Table 1). Such a discrepancy could result from a difference of mRNA stability regulation, as the endogenous transcript, but not the reporter transcript, bears a 1700-nt long 3′ UTR.
Figure 3.
Activities of FGF1 promoters during myoblast differentiation and muscle regeneration. (A) Activities of FGF1 promoters A and B during C2C12 myoblast differentiation. Luciferase activities were measured on C2C12 cells transfected with plasmids containing the Firefly luciferase expressed under the control of the FGF1 promoter A or B. Results are expressed relatively to the proliferation state. The graphs show the mean ± SEM of five independent experiments. (B) Activities of FGF1 promoter A and CMV promoter during mouse muscle regeneration. Plasmids expressing the Firefly luciferase under the control of FGF1 promoter A or CMV promoter were injected into the Tibialis anterior muscles of wild-type mice. Luciferase activities were measured on cardiotoxin-treated (+) or untreated (−) muscles at Day 5 after CTX treatment. Results are expressed relatively to the level of untreated muscle and represent mean ± SEM. n, number of muscles, *P < 0.05. (C) RT qPCR quantification of endogenous FGF-1 mRNA and of the transcript A standardized to mRL8 RNA in untreated or cardiotoxin treated muscle extracts (Days 3 and 7). Results are expressed relative to the level of untreated muscle and represent mean ± SEM of two independent experiments.
We then studied the promoter A activity and the FGF1 endogenous mRNA levels in vivo, during muscle regeneration. Plasmids expressing LucF under the control of the promoter A or the ubiquitous cytomegalovirus (CMV) promoter were injected into cardiotoxin-treated or untreated muscles of wild-type mice. We observed a significant 1.5-fold increase of promoter A activity 5 days after CTX-treatment relatively to untreated muscles, while no change of the CMV promoter activity could be detected (Figure 3B). RT–qPCR analysis of the endogenous transcript A in CTX-muscle indicated a 2.4-fold increase at Day 3, which remained stable until Day 7, whereas the total FGF1 mRNA did not significantly increase (Figure 3C).
These results, although showing differences between the levels of FGF1 mRNAs and promoter activation observed in vivo and in vitro, were concordant by revealing the induction of the FGF1 promoter A in both differentiating myoblast and regenerating muscle.
Cap-dependent translation decreases during myoblast differentiation
To determine whether FGF1 expression during myoblast differentiation involved cap- or IRES- dependent translation, we looked at the status of global cap-dependent translation during myoblast differentiation by analyzing 4E-BP1 and eIF4E expression, and monitoring eIF4E sequestration by 4E-BP1.
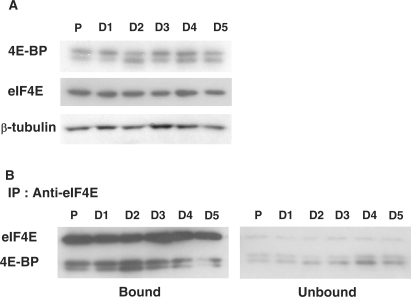
Western blot experiments performed on protein extracts from proliferating and differentiating myoblasts showed that the expression level of eIF4E and 4E-BP1 did not vary whatever the differentiation state (Figure 4A). However, changes in 4E-BP1 gel mobility were visible with a prominent modification at Day 2 of differentiation. 4E-BP1 protein is known to exhibit a complex gel mobility pattern due to post-translational modifications including different degrees of phosphorylations. Other post-translational modifications that can render 4E-BP1 data analysis even more complex are that the phosphorylated protein can be ubiquitinated (29). Furthermore, phosphorylation at sites initially considered as important for 4E-BP1 dissociation from eIF4E may not affect directly 4E-BP1 affinity for eIF4E, but may serve instead as priming events for another type of a yet unknown post-translational modification that is actually required for 4E-BP1 dissociation from eIF4E (30). Therefore, we directly tested whether modifications in 4E-BP1 electrophoretic properties were correlated to changes in 4E-BP1 affinity for eIF4E by co-immunoprecipitation experiments. The data revealed a transient increase in the formation of the 4E-BP1/eIF4E complex from Day 1 to Day 3 and peaking at Day 2 of differentiation (Figure 4B). These results indicated that eIF4E was transiently sequestered by 4E-BP1 through the process of myoblast differentiation.
Figure 4.
Regulation of the cap-dependent translation during myoblast differentiation. (A) Western blot analysis of eIF4E and its inhibitor 4E-BP during myoblast proliferation (P) and differentiation (D), using anti-eIF4E and anti-4E-BP antibodies, respectively. β-tubulin was used as a normalization control. These data correspond to a representative experiment (repeated at least three times). (B) Co-immunoprecipitation of eIF4E and 4E-BP during myoblast proliferation (P) and differentiation (D). Immunoprecipitation was performed on C2C12 cell extracts with an antibody against eIF4E. The bound and the unbound fractions were applied to Tris–Tricine SDS–PAGE followed by Western transfer and immunodetection with antibodies as indicated. These data correspond to a representative experiment (repeated three times).
FGF1 mRNA translation is induced through an IRES A-dependent mechanism during myoblast differentiation and muscle regeneration
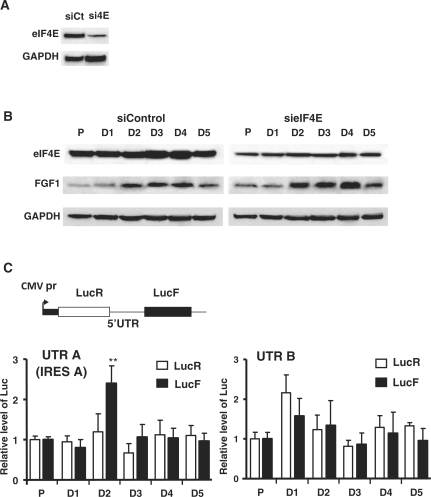
We have shown above that FGF1 protein induction starts at Day 2 of myoblast differentiation, concomitantly with a peak of promoter A activity and a transient decrease of cap-dependent translation. The presence of an IRES in transcript A, appearing as mainly responsible for FGF1 induction, suggested that FGF1 mRNA translation could occur by an IRES-dependent mechanism (22). This hypothesis was assessed by measuring FGF1 protein expression in differentiating C2C12 cells treated with an eIF4E-targeted siRNA (sieIF4E) or a control siRNA (siControl) (Figure 5A and B). The eIF4E knock-down did not affect the FGF1 induction at Day 2, suggesting that its expression is mediated by a cap-independent mechanism (Figure 5B, right panel).
Figure 5.
IRES-dependent expression of FGF1 during myoblast differentiation. (A) Control of eIF4E knock-down. Proliferating C1C12 cells were transfected with an siRNA against eIF4E (sieIF4E) and siRNA control (sicontrol), in conditions described in ‘Materials and Methods’ section, and submitted to serum starvation. eIF4E level following siRNA treatments was analyzed on the same gel by western blot at Day 1 of differentiation. (B) Effect of eIF4E knock-down on FGF1 expression during myoblast differentiation. C2C12 cells were transfected with sieIF4E or sicontrol as above. eIF4E, FGF1 and as a control GAPDH were detected by Western blot on proliferating (P) and differentiating myoblasts (D). These data correspond to a representative experiment (repeated three times). (C) Measure of IRES activities in the 5′UTR A and B of FGF1. The bicistronic construct used to evaluate IRES activities is schematized. The bicistronic cassette expresses, under the control of the CMV (Cytomegalovirus) promoter, Renilla (LucR) and Firefly (LucF) luciferase reporter genes in a cap- and IRES-dependent manner, respectively. The bicistronic cassette contains the FGF1 mRNA 5′ UTRs A or B. Renilla and Firefly luciferase activities were measured on extracts of C2C12 cells transfected with the indicated bicistronic expression vectors. Relative LucR (cap-dependent) and LucF (IRES-dependent) activities are shown for the different differentiation days (Days 1–5), normalized to the activities obtained in proliferation (P). The graphs show the mean ± SEM of five independent experiments; **P < 0.01.
IRES activities in the 5′UTR of FGF1 mRNAs were measured using bicistronic constructs expressing the Renilla (LucR) and Firefly luciferases in a cap- and IRES-dependent manner respectively (Figure 5C) (22). Consequently, the ratio of LucF to LucR activities is expected to reflect the relative level of IRES- to cap-dependent translation. We observed a transient increase of LucF activity (2.5-fold) and of the ratio LucF/LucR peaking at Day 2 after the induction of differentiation, with the construct containing the transcript A 5′UTR, thus the IRES A (Figure 5C and Supplementary Table II). No relative increase of IRES activities was detected in the 5′UTRs of transcript B or D, while the UTR C containing the IRES C also showed a small peak of activity (but not statistically significant) at Day 2 of FGF1 during myoblast differentiation (Supplementary Table II). In addition, the equivalent amount of lucR and lucF RNA confirmed that all the mRNAs were bicistronic (Supplementary Figure S1 and data not shown). These data showed that the IRES A is the only one to be significantly activated at Day 2 of differentiation.
In order to check IRES A involvement in FGF1 induction during myogenesis in vivo, FGF1-based bicistronic rAAVs (adeno-associated virus) containing the expression cassette described in Figure 5C were injected into CTX-treated or untreated muscles of wild-type mice. The IRES-driven LucF expression was followed on live animals, using a CCD bioluminescence camera. LucF was measured during 7 days and quantified at Days 3 and 7 after CTX-treatment. A weak increase of the IRES A activity was observed at Day 3, which became significant at Day 7 with a 2.5-fold increase measured in CTX-treated-muscles relatively to untreated muscles (Figure 6A and B). Measurement of LucR and LucF activities in muscle extracts at Day 5 after CTX-treatment confirmed such a significant augmentation of the FGF1 IRES A activity (Figure 6C, left panel), whereas the activity of the EMCV (EncephaloMyoCarditis Virus) IRES, used as a control, remained unchanged (Figure 6C, right panel).
Figure 6.
Regulation of FGF1 IRES A-driven translation activity during muscle regeneration. (A) Kinetics of the expression of luciferase Firefly under the control of the IRES A during muscle regeneration. The luciferase Firefly activity was measured in mice transduced with a bicistronic AAV vector containing the gene-encoding LucF under the control of the IRES A of FGF1 (1010 total particles/muscle). Mice were imaged using a bioluminescent camera after injection of D-luciferin intraperitonealy. The Firefly luciferase activities are shown in pseudocolors at Days 3 and 7 after cardiotoxin treatment. The color scale illustrates the variation of luciferase activities from maximum (red) to minimum (dark blue). (B) Luminescent measurement of luciferase Firefly activities with CCD camera in the experiments described in (A). Each dot of the graph represents one muscle and the mean is indicated by a short line. (C) Specific activation of the FGF1 IRES A during muscle regeneration. Plasmids expressing the Renilla and Firefly luciferases through a cap- and IRES-dependent mechanisms respectively were injected into the Tibialis anterior muscles of wild-type mice. Luciferase activities were measured 5 days after intra-muscular injection of cardiotoxin. IRES activities are given as the LucF/LucR ratio relatively to the level of untreated muscles and represent mean ± SEM. n, number of muscles, ***P < 0.001.
These results highlighted the specific activation of the IRES A of FGF1 during myoblast differentiation and muscle regeneration. Furthermore they provided new insight into the mechanisms of FGF1 synthesis by showing that it is expressed by IRES-dependent mechanism in the early stage of myoblast differentiation, when the cap-dependent translation is inhibited.
The FGF1 IRES A is activated by a cis-acting element present in the promoter A
Our data showed that the IRES A activation is concomitant with the activation of the FGF1 promoter A. This observation, together with previous data indicating that the IRES activity may be influenced by the promoter (Y. Martineau, PhD thesis 2004), raised the hypothesis of a mechanism of transcription-translation coupling.
In order to investigate the physiological relevance of such a correlation and the putative functional link between promoter and IRES, we replaced the CMV promoter by the FGF1 promoter A upstream from the bicistronic cassette containing either the FGF1 IRES A or the EMCV IRES (Figure 7A). The bicistronic constructs with either FGF1 or CMV promoter were used to transfect C2C12 cells as above. IRES activities are presented as the LucF/LucR ratio, thus normalized to the mRNA level reflected by LucR. As shown in Figure 7B, the FGF1 IRES activity at Day 2 was 15-fold superior for the construct containing the FGF1 promoter A compared to the one obtained with the CMV promoter. In contrast, the EMCV IRES activity was similar in the presence of either promoter A or CMV promoter (Figure 7C).
Figure 7.
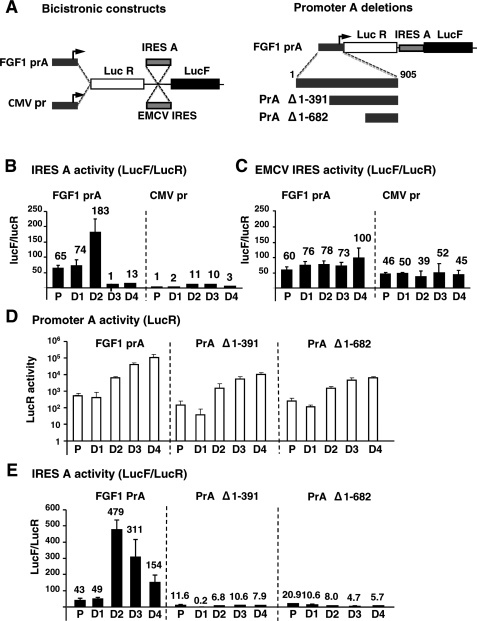
Effect of the FGF1 promoter A on IRES A activity. (A) Left: schema of the bicistronic constructs containing on the one hand either the FGF1 Promoter A or the CMV promoter and on the other hand either the FGF1 IRES A or the EMCV IRES. Right: Different promoter A deletions were performed (Δ1-391 and Δ1-682) and are schematized below the bicistronic construct. This latter series of constructs contains a more recent version of firefly luciferase, Luc2CP, which displays higher expression than the classical firefly luciferase. (B) and (C). Activities of the FGF1 IRES A (B) and the EMCV IRES (C) obtained after transfection of C2C12 cells with the bicistronic constructs schematized in A (left). IRES activities obtained in proliferating and differentiating myoblasts in the presence of promoter A (left panels) or CMV promoter (right panels) are represented as LucF/LucR ratios. The activities are indicated above each histogram. Experiments were repeated at least three times. Results represent means ± standard errors from a representative experiment done in triplicate. (D) Transcriptional activities of the complete and deleted FGF1 promoter A in proliferating and differentiating myoblasts. Myoblasts were transfected with the bicistronic constructs described in Figure 7A (right) and the LucR activities were measured. (E) Activities of the FGF1 IRES A in the different constructs containing the complete or deleted FGF1 promoter A. IRES activities are given as the LucF/LucR ratio. The activities are indicated above each histogram. Experiments were repeated at least three times. Results represent means ± standard errors from a representative experiment done in triplicate.
These data also showed that the peak of IRES A activation at Day 2 of differentiation occurs with both promoters and is specific. However, the presence of the FGF1 promoter renders the FGF1 IRES as active as the EMCV IRES in proliferating cells and largely superior to the EMCV IRES in differentiating cells.
In order to map the region of the FGF1 promoter A responsible for the IRES activation, the promoter was deleted of nt 1–392 or nt 1–682 (Figure 7A). The LucR activity of the corresponding constructs clearly showed that the region between nt 1 and 392 contains an element necessary for transcriptional activation at the FGF1 promoter A at Day 2 and later (Figure 7D). Furthermore, the IRES A-dependent translation (LucF/LucR ratio) peaked at Day 2 with the complete promoter, but not with the deleted constructs (Figure 6E).
In conclusion, our results clearly show that FGF1 IRES A-dependent translation is strongly activated in the presence of the promoter A, and that this activation requires a cis-acting element localized between nt 1 and 392 of the promoter.
DISCUSSION
In this study, we have investigated the molecular mechanisms implicated in the induction of FGF1 expression during myogenesis. We demonstrate that FGF1 up-regulation is required for myoblast differentiation and results from a concomitant and specific activation of both FGF1 promoter A and IRES A, when the cap-dependent translation is blocked, in differentiating myoblasts as well as in regenerating mouse muscle. Furthermore, our data show that the IRES A-dependent translation is strongly activated in the presence of the FGF1 promoter A, revealing a novel mechanism of coupled translation and transcription.
FGF1 has a double role in myogenesis
FGF1 induction during muscle regeneration is concomitant with the apparition of myotubes. Furthermore, our data clearly show that FGF1 knock-down attenuates the myogenin induction peaking at Day 2 and generates formation of abnormal myotubes. This observation could seem discordant with the known function of FGFs as myoblast differentiation inhibitors (3,31). However FGF1, in contrast to other members of the FGF family, is expressed in dystrophic muscle, suggesting a positive role in the regeneration of skeletal muscle fiber (5,6). The double role of FGF1 as a proliferation activator as well as a differentiation inducer may result from its different functions as an extracellular or intracellular factor (4). Extracellular FGF1 would activate myoblast proliferation by activating the FGF-signaling pathway mediated by FGFR1 receptor upregulated in proliferating myoblasts, (32). In contrast, intracellular FGF1 would act on differentiation by an intracrine pathway. Alternatively, the different roles of FGF1 could be due to differential expression of FGF receptors (33). FGFR1 and FGFR4 are the most prominent FGF receptors in muscle cells. FGFR4 has been recently shown to be strongly up-regulated during myoblast–myotube transition (34). Interestingly, Fgfr4–/– mice show impaired muscle regeneration with slowed maturation of regenerating fibers (34).
Translational regulation during myoblast differentiation
It has been well established that cap-dependent translation is tightly regulated during the cell cycle (35,36). However, little is known about translational control of gene expression during cell differentiation. Our work shows that cap-dependent translation is repressed during myoblast differentiation. This suggests that, at this stage, the global translation is blocked in cells, whereas a small number of mRNAs may be translated through cap-independent mechanisms. This is the case for FGF1 whose induction is not affected by eIF4E sequestration, concomitant with the activation of its IRES A. Thus, our results together with previous studies show that IRESs play a key role in gene expression during differentiation of different cell types and during development (37–40).
The role of IRES in FGF1 expression was further confirmed in vivo during muscle regeneration. The importance of IRES-dependent translation during muscle regeneration has also been shown for utrophin A, a structural protein of the skeletal muscle fiber, mainly expressed at the neuromuscular junction (41). In contrast to utrophin A, FGF1 has no structural role, but a regulating function during the myogenesis process. The importance of the IRES-mediated translational control for two proteins with such different roles but both involved in the muscle fiber regenerative response suggest that the IRES-dependent process could drive expression of a subclass of mRNAs whose coordinated induction would ensure the success of the muscle regeneration process.
Coupling between transcription and IRES-dependent translation
Among the four FGF1 alternative promoters, A and B have been reported to be tissue-specific whereas C and D are inducible (7,42). Our results reveal that promoter A is active in skeletal muscle, and demonstrate its inducibility during myoblast differentiation. Interestingly, the activity of IRES A, contained in the transcript generated by promoter A, specifically increases during myoblast differentiation and muscle regeneration.
Coordinated activation of IRES with the corresponding promoter in conditions of cap-dependent translation blockade has been reported for the Drosophila insulin-like receptor, underlining the key role of IRESs in the activation of gene expression when global translation is silenced (43). The coupling of translation with transcription shown in our study goes further by suggesting the existence of a molecular mechanism of IRES A activation depending on a cis-acting element in the promoter. One could argue that the higher IRES A activity in the presence of promoter A could result from the weakness of this promoter, whereas the strong CMV promoter would result in mRNA overexpression and decreased IRES A activity due to titration of a limiting ITAF. This was proposed in a previous study showing that the c-myc IRES is higher with the SV40 promoter than with the CMV promoter (44). However, in our study, the strength of the FGF1 promoter A is similar to that of the CMV promoter (data not shown), and the deleted versions of promoter A, which render the promoter very weak, are not related to any IRES activation (Figure 7). This clearly shows that the FGF1 promoter A has a positive effect on FGF1 IRES-driven translation. Our data suggest the existence of a translation–transcription coupling mechanism requiring an element in the distal part of promoter A, upstream from nt 392.
Such coupling of transcription with translation would appear as a novel mechanism completing the complex network of coupled interactions in gene expression proposed by Maniatis and Reed (45). That model proposes coupling of transcription with subsequent steps of mRNA processing and export, in gene expression factories anchored to the nuclear substructure, where DNA is reeled through the RNA polymerase as the nascent mRNA is extruded through its exit channel. Such gene expression factories contain all machineries involved in transcription, capping, splicing and polyadenylation. Our data suggest that IRES-dependent translation could also be controlled in these gene expression factories, due to a co-transcriptional ‘loading’ of ITAFs on nascent mRNA. Consistent with this hypothesis, most ITAFs identified so far are nuclear or nucleocytoplasmic shuttling proteins implicated in a staggering array of cellular activities, ranging from transcription and pre-mRNA processing in the nucleus, to cytoplasmic mRNA translation and turnover (46,47). Interestingly, it has been shown that the same factor, hnRNPK, controls the promoter and the IRES activities of c-myc (48–50).
Such a direct coupling of transcription and IRES-dependent translation despite the compartmentalization of the cell nucleus might allow cells to respond more quickly to environmental signals.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Agence Nationale pour la Recherche; Association Française contre les Myopathies; Association pour la Recherche sur le Cancer; Ligue Contre le Cancer, Fondation de l’Avenir; Conseil Régional Midi-Pyrénées; Association Française contre les Myopathies fellowship to N.A. Funding for open access charge: Institut National de la Santé de la Recherche Rédicale (Inserm).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr I. M. Chiu for providing plasmids. We are also grateful to L. Van den Berghe and F. Sage for technical assistance and Dr L. Prado-Lourenço for helpful discussions. We also thank Y. Barreira and C. Nevoit (IFR150 animal facility) and J. J. Maoret (IFR150 plateform of molecular biology).
Footnotes
The authors wish it to be known that, in their opinion, the second, third and fourth authors should be regarded as joint Second Authors.
REFERENCES
- 1.Auguste P, Javerzat S, Bikfalvi A. Regulation of vascular development by fibroblast growth factors. Cell Tissue Res. 2003;314:157–166. doi: 10.1007/s00441-003-0750-0. [DOI] [PubMed] [Google Scholar]
- 2.Dawid IB, Taira M, Good PJ, Rebagliati MR. The role of growth factors in embryonic induction in Xenopus laevis. Mol. Reprod. Dev. 1992;32:136–144. doi: 10.1002/mrd.1080320209. [DOI] [PubMed] [Google Scholar]
- 3.Clegg CH, Linkhart TA, Olwin BB, Hauschka SD. Growth factor control of skeletal muscle differentiation: commitment to terminal differentiation occurs in G1 phase and is repressed by fibroblast growth factor. J. Cell Biol. 1987;105:949–956. doi: 10.1083/jcb.105.2.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Uruno T, Oki J, Ozawa K, Miyakawa K, Ueno H, Imamura T. Distinct regulation of myoblast differentiation by intracellular and extracellular fibroblast growth factor-1. Growth Factors. 1999;17:93–113. doi: 10.3109/08977199909103519. [DOI] [PubMed] [Google Scholar]
- 5.Oliver L, Raulais D, Vigny M. Acidic fibroblast growth factor (aFGF) in developing normal and dystrophic (mdx) mouse muscles. Distribution in degenerating and regenerating mdx myofibres. Growth Factors. 1992;7:97–106. doi: 10.3109/08977199209046399. [DOI] [PubMed] [Google Scholar]
- 6.Saito A, Higuchi I, Nakagawa M, Saito M, Uchida Y, Inose M, Kasai T, Niiyama T, Fukunaga H, Arimura K, et al. An overexpression of fibroblast growth factor (FGF) and FGF receptor 4 in a severe clinical phenotype of facioscapulohumeral muscular dystrophy. Muscle Nerve. 2000;23:490–497. doi: 10.1002/(sici)1097-4598(200004)23:4<490::aid-mus6>3.0.co;2-k. [DOI] [PubMed] [Google Scholar]
- 7.Madiai F, Hackshaw KV, Chiu IM. Characterization of the entire transcription unit of the mouse fibroblast growth factor 1 (FGF-1) gene. Tissue-specific expression of the FGF-1.A mRNA. J. Biol. Chem. 1999;274:11937–11944. doi: 10.1074/jbc.274.17.11937. [DOI] [PubMed] [Google Scholar]
- 8.Myers RL, Chedid M, Tronick SR, Chiu IM. Different fibroblast growth factor 1 (FGF-1) transcripts in neural tissues, glioblastomas and kidney carcinoma cell lines. Oncogene. 1995;11:785789. [PubMed] [Google Scholar]
- 9.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Richter JD, Sonenberg N. Regulation of cap-dependent translation by eIF4E inhibitory proteins. Nature. 2005;433:477–480. doi: 10.1038/nature03205. [DOI] [PubMed] [Google Scholar]
- 11.Svitkin YV, Herdy B, Costa-Mattioli M, Gingras AC, Raught B, Sonenberg N. Eukaryotic translation initiation factor 4E availability controls the switch between cap-dependent and internal ribosomal entry site-mediated translation. Mol. Cell Biol. 2005;25:10556–10565. doi: 10.1128/MCB.25.23.10556-10565.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holcik M, Sonenberg N. Translational control in stress and apoptosis. Nat. Rev. Mol. Cell Biol. 2005;6:318–327. doi: 10.1038/nrm1618. [DOI] [PubMed] [Google Scholar]
- 13.Pause A, Belsham GJ, Gingras AC, Donze O, Lin TA, Lawrence JC, Jr, Sonenberg N. Insulin-dependent stimulation of protein synthesis by phosphorylation of a regulator of 5'-cap function. Nature. 1994;371:762–767. doi: 10.1038/371762a0. [DOI] [PubMed] [Google Scholar]
- 14.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 15.Spriggs KA, Bushell M, Mitchell SA, Willis AE. Internal ribosome entry segment-mediated translation during apoptosis: the role of IRES-trans-acting factors. Cell Death Differ. 2005;12:585–591. doi: 10.1038/sj.cdd.4401642. [DOI] [PubMed] [Google Scholar]
- 16.Vagner S, Galy B, Pyronnet S. Irresistible IRES. Attracting the translation machinery to internal ribosome entry sites. EMBO Rep. 2001;2:893–898. doi: 10.1093/embo-reports/kve208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pyronnet S, Pradayrol L, Sonenberg N. A cell cycle-dependent internal ribosome entry site. Mol. Cell. 2000;5:607–616. doi: 10.1016/s1097-2765(00)80240-3. [DOI] [PubMed] [Google Scholar]
- 18.Audigier S, Guiramand J, Prado-Lourenco L, Conte C, Gonzalez-Herrera IG, Cohen-Solal C, Recasens M, Prats AC. Potent activation of FGF-2 IRES-dependent mechanism of translation during brain development. RNA. 2008;14:1852–1864. doi: 10.1261/rna.790608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conte C, Riant E, Toutain C, Pujol F, Arnal JF, Lenfant F, Prats AC. FGF2 translationally induced by hypoxia is involved in negative and positive feedback loops with HIF-1alpha. PLoS ONE. 2008;3 doi: 10.1371/journal.pone.0003078. e3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gonzalez-Herrera IG, Prado-Lourenco L, Pileur F, Conte C, Morin A, Cabon F, Prats H, Vagner S, Bayard F, Audigier S, et al. Testosterone regulates FGF-2 expression during testis maturation by an IRES-dependent translational mechanism. FASEB J. 2006;20:476–478. doi: 10.1096/fj.04-3314fje. [DOI] [PubMed] [Google Scholar]
- 21.Teshima-Kondo S, Kondo K, Prado-Lourenco L, Gonzalez-Herrera IG, Rokutan K, Bayard F, Arnal JF, Prats AC. Hyperglycemia upregulates translation of the fibroblast growth factor 2 mRNA in mouse aorta via internal ribosome entry site. FASEB J. 2004;18:1583–1585. doi: 10.1096/fj.03-1118fje. [DOI] [PubMed] [Google Scholar]
- 22.Martineau Y, Le Bec C, Monbrun L, Allo V, Chiu IM, Danos O, Moine H, Prats H, Prats AC. Internal ribosome entry site structural motifs conserved among mammalian fibroblast growth factor 1 alternatively spliced mRNAs. Mol. Cell Biol. 2004;24:7622–7635. doi: 10.1128/MCB.24.17.7622-7635.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sullivan DE, Storch TG. Tissue- and development-specific expression of HBGF-1 mRNA. Biochim Biophys Acta. 1991;1090:17–21. doi: 10.1016/0167-4781(91)90031-g. [DOI] [PubMed] [Google Scholar]
- 24.Creancier L, Morello D, Mercier P, Prats AC. Fibroblast growth factor 2 internal ribosome entry site (IRES) activity ex vivo and in transgenic mice reveals a stringent tissue-specific regulation. J. Cell Biol. 2000;150:275–281. doi: 10.1083/jcb.150.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan Z, Choi S, Liu X, Zhang M, Schageman JJ, Lee SY, Hart R, Lin L, Thurmond FA, Williams RS. Highly coordinated gene regulation in mouse skeletal muscle regeneration. J. Biol. Chem. 2003;278:8826–8836. doi: 10.1074/jbc.M209879200. [DOI] [PubMed] [Google Scholar]
- 26.Delluc-Clavieres A, Le Bec C, Van den Berghe L, Conte C, Allo V, Danos O, Prats AC. Efficient gene transfer in skeletal muscle with AAV-derived bicistronic vector using the FGF-1 IRES. Gene Ther. 2008;15:1090–1098. doi: 10.1038/gt.2008.49. [DOI] [PubMed] [Google Scholar]
- 27.Charge SB, Rudnicki MA. Cellular and molecular regulation of muscle regeneration. Physiol. Rev. 2004;84:209–238. doi: 10.1152/physrev.00019.2003. [DOI] [PubMed] [Google Scholar]
- 28.Yaffe D, Saxel O. Serial passaging and differentiation of myogenic cells isolated from dystrophic mouse muscle. Nature. 1977;270:725–727. doi: 10.1038/270725a0. [DOI] [PubMed] [Google Scholar]
- 29.Elia A, Constantinou C, Clemens MJ. Effects of protein phosphorylation on ubiquitination and stability of the translational inhibitor protein 4E-BP1. Oncogene. 2008;27:811–822. doi: 10.1038/sj.onc.1210678. [DOI] [PubMed] [Google Scholar]
- 30.Oulhen N, Boulben S, Bidinosti M, Morales J, Cormier P, Cosson B. A variant mimicking hyperphosphorylated 4E-BP inhibits protein synthesis in a sea urchin cell-free, cap-dependent translation system. PLoS ONE. 2009;4 doi: 10.1371/journal.pone.0005070. e5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Husmann I, Soulet L, Gautron J, Martelly I, Barritault D. Growth factors in skeletal muscle regeneration. Cytokine Growth Factor Rev. 1996;7:249–258. doi: 10.1016/s1359-6101(96)00029-9. [DOI] [PubMed] [Google Scholar]
- 32.Scata KA, Bernard DW, Fox J, Swain JL. FGF receptor availability regulates skeletal myogenesis. Exp. Cell Res. 1999;250:10–21. doi: 10.1006/excr.1999.4506. [DOI] [PubMed] [Google Scholar]
- 33.Dusterhoft S, Pette D. Evidence that acidic fibroblast growth factor promotes maturation of rat satellite-cell-derived myotubes in vitro. Differentiation. 1999;65:161–169. doi: 10.1046/j.1432-0436.1999.6530161.x. [DOI] [PubMed] [Google Scholar]
- 34.Zhao P, Caretti G, Mitchell S, McKeehan WL, Boskey AL, Pachman LM, Sartorelli V, Hoffman EP. Fgfr4 is required for effective muscle regeneration in vivo. Delineation of a MyoD-Tead2-Fgfr4 transcriptional pathway. J. Biol. Chem. 2006;281:429–438. doi: 10.1074/jbc.M507440200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sachs AB. Cell cycle-dependent translation initiation: IRES elements prevail. Cell. 2000;101:243–245. doi: 10.1016/s0092-8674(00)80834-x. [DOI] [PubMed] [Google Scholar]
- 36.Cormier P, Pyronnet S, Salaun P, Mulner-Lorillon O, Sonenberg N. Cap-dependent translation and control of the cell cycle. Prog. Cell Cycle Res. 2003;5:469–475. [PubMed] [Google Scholar]
- 37.Bernstein J, Shefler I, Elroy-Stein O. The translational repression mediated by the platelet-derived growth factor 2/c-sis mRNA leader is relieved during megakaryocytic differentiation. J. Biol. Chem. 1995;270:10559–10565. doi: 10.1074/jbc.270.18.10559. [DOI] [PubMed] [Google Scholar]
- 38.Bernstein J, Sella O, Le SY, Elroy-Stein O. PDGF2/c-sis mRNA leader contains a differentiation-linked internal ribosomal entry site (D-IRES) J. Biol. Chem. 1997;272:9356–9362. doi: 10.1074/jbc.272.14.9356. [DOI] [PubMed] [Google Scholar]
- 39.Ye X, Fong P, Iizuka N, Choate D, Cavener DR. Ultrabithorax and Antennapedia 5′ untranslated regions promote developmentally regulated internal translation initiation. Mol. Cell Biol. 1997;17:1714–1721. doi: 10.1128/mcb.17.3.1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pozner A, Goldenberg D, Negreanu V, Le SY, Elroy-Stein O, Levanon D, Groner Y. Transcription-coupled translation control of AML1/RUNX1 is mediated by cap- and internal ribosome entry site-dependent mechanisms. Mol. Cell Biol. 2000;20:2297–2307. doi: 10.1128/mcb.20.7.2297-2307.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miura P, Thompson J, Chakkalakal JV, Holcik M, Jasmin BJ. The utrophin A 5′-untranslated region confers internal ribosome entry site-mediated translational control during regeneration of skeletal muscle fibers. J. Biol. Chem. 2005;280:32997–33005. doi: 10.1074/jbc.M503994200. [DOI] [PubMed] [Google Scholar]
- 42.Chotani MA, Chiu IM. Differential regulation of human fibroblast growth factor 1 transcripts provides a distinct mechanism of cell-specific growth factor expression. Cell Growth Differ. 1997;8:999–1013. [PubMed] [Google Scholar]
- 43.Marr MT, 2nd, D'Alessio JA, Puig O, Tjian R. IRES-mediated functional coupling of transcription and translation amplifies insulin receptor feedback. Genes Dev. 2007;21:175–183. doi: 10.1101/gad.1506407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stoneley M, Subkhankulova T, Le Quesne JP, Coldwell MJ, Jopling CL, Belsham GJ, Willis AE. Analysis of the c-myc IRES; a potential role for cell-type specific trans-acting factors and the nuclear compartment. Nucleic Acids Res. 2000;28:687–694. doi: 10.1093/nar/28.3.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 46.Stoneley M, Willis AE. Cellular internal ribosome entry segments: structures, trans-acting factors and regulation of gene expression. Oncogene. 2004;23:3200–3207. doi: 10.1038/sj.onc.1207551. [DOI] [PubMed] [Google Scholar]
- 47.Komar AA, Hatzoglou M. Internal ribosome entry sites in cellular mRNAs: The mystery of their existence. J. Biol. Chem. 2005;280:23425–23428. doi: 10.1074/jbc.R400041200. [DOI] [PubMed] [Google Scholar]
- 48.Michelotti EF, Michelotti GA, Aronsohn AI, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol. Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomonaga T, Levens D. Activating transcription from single stranded DNA. Proc. Natl Acad. Sci. USA. 1996;93:5830–5835. doi: 10.1073/pnas.93.12.5830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Evans JR, Mitchell SA, Spriggs KA, Ostrowski J, Bomsztyk K, Ostarek D, Willis AE. Members of the poly (rC) binding protein family stimulate the activity of the c-myc internal ribosome entry segment in vitro and in vivo. Oncogene. 2003;22:8012–8020. doi: 10.1038/sj.onc.1206645. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.