Abstract
We have screened peptoid microarrays to identify specific ligands for the RNA hairpin precursor of miR-21, a microRNA involved in cancer and heart disease. Microarrays were printed by spotting a library of 7680 N-substituted oligoglycines (peptoids) onto glass slides. Two compounds on the array specifically bind RNA having the sequence and predicted secondary structure of the miR-21 precursor hairpin and have specific affinity for the target in solution. Their binding induces a conformational change around the hairpin loop, and the most specific compound recognizes the loop sequence and a bulged uridine in the proximal duplex. Functional groups contributing affinity and specificity were identified, and by varying a critical methylpyridine group, a compound with a dissociation constant of 1.9 μM for the miR-21 precursor hairpin and a 20-fold discrimination against a closely-related hairpin was created. This work describes a systematic approach to discovery of ligands for specific pre-defined novel RNA structures. It demonstrates discovery of new ligands for an RNA for which no specific lead compounds were previously known by screening a microarray of small molecules.
INTRODUCTION
The discovery of microRNAs (miRNAs) and their essential role in translational regulation has brought to light a promising new class of RNA therapeutic targets. MiRNAs are short, naturally occurring RNA molecules that regulate translation of messenger RNAs by binding to sequences in their 3′-untranslated regions (1). They are important regulators of development and cellular responses to stress (2,3). Consistent with their ubiquitous role in regulating translation, aberrant expression of specific miRNAs is often associated with disease of disregulated translation.
Cancer and cardiovascular diseases are among the disease for which this association is well established. Signature profiles of miRNA expression accompany the cancerous state, and a miRNA that is dramatically up-regulated in the expression signatures of many solid tumors is miR-21 (4). This miRNA targets tumor suppressor genes, including pro-apoptotic genes such as PDCD4 (programmed cell death 4) (5,6). By suppressing translation of pro-apoptotic genes, miR-21 contributes to an essential feature of the cancer phenotype, a diminished ability to undergo programmed cell death. Inhibition of miR-21 function renders cancer cells vulnerable to self-destruction (7). Increased expression of miR-21 is also a characteristic of the miRNA profile associated with pathological remodeling of heart tissue in response to persistent stress (8–10). This effect is characterized by increased cardiac size and re-expression of fetal genes, and miR-21 appears to regulate these responses. Thus, increased expression of miR-21 is associated with cancer and heart disease, implicating it as a potential therapeutic target in both of these conditions (11,12).
Like other miRNAs, miR-21 is produced by excision from a longer primary transcript, where it is found in the stem of a hairpin structure (1). Endonucleases cleave the hairpin from the primary transcript and remove the apical loop. Cleavage from the primary transcript by the ribonuclease Drosha is emerging as an important point at which miRNA expression is regulated. Expression of miR-21 is promoted by factors that recruit the microprocessor, the complex of Drosha and associated proteins, to its primary transcript (13). Conversely, the RNA-binding protein Lin-28 mediates diminished processing of Let-7 family pri-miRNAs by association with the loop of the precursor hairpin (14–16). This natural regulation of miRNA processing suggests a strategy for manipulating miRNA levels: binding of a small molecule within or adjacent to the hairpin loop of a miRNA precursor could interfere with its maturation.
However, discovery of small molecule ligands for specific, pre-defined RNAs remains a significant challenge. As a result, specific ligands are known for only a small number of disease-related RNAs (17). Structure-based methods have been possible in a few cases (18–21), but the lack of high-resolution structural data for most RNAs of interest favors screening of diverse chemical collections and combinatorial libraries for specific RNA-binding molecules (22–25). In the study presented here, we have screened a combinatorial library of N-substituted oligoglycines, commonly called peptoids (26), for molecules with specific affinity for the hairpin loop of the miR-21 precursor. Peptoids are peptidomimetic compounds that are cell permeable (27), protease resistant, and easily synthesized in highly diverse libraries by solid phase ‘split-pool’ synthesis (28–31).
We screened the peptoid library in a microarray format. Microarrays of surface-bound small molecules are increasingly important tools for high-throughput screening and discovery of new ligands (32). They have been used to discover new protein–small molecule interactions (33–35) and to profile the specificities of known RNA-binding antibiotics (36–38). The microarray format offers potential benefits as a means to discover new ligands. Thousands of compounds can be screened simultaneously on microarrays with low requirements for costly RNA samples, and many replicate arrays can be printed from a single chemical library, allowing the same library to be screened against multiple targets or in multiple screening trials with the same target. We have used peptoid microarrays as the foundation for a system of RNA ligand discovery in which lead compounds with specific affinity are identified by screening, and the positive, specific molecular interactions discovered are utilized to create improved ligands.
MATERIALS AND METHODS
Abbreviations
DMF: dimethylformamide; NMM: N-methyl morpholine; DIC: diisopropylcarbodiimide; HBTU: O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate; HOBT: 1-hydroxybenzotriazole; NMP: N-methylpyrrolidinone; DMSO: dimethylsulfoxide; EDTA: ethylenediaminetetraacetic acid; PBS: phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4)
General procedures
Library synthesis and all peptoid syntheses were carried out in fritted glass peptide synthesis reaction vessels (Chemglass). Matrix assisted laser desorption ionization (MALDI) mass spectrometry (Voyager DE Pro, ABI) was done using α-cyano-4-hydroxycinnamic acid as matrix. Analytical and preparative HPLC were carried out on C18 columns (Vydac), eluting with a gradient from 100% H2O to 20% acetonitrile (with constant 0.1% trifluoroacetic acid). Except where noted, all reagents were obtained from Sigma.
Synthesis of peptoid library
Polystyrene macrobeads (2.0 g; 500–560 μm; substitution: 0.56 mmol/g, Rapp Polymere) were swollen in dimethylformamide (DMF) at room temperature overnight. The beads were treated twice with 20% (v/v) piperidine in DMF at room temperature with shaking (230 r.p.m.) for 15 min (2 × 20 ml). The reaction vessel was drained and the beads were thoroughly washed with DMF (6 × 20 ml). Cysteine, glycine and arginine residues were coupled using standard manual peptide synthesis protocols. To couple the cysteine, 10 ml 200 mM Fmoc-Cys(Trt)-OH solution was added to the beads followed by 10 ml 200 mM solution of O-(benzotriazol-1-yl)-N,N,N′,N′-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxybenzotriazole (HOBT) in DMF containing 400 mM N-methyl morpholine (NMM). The reaction vessel was shaken at room temperature for 1 h. The vessel was drained and washed with DMF (6 × 20 ml). It was treated twice with 20% piperidine in DMF for 15 min (2 × 20 ml), drained and washed thoroughly with DMF. The same protocol was repeated to couple the two glycine residues and an arginine residue using Fmoc-Gly-OH and Fmoc-Arg(Pbf)-OH. A small portion of the beads was set aside after coupling arginine for spotting of control features following deprotection and cleavage from the support.
Combinatorial peptoid synthesis was performed on these beads following deprotection of the terminal amine by treatment with 20% piperidine in DMF. The beads were distributed equally into 21 peptide synthesis reaction vessels. Monomer addition was carried out in two steps (29). In step 1, acylation was carried out using chloroacetic acid and di-isopropylcarbodiimide (DIC). To each vessel was added 1.5 ml of 2 M chloroacetic acid in anhydrous DMF and 1.5 ml 2M DIC in anhydrous DMF. The reaction vessels were shaken at 37°C for 10 min. The vessels were drained, and the beads were washed with DMF (6 × 5 ml). In step 2, displacement of the chloride by the primary amine was carried out. The beads in each of the vessels were treated with 1 of 21 primary amines at 2 M concentrations in N-methylpyrrolidinone (NMP) at 37°C with shaking. The amines used were tert-butyl N-(4-aminobutyl)carbamate (TCI America), 1-amino-2-methylpropane, 3-amino-1-propene, glycine-t-butyl ester, 2-aminomethylfuran, 2-(aminomethyl)tetrahydofuran, O-t-butyl-2-aminoethanol (CSPS Pharmaceuticals, San Diego, USA), 3-(2-aminoethyl)indole, R-(+)-1-phenylethylamine, 2,4-dimethoxybenzylamine, 2,2-diphenylethylamine, 2-(1-cyclohexenyl)ethylamine, 4-aminomethyl-boc-piperidine (Fluka), cyclobutylamine, 3,4-(methylenedioxy)benzylamine (Acros), 2-(4-imidazolyl)ethylamine, 4-(2-aminoethyl)phenol, 4-aminoethylmorpholine, 4-aminomethylpyridine, 1-(3-aminopropyl)-2-pyrrolidinone and 2-(2-aminoethylamino)-5-nitropyridine. These amines are illustrated in their unprotected forms in Figure 1B. The vessels were drained and washed thoroughly with DMF (6 × 5 ml). A small portion of the beads were set aside after the first cycle of peptoid coupling to spot as control features on the arrays. The beads in each of the reaction vessels were pooled into a 250 ml peptide synthesis vessel, drained, suspended in 50 ml dichloromethane/DMF (2:1) and thoroughly mixed. The beads were then distributed equally into 21 reaction vessels and the acylation and displacement procedures were repeated for two more monomer residues. After completion of the library synthesis, the beads were washed with dichloromethane (5 × 20 ml) and finally with absolute ethanol (5 × 20 ml). The beads were manually sorted into 96-well polystyrene plates, one bead per well. To remove side chain protective groups from amine, carboxyl and hydroxyl side chains and to cleave the library elements from the beads, 30 μl of a cleavage cocktail consisting of 47.5% TFA, 47.5% dichloroethane, 2.5% tri-isopropylsilane and 2.5% water was added to each well. The plates were sealed and incubated at room temperature for 6 h, after which the cleavage cocktail was allowed to evaporate in a fume hood overnight. The peptoids were dissolved in 50 μl of acetonitrile/water (1:1) and transferred quantitatively into a duplicate set of 384-well polystyrene plates. Solvent was evaporated from one set of plates and the residue in each well was dissolved in 50 µl dimethylsulfoxide (DMSO). The remaining set of plates was reserved for mass spectral analysis of ‘hit’ compounds.
Figure 1.
Composition of peptoid microarrays. (A) Spots included a combinatorial library of peptoids with three variable peptoid monomers followed by an arginine and a linker. Control spots included linker only and linker with one peptoid monomer. Each spot contained one compound. (B) Amines used in library synthesis, shown with the substituents as they are after deprotection to illustrate the composition of the final library. The amine groups shown in blue are incorporated into the amide backbone during peptoid synthesis.
Library characterization
Seventeen individual library elements (i.e. products from individual beads) were randomly chosen from the peptoid library and analyzed by MALDI mass spectrometry. The mass spectra indicated a single major component in each of the products. Each of the selected library elements was sequenced by MS/MS. Of the 21, 16 possible side chains were represented at least once, with the most common frequency of occurrence being two (nine of the side chains). The highest frequency of occurrence in this sample is seven times, for one side chain. This distribution is consistent with the random incorporation of submonomers expected from split-pool synthesis.
Printing of microarrays
Microarrays were printed on 1″ × 3″ glass microscope slides. Prior to spotting, the slides were cleaned and coated with 3-glycidoxypropyltrimethoxysilane and polyethylene glycol as described by Maskos and Southern (39). One face of each polyethylene glycol functionalized slide was treated with 20 mM N-(p-maleimidophenyl) isocyanate (Pierce) in dry DMSO in a custom-made reaction chamber (35). After overnight incubation at room temperature, the slides were washed several times with DMSO and acetonitrile. Cysteine-terminated peptoids dissolved in DMSO were printed to these slides at a density of 500 spots/cm2 with an ArrayIT robotic printer (Telechem International, Inc.). After spotting, the slides were left on the printer at room temperature for 16 h. To block unreacted maleimide groups, the slides were then immersed in a 1% (v/v) solution of 2-mercaptoethanol in DMF for 2 h. All the slides were then washed in DMF, acetonitrile and iso-propanol, dried and stored in darkness under vacuum until used.
Large scale synthesis of peptoids
Synthesis was performed on 100 mg Knorr Amide MBHA resin (substitution: 0.78 mmol/g; Nova Biochem). The beads were swollen in DMF for 2 h, drained, treated twice with 20% piperidine in DMF for 15 min (2 × 5 ml) and washed thoroughly with DMF (6 × 5 ml). Two milliliters of 200 mM solution of Fmoc-gly-OH in DMF was added followed by 2 ml of 200 mM solution of HBTU/HOBT in DMF containing 400 mM NMM. The reaction vessel was shaken at room temperature for 1 h. The beads were washed with DMF (6 × 5.0 ml). The beads were treated twice with 20% piperidine in DMF for 15 min (2 × 5 ml) drained and washed with DMF (6 × 5.0 ml). Another glycine residue and an arginine residue were coupled in the same manner. Two milliliters of a 2M solution of the chloroacetic acid in anhydrous DMF was added to the resin-bound amine at 37°C followed by 2 ml of 2.0 M DIC in anhydrous DMF. The resin was mixed for 10 min, drained and then washed with DMF (6 × 2.0 ml). The chloride was displaced by addition of 2 ml of a 2.0 M solution of the furfuryl amine in NMP, allowed to react for 60 min at 37°C. The resin was drained and then washed with DMF (6 × 2.0 ml). The monomer addition steps were repeated for O-t-butyl-2-aminoethanol and 4-(aminomethyl) pyridine. The beads were washed with dichloromethane (5 × 5 ml). The peptoid was released from the resin with concomitant removal of the protecting groups by treating the beads with 5 ml of 95% TFA, 2.5% tri-isopropylsilane and 2.5% water. The cleavage cocktail was concentrated by blowing argon over the solution. The concentrated filtrate was dissolved in 5 ml of 1:1 acetonitrile/water and lyophilized. The residue was purified by HPLC.
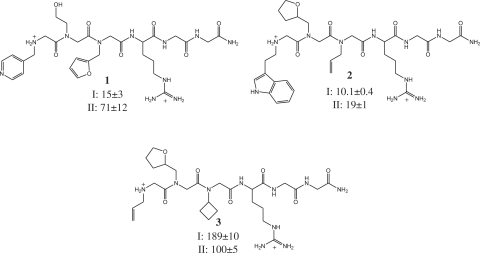
Compound 2 was synthesized in a similar manner using allyl amine, tetrahydofurfuryl amine and tryptamine in the chloride displacement steps, and 3 was synthesized in a similar manner using cyclobutylamine, tetrahydrofurfurylamine amine and allylamine. Methyl side chains were incorporated into derivatives of 1 by using a 2 M solution of methylamine in THF in the chloride displacement step of peptoid synthesis.
General RNA sample preparation
RNA samples were obtained purified from Dharmacon with the 2′-ACE protective groups left in place. Protective groups were removed immediately prior to use according to manufacturer's protocol. The deprotected RNA was precipitated with ethanol and sodium acetate prior to use.
Tm measurement by UV-monitored thermal denaturation
RNA melting points were measured with a Cary 100Bio UV spectrophotometer (Varian). RNA samples were dissolved in phosphate buffer saline (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 2 mM KH2PO4, pH 7.4) (PBS) buffer and overlayed with mineral oil in 1 cm path length quartz cells. Absorbance at 260 nm was measured as the temperature varied from 14 to 95°C at a rate of 2.0°C/min. Melting points were determined from the first derivative of the melting curves. Each temperature ramp was repeated three times and the mean and SD of the replicates is reported.
Array probing with RNA
Samples for screening and electrophoretic analysis were obtained with the Dy547 label (Fluorescence properties similar to Cy3) at the 5′-end. Deprotected RNA precipitates were re-dissolved in PBS to a concentration of 5 µM. The resulting solution was heated briefly to 95°C, then allowed to cool slowly to room temperature. The cooled solution (80 µl) was applied to an array and covered with a glass coverslip. After 1 h, the array was washed four times briefly with PBS followed by a brief wash in deionized water and centrifugal drying. The array was scanned on a GenePix 4100A scanner (Axon Instruments) with laser excitation at 532 nm and a 550–600 nm emission filter (Cy3 emission maximum at 570 nm). Results were analyzed with GenePix Pro software. Each RNA sample was applied to three replicate arrays. To be considered positive, an array feature (spot) was required to have fluorescence signal above background in each replicate treated with RNA I, and features with a high fluorescence prior to application of RNA were disregarded. Features with fluorescence signal above background in any one of three replicates with control RNA II were disregarded. The two positive hit compounds and a negative control compound were identified by MS/MS analysis (ABI 4700 proteomics analyzer) of the stock solutions of the library members spotted to the positive features.
Fluorescence binding assay
RNA samples with 2-aminopurine (2-ap) substituted for adenine were deprotected according to vendor protocol, precipitated with ethanol and sodium acetate, and dissolved in PBS to a concentration of 5 µM. The resulting solution was heated briefly to 95°C, then allowed to cool slowly to 25°C. The RNA was diluted to 1 µM with the assay buffer: PBS, 10 mM KCl, 50 mM Tris, pH 8.5, 1 mM MgCl2 (same as in-line cleavage conditions) or 10 mM potassium phosphate, pH 7.2, 140 mM potassium acetate and 2 mM MgCl2. Ligand solutions were prepared as serial dilutions in assay buffer, and equal volumes of ligand and RNA were mixed prior to delivery to wells of a standard black 96-well plate. The final RNA concentration in each well was 0.5 µM. Background fluorescence from the ligand was assessed in wells containing ligand at each final concentration in the absence of RNA. Fluorescence was measured on a SpectraMax M5 plate reader (Molecular Devices), with excitation at 320 nm, emission measured at 390 nm and a 325 nm cut-off filter. Equilibrium was confirmed by comparison of data sets separated by 30 min. The dissociation constant was determined by fitting a sigmoidal dose–response curve to the mean fluorescence of triplicate measurements after subtraction of background fluorescence due to ligand. The error bars on graphs represent 1 SD from the mean, in many cases smaller than the data marker. Three independent determinations of each dissociation constant were made, and the mean and standard deviation are reported.
Isothermal titration calorimetry
Isothermal titration calorimetry (ITC) measurements were made at 25°C with a MicroCal VP-ITC (MicroCal, Inc.). A 1 mM solution of compound 2 in PBS was injected in 8 µl aliquots into the sample cell containing 1.4 ml of 70 µM RNA solutions in PBS or buffer alone. Injection was over 16 s, with 240 s between injections. The area under the curve of microcalories per second versus seconds was integrated for each injection (using Origin version 7.0 software, MicroCal, Inc.) to derive the heat associated with each injection. The heat associated with ligand solvation, measured by injection of ligand into buffer alone, was subtracted from the corresponding heat from injection of the ligand into the RNA solution to determine the heat due to association of the ligand with the RNA. A single-site binding model was used to fit the binding curves from which thermodynamic parameters were obtained.
Analysis of Mg2+-induced RNA cleavage
The precipitate of deprotected RNA (5′-labeled with Dy547) was dissolved in 0.1X PBS to a final concentration of 15 µM, then heated briefly to 95°C and allowed to cool slowly (over ∼2 h) to room temperature. The resulting RNA was incubated, at a final concentration of 3 µM, in 10 mM KCl, 50 mM Tris, pH 8.5. MgCl2 and peptoid were added to specified concentrations as stock solutions. Each reaction was done in triplicate. Reaction mixtures were incubated at room temperature in darkness for 10 days, then precipitated with ethanol, sodium acetate and 5 µg of yeast phenylalanine tRNA. Alkaline hydrolysis was carried out in 10 mM NaHCO3, pH 9.0, at 95°C for 4 min. Ribonuclease T1 digestion was carried out with 0.007 U of ribonuclease T1 (Ambion) in 20 mM Tris, pH 7.15, 50 mM NaCl, 0.1 mM MgCl2, at room temperature for 20 min. The reaction was stopped by addition of 0.2 volumes of 5 mM ethylenediaminetetraacetic acid (EDTA) and precipitation with ethanol, sodium acetate and 5 µg of yeast phenylalanine tRNA. The precipitates were heated briefly at 95°C in formamide prior to analysis by electrophoresis on a polyacrylamide gel (20%, 19:1 crosslinking, 8 M urea). The gel was visualized by fluorescence (532 nm excitation) with a Typhoon 9200 scanner (Amersham Biosciences) and analyzed with ImageQuant software. Fraction cleavage at specific positions was calculated from the background-corrected fluorescence in the band associated with that position divided by the background-corrected total fluorescence in the gel lane. Triplicate measurements were averaged and the dissociation constant calculated by fitting a sigmoidal dose–response curve to the data using GraphPad Prism.
RESULTS
Microarray synthesis and screening
Microarrays were created from a combinatorial library of peptoids (Figure 1A). The library was constructed from a chemically diverse set of 21 monomers (Figure 1B). An arginine was present at the C-terminal side of each peptoid trimer in the library. Arginine binds with low affinity (Kd > 2 mM) to many RNA molecules, including HIV TAR RNA (40) and group I introns (41), and was included to provide a ‘toe-hold’ for additional specific binding interactions. A linker comprising two glycines and ending in a cysteine enabled covalent attachment of the library elements to maleimide-derivatized glass slides (35,42). The library was robotically spotted onto microarrays, each with a total of 8736 spots.
The arrays were screened to identify ligands specific for an RNA hairpin corresponding to the primary transcript of miR-21 (Figure 2A, RNA I). MiR-21hp with a fluorescent label at its 5′-end was applied to a peptoid array, and after washing, spots to which it bound were identified. Features with a ratio of signal to local background (SNR) ≥1.2, the threshold for visibly discernable fluorescence on the scanned image, were considered to have signal above background. To be considered positive, an array feature was required to have fluorescence signal above background in each of three replicate arrays treated with labeled RNA I. The requirement for reproducibility on three arrays was imposed to minimize false positives due to washing artifacts (e.g. water spots) or other irregularities on the array (e.g. scratches). None of the control features (arginine and linker only) or features with a single peptoid residue had signal above background. 891 of the library features (12%) were fluorescent above background. The SNR of these features ranged from 1.2 to 468, with a mean SNR of 7.9 and a median value of 2.2.
Figure 2.
Screening peptoid microarrays with labeled RNA. (A) RNAs used to probe the arrays. A fluorescent dye moiety, Dy547, was appended to the 5′-end of each RNA for microarray screening. The adenosines in large blue type were replaced with 2-ap in subsequent experiments to determine dissociation constants by fluorescence. RNA I (miR-21hp) corresponds to the hairpin from the primary transcript of miR-21. RNA II (control) was used as a control for specificity. The secondary structures shown for RNAs I and II are predicted by the mfold program (54), and the melting temperatures (Tm) shown are invariant between 1 and 10 µM, confirming unimolecular structures. (B–E) Fluorescence scans of microarray regions containing lead compounds. A grid of circles is overlayed on the scan to indicate the pattern of spotting. The grid was aligned with fluorescent spots created by printing a fluorescein-labeled peptoid at known positions. Each circle corresponds to an area with diameter of 270 µm. (B) and (C) Region of peptoid microarray containing the feature (indicated by the arrow) corresponding to 1 after treatment with (B) labeled RNA I or (C) labeled RNA II. (D) and (E) Region of the peptoid microarray containing the feature (indicated by the arrow) corresponding to compound 2 after treatment with (D) labeled RNA I or (E) labeled RNA II.
A dye-labeled control hairpin (Figure 2A, RNA II) was tested to eliminate from consideration peptoids with a high nonspecific affinity for RNA hairpins or for the dye molecule and to narrow the search for peptoids that recognize distinctive features of the miR-21 precursor hairpin. This control hairpin was similar to the target hairpin, but differed from it in several respects to allow for the possibility of recognizing various features of the target. The most distinctive features of the target RNA's predicted structure were judged to be the adjacent G•U base pairs, the bulged U, and the loop sequence. Therefore, in the control hairpin, the G•U pairs were changed to G•C pairs, the bulged U was omitted and the loop sequence was scrambled, maintaining the base composition but changing the sequence order.
Most of the 891 features that were fluorescent above background after treatment with hairpin I were also fluorescent on one or more of three replicate arrays treated with RNA II. Examples of such features can be seen in Figure 2B–E, identifiable as fluorescent features not indicated with an arrow. These features were considered to be nonspecific in their affinity for the labeled RNA, though some might have specificity for the target that was not distinguished at the level of stringency of the binding and washing conditions used. Sequencing some of these nominally nonspecific compounds by MS/MS revealed them to be rich in monomers h and i (Figure 1B).
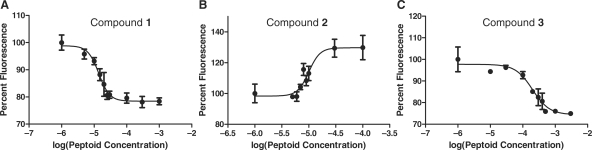
Two spots were identified that reproducibly had specific affinity for miR-21hp (Figure 2). The SNR of the feature indicated by the arrow in Figure 2B and C was 1.6, 1.7 and 1.3 in three replicate experiments with RNA I (2B); whereas it was 1.1, 0.83 and 1.0 in replicate experiments with RNA II (2C). The SNR of the feature indicated by the arrow in Figure 2D and E was 8.3, 3.7 and 14 in three replicate experiments with RNA I (2D); whereas it was 0.9, 0.9 and 1.0 in replicate experiments with RNA II (2E). These features were revealed by mass spectral analysis to be compounds 1 and 2 (Figure 3). Compound 3, identified from a spot to which neither RNA bound, was made and tested in solution binding assays for comparison. Binding of ligands to the RNA hairpins in solution was assessed using the base analogue 2-ap as a fluorescent probe (43,44). The fluorescent base was substituted for the adenine indicated in Figure 2, and the peptoid-dependent change in fluorescence was used to determine the dissociation constant (Figure 4). Association of peptoids with the RNA typically results in a 20–30% decrease in 2-ap fluorescence. In one case, the complex of miR-21hp (RNA I) with compound 2, binding of the peptoid results in an increase in fluorescence by 30% (Figure 4B). No change in fluorescence is observed with peptoid concentrations up to 1 mM when 2-ap is substituted for A7 (data not shown). Dissociation constants determined by this method are shown in Figure 3. Binding of compounds 1 and 2 is indeed specific to RNA I. As anticipated, 3 does not have specific affinity for RNA I. It has highest affinity for RNA II and relatively low affinity for both test RNAs. The affinities of 1 and 2 for RNA I are similar, but 1 has higher specificity for the target RNA.
Figure 3.
Compounds identified in microarray screen. Compound 1 corresponds to the indicated array feature in Figure 2B. Compound 2 corresponds to the indicated array feature in Figure 2D. Compound 3 was identified from an array feature to which neither RNA bound. Solution-phase dissociation constants (µM) measured by monitoring 2-ap fluorescence are shown for RNAs I and II. Reported dissociation constants are the mean ± SD for three independent determinations.
Figure 4.
Representative binding curves of RNA I with compounds identified in the microarray screen. Fluorescence of 2-ap, substituted at the position indicated in bold in Figure 2, was monitored as a function of peptoid concentration. Each point is the mean of three measurements, and error bars represent the standard deviation in the mean. The data were fit to a logistic dose–response curve from which the dissociation constant was determined. (A) Compound 1 (B) Compound 2 (C) Compound 3.
Fluorescence of 2-ap is commonly used to measure affinities of ligands for nucleic acids, but this technique relies on the assumption that the substitution does not perturb the binding interaction. To test this assumption for the system of this study, we have compared the results of this assay with dissociation constants determined by ITC (see Supplementary Data) and probing with Mg2+-induced hydrolytic cleavage (see below and Supplementary Data). The agreement of these methods confirms that substitution of 2-ap for adenine does not perturb the binding interaction.
Effects of compounds 1 and 2 on Mg2+-induced cleavage of RNA I
Effects of peptoids 1 and 2 on Mg2+-induced hydrolytic cleavage of RNA I were used to characterize their binding (Figure 5). Hydrolytic cleavage of the RNA backbone occurs principally through nucleophilic attack of a 2′-hydroxyl on the adjacent phosphodiester, displacing the 5′-hydroxyl of the following nucleotide. For this displacement to occur, the attacking 2′-hydroxyl must be in-line with the scissile phosphorus–oxygen bond. Thus, in-line cleavage is a useful probe of conformational changes such as those that are frequently induced by ligand binding (45). The reaction is stimulated by divalent metal ions such as Mg2+.
Figure 5.
Analysis of peptoid binding to RNA I by Mg2+-induced cleavage. (A) Fluorescent scan of polyacrylamide gel for analysis of 5′-end labeled RNA after treatment in the presence (+) or absence (−) of 1 mM MgCl2. Samples were treated with 30, 300 or 3000 µM 1 or 2 as indicated. OH and T1 are alkaline hydrolysis ladder and ribonuclease T1 digestion, respectively. (B) Cleavage patterns in the presence and absence of peptoid. The lengths of the arrows are semi-quantitative indicators of relative cleavage efficiency. (C) Fraction of total RNA I cleaved after C19 as a function of the log molar concentration of 1 and 2. Error bars, in most cases smaller than the data markers, are SDs in means of three measurements. Dissociation constants of 251 ± 52 and 164 ± 13 µM (mean ± SD for three independent determinations) were determined for binding of RNA I to 1 and 2, respectively.
In the absence of peptoid, Mg2+-induced cleavage occurs predominantly within the 3′-side of the hairpin loop and, with much lower efficiency, after U21 (Figure 5A and B). This pattern is consistent with the secondary structure predicted for the hairpin. The pattern of cleavage changes significantly when carried out in the presence of peptoids 1 and 2. With each, the most pronounced effect is an increase in cleavage after C19. Reactivity also increases after G10 and U11 and to a small extent after U16. There is a discernible decrease in reactivity after U21. These changes indicate binding of the peptoids proximal to or within the hairpin loop with concurrent alteration of the backbone conformation. The EC50 for enhancement of cleavage at C19 (Figure 5C) provides apparent dissociation constants for 1 and 2. Under the conditions of the cleavage assay (1 mM Mg2+, pH 8.5), these dissociation constants are higher than measured with 2-ap fluorescence under the screening conditions. However, dissociation constants measured by fluorescence and in-line cleavage under equivalent conditions gave equivalent binding constants (See Supplementary Data).
RNA determinants of compound 1 affinity
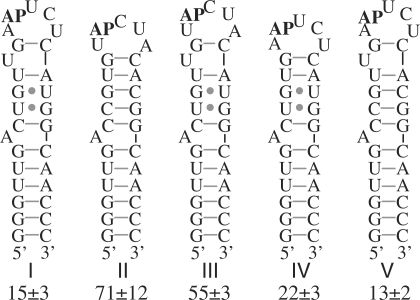
Further characterization was focused on compound 1, because it has higher specificity for the target RNA than 2. Dissociation constants of 1 for RNAs III, IV and V were measured to dissect the contributions of the differences between RNAs I and II to the difference in their affinities (Figure 6). The largest effect on affinity (3- to 4-fold) resulted from scrambling the loop sequence. The putatively bulged U apparently plays a small but measurable role in recognition, its omission increasing the dissociation constant by a factor of ∼1.5. No apparent effect on affinity results from changing the G•U pairs to G•C pairs. The recognition of both the loop sequence and the bulge illustrates the rationale for varying several features of the RNA hairpin in the design of the control hairpin used for screening.
Figure 6.
Affinities of 1 for RNA hairpins related to RNA I. Dissociation constants (µM) were measured by monitoring fluorescence of 2-ap substituted at the position indicated (AP). Means of three determinations are reported ± SD.
Contributions of individual monomers to affinity and specificity of compound 1
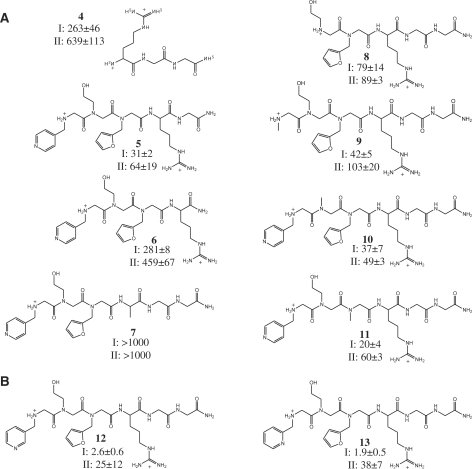
To analyze the contributions made by different parts of compound 1 to affinity and specificity, we measured affinities of derivatives that were truncated or in which a side chain was replaced with a methyl group (Figure 7A). The arg–gly–gly peptide alone has low but measurable affinity, with some specificity for the targeted hairpin (compound 4). As expected, the positively charged side chain of the arginine is required for binding (compound 7). Unexpectedly, the glycines included as a linker to the array surface, especially the glycine adjacent to the arginine, contribute to the affinity and specificity for RNA I (compounds 5 and 6). Of the variable components of the library, the amino-terminal residue and its pyridine side chain are clearly important contributors to affinity and specificity (compounds 8 and 9). Replacement of the hydroxyethyl side chain with a methyl group (compound 10) also significantly lowers affinity, indicating its involvement in binding. The furfuryl side chain is of lower significance for binding, its replacement with a methyl group (compound 11) having little or no effect on binding.
Figure 7.
Affinities for derivatives of 1 for RNAs I and II. Dissociation constants (µM) were determined using 2-ap fluorescence and are reported as mean ± SD for three independent measurements. (A) Compounds used to characterize contributions of functional groups of 1 to affinity and specificity. (B) Affinities of methyl pyridine positional isomers 12 and 13 for RNAs I and II.
Improved affinity and specificity by conservative modification of initial lead compound
A benefit of the peptoid scaffold for ligand development is the ease with which variations can be made toward optimizing lead compounds. Once elements of a lead compound that contribute specific positive interactions are identified, the approach of incrementally varying those elements to improve the interactions can be applied readily. In a first application of this approach to lead compound 1, we focused on the pyridine side chain at the N-terminus, which appeared to be partly responsible for specific affinity. Variants were synthesized in which the 4-substituted pyridine was replaced by 2- and 3-substituted pyridines (Figure 7, compounds 12 and 13, respectively). Each of these derivatives had higher affinity for the target RNA and higher specificity against binding to the control hairpin than the parent lead. The best of these compounds was 13, with a dissociation constant of 1.9 µM and an ∼20-fold discrimination against binding the control RNA. The affinity of this compound for the target and control RNAs was also measured in a buffer containing 2 mM Mg2+ (10 mM potassium phosphate, pH 7.2, 140 mM potassium acetate and 2 mM MgCl2), to approximate a biological background of divalent cations. Under these conditions, the Kd increased somewhat, as expected, to 8.0 ± 2.8 µM, but the discrimination against the control RNA (II) remained at ∼20-fold, with the Kd for that RNA being 156 ± 33 µM.
DISCUSSION
This work demonstrates the utility of peptoid microarrays for discovering new ligands for specific RNAs. Previous work has employed the microarray format as a quantitative analytical tool to profile the relative affinities and specificities of known RNA-binding antibiotics (36–38) and to find improvements to peptide sequences that bind to an RNA required for packaging HIV (46). Recently, Labuda and co-workers reported a screen of peptoids in a microarray format to find improved inhibitors of a group I intron (47). These efforts used libraries of 4–120 different RNA-binding compounds. To discover new ligands for a novel, pre-defined RNA target, we have used the microarray platform as a qualitative tool to screen a relatively large library of 7680 different compounds. As a qualitative screen, it is akin to bead-based library screens, in which a library of compounds on beads is screened for binding to a fluorescently labeled biomolecule, and hit compounds are identified by visual brightness of individual beads with the use of a fluorescent microscope (25,48). However, having the library elements arrayed on a surface allows systematic consideration of each, which is difficult in the bead-based format. Also, the ability to perform replicate experiments allows confirmation of reproducibility and specificity prior to hit identification and solution-phase testing.
The miR-21 precursor hairpin is a novel target, without previously known small molecule ligands, and very few sequence-specific ligands are known for any RNA hairpin loop. Thus, we had no a priori expectations for the affinities that library compounds with specificity for the target RNA would have. For this initial test of the approach, our goal was to discover positive and specific molecular interactions that can be subsequently utilized toward making ligands of high affinity and specificity. We chose the peptoid scaffold for our library, in part, because these subsequent steps are facilitated by the ease with which peptoids can be modified. We attempted to bias our library toward a baseline affinity of Kd ∼1–10 mM by including arginine in every library element. The affinity of the diglycine linker was fortuitous, since we had no prior expectation that it would contribute to binding. Together, the arginine and glycine components of the library contribute 4.9 kcal/mol (calculated from the Kd) to the free energy of association of the library elements to the target RNA, with a weak specificity for the target RNA over the control.
Most of the array features gave no indication of binding the labeled target RNA, but 12% of them were fluorescent after its application, some with SNR greater than 100. Most of these features displayed similar signal to noise with the control hairpin. The compounds rich in alkyl amines that were predominant among the peptoids with high signal to noise are likely to have high affinity for the RNA because the amines are substantially protonated, creating an electrostatic attraction for the polyanionic RNA. While some of these compounds may have specificity for the target RNA that could be revealed by screening under more stringent conditions and by rigorous quantitation of the fluorescent signal, a large nonspecific component of binding is likely to interfere with optimization of the specific interactions. Therefore, we focused our attention on hit compounds that reproducibly showed no interaction with the control hairpin.
Both of the compounds to which this requirement narrowed our focus had affinity for the target RNA in solution 15- to 20-fold higher than the affinity of a compound that did not bind the labeled target on the array (compound 3). The affinity of compound 3 is comparable to that of the arginine and linker alone (4). Thus, the array screen was successful in identifying compounds from the library that are better ligands than one that shows no interaction on the array. The variable peptoid components of the lead compounds identified contribute ∼20-fold to the affinity over the arginine and glycines alone, corresponding to ∼1.6 kcal/mol of additional free energy of binding. Furthermore, peptoids 1 and 2 share common features that are not found in other library elements: an aromatic heterocycle at the N-terminus and an oxygen three atomic centers from the backbone in the second position. These similarities suggest involvement of specific molecular interactions in the binding of the RNA, supporting the conclusion that the microarray screen successfully identified library elements that form positive, specific interactions with the target. Though the signal to noise ratio of compound 1, near the threshold of being visible, was significantly lower than that of compound 2, the solution phase affinities of these two compounds for the target were similar, indicating that fluorescent signal on the array is influenced by factors other than solution-phase affinity, such as linker effects, well known in the interactions of surface-bound ligands with RNA (36,49).
Though the specificities of the initial lead compounds are modest, the 4.7-fold specificity of 1 is significant in light of the similarity between the target and control hairpins and the paucity of ligands reported in the literature with appreciable sequence specificity for RNA hairpin loops. This difference is all the more significant in light of the fact that it is primarily due to the sequence order within the five-nucleotide loop, the loop sequence composition being held constant, with little dependence on other differences between the target and control hairpins. Furthermore, much of this specificity can be attributed to distinct functional elements of the ligand. The terminal pyridine and the hydroxyethyl side chain contribute the majority of the free energy of association that is specific to the target RNA.
Although these groups contribute specific interactions to the complex, they do not necessarily optimize those interactions. The potential for enhancing these contacts suggests an avenue for improvement of the lead compound, by screening a series of derivatives in which a critical group is incrementally varied. This approach was immediately productive when applied to the terminal pyridine of 1. By simply varying the position at which the pyridine is linked to the backbone, the affinity and specificity were both increased. The affinities of the resulting compounds for the target RNA are comparable to the affinity of DGCR8, the double-stranded RNA-binding component of the microprocessor complex, for primary miRNAs (50). This is true for compound 13 in the presence of physiologically relevant concentrations of mono- and divalent cations. Development of further optimized ligands by testing conservative variations of the hydroxyethyl side chain is in progress.
The process followed to discover ligand 13 illustrates a systematic approach to the development of ligands for specific, pre-defined RNA molecules. The screen of a library of potential ligands is the first step, aimed at discovery of specific interactions. The library is designed so that its members not only have desirable qualities for biologically active compounds (cell permeability, limited size, biological stability), but are also easily modified in the subsequent steps, when the structural elements of the lead compounds that contribute specific affinity are identified and varied to find improvements.
Work is underway to test the effects of the ligands found here on miRNA processing in cell culture and in cell-free miRNA processing assays and to explore the broader biological effects of these compounds. Though cleavage of miRNA precursors by Drosha does not depend on specific loop sequences, mutations that stabilize an internal conformation of the loop diminish or abolish cleavage (51,52). Thus, a ligand that recognizes, thereby stabilizing, a hairpin conformation might similarly inhibit processing. This inhibition could be realized even if the bound conformation is different from the most stable conformation in the absence of ligand. The dependence of binding on loop sequence, the binding-induced change of 2-ap fluorescence when the fluorescent probe is in the hairpin but not in the stem, and the conformational changes within the loop revealed by Mg2+-mediated cleavage indicate that the binding site for the compounds identified in this study includes the loop. A small molecule that binds to a miRNA precursor might also interfere with endogenous factors that interact with the hairpin loop to stimulate cleavage by Drosha (13) or with processing steps that follow Drosha cleavage, including removal of the loop or intracellular localization (53). In any of these cases, a compound that specifically interferes with maturation of miR-21 will be a useful tool for understanding miRNA regulation and could be of therapeutic value.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Heart, Lung, and Blood Institute (contract number N01-HV-2185); University of Texas Southwestern Division of Translational Research. Funding for open access charge: Division of Translational Research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. Reddy Moola for expert assistance with ITC. Peptoid libraries were printed by the Division of Translational Research Array Spotting Core.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Ambros V. The function of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 3.Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- 4.Volinia S, Calin G, Liu C-G, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Ferracin M, Prueitt RL, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Asangani IA, Rasheed SAK, Nikolova DA, Leupold JH, Colburn NH, Post S, Allgayer H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27:2128–2136. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 6.Zhu S, Wu H, Wu F, Nie D, Sheng S, Mo YY. MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res. 2008;18:350–359. doi: 10.1038/cr.2008.24. [DOI] [PubMed] [Google Scholar]
- 7.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 8.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. MicroRNAs are aberrantly expressed in hypertrophic heart. Am. J. Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen J-F, Newman M, Rojas M, Hammond SM, Wang Q-Z. Expression of microRNAs is dynamically regulated during cardiomyocyte hypertrophy. J. Mol. Cell Cardiol. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-reponsive microRNAs that can evoke cardiac hypertrophy and heart failure. Proc. Natl Acad. Sci. USA. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.van Rooij E, Marshall WS, Olson EN. Toward microRNA-based therapeutics for heart disease. Circ. Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–62. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heo I, Joo C, Cho J, Ha M, Han J, Kim VN. Lin28 mediates the terminal uridylation of let-7 precursor microRNA. Mol. Cell. 2008;32:276–284. doi: 10.1016/j.molcel.2008.09.014. [DOI] [PubMed] [Google Scholar]
- 15.Newman MA, Thomson JM, Hammond SM. Lin-28 interaction with the Let-7 precursor loop mediates regulated microRNA processing. RNA. 2008;14:1539–1549. doi: 10.1261/rna.1155108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piskounova E, Viswanathan S, Janas M, LaPierre RJ, Daley GQ, Sliz P, Gegory RI. Determinants of microRNA processing inhibition by the developmentally regulated RNA-binding protein Lin28. J. Biol. Chem. 2008;283:21310–21314. doi: 10.1074/jbc.C800108200. [DOI] [PubMed] [Google Scholar]
- 17.Thomas JR, Hergenrother PJ. Targeting RNA with small molecules. Chem. Rev. 2008;108:1171–1224. doi: 10.1021/cr0681546. [DOI] [PubMed] [Google Scholar]
- 18.Athanassiou Z, Patora K, Dias RLA, Moehle RLA, Robinson JA, Varani G. Structure-guided peptidomimetic design leads to nanomolar β-hairpin inhibitors of the Tat-TAR interaction of bovine immunodeficiency virus. Biochemistry. 2007;46:741–751. doi: 10.1021/bi0619371. [DOI] [PubMed] [Google Scholar]
- 19.Foloppe N, Chen IJ, Davis B, Hold A, Morley D, Howes R. A structure-based strategy to identify new molecular scaffolds targeting the bacterial ribosomal A-site. Bioorg. Med. Chem. 2004;12:935–947. doi: 10.1016/j.bmc.2003.12.023. [DOI] [PubMed] [Google Scholar]
- 20.Lind KE, Du Z, Fujinaga K, Peterlin BM, James TL. Structure-based computational database screening, in vitro assay, and NMR assessment of compounds that target TAR RNA. Chem. Biol. 2002;9:185–193. doi: 10.1016/s1074-5521(02)00106-0. [DOI] [PubMed] [Google Scholar]
- 21.Yan Z, Sikri S, Beveridge DL, Baranger AM. Identification of an aminoacridine derivative that binds to RNA tetraloops. J. Med. Chem. 2007;50:4096–4104. doi: 10.1021/jm070305p. [DOI] [PubMed] [Google Scholar]
- 22.Mei HY, Cui M, Heldsinger A, Lemrow SM, Loo JA, Sannes-Lowery KA, Sharmeen L, Czarnik AW. Inhibitors of protein-RNA complexation that target the RNA: Specific recognition of human immunodeficiency virus type I TAR RNA by small organic molecules. Biochemistry. 1998;37:14204–14212. doi: 10.1021/bi981308u. [DOI] [PubMed] [Google Scholar]
- 23.Swayze EE, Jefferson EA, Sannes-Lowery KA, Blyn LB, Risen LM, Arakawa S, Osgood SA, Hofstadtler SA, Griffey RH. SAR by MS: a ligand based technique for drug lead discovery against structured RNA targets. J. Med. Chem. 2002;45:3816–3823. doi: 10.1021/jm0255466. [DOI] [PubMed] [Google Scholar]
- 24.Yu L, Oost TK, Scherykantz JM, Yang J, Janowick D, Fesik SW. Discovery of aminoglycoside mimetics by NMR-based screening of Escherichia coli A-site RNA. J. Am. Chem. Soc. 2003;125:4444–4445. doi: 10.1021/ja021354o. [DOI] [PubMed] [Google Scholar]
- 25.Hwang S, Tamilarasu N, Kibler K, Cao H, Ali A, Ping Y-H, Jaeang K-T, Rana TM. Discovery of a small molecule Tat-trans-activation-responsive RNA antagonist that potently inhibits human immunodeficiency virus-1 replication. J. Biol. Chem. 2003;278:39092–39103. doi: 10.1074/jbc.M301749200. [DOI] [PubMed] [Google Scholar]
- 26.Zuckermann RN, Kerr JM, Kent SBH, Moos WH. Efficient method for the preparation of oligo N-substituted glycines by submonomer solid-phase synthesis. J. Am. Chem. Soc. 1992;114:10646–10647. [Google Scholar]
- 27.Kwon YU, Kodadek T. Quantitative evaluation of the relative cell permeability of peptoids and peptides. J. Am. Chem. Soc. 2007;129:1508–1509. doi: 10.1021/ja0668623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alluri P, Reddy MM, Bacchawat-Sikder K, Olivos HJ, Kodadek T. Isolation of protein ligands from large peptoid libraries. J. Am. Chem. Soc. 2003;125:13995–14004. doi: 10.1021/ja036417x. [DOI] [PubMed] [Google Scholar]
- 29.Burkoth TS, Fafarman AT, Charych DH, Connolly MD, Zuckermann RN. Incorporation of unprotected heterocyclic side chains into peptoid oligomers via solid-phase submonomer synthesis. J. Am. Chem. Soc. 2003;125:8841–8845. doi: 10.1021/ja0352101. [DOI] [PubMed] [Google Scholar]
- 30.Lam KS, Salmon SE, Hersh EM, Hruby VJ, Kazmierski WM, Knapp RJ. A new type of synthetic peptide library for identifying ligand-binding activity. Nature. 1991;354:82–84. doi: 10.1038/354082a0. [DOI] [PubMed] [Google Scholar]
- 31.Hamy F, Felder ER, Heizmann G, Lazdins J, Aboul-Ela FA, Varani G, Karn J, Klimkait T. An inhibitor of the Tat/TAR interaction that effectively suppresses HIV-1 replication. Proc. Natl Acad. Sci. USA. 1997;94:3548–3553. doi: 10.1073/pnas.94.8.3548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uttamchandani M, Walsh DP, Yao QS, Chang T-T. Small molecule microarrays: recent advances and applications. Curr. Opinion Chem. Biol. 2005;9:4–13. doi: 10.1016/j.cbpa.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 33.Koehler AN, Shamji AF, Schreiber SL. Discovery of an inhibitor of a transcription factor using small molecule microarrays and diversity-oriented synthesis. J. Am. Chem. Soc. 2002;125:8420–8421. doi: 10.1021/ja0352698. [DOI] [PubMed] [Google Scholar]
- 34.Kuruvilla FG, Shamji AF, Sternson SM, Hergenrother PJ, Schreiber SL. Dissecting glucose signaling with diversity-oriented synthesis and small-molecule microarrays. Nature. 2002;416:653–657. doi: 10.1038/416653a. [DOI] [PubMed] [Google Scholar]
- 35.Reddy MM, Kodadek T. Protein ‘fingerprinting’ in complex mixtures with peptoid microarrays. Proc. Natl Acad. Sci. USA. 2005;102:12672–12677. doi: 10.1073/pnas.0501208102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bryan MC, Wong C-H. Aminoglycoside arrays for the high-throughput analysis of small-molecule-RNA interactions. Tet. Lett. 2004;45:3639–3642. [Google Scholar]
- 37.Childs-Disney JL, Wu M, Pushechnikov A, Aminova O, Disney MD. A small molecule microarray platform to select RNA internal loop-ligand interactions. ACS Chem. Biol. 2007;2:745–754. doi: 10.1021/cb700174r. [DOI] [PubMed] [Google Scholar]
- 38.Disney MD, Seeberger PH. Aminoglycoside microarrays to explore interactions of antibiotics with RNAs and proteins. Chem. Eur. J. 2004;10:3308–3314. doi: 10.1002/chem.200306017. [DOI] [PubMed] [Google Scholar]
- 39.Maskos U, Southern E. Oligonucleotide hybridizations on glass supports: a novel linker for oligonucleotide synthesis and hybridization properties of oligonucleotides synthesized in situ. Nucleic Acids Res. 1992;20:1679–1684. doi: 10.1093/nar/20.7.1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tao J, Frankel AD. Specific binding of arginine to TAR RNA. Proc. Natl Acad. Sci. USA. 1992;89:2723–2726. doi: 10.1073/pnas.89.7.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hicke BJ, Christian EL, Yarus M. Stereoselective arginine binding is a phylogenetically conserved property of group I self-splicing RNAs. EMBO J. 1989;8:3843–3851. doi: 10.1002/j.1460-2075.1989.tb08562.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Macbeath G, Koehler AN, Schreiber SL. Printing small molecules as microarrays and detecting protein-ligand interactions en masse. J. Am. Chem. Soc. 1999;121:7967–7968. [Google Scholar]
- 43.Bradrick TD, Marino JP. Ligand-induced changes in 2-aminopurine fluorecence as a probe for small molecule binding to HIV-1 TAR RNA. RNA. 2004;10:1459–1468. doi: 10.1261/rna.7620304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Means JA, Katz S, Nayek A, Anupam R, Hines JV, Bergmeier SC. Structure-activity studies of oxazolidinone analogs as RNA-binding agents. Bioorg. Med. Chem. Lett. 2006;16:3600–3604. doi: 10.1016/j.bmcl.2006.03.068. [DOI] [PubMed] [Google Scholar]
- 45.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dietz J, Kock J, Kaur A, Raja C, Stein S, Grez M, Pustowka A, Mensch S, Ferner J, Moller L, et al. Inhibition of HIV-1 by a peptide ligand of the genomic RNA packaging signal psi. Chem. Med. Chem. 2008;3:749–755. doi: 10.1002/cmdc.200700194. [DOI] [PubMed] [Google Scholar]
- 47.Labuda LP, Pushechnikov A, Disney MD. Small molecule microarrays of RNA-focused peptoids help identify inhibitors of a pathogenic group I intron. ACS Chem. Biol. 2009;4:299–307. doi: 10.1021/cb800313m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lim HS, Archer CT, Kodadek T. Identification of a peptoid inhibitor of the proteasome 19S regulatory particle. J. Am. Chem. Soc. 2007;129:7750–7751. doi: 10.1021/ja072027p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Carlson CB, Beal PA. Point of attachment and sequence of immobilized peptide-acridine conjugates control affinity for nucleic acids. J. Am. Chem. Soc. 2002;124:8510–8511. doi: 10.1021/ja026029f. [DOI] [PubMed] [Google Scholar]
- 50.Sohn SY, Bae WJ, Kim JJ, Yeom KH, Kim VN, Cho Y. Crystal structure of human DGCR8 core. Nat. Struct. Mol. Biol. 2007;14:847–853. doi: 10.1038/nsmb1294. [DOI] [PubMed] [Google Scholar]
- 51.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 52.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme DROSHA. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krishnamurthy M, Simon K, Orendt AM, Beal PA. Macrocyclic helix-threading peptides for targeting RNA. Angew. Chem. Int. Ed. 2007;46:7044–7047. doi: 10.1002/anie.200702247. [DOI] [PubMed] [Google Scholar]
- 54.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.