Abstract
Restriction-modification (R-M) system Ecl18kI is representative of R-M systems whose coordinated transcription is achieved through a separate DNA-binding domain of the methyltransferase. M.Ecl18kI recognizes an operator sequence located in the noncoding region that separates the divergently transcribed R and M genes. Here we show that, contrary to previous predictions, the two ecl18kI promoters are not divergent, but actually face one another. The binding of M.Ecl18kI to its operator prevents RNA polymerase (RNAP) binding to the M promoter by steric exclusion, but has no direct effect on RNAP interaction with the R promoter. The start point for R transcription is located outside of the intergenic region, opposite the initiation codon of the M gene. Regulated transcription of the potentially toxic ecl18kI R gene is accomplished (i) at the stage of promoter complex formation, through direct competition from complexes formed at the M promoter, and (ii) at the stage of promoter clearance, since R promoter-bound RNAP escapes the promoter more slowly than RNAP bound to the M promoter.
INTRODUCTION
Enterobacter cloacae type II restriction-modification (R-M) system Ecl18kI is naturally carried on plasmid pECL18 (1). Ecl18kI consists of two divergently transcribed genes, one coding for restriction endonuclease (R), and another coding for methyltransferase (M). The two genes are separated by an intergenic region of 109 bp. Ecl18kI is virtually identical to the better-studied SsoII system from Shigella sonnei: the two systems differ from each other in only 8 nt positions over their entire length; their intergenic regions are identical. In both systems, the intergenic region contains an inverted repeat AGGACAATTTGTCCT separated by a T at the center of symmetry. Coordinated expression of the ssoII genes is achieved through specific binding of the methyltransferase to this inverted repeat, called the operator (2). The operator is distinct from the SsoII recognition site 5′-CCNGG-3′ and is recognized by a helix–turn–helix motif located at the N-terminus of the methyltransferase polypeptide (2). DNase I footprinting experiments demonstrate that M.SsoII protects a 48–52-bp region immediately upstream of its coding sequence (2). The region includes predicted consensus elements of the ssoII.M promoter, suggesting that the binding of M.SsoII autogeneously regulates its own synthesis by sterically excluding RNAP from the promoter.
While transcription regulation of SsoII was never explicitly studied, previous work assumed that both divergent ssoII promoters are located in the 109-bp intergenic spacer (2). Such an assumption, however, created a problem, since it was not clear how the methyltransferase interaction with the operator could stimulate transcription of ssoII.R, an essential step during establishment of a plasmid containing the ssoII genes in a naive host. In fact, bioinformatically predicted consensus promoter elements of the putative ssoII.R promoter overlapped with the M.SsoII binding site (2). Such an arrangement is expected to repress rather than activate ssoII.R transcription by bound M.SsoII, in the same way as in the case of ssoII.M transcription.
We felt that transcription regulation of ssoII genes needs to be revisited. For historical reasons, we studied transcription regulation in the Ecl18kI system. However, since the intergenic regions and adjacent DNA sequences in both Ecl18kI and SsoII are identical, there is no doubt that transcription regulation in both systems is identical too. The results reported here reveal that only the methyltransferase promoter was predicted correctly in the previous work. The restriction endonuclease promoter is located outside of the intergenic region, inside the coding sequence of the methyltransferase gene. Methyltransferase binding to the operator has little or no direct effect on transcription from the restriction endonuclease promoter, which becomes activated due to decreased competition from the methyltransferase promoter. The competition is caused by two effects: first, RNAP bound to the methyltransferase promoter decreases, but does not prevent, RNAP interaction with the restriction endonuclease promoter. Second, at conditions when both promoters are occupied by RNAP, transcription initiation from methyltransferase promoter occurs faster than from restriction endonuclease promoter.
MATERIALS AND METHODS
Bacterial strains and plasmids
Escherichia coli strains RR1 (Δ(gpt-proA)62 leuB6 thi-1 lacY1 hsdSB20 rpsL20 (Strr) ara-14 galK2 xyl-5 mtl-1 supE44 mcrBB) (3), K802 (F−, lac-6 (del), lacY1, glnV44 (AS), galT22, galK2 (Oc), LAM, e14, rfbD1, metB1, mcrB, hsdR2, hsdM+, supE, mcrA) (4), M15[pREP4] (F−, Δ(pro-lac), thi, ϕ80d, lacZ, ΔM15, ara, rpsL, recA) (5,6) were used throughout this work. Bacteria were grown in standard LB medium (7).
The plasmid vectors used were pUC18/19 (8), pQE30 (9,10), and pFD51 (11).
The entire Ecl18kI system was cloned into the polylinker of pUC19. The resultant plasmid was named pBcn(m+r+). Plasmid pBcn(m+r–) was obtained from the pBcn(m+r+) by digestion with BglII (cuts once within the ecl18kI.R gene), followed by fill-in with the Klenow enzyme and religation. Plasmid pBcn(m–r–) was prepared by excision of an EcoRV fragment containing part of the ecl18kI.M gene from pBcn(m+r–).
Plasmid pRmGalK was prepared by cloning a fragment of ecl18kI DNA containing the entire Ecl18kI intergenic region and beginnings of Ecl18kI structural genes into promoterless plasmid pFD51; in pRmGalK the galK gene of pFD51 is controlled by ecl18kI.R promoter (Pr); the ecl18kI.M promoter (Pm) is also present and initiates transcription in the opposite direction. In plasmid pMrGalK, the galK gene of pFD51 is controlled by Pm; Pr is also present and initiates transcription in the opposite direction. In pResBSGalK, the galK gene of pFD51 is controlled by Pr; Pm is absent.
Plasmid p18Km(m+r–) was obtained from the p18Km plasmid carrying the ecl18kI genes (1) by digestion with BglII (cuts once within the ecl18kI.R gene), followed by fill-in with the Klenow enzyme and religation. This plasmid is compatible with plasmids described above and was used as a source of Ecl18kI methyltransferase M.Ecl18kI.
Proteins
RNAP and N-terminally hexahistidine-tagged M.Ecl18kI were purified as described in Refs. 14 and 22, respectively.
Total RNA preparation and primer extension reactions
RNA was purified from E. coli K802 harboring indicated plasmids using SV Total RNA Isolation System (Promega). Purified samples were treated with DNase I and then repurified using the same kit. Total RNA (3 μg) was reverse-transcribed with 200 U RevertAid M-MuLV Reverse Transcriptase (Fermentas) in the presence of 1 pmol of γ-32P-labeled primer. Primer extension reactions were carried out at 42°C for 60 min and terminated by a 5-min incubation at 70°C. Samples were extracted with chloroform, ethanol-precipitated, dissolved in formamide-containing loading buffer and loaded on 7% sequencing gels. As markers, the products of DNA sequencing reactions performed with fmol DNA Cycle Sequencing System (Promega) using ecl18kI DNA as template and the same primer were used. Reaction products were revealed by autoradiography. 5′RACE was performed exactly as described in refs. (12) and (13).
Quantitative real-time PCR (qRT-PCR) analysis
Three micrograms of total RNA was used to synthesize first-strand cDNA using RevertAid M-MuLV Reverse Transcriptase (Fermentas). One microliter of the reverse transcription reactions was used as template for RT-PCR. Three sets of primers were used to assay transcript levels of plasmid-borne ecl18kIM, ecl18kIR and bla. Ecl18kIM-specific primers RTrevM (cggtgtttgggtaaggtggt) and RTforM (cgcaacaatcaaagaaa) annealed to ecl18kIM codons 67–62 and 6–11, respectively. Ecl18kIR-specific primers RTforR (cgccatttgctttggtttat) and RTrevR (tgttgtgaattctttttcaaatgg) annealed to ecl18kIR codons 23–29 and 73–66, respectively. The expected sizes of PCR products were 188, 151, and 189 bp for ecl18kIM, ecl18kIR and bla, respectively. Each RT-PCR mixture (25 µl) contained 10 µl SYBR Green I PCR Master Mix (2.5 × PCR buffer, Taq DNA-polymerase, dNTP, glycerol, Tween-20, SYBR Green I), 12 µl distilled H2O, 1 µl of 40 µM forward primer, 1 µl of 40 µM reverse primer and 1 µl of cDNA template. Amplifications were carried out using ANK32 CyclerSystem (Syntol, Russia). The RT-PCR conditions were 95°C for 5 min (1 cycle), and 45 cycles of 95°C for 15 s and 62°C for 45 s. Reaction products were analyzed using 2% agarose gel electrophoresis to confirm that the signals detected originated from products of expected lengths.
In vitro transcription reactions
As templates for in vitro transcription, a PCR-amplified 290-bp-long ecl18kI fragment containing both ecl18kI promoters amplified from plasmid pBcn(m+r+) or a 295-bp-long fragment containing only the ecl18kI.R promoter and the Ecl18kI.M operator (amplified from plasmid pResBSGalK) were used. Transcription reactions were set in 14 μl and contained 50 ng of transcription templates and various amounts of E. coli RNAP σ70 holoenzyme in a buffer containing 20 mM Tris–HCl, pH 8.0, 100 mM MgCl2, 0.1 mM EDTA, 1 mM DTT, 50 μg BSA, 5% glycerol. After a 5–10-min incubation at 37°C, the reactions were supplemented with 2 μl of nucleotide hot mix [2 mM ATP, GTP and CTP, 500 μM UTP and 0.5 μCi [α-32P]-UTP (3000 Ci/mmol) and 20 μg heparin] and incubated for additional 10 min at the same temperature. In the order of additions experiments, the first protein was incubated with DNA for 10 min followed by the addition of the second protein and an additional 10-min incubation prior to the addition of the hot mix. Reactions were terminated by the addition of 15 μl of formamide-containing loading buffer and loaded on a 7% sequencing gels.
Footprinting
KMnO4 probing and DNase I footprinting were conducted under conditions used for in vitro transcription using 32P-labeled promoter fragments as described (14). G+A sequencing reactions were carried out as described (15). Samples were applied on 6% sequencing gels and reaction products revealed using autoradiography.
RESULTS
Transcription of ecl18kI genes in vivo
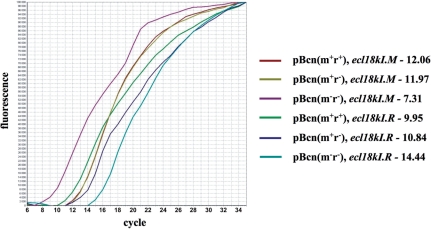
The entire Ecl18kI system was cloned into E. coli plasmid pUC19. The resultant plasmid was named pBcn(m+r+). Escherichia coli cells harboring pBcn(m+r+) grew well and restricted the growth of λvir phage (data not shown), indicating that both ecl18kI genes are expressed. To measure the abundance of ecl18kI transcripts in the presence or in the absence of M, RT-PCR analysis of RNA prepared from cells harboring pBcn(m+r+) or its two derivatives was performed. Plasmid pBcn(m+r–) has a frame-shift mutation in the beginning of the R gene and does not produce restriction endonuclease. Plasmid pBcn(m–r–) has both ecl18kI genes disrupted (see ‘Materials and Methods’ section). After reverse transcription of total cellular RNA with R or M-specific primers, the amounts of reverse-transcribed DNA were determined by registering threshold cycles in RT-PCR reactions for each of the genes monitored. As a control, the amount of transcripts from ampicillin-resistance gene bla present in pBcn(m+r+) and its derivatives was determined alongside determinations of the amounts of ecl18kI transcripts and found to be very similar or identical in all cases (threshold cycles of 7.5 ± 0.4). A typical result is presented in Figure 1 and can be summarized as follows. For M transcripts, threshold cycle values for RNA purified from cells harboring pBcn(m+r+) and pBcn(m+r–) were similar (12.0 ± 0.3). Threshold cycle values for M transcripts from cells harboring pBcn(m–r–) were 7.3 ± 0.5. Thus, the presence of M leads to ∼25 (32-fold) decrease in steady-state levels of M transcripts. For R transcripts, threshold cycle values for RNA purified from cells harboring pBcn(m+r+) and pBcn(m+r–) were 10.4 ± 0.4. The value was 14.4 ± 0.2 for RNA from pBcn(m–r–)-harboring cells. Comparable threshold cycle values for pBcn(m+r+) and pBcn(m+r–) exclude the effect of transcription polarity as the cause of decreased amounts of R transcripts in cells harboring pBcn(m–r–). Thus, the presence of M leads to ∼24 (16-fold) increase in steady-state levels of R transcripts.
Figure 1.
Real-time PCR analysis of ecl18kI transcripts. A typical result of RT-PCR analysis of total RNA prepared from cells harboring the indicated plasmids with ecl18kI genes specific primer pairs is shown. Values of threshold cycles for each transcript analyzed in the experiment shown are indicated.
The reverse-transcription stage of the RT-PCR experiment was carried in the presence of large excess of gene-specific primers. Therefore, some conclusions about the relative abundance of individual ecl18kI transcripts can be made. It appears that during stable maintenance of a plasmid containing functional Ecl18kI system (a condition of our experiment) the amount of R transcripts exceeds the amount of M transcripts (threshold cycle values of ∼10 and ∼12, correspondingly), though not by a large margin. Since the RT-PCR experiment measures the steady-state levels of transcripts, which depend both on the rate of transcript synthesis and its decay, no firm conclusions about transcription activity of ecl18kI promoters can be made.
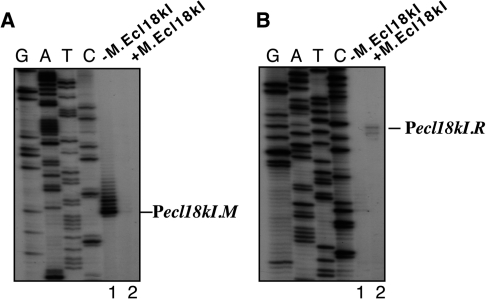
Transcription initiation start points for ecl18kI genes were next established. Primer extension analysis of RNA prepared from cells harboring pBcn(m+r+) with several ecl18kI specific primers revealed no primer extension products, possibly due to low levels of ecl18kI transcripts in the cell. The ecl18kI intergenic region and its flanking sequences were cloned, in both orientations, in front of promoterless galK gene of the plasmid pFD51. The resulting plasmids were introduced in galK− E. coli cells with or without a compatible plasmid that harbored intact M gene and its upstream sequences but lacked the R gene. Cells harboring a pFD51 derivative in which galK was under the control of Pm formed red colonies on McConkey agar plates, but only in the absence of compatible plasmid producing the methyltransferase (data not shown). RNA purified from these cells was subjected to primer extension analysis using a primer specific for the galK gene. A single, major, primer extension product was revealed in the absence of M (Figure 2A, lane 1). As can be seen from Figure 3, the position corresponding to primer extension product endpoint was preceded by an appropriately positioned −10 element cataat (consensus sequence TATAAT) and, 17 bases upstream, a −35 element ttgaaa (consensus sequence TTGACA). The two elements matched those bioinformatically predicted for SsoII promoter Pm in the earlier study (2). In addition to the major primer extension product, a series of longer primer extension products extending as much as 12 nt upstream of the major product with 1-nt increments were observed. The abundance of these products decreased for longer products. The initial transcribed sequence of Pm contains a run of four adenosines in positions +2 to +5. While this was not rigorously investigated, the longer primer extension products most likely result from transcript slippage during transcription through the run of adenosines. Transcript slippage is known to occur on some promoters containing runs of three or more identical nucleotides immediately downstream of the transcription start point and is a subject of biological regulation in some systems [see (16) and references therein]. Transcript slippage was also observed during transcription initiation in the EcoRV R-M system (17). The significance of transcript slippage, if any, for regulation of methyltransferase synthesis in the case of Ecl18kI remains to be determined. No primer extension product was detected when RNA from cells containing M encoded by a separate plasmid was analyzed (Figure 2A, lane 2), indicating that M represses transcription from its promoter, as expected.
Figure 2.
Expression of the ecl18kI genes in vivo. RNA was purified from E. coli cells harboring plasmids with ecl18kI.M (A) or ecl18kI.R (B) promoters fused to promotorless galK in the absence (lanes 1) or presence (lanes 2) of a compatible plasmid expressing M.Ecl18kI and subjected to primer extension with a galK specific primer.
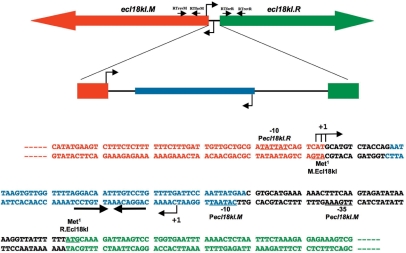
Figure 3.
Genetic organization of restriction-modification system Ecl18kI. The ecl18kI genes with the M.Ecl18kI binding site located in the intergenic region are schematically shown on the top. Arrows show the direction of transcription. The primers used in real-time PCR experiments (Figure 1) are shown as horizontal arrows below schematic representations of the ecl18kI genes; the primer names are indicated and their positions approximately match the sites of ecl18kI genes to which they anneal to. The DNA sequence of the intergenic region and of beginnings of the ecl18kI.R and ecl18kI.M genes is expanded below (both DNA strands are shown). The coding regions are indicated with colours that match those at the top of the figure, the beginnings of ORFs are highlighted in bold typeface, and marked as ‘Met1’. The Pm and Pr start sites are indicated by leftward and rightward arrows below and above the sequence. Conserved promoter elements are labeled and underlined. The M.Ecl18kI binding site is indicated by blue colour; a set of inverted repeats is shown by convergent arrows.
Primer extension reactions designed to reveal transcripts initiated from Pr revealed, in the presence of M, two products (Figure 2B, lane 2) located 109 and 111 bp upstream of the R gene initiation codon (Figure 3). These products were undetectable in the absence of M (Figure 2B, lane 1). Upstream of primer extension product end points there is an appropriately positioned sequence tattat with similarity to consensus −10 promoter element sequence TATAAT (Figure 3). However, a sequence similar to the −35 promoter element consensus is absent. The 5′ ends of RNAs corresponding to primer extension products are located upstream of the start point of bioinformatically predicted restriction endonuclease promoter of SsoII (2) and overlap with the translation start codon of the Ecl18kI methyltransferase gene. No primer extension products corresponding to transcripts initiated from previously predicted SsoII Pr (2) could be detected (data not shown).
We used 5′-RACE as an alternative approach to confirm Pr start point(s). A single TAP-stimulated PCR amplification product was revealed during the analysis of RNA purified from cells containing pBcn(m+r+) (data not shown). Upon sequencing, sequences corresponding to three start points, two matching those revealed by primer extension (above), and another one located 112 bp upstream of the restriction endonuclease gene initiating codon, were found (see Figure 3). We conclude that primer extension experiments indeed reveal a promoter that is distinct from the one predicted in the earlier work on the SsoII system (2). Conversely, the predicted endonuclease promoter either does not exist or is inactive in our conditions. The fact that the Pr is located outside of the ecl18kI intergenic region indicates that the mechanism of ecl18kI genes regulation must be distinct from that proposed previously.
Transcription of ecl18kI genes in vitro
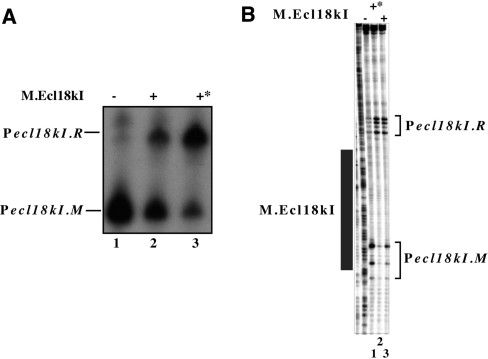
Transcription from ecl18kI promoters was next investigated in vitro. A fragment of ecl18kI DNA containing both promoters was combined with E. coli RNAP σ70 holoenzyme in the presence or in the absence of purified His-tagged methyltransferase, and a single round of transcription was allowed by the addition of NTPs and heparin. The results revealed a single transcription product in the absence of M (Figure 4A, lane 1). By primer extension, this product corresponded to Pm-initiated transcript (data not shown). In the presence of M, added either before or after RNAP, the abundance of this transcript decreased and another, longer, transcript became prominent (Figure 4A, lanes 2 and 3). The start of this transcript was also mapped and found to match Pr transcription start points (data not shown). Thus, M decreases transcription from Pm and activates transcription from Pr in vitro.
Figure 4.
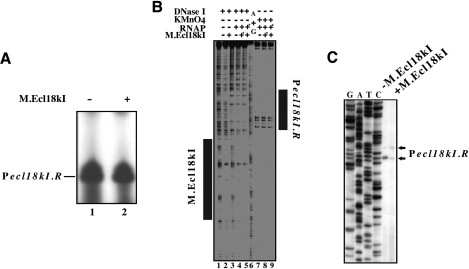
In vitro transcription from and open complex formation on ecl18kI promoters. Escherichia coli RNAP σ70 holoenzyme was combined with a DNA fragment containing the entire ecl18kI intergenic region and flanking sequences in the presence, where indicated, of M.Ecl18kI (the latter was added before RNAP in lanes labelled with an asterisk). In (A), a single round of transcription was allowed to occur by supplementing the reactions with a mixture of NTPs and heparin. In (B), promoter complexes were probed with KMnO4. Reaction products were resolved by denaturing polyacrylamide gel electrophoresis and revealed by autoradiography. Area protected from DNase I cleavage by M.Ecl18kI is shown as a dark rectangle at the right hand side of the gel of (B).
KMnO4 probing revealed that RNAP alone formed open promoter complexes on Pm: KMnO4-sensitivity of thymidines at positions −7 and −10 and strong sensitivity of thymidine at position −1 with respect to Pm transcription start point was observed (Figure 4B, lane 1). Weak KMnO4-sensitive bands reporting low-level open complex formation at Pr were also evident. The addition of M, either before or after the addition of RNAP, led to strong decrease of open complex formation on Pm (Figure 4B, lanes 2 and 3, correspondingly). In contrast, formation of open complex at Pr was stimulated.
The results presented above do not allow one to determine whether transcription from Pr is activated directly (for example through a protein–protein contact between operator-bound M and RNAP), or activation is indirect, i.e. is a simple consequence of removing an RNAP molecule bound to Pm. To address this question, in vitro transcription and KMnO4 probing on a mutant fragment that contained Pr and the M operator but lacked Pm, which was destroyed by a mutation that removed its −35 element, were performed. The results of the in vitro transcription experiment revealed robust activity from Pr that was unaffected by M (Figure 5A, compare lanes 1 and 2). No transcription from Pm was evident, as expected. KMnO4 probing revealed that Pr open complexes were formed on the mutant template both in the presence and in the absence of M (Figure 5B, lanes 7–9). No open complex formation on Pm was observed, as expected. As judged by DNase I footprinting, M bound the mutant template normally, and protected DNA from position +15 to +67 with respect to the Pr transcription start point (Figure 5B, lane 2).
Figure 5.
Pecl18kI.R transcription and promoter complex formation in the absence of Pecl18kI.M. (A and B) Escherichia coli RNAP σ70 holoenzyme was combined with a DNA fragment containing the entire ecl18kI intergenic region and flanking sequences in the presence, where indicated, of M.Ecl18kI. In (A), a single round of transcription was allowed to occur by supplementing the reactions with a mixture of NTPs and heparin. In (B), promoter complexes were footprinted with DNase I or probed with KMnO4. Components labeled with asterisks were added to DNA first. Reaction products were resolved by denaturing polyacrylamide gel electrophoresis and revealed by autoradiography. Areas protected from DNase I cleavage by M.Ecl18kI and RNAP bound to Pecl18kI.R are shown as dark rectangles. (C) RNA was purified from E. coli cells harboring a plasmid with Pecl18kI.R fused to promotorless galK (and disrupted Pecl18kI.M) in the absence (lane 1) or presence (lane 2) of a compatible plasmid expressing M.Ecl18kI and subjected to primer extension with a galK-specific primer.
The absence of direct effect of M on transcription from Pm could also be demonstrated in vivo (Figure 5C). Primer extension analysis of RNA initiated from plasmid-borne Pr–galK fusion plasmid with disrupted Pm was performed. As can be seen, there was no increase in abundance of Pr-initiated transcripts when M was provided from a compatible plasmid (in fact, a small decrease was observed, which may have been due to interference of bound M with transcription initiated from Pr).
From these experiments, we conclude that Pr is a strong promoter whose activity is masked by RNAP binding to an even stronger Pm. Clearly, the Ecl18kI methyltransferase activates Pr indirectly, by preventing promoter complex formation on Pm.
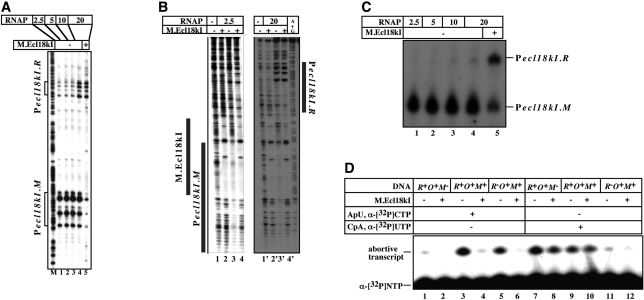
In vitro experiments presented above were conducted at conditions when RNAP was present in equimolar amounts with DNA fragments containing ecl18kI promoters. The results of these experiments suggest that formation of the Pr and Pm promoter complexes is mutually exclusive. However, experiments conducted in the presence of higher concentrations of RNAP suggest that this is not necessarily so. As can be seen from Figure 6A, when the ratio of RNAP and promoter DNA reached 20:1, KMnO4-sensitive bands indicative of open promoter complexes on both Pm and Pr were observed in the absence of M (Figure 6A, lane 4). The effect appeared to be cooperative, since in the presence of intermediate concentrations of RNAP no gradual increase of Pr complexes was detected. We therefore conclude that both ecl18kI promoters can be simultaneously occupied by RNAP. The conclusion was also supported by the results of gel-retardation experiments (data not shown) and DNase I footprinting conducted at two different concentrations of RNAP (Figure 6B). As can be seen from the figure, at high RNAP concentrations, complexes at both promoters were formed (Figure 6B, lane 3′). In the presence of high concentrations of RNAP, the addition of M decreased open complex formation on Pm, as expected, but did not lead to further stimulation of complex formation Pr (Figure 6A, lane 5).
Figure 6.
In vitro transcription and promoter complex formation from ecl18kI promoters in the presence of increasing concentrations of RNAP. (A–C) Increasing amounts of E. coli RNAP σ70 holoenzyme were combined with a DNA fragment containing the entire ecl18kI intergenic region and the flanking sequences in the absence (lanes 1–4) or in the presence (lane 5) of M.Ecl18kI. In (A), promoter complexes were probed with KMnO4; in (B), promoter complexes were footprinted with DNase I; in (C), a single round of transcription was allowed to occur by supplementing the reactions with a mixture of NTPs and heparin. Reaction products were resolved by denaturing polyacrylamide gel electrophoresis and revealed by autoradiography. (D) RNAP was combined [at high ratio corresponding to lanes 4 and 5 in (A)] with indicated wild-type or mutant ecl18kI intergenic DNA fragments and abortive transcription initiation reactions characteristic of Pecl18kI.R or Pecl18kI.M were initiated in the presence of in the absence of M.Ecl18kI. Reaction products were resolved by denaturing polyacrylamide gel electrophoresis and revealed by autoradiography.
Surprisingly, the results of single-round transcription did not follow the results of KMnO4 probing. As can be seen from Figure 6C, increasing concentrations of RNAP led only to a marginal increase in the amounts of Pr transcripts in absence of M the (compare lanes 1 and 4). In contrast and as expected, the addition of M strongly increased the amount of Pr transcripts (and decreased the amount of Pm transcripts, lane 5). Thus, in the presence of excess RNAP, promoter complexes on Pr form in the absence of M but no full-sized transcripts from these complexes is produced.
To determine if promoter complexes formed on Pr in the absence of M are transcriptionally active, abortive transcription initiation reactions were performed. To reveal transcription initiation from Pr, the synthesis of CpApU from the CpA primer and radioactive UTP was monitored; to reveal transcription from Pm, the synthesis of ApUpC from the ApU primer and radioactive CTP was followed. The reactions contained enough RNAP to allow promoter complex formation on both promoters in the absence of M. In addition to the wild-type ecl18kI template containing both promoters and extending from −60 to +132 relative to Pr start point (–81 to +110 relative to Pm start point) templates containing mutations disrupting either Pr (–81 to +36 relative to Pm start point, +17 to +132 relative to Pr start point) or Pm (–60 to +53 relative to Pr start point, −2 to +111 relative to Pm start point) were also used as controls. As can be seen from Figure 6D, on template containing both promoters (R+O+M+), both abortive trinucleotide diphosphates were synthesized by RNAP alone (Figure 6D, lanes 3 and 9). In the presence of M, the synthesis of ApUpC was strongly inhibited (compare lanes 3 and 4), while the CpApU synthesis was unaffected (compare lanes 9 and 10). Reactions with mutant templates (labeled R−O+M+ or R+O+M– on Figure 6D) confirmed that each trinucleotide was synthesized from the expected promoter since disruption of Pr abolished the synthesis of CpApU but not ApUpC (compare lanes 11 and 5), while disruption of Pm abolished the synthesis of ApUpC but not of CpApU (compare lanes 1 and 7). Thus, when RNAP is in excess, active promoter complexes on both ecl18kI promoters form in the absence of M, but full-sized transcripts from Pm only are produced.
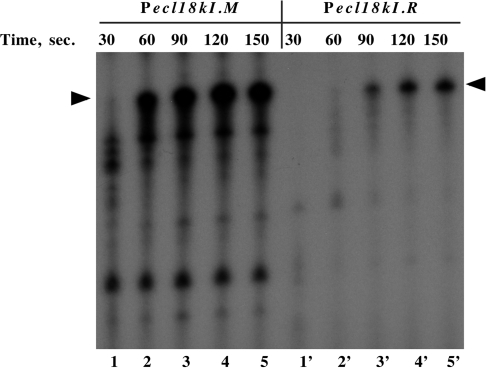
An arrangement of promoters similar to that found in Ecl18kI has been described in immunity regions of several lambdoid phages (18). There, preferential transcription from one promoter was explained by a ‘sitting duck’ model, which posits that promoter from which RNAP escapes faster ‘wins’ over a promoter from which the escape is slow (19). We determined the time course of RNAP escape from Pr and Pm in the absence of the interfering promoter (Pm or Pr, respectively). The results are shown in Figure 7. As can be seen, in a single-round transcription experiment, the Pm transcript accumulated considerably faster than the Pr transcript (for example, at 60-s time point, no full-sized Pr transcript was produced, while substantial amounts of Pm transcript was already synthesized, compare lane 2′ with lane 2). We propose that, in accordance with the ‘sitting duck’ model, faster promoter clearance from Pm leads to disruption of open complexes on Pr, which require more time to escape to elongation. As a result, more Pm transcripts are produced even when RNAP forms complexes on both Ecl18kI promoters.
Figure 7.
The time course of accumulation of full-sized ecl18kI transcripts in vitro. Escherichia coli RNAP σ70 holoenzyme was combined with DNA fragments containing the indicated promoters and a single round of transcription was initiated by the addition of NTPs and heparin. PCR fragments used as templates for in vitro transcription allowed the generation of similarly sized full-sized transcripts from both ecl18kI promoters. Reaction aliquots were withdrawn and transcription terminated at times indicated. Reaction products were resolved by denaturing PAGE and revealed by autoradiography.
DISCUSSION
In every type II R-M system studied today, special mechanisms exist that ensure coordinated expression of restriction endonuclease and methyltransferase genes. The functional requirements for such regulation are 2-fold. First, during establishment of an R-M system in a naive host, methyltransferase needs to be expressed first and in amounts sufficient for complete modification of recognition sites in the host genome before restriction endonuclease activity appears. Second, during stable maintenance of an R-M system, expression of methyltransferase needs to be decreased to prevent inadvertent methylation (and therefore protection) of incoming foreign DNA, for example phage DNA. Regulation of R-M genes expression at the level of transcription appears to be the most economical from the point of view of expenditure of cellular resources.
Genetic architecture of various R-M systems imposes important constraints on molecular mechanisms that ensure coordinated gene expression. R-M system Ecl18kI studied in this work is characterized by divergent orientation of structural genes and a presence of a predicted helix–turn–helix domain in the N-terminal portion of methyltransferase. The intergenic region separating the two structural genes contains a palindromic sequence (an operator) to which the N-terminal domain of methyltransferase can bind to. Methyltransferase interaction with the operator provides a negative-feedback loop that autogeneously decreases transcription of the M gene and a positive-feedback loop that increases transcription of the R gene. R-M systems MspI (20) and EcoRII (21) are organized similarly to SsoII. In the case of MspI, transcription start points have been mapped (20) and it was shown that (i) the M gene transcript start point overlaps with the methylase binding site, suggesting that methylase inhibits M gene transcription by steric exclusion and (ii) the R gene transcript start point is located ∼50 bp downstream of the methylase binding site, suggesting direct activation of R gene transcription by the bound methylase.
The SsoII transcription start points have not been mapped. While it was a priori clear that negative regulation of the SsoII M gene transcription occurred most likely at the level of steric occlusion of the promoter by operator-bound methylase, the mechanism of activation of SsoII Pr remained less clear, especially since this promoter was predicted to be very close to Pm. The principal result of our work is the demonstration that Pr is located outside the intergenic region, in the beginning of the M gene. As a result, the two promoters are facing each other, their transcriptions start points separated by 50 bp of DNA. The Pr promoter identified here could not have been revealed by bioinformatic analysis alone due to its unusual location (outside the ecl18kI intergenic region and within the M gene) and low similarity to existing bioinformatic models of σ70 holoenzyme promoters. In fact, Pr should be considered a novel promoter as it lacks clear similarity to either −10/–35 or extended −10 promoters and yet, in the absence of interference from Pm, it acts as a strong promoter both in vitro and in vivo. The result underscores the fact that there may be a large number of promoters and RNAP-binding sites in bacterial genomes that escape identification by bioinformatic means and yet perform important functions. Further, it appears that molecular mechanisms of regulation of restriction endonuclease genes in Ecl18kI (SsoII) and MspI R-M systems are quite different despite the common architecture of these systems.
Our results suggest that at low RNAP concentrations, RNAP binding to Pm decreases the interaction with Pr, probably due to direct interference between front ends of RNAP molecules bound to either one of the two promoters. The binding of Ecl18kI methyltransferase directly interferes with promoter complex formation on Pm but per se has no effect on RNAP interaction with Pr. However, decreased complex formation on Pm allows the Pr complex to form more efficiently. Apparently, transcription from Pr proceeds through the methyltrasferase–operator complex. The fate of this complex during the passage of Pr-initiated transcription elongation complex is unknown. It is attractive to speculate that such transcription dislodges methyltransferase from the complex, thus allowing promoter competition to be reenacted after each transcription initiation event from Pr, and thus providing a mechanism to fine-tune the amount of Pm and Pr transcripts in the cell. Pr is located more than 100 bp away from the R gene starting codon. It is possible that this long leader contributes to the regulation of Ecl18kI gene expression since Ecl18kI mRNAs may hybridize to one another over the full distance between their respective start sites. Such hybridization would block the M translation initiation region, but not the R initiation region, which may help in the transition from ‘methylate only’ stage when a plasmid containing Ecl18kI genes establishes in naive host to ‘begin restriction’ stage during stable maintenance of the plasmid.
At high RNAP concentrations, open complexes on both ecl18kI promoters form even in the absence of M. However, no full-sized Pr transcripts are produced. A likely explanation is that in the presence of transcription substrates, promoter complexes on Pr are disrupted by transcription elongation complexes initiated from Pm, which is a faster-clearing promoter. Both mechanisms of Pr activation by M (through increase of Pr open complex formation and through increase of productive transcription from Pr open complexes) may be operational in vivo and controlled not only by RNAP availability but also by DNA topology. The relative contribution of both mechanisms requires further investigation and is a subject of ongoing work in our laboratories.
There is an interesting question about the relationship between the HTH regulatory DNA-binding domains of methyltransferases such as M.Ecl18kI, M.MspI and M.EcoRII and C-proteins, small DNA-binding proteins that regulate transcription in many type II R-M systems (22). All C-proteins are evolutionarily related to each other and are also distant relatives of phage regulators such as the λ repressor. Recently, a comprehensive bioinformatics survey identified several hundred C-proteins that could be clustered into 10 families (23). In all cases that have been studied, C-proteins recognize duplicated palindromic binding sites called C-boxes. Binding of a C-protein dimer to the high-affinity site activates the R gene promoter. Binding to the low-affinity site represses it. Sequence analysis indicates that the HTH domain of M.Ecl18kI is distinct from C-proteins, that is, even most distant C-proteins are more closely related to each other than to M.Ecl18kI DNA-binding domain. Likewise, the structure of M.Ecl18kI operator is distinct from any known or predicted C-box. The operator contains a single rather than a duplicated palindrome and the inverted repeat in the operator is much longer than those in C-boxes. Our recent attempts to observe operator binding with recombinant HTH domain of M.Ecl18kI were unsuccessful (24), indicating that sequences in the methyltrasnferase part of M.Ecl18kI are required for DNA binding (the reverse is not true since M.Ecl18kI lacking the HTH domain is fully capable of site methylation). Structural characterization of M.Ecl18kI, alone and in complex with the operator, will be necessary to clarify these issues.
FUNDING
NIH RO1 grant GM59295; NIH FIRCA research grant R03 TW07145; and Presidium of Russian Academy of Sciences Molecular and Cellular Biology Program grant (to K.S.). Funding for open access charge: National Institutes of Health GM R01 59295 grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr. Ekaterina Semenova for assistance and advice with 5′-RACE and Valery Sorokin for help with bioinformatic analysis of M.Ecl18kI DNA binding domain.
REFERENCES
- 1.Den'mukhametov MM, Zakharova MV, Kravets AN, Pertsev AV, Sineva EV, Repik AV, Beletskaia IV, Gromova ES, Solonin AS. Characteristics of a plasmid bearing a gene of a restriction modification type II system—the SsoII isoschizomer. Mol. Biol. 1997;31:831–838. [PubMed] [Google Scholar]
- 2.Karyagina A, Shilov I, Tashlitskii V, Khodoun M, Vasil’ev S, Lau PCK, Nikolskaya I. Specific binding of SsoII DNA methyltransferase to its promoter region provides the regulation of SsoII restriction-modification gene expression. Nucleic Acids Res. 1997;25:2114–2120. doi: 10.1093/nar/25.11.2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyer HW, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 4.Wood WB. Host specificity of DNA produced by Escherichia coli: Bacterial mutations affecting the restriction and modification of DNA. J. Mol. Biol. 1966;16:118–133. doi: 10.1016/s0022-2836(66)80267-x. [DOI] [PubMed] [Google Scholar]
- 5.Ruther U. A recA lacZ transformation host. Nucleic Acids Res. 1981;9:4087–4098. doi: 10.1093/nar/9.16.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Groger RK, Morrow DM, Tykocinski ML. Directional antisense and sense cDNA cloning using Epstein-Barr virus episomal expression vectors. Gene. 1989;81:285–294. doi: 10.1016/0378-1119(89)90189-3. [DOI] [PubMed] [Google Scholar]
- 7.Sambrook J, Frith EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 8.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 9.Bujard H, Gentz R, Lanzer M, Stueber D, Mueller M, Ibrahimi I, Haeuptle MT, Dobberstein B. A T5 promoter-based transcription-translation system for the analysis of proteins in vivo and in vitro. Methods Enzymol. 1987;155:416–433. doi: 10.1016/0076-6879(87)55028-5. [DOI] [PubMed] [Google Scholar]
- 10.Stuber D, Matile H, Garotta G. System for high-level production in Escherichia coli and rapid purification of recombinant proteins: application to epitope mapping, preparation of antibodies, and structure-function analysis. In: Lefkovits I, Pernis B, editors. Immunological Methods, IV. New York: Academic Press; 1990. pp. 121–152. [Google Scholar]
- 11.Rak B, von Reutern M. Insertion element IS5 contains a third gene. EMBO J. 1984;3:807–811. doi: 10.1002/j.1460-2075.1984.tb01889.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Semenova E, Djordjevic M, Shraiman B, Severinov K. The tale of two polymerases: transcription profiling and gene expression strategy of bacteriophage Xp10. Mol. Microbiol. 2005;55:764–777. doi: 10.1111/j.1365-2958.2004.04442.x. [DOI] [PubMed] [Google Scholar]
- 13.Minakhin L, Semenova E, Liu J, Vasilov A, Severinova E, Gabisonia T, Inman R, Mushegian A, Severinov K. Genome and gene expression of Bacillus anthracis bacteriophage Fah. J. Mol. Biol. 2005;354:1–15. doi: 10.1016/j.jmb.2005.09.052. [DOI] [PubMed] [Google Scholar]
- 14.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual promoter complexes. Proc. Natl Acad. Sci. USA. 1997;94:13481–13486. doi: 10.1073/pnas.94.25.13481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Metzger W, Heumann H. Footprinting with exonuclease III. In: Kneale G, editor. DNA-Protein Interaction: Principles and Protocols, Vol. 30. Methods in Molcular Biology. Totowa, NJ: Humana Press Inc.; 1994. pp. 11–20. [DOI] [PubMed] [Google Scholar]
- 16.Turnbough CL. Regulation of bacterial gene expression by the NTP substrates of transcription initiation. Mol. Microbiol. 2008;69:10–14. doi: 10.1111/j.1365-2958.2008.06272.x. [DOI] [PubMed] [Google Scholar]
- 17.Semenova E, Minakhin L, Vasilov A, Solonin A, Heyduk T, Zakharova M, Severinov K. Transcription regulation of the EcoRV restriction-modification system. Nucleic Acids Res. 2005;33:6942–6951. doi: 10.1093/nar/gki998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dodd IB, Kalionis B, Egan JB. Control of gene expression in the temperate coliphage 186. VIII. Control of lysis and lysogeny by a transcriptional switch involving face-to-face promoters. J. Mol. Biol. 1990;214:27–37. doi: 10.1016/0022-2836(90)90144-B. [DOI] [PubMed] [Google Scholar]
- 19.Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol. Cell. 2004;14:647–656. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Som S, Friedman S. Characterization of the intergenic region which regulates the MspI restriction-modification system. J. Bacteriol. 1997;179:964–967. doi: 10.1128/jb.179.3.964-967.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Som S, Friedman S. Autogenous regulation of the EcoRII methylase gene at the transcriptional level: effect of 5-azacytidine. EMBO J. 1993;12:4297–4303. doi: 10.1002/j.1460-2075.1993.tb06114.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tao T, Bourne JC, Blumenthal RM. A family of regulatory genes associated with type II restriction-modification systems. J. Bacteriol. 1991;173:1367–1375. doi: 10.1128/jb.173.4.1367-1375.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sorokin V, Severinov K, Gelfand MS. Systematic prediction of control proteins and their DNA binding sites. Nucleic Acids Res. 2009;37:441–451. doi: 10.1093/nar/gkn931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fedotova EA, Protsenko AS, Zakharova MV, Lavrova NV, Alekseevsky AV, Oretskaya TS, Karyagina AS, Solonin AS, Kubareva EA. SsoII-like DNA-methyltransferase Ecl18kI: interaction between regulatory and methylating functions. Biochemistry. 2009;74:85–91. doi: 10.1134/s0006297909010131. [DOI] [PubMed] [Google Scholar]