Abstract
Escherichia coli ribosomal protein S1 is required for the translation initiation of messenger RNAs, in particular when their Shine–Dalgarno sequence is degenerated. Closely related forms of the protein, composed of the same number of domains (six), are found in all Gram-negative bacteria. More distant proteins, generally formed of fewer domains, have been identified, by sequence similarities, in Gram-positive bacteria and are also termed ‘S1 proteins’. However in the absence of functional information, it is generally difficult to ascertain their relationship with Gram-negative S1. In this article, we report the solution structure of the fourth and sixth domains of the E. coli protein S1 and show that it is possible to characterize their β-barrel by a consensus sequence that allows a precise identification of all domains in Gram-negative and Gram-positive S1 proteins. In addition, we show that it is possible to discriminate between five domain types corresponding to the domains 1, 2, 3, 4–5 and 6 of E. coli S1 on the basis of their sequence. This enabled us to identify the nature of the domains present in Gram-positive proteins and, subsequently, to probe the filiations between all forms of S1.
INTRODUCTION
Prokaryotic cells (and chloroplasts) translation seldom starts at the first AUG codon. The small ribosomal subunit identifies the initiation codons among synonymous triplets by the means of specific signals in their vicinity. The most often encountered signal, recognized by the translation system of all bacteria, is the Shine–Dalgarno sequence (with consensus AAGGAG), located 5–13 nt upstream from the initiator AUG (1,2). However, in addition to this universal mechanism, specific systems co-exist. In particular, the translation initiation of most, if not all, Escherichia coli messenger RNAs (mRNAs) depends on the presence of the ribosomal protein S1 (3) that was described to recognize an A/U rich sequence upstream of the initiation codon (4). This protein is found in Gram-negative bacteria. More or less distantly related forms have been observed in Gram-positive bacteria, but the nature of these proteins, their roles and their relationship with the Gram-negative S1 remain unclear.
Gram-negative proteins S1 are formed of six similar domains. These domains have been shown to play different roles in the E. coli protein S1. The two first are involved in the binding of the protein to the ribosome, while the four following are involved in the interactions with mRNAs (5). As the last domain has been shown to be dispensable for translation initiation (6), its function is still unknown. The fragment of S1 formed of the domains 3, 4 and 5 (F3–5 fragment) also enhances the activity of the ribonuclease RegB of the bacteriophage T4, whose function is to inactivate several of the phage early messengers, when their translation in no more required, by cleaving them in their Shine–Dalgarno sequence. The domains 3, 4 and 5 are simultaneously involved in the binding of RNAs (7), but there is a structural and functional asymmetry between the domain 3 on the one hand and the domains 4 and 5 on the other hand. The domains 4 and 5 are structurally associated in the F3–5 fragment, while the domain 3 is free to move, in the absence of RNA at least (7). The fragment 45 has a function by itself (in RegB activation) while the domain 3 is only active when linked to the two others (8).
The domains of the protein S1 are more or less closely related to a set of domains found in many proteins involved in the RNA metabolism in all kinds of organisms. Several structures of such domains have been determined (9), showing that they belong to the OBfold (oligonucleotide–oligosaccharide-binding fold) family. However, to date, there is no known structure of a protein S1 domain. To gain insight into the properties of these domains and with the objective to find structural features that could be related to their functional differences, we determined the structure of the domains 4 and 6 of the E. coli protein S1 and investigated the interactions between the domain 6 and two RNAs [poly(A) and poly(U)]. We show that there is no structural difference between the domains. RNA binding on the domain 6 induces fewer modifications at the surface of the β-barrel than in the case of the domains 3, 4 or 5, and involves the C-terminal flexible segment of the protein, but the interaction occurs on the same side of the β-barrel as for the other domains. However, comparison of the structures of the two domains allowed us to identify consensus sequences characteristic of the five β-strands of the domains of all Gram-negative and, apparently, Gram-positive proteins S1. Using these sequences we were able to precisely align the sequences of the domains of the Gram-negative proteins S1 and consequently to show that it is possible to discriminate between the different domains (at positions 1, 2, 3, 4, 5 and 6) by using only the sequences. Accordingly we also were able to identify the nature of all domains of the Gram-positive proteins S1 and to probe the relationship between the proteins S1 of the two groups of bacteria.
MATERIALS AND METHODS
Proteins production and purification
Typically, 25 ml overnight cultures of the E. coli strain BL21(DE3) transformed with the adequate plasmid (8) were used to inoculate 1 l of M9 minimal medium supplemented with 1.0 g l–1 15NH4Cl and either 4 g l–1 glucose (15N U-labeled protein samples) or 2 g l–1 13C-glucose (15N13C U-labeled protein samples). Protein expression was induced at OD600 = 0.6 using 1 mmol l–1 isopropyl-β-d-thiogalactopyranoside. Cells were harvested 3–4 h later, disrupted by sonication and proteins were purified either on a Talon resin (Clontech) or on a Fast Flow Ni-Histrap column (GE Healtcare) using the manufacturer's recommended protocols. The proteins were dialyzed against nuclear magnetic resonance (NMR) buffer [50 mmol l–1 phosphate, pH 6.8, 200 mmol l–1 NaCl, 20 mmol l–1 dithiothreitol (DTT)] and concentrated up to 0.7–0.8 mmol l–1.
NMR spectroscopy
All NMR experiments were realized on a Bruker DRX 600 spectrometer equipped first with a 5-mm TXI triple resonance Z-gradient probe (F6 study) and then with a 5-mm TXI triple resonance Z-gradient cryoprobe (F4 study). Data were processed with GIFA (10) or XWINNMR 3.0 (Bruker) and analyzed with XEASY (11) or Sparky software (University of California San Francisco, Thomas L. Goddard). All spectra were recorded at 303 K.
The resonance frequency assignment of the backbone atoms (HN, N, C′, Cα and Cβ) was obtained by recording and analyzing HNCO, HNCA, HN(CO)CA, HNCACB and CBCA(CO)NH triple-resonance experiments recorded on 15N13C U-labeled samples in 90/10% H2O/D2O. The Hα and aliphatic side-chain 1H and 13C resonance frequencies were further assigned by the means of 3D TOCSY-HSQC (60 ms spin-lock time, 15N U-labeled samples in 90/10% H2O/D2O) and HCCH-TOCSY (12 ms spin-lock time, 15N13C U-labeled samples in 100% D2O) experiments. Finally, the aromatic 1H resonance frequencies were assigned using NOESY and COSY spectra (15N U-labeled samples in 100% D2O). Distance constraints were obtained from the analysis of 15N-NOESY-HSQC (80 ms mixing-time, 15N U-labeled samples in 90/10% H2O/D2O), 13Caliphatic-NOESY-HSQC (80 ms mixing-time, 15N13C U-labeled samples in 90/10% H2O/D2O for the F4 fragment or 100% D2O for the F6 fragment) and a series of 2D-NOESY (25–100 ms mixing-times experiments, 15N U-labeled samples in 100% D2O).
Domain 6/poly(A) and poly(U) interaction experiments were analyzed by recording 15N–HQSC spectra on 0.5 mmol l–1 15N U-labeled samples of the domain 6 in the presence of increasing amounts of poly(A) and poly(U) (Amersham Pharmacia Biotech.). Each titration point was obtained by transferring the sample from the NMR tube into a 1.5 ml vial containing the desired amount of dry polyribonucleotide and transferring it back into the NMR tube. For each series, five HSQC spectra were recorded corresponding to RNA/protein ratios of 0, 1, 5, 10 and 20, where RNA quantities are expressed in nucleotide (monomer) quantities. In all interaction experiments, all recording parameters were kept rigorously constant, the only adjustment concerning the probe tuning and the field shimming. All HSQC were recorded with 200 15N increments in order to obtain a sufficient resolution.
Structure calculations
The structures of domains 4 and 6 were calculated using the INCA software (12), which mainly uses unassigned NOE cross peaks to perform the structure determination. Typically, the software carried out 22 calculation cycles, each corresponding to an automatic assignment, a structure calculation and an analysis step. The assignment step converts each NOE cross peak to a list of possible constraints based on the distances observed in the structures calculated during the preceding cycle. The calculation step carries out the calculation of 500 structures from the constraint lists by simulated annealing. Subsequently, the best 20 structures are selected during the analysis step. The program input consisted of three lists of unassigned NOE correlations picked from the 80 ms 13C- and 15N-NOESY–HSQC and the 60 ms 2D-NOESY spectra, with the corresponding lists of 1H, 15N and 13C chemical shifts, with a list of constraints derived from manually assigned NOEs (mainly those characteristic of the secondary structure elements and topology) and the identified hydrogen bonds and finally with a list of phi and psi dihedral angle constraints derived from the TALOS analysis of the backbone chemical shifts (13). The coordinates of 12 structures and the restraint files have been deposited at the RCSB with accession code 2KHI (domain 4) and 2KHJ (domain 6).
Sequence analysis
Using the NCBI search engine (www.ncbi.nlm.nih.gov/sites/entrez), we retrieved the sequences of nine S1 proteins belonging to the main groups of Gram-negative bacteria, namely those of the proteobacter α Sinorhizobium melitoti (Sm, P14129), proteobacter β Neisseria meningitidis (Nm, CAM08662), proteobacter γ E. coli (Ec, ABJOO328), proteobacter δ Anaeromyxobacter sp. (A, YP_002134703), proteobacter ε Arcobacter butzleri (Ab, YP_001490936), aquificae Aquifex aeolicus (Aa, AAC07419), chlamydiaec Chlamydophila pneumonia (Cp, Q9Z8M3), bacteroides Bacteroides fragilis (Bf, CAH06700) and that of the spirochetes Borrelia burgdorferi (Bb, NP212261). All these proteins have similar length (between 550 for the S1 of A. butzleri and 597 for the S1 of B. fragilis) and are formed of the same number of S1 domains (six). We also retrieved 17 sequences noted as ‘S1 proteins’ representative of the main Gram-positive eubacteria subdivisions as defined by Olsen and collaborators (14) that possess very different sizes (from 111 for the tenericutes Spiroplasma kunkelii to 827 for the fusobacteria Fusobacterium nucleatum) and a variable number of S1 domains (1–6). We did not found any S1 sequences for Archea and thermodesulfobacteria.
The sequences of the Gram-negative S1 proteins were aligned in two steps. The β-strands were first manually aligned by using characteristic residues from the hydrophobic core of the S1 domains (see ‘Results’ section). The loop regions were then automatically aligned by clustalW2 (15). The phylogenic trees were built using the parsimony method (protpars algorithm of the PHYLYP 3.68 package). The contribution of the different S1 regions to the functional specialization of the domains was estimated by comparing the evolutionary trees obtained from the complete sequences and from partially masked sequences, the masks resulting of the use of a weighting file containing 0 (masked) and 1 (unmasked) coefficients applied to the desired amino acids. The hmm profile library of the domains of the Gram-negative S1 proteins was built by using the hmmbuild algorithm of the HHMER suite (16). The agreement between all other S1 domain sequences and the hmm profiles of the library was then tested by using the hmmpfam algorithm of the same suite.
RESULTS
Escherichia coli domain 4 and 6 structure determination
We used the INCA software (12) that performs simultaneous NOE cross peak interpretation and structure determination. The success of such an automatic procedure critically relies on the availability of an essentially complete list of atom resonance frequencies for each used NOESY spectrum (17). We obtained 94% of the proton frequencies in domain 4 spectra and more than 95% in domain 6 spectra. In the case of domain 4, we missed all Met–CεH3, the side chain protons of Lys314 and Lys347, the aromatic proton of His305, His317, His361 and Trp357, and the Trp311-Hε1,Hζ3 in all spectra. We also missed Asn315-NγH2 and Gln348-NδH2 in 15N-NOESY-HSQC. In the case of domain 6, we missed all Met-CεH3 Glu527-Hβ and Phe505-Hζ protons in all NOESY. We also missed Lys449–Hε and Lys450-Hγ,δ in 15N-NOESY-HSQC and 2D–NOESY but assigned them in 13C–NOESY-HSQC.
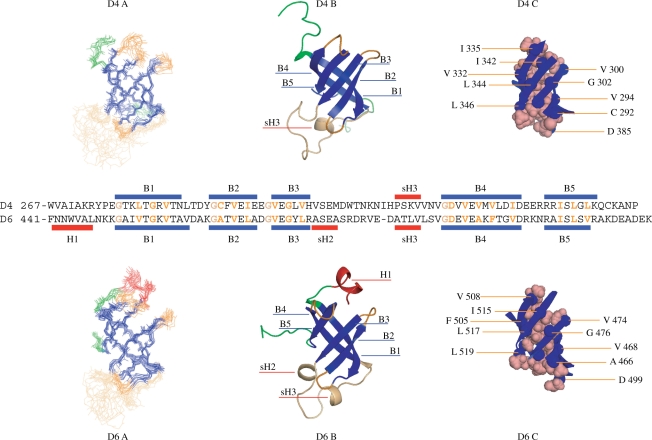
The structures were calculated by using a set of already assigned NOEs characteristic of the secondary structure elements, the (φ,ψ) dihedral angles, and the lists of nonassigned correlations manually peaked in the NOESY spectra (1685 for domain 4, 1326 for domain 6) (Table 1). These correlations were converted during the structure calculation process in 1176 (domain 4) and 921 (domain 6) nontrivial, non redundant constraints among which 880 (domain 4) and 740 (domain 6) were unambiguously assigned. Some of these constraints completed the net of backbone-backbone proximities defining the protein secondary structures. Twelve structures are represented for each domain in the Figure 1. None of these structures presents a distance violation larger than 0.5 Å and a dihedral angle violation larger than 10°. Their covalent geometry is nearly perfect and 96% (domain 4) and 97% (domain 6) of the residues of the structured regions are in the most and additionally allowed regions of the Ramachandran diagram (Table 1). The structure of domain 4 (rmsd of 0.7 Å, calculated on the backbone atoms of the structured regions) is better defined than that of the domain 6 (0.8 Å), due to a greater number of constraints.
Table 1.
Structural statistics of the S1 domains 4 and 6
| Domain 4 (12 structures) | Domain 6 (12 structures) | |
|---|---|---|
| Experimental restraints | ||
| A priori assigned restraints | 144 | 96 |
| Sequential (HN-HN, Hα-HN) | 51 | 38 |
| Non-sequential (HN-HN, Hα-Hα, HN-Hα) | 57 | 40 |
| Hydrogen bonds (two restraints by bond) | 36 | 18 |
| A posteriori assigned restraints | ||
| Peak number | 1685 | 1326 |
| Assigned peaks | 1606 | 1244 |
| Constraint number | 1176 | 921 |
| Non-ambiguous restraints | 889 | 740 |
| Intraresidual | 432 | 411 |
| Sequential | 223 | 130 |
| Non-sequential | 234 | 199 |
| Ambiguous constraints | 287 | 181 |
| Phi-Psi dihedral angle constraints | 62 | 120 |
| Restraints violations | ||
| NOE violations > 0.5 Å | 0 | 0 |
| Dihedral angle violations > 10° | 0 | 0 |
| Structural coordinates rmsd | ||
| Bond rmsd (maximal deviation) | 0.016 Å (<0.1 Å) | 0.016 (<0.1 Å) |
| Angle rmsd (maximal deviation) | 3.34° (29°) | 3.31° (23°) |
| Improper rmsd (maximal deviation) | 2.13° (12°) | 2.44° (19°) |
| Ramachadran plot | Residues 276–305 and 325–345 | Residues 12–40 and 59–80 |
| Most allowed (most and additional allowed) | 83.9 (96.2) | 90.5 (97.4) |
| Structure precision | Residues 276–305 and 325–345 | Residues 12–40 and 59–80 |
| Mean ± variance | 0.72 ± 0.04 | 0.84 ± 0.05 |
Figure 1.
Comparison of the domain 4 and 6 structures. In (A) the backbone traces of 12 structures are superimposed. β-strands are in blue, loops in orange, ends in green (only their structured part have been represented here). The domain 6 possesses a short α-helix (colored in red) at its N-terminus. In (B) is represented a schematic (ribbon) view of one model using the same color code. In (C) are represented the residues involved in the packing of the β-strands forming the β-barrel. They are indicated in orange in the aligned sequences of the domains. On this alignment, we also indicated the glycines found at the strand extremities, conserved in all domains but not involved in the packing.
As awaited, both structures consist of the five-stranded β-barrel characteristic of the S1 domain structures (9). Their geometry is very similar; the rmsd between the Cα of the β-barrel and of the short loops connecting the strands B1 and B2, B2 and B3, and B4 and B5 is about 1.2 Å. The long loop between the strands B3 and B4 is mainly disorganized, but presents a propency to form a helix turn at each of its extremities. The β-barrels are stabilized through a set of similar hydrophobic interactions. In both barrels, three residues are involved in the case of the strands B1 (L/V-x-G-x-V), B2 (C/A-x-V-x-I/L), B3 (V-x-G-x-V/L) and B5 (I-x-L-x-L/V). Four hydrophobic residues and, more surprisingly, an aspartate are found in the case of the strand B4 (D-x-V-x-V/A-x-V/F-xx-I/V). Similarly, a set of conserved glycines is found at or near the extremities of the strands B1, B2, B3 and B4, which does not participate to the packing of the barrels, but seems important for the connections between the strands. The main difference between the two domains resides in the space between the parallel B3 and B5 strands (slightly wider in domain 6 than in domain 4) and the presence of a two-turns α-helix at the N-terminus extremity of the domain 6. However, the pertinence of these observations is difficult to assess. The absence of the adjacent domains, in particular, is likely to influence the structures of the extremities.
Domain 6 interactions with poly(U) and poly(A) RNAs
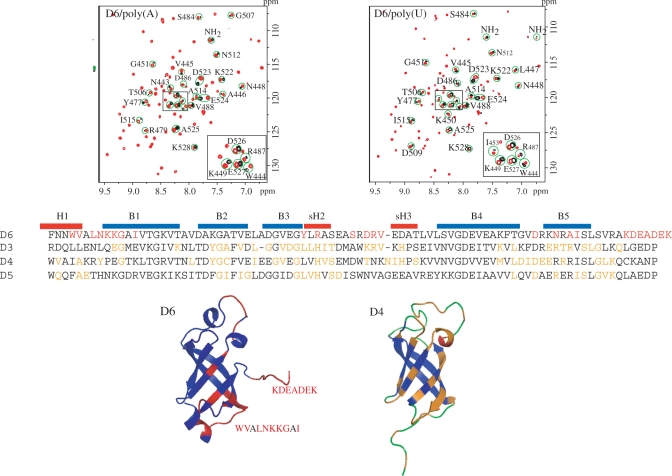
We recently characterized the interactions of an S1 fragment composed of the domains 3, 4 and 5 with three different RNAs: two substrates of RegB (a T4 phage ribonuclease whose activity is enhanced by S1) and one translation initiation region (7). We showed that the three RNAs are similarly recognized by S1 and that the interaction surface is formed on the same region of the three domains (Figure 2). We wanted to determine if the domain 6 also interacts with RNAs and if the interaction involves the same region as in the domains 3, 4 and 5. The domain 6 is dispensable for both RegB cleavage rate acceleration and translation initiation, raising the question of the choice of an RNA fragment to test the interaction. The protein S1 is known to bind with high affinity to poly(A) and poly(U) ribonucleotides (5). Accordingly we chose these two molecules.
Figure 2.
Identification of the domain 6 residues affected by poly(A) and poly(U) binding. The differences map were obtained by subtracting the domain 6 15N-HSQCs recorded in the presence of poly(A) or poly(U) from the reference spectrum. A slight imbalance between the two spectra was introduced in the subtraction to conserve the trace of the whole HSQC spectrum (in red). The affected residues are in red in the domain 6 structure and sequence. The residues of the domains 3, 4 and 5 previously identified as involved in RNA binding are in orange (7).
The results of the titration experiments are reported in the Figure 2. In both cases, the system is in the rapid exchange regime. There are many differences between the reference spectrum of the domain 6 and those recorded in the presence of poly(A) or poly(U) (at 20:1 nucleotide:protein ratio). The comparison of the two difference maps clearly shows that the effects induced by the two RNAs are very similar. The localization of the affected amino acids on the protein structure reveals that most of them (15/25) are located in the N- and C-terminal extremities of the domains. The others are in the strand B3 (Tyr477, Arg479), in the following long loop (Ser484, Asp486, Arg487 and Val488), and at the apex of the hairpin formed by the strands B4 and B5 (Asp509, Asn512, Ala514 and Ile515). The significance of the perturbations observed in the domain extremities is difficult to interpret in the absence of the flanking elements (the domain 5 at the N-terminus, an extension of 40 amino acids at the C-terminus). However, it is worth noticing that the 40 amino acid extension is not structured and does not interact with the rest of the protein (8). Accordingly, its removal is likely to have no or little influence on the properties of the domain. In addition, Bernstein et al. (18) showed that a mutation of Ala530 (two residues downstream the end of our fragment) influences the ability of S1 to initiate the translation of foreign messengers in E. coli, comforting the idea that this extremity could interact with RNAs. All other affected amino acids, except Ser484, occupy positions also affected in the domains 3, 4 and 5 in the presence of different RNAs (7). In addition, the affected Tyr477, in the middle of the strand B3, is another of the residues identified by Bernstein et al. All these results strongly suggest that the domain 6 binds RNAs. The binding area encompasses residues at the surface of the β-barrel also found in the case of the domain 3, 4 and 5, but is smaller. Finally, the C-terminal, unstructured, region could play a role in the domain 6/RNA interaction.
S1 domain specialization
One of our long-term purposes is to analyze the functional specialization of the domains of the S1 proteins. In particular, we wondered if the positions of the domains in the long Gram-negative S1 proteins correspond to different domain characteristics, or if some of them (for example, the positions 1 and 2, or 3, 4 and 5) could be equivalent. Similarly, in the case of a functional specialization of the domains, we wondered if the different functions would always correspond to the same positions or if swapping could occur.
To approach this question, we built a phylogenic tree derived from the alignment of S1 protein sequences representative of the different Gram-negative bacteria groups. For this, it was crucial to dispose of a reliable sequence alignment. We took advantage of the characteristic sequences of the β-strands identified by analyzing the structures of the E. coli domains 4 and 6. Using these sequences as a starting point, we could define a consensus sequence for each strand. As shown in the Supplementary Figure 1, the consensus sequences characteristic of the strands B1 (V/I/L–x–G–x-V/I), B2 (φ-x-V/L/I-x-φ) and B4 (D/Q/E-x-V/I/L-x-V/A/F-x-V/I/L-x-x-φ) are well conserved throughout the sequences of the six domains. In the case of the strands B3 and B5, the consensus sequences (B3: V/I-x-G-x-L/V/I; B5: I/V/L-x-L-x-L/I/V/M) are well conserved for the domains 3 to 6, but are more degenerated in the domains 1 and 2. In the case of the strand B3, the Gly is replaced by an Ala in the domain 2 and the first hydrophobic residue is missing in the domain 1 sequences excepted those of S. melitoti and A. butzleri. In the case of the strand B5, the first hydrophobic residue is missing in domain 1 and 2 sequences excepted those of B. burgdorferi. However, the remaining elements and the other residues of the strands were sufficient to align them. In a second step, the interstrand regions were aligned using clustalW2 (15).
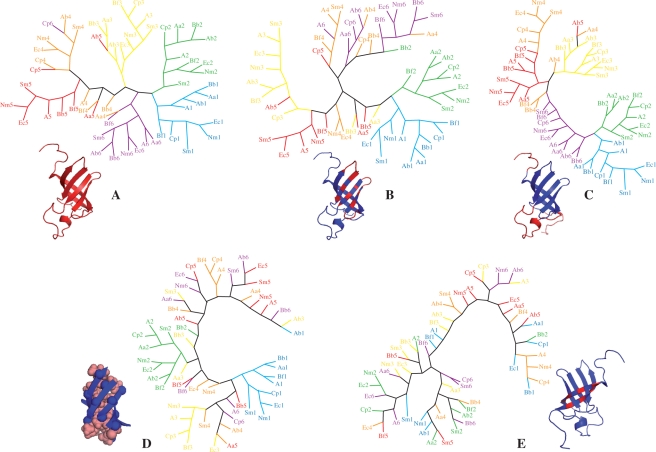
The tree calculated by using the complete sequences of the domains (amino acid set A) is represented in the Figure 3A. This tree shows a clear segregation of the domains as a function of their position in the proteins. The segregation is complete or nearly complete for the domains 1, 2, 3 and 6, while the domains 4 and 5 are more imbricate. This readily indicates that two domains occupying a similar position in two different S1 proteins are evolutionary closer that two domains occupying different positions in the same S1 protein, suggesting that the evolution of the domains at each position is specifically constrained, and accordingly, that each position could correspond to a specific function. This also suggests that there is no possibility of swapping the different positions.
Figure 3.
Phylogenic trees built from the alignment of domain sequences of the Gram-negative S1 proteins (see ‘Materials and Methods’ section). Each domain sequence is represented by the initials of the bacteria followed by the number of the domain (e.g. Ec5 stands for Escherichia coli domain 5). The sequences of the domains 1 are in cyan, 2 in green, 3 in yellow, 4 in orange, 5 in red and 6 in magenta. (A) Tree built by using all amino acids of the domains. (B) Tree built by using the exposed residues of the ‘front side’ of the β-barrel (i.e. strands B2, B3 and B5 forming the RNA-binding area) and of the short loops connecting them. (C) The tree built by using the amino acids of the two linkers and of the long loop connecting the strands B3 and B4. (D) Tree built by using the residues of the β-barrel hydrophobic core. (E) Tree built by using the exposed residues of the ‘back side’ of the β-barrel (strands B1 and B4). In all cases, the positions of the residues used for the tree building are represented in red on the structure of the domain 4.
We also tried to determine whether it was possible to relate the global difference between the domains occupying different positions in the S1 protein, evidenced in the previous tree, to a given structural region of the same domains. For this, we built four supplementary evolutionary trees by using subsets of amino acids corresponding to different parts of the domains (Figure 3B–E). We probed the amino acids at the surface of the strands B1, B3 and B5 and in the loops preceding them (set B). They correspond to the RNA-binding area we identified previously on domains 3, 4, 5 (7) and in this study on domain 6. We also probed the amino acids of the long loop between the strands B3 and B4 and of the two extremities of the domains (set C), which are likely involved in the interdomain interactions in the case of the domains 3 and 4 (7). We finally probed the amino acids of the domain hydrophobic core (set D) and those exposed at the surface of the strands B1 and B4 (set E), that constitute the ‘back’ of the domains with respect to the RNA interaction area.
Both the set B (RNA interaction surface) and C (long loop and extremities) lead to segregation of the domains as a function of their position. In the tree built with set B, the domains 1, 2 and 3 belong independent branches. Five domains 5 are gathered, while the four others are dispersed. The domains 4 and 6 are imbricate in a last branch. In the case of the set C, the domains 1, 2, 3, 5 and 6 constitute distinct clusters. Five domains 4 are gathered at the extremity of the branch formed by the domains 5, the others being dispersed. On the opposite, there is little or no segregation in the trees built from the set D (hydrophobic core) and E (back of the β-barrel). In the case of the hydrophobic core, we observe two branches corresponding to the domains 1 and 2. In the case of the back of the β-barrel, there is no segregation at all. However, the small number of residues constituting this last set (seven residues) is likely to bias this result.
According to all these observations, the domains 1 and 2 seem to be the most specific, as they form independent branches when considering the RNA-binding area (set B), the long loop and the extremities (set C) and the hydrophobic core (set D). It is tempting to rely this to the fact that these domains play a particular role, as they are responsible for ribosome binding, while the others are involved in the interactions with mRNAs. However, an interesting point is that the domains 1 and 2 are also segregating from each other, strongly suggesting the idea that they have different roles in the ribosome binding. The other domains seem more homogeneous from a structural point of view (they are indiscernible in the tree built using the hydrophobic core). Among them, the domains 3 occupy a place apart, as they segregate in the trees built using the sets A (all residues), B (RNA-binding area) and C (long loop and extremities), while the domains 4 and 5 are associated in the tree built with all residues and the domains 4 and 6 are associated in the tree built with the residues of the RNA-binding surface.
S1 protein evolution
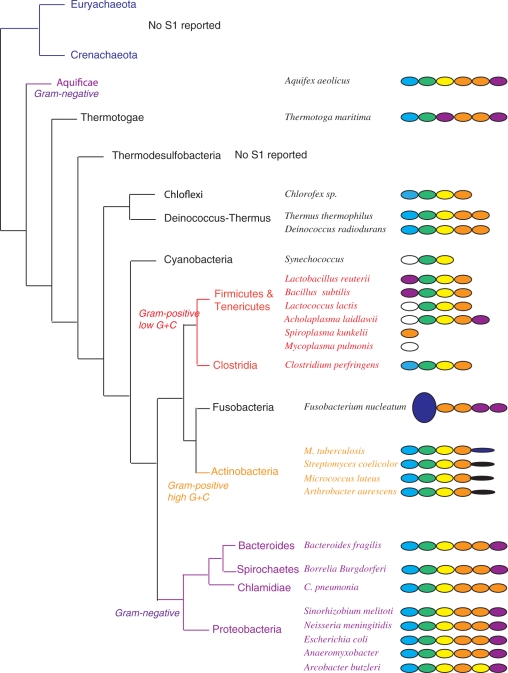
A second purpose was to probe the relationship between the S1 proteins belonging to the Gram-negative and Gram-positive bacteria. Considering the possibility to identify the domains of the Gram-negative S1 proteins by the means of the consensus sequences of the β-barrel and to discriminate between the domain types (domains 1, 2, 3, 4–5 and 6), we wondered whether this was transposable to the Gram-positive forms of the protein. Indeed, this would allow us to establish the number and the nature of the domains present in the Gram-positive S1 proteins, and therefore to verify whether the order of the domains is conserved in all S1 proteins and which domains are missing in the shorter forms. We chose to analyze a set of 17 sequences corresponding to the main divisions of Gram-positive bacteria proposed by Olsen and collaborators (14).
We were able to locate the S1 domains in all Gram-positive S1 proteins by using the β-barrel consensus sequences (Supplementary Figure 1). In all but one case, the number of domains we found is identical to that indicated in the sequence records. We identified six domains in the sequence of Thermotoga maritima S1 instead of the five reported in the CAB08883 file. Each Gram-positive domain sequence was then tested against an hmm profile library built from the alignments of the sequences of the Gram-negative S1 domains (Figure 4 and Supplementary Figure 2). Seven profiles were considered, one for each domain position (i.e. one from the domain 1 alignment, one from the domain 2…) and one from the simultaneous alignment of the domains 4 and 5, to take into account the fact that they are not discernable in the phylogenic tree built using the whole sequences. We validated this library by testing several domains from Gram-negative S1 proteins not used to build the profile. We also determined a significance threshold value (about 10) by testing several S1 domain sequences not belonging an S1 protein.
Figure 4.
Schematic representation of the organization of protein S1 found in a set of bacteria representative of the classification proposed by Olsen and collaborators (14). The domains identified as domain 1 are in blue, as domain 2 in green, as domain 3 in yellow, as domains 4/5 in orange and as domain 6 in magenta. The domains possessing the consensus sequences characteristic of the β-barrel of the domains of protein S1 but presenting a score against any profile below 10 are in white. The domains not identified as S1 domains are in deep blue.
In all records we retrieved but those of Mycoplasma pulmonis, S. kunkelii (tenericutes) and F. nucleatum, (see below), the two first domains are referenced as similar to the E. coli S1 domains 1 and 2, respectively. We confirmed this result for the second domain. We also confirmed it for the first domain in the case of the actinobacter M. tuberculosis, S. coelicolor, M. luteus, A. aurescens, the deinococcus-thermus Thermus thermophilus, D. radiodurans, the clostridia C. perfirngens and the thermotogae T. maritima. However, in the case of the firmicutes Lactobacillus reuterii, Bacillus subtilis, Lactococcus lactis, the tenericutes Archoplasma laidlawii, the cyanobacteria Synechococcus sp. and the chloroflexi Chloroflex sp., the score obtained by the first domain against any profile is negative or very low (below 10) and the best value (except in the case of Chloroflex sp.) does not correspond to the domain 1 profile. In all these sequences, the identification proposed in the data banks for the first domain thus seems questionable. Finally, the ‘S1 protein’ of S. kunkelii and M. pulmonis is formed of a unique domain, which seems to be a domain 4 in the case of S. kunkelii and not an S1 protein domain in the case M. pulmonis. The F. nucleatum (Fusobacteria) protein S1 is the fusion between protein LytB (residues 1–286) and four S1 domains (residues 450–800). None of these four domains is a domain 1 or 2 and no other S1 domains could be identified in the region 290–450.
Most of the other domains (23/34) are designated as ‘S1-like’ in the records. In fact, all of them correspond to S1 protein domains and are in all but three cases ordered as in the Gram-negative proteins. The exceptions concern the third domain of the T. maritima, the fifth domain of A. laidlawii and the penultimate domain of the F. nucleatum proteins that were identified as domain 6. However, in the cases of T. maritima and F. nucleatum, the scores between the domain 6 and 4–5 profiles are very close. In addition, it should be noticed that some domain swaps also occur in the Gram-negative proteins. All these results tend to show that all S1 proteins (excepted those formed of one unique domain and maybe that of F. nucleatum, for which nothing definitive can be told) are related and that the shorter forms are due to the loss of one (T. thermophilus, D. radiodurans, A. laidlawii), two (L. reuterii, B. subtilis, L. lactis, Clostridium perfringens, Chloroflex sp. and all the actinobacteria) or even three (Synechococcus sp.) domains at their C-terminal extremities.
DISCUSSION
A partial functional specialization of the domains of the E. coli protein S1 is attested by many studies. It has long been known that the two first domains are responsible for the ribosome binding (5), while the four following are involved in the interactions with mRNAs. It was further shown that the sixth domain is dispensable for translation initiation (6). Similar observations were reported in the case of the other functions of S1. The two first domains are responsible for the binding of S1 to the Qβ phage RNA replicase, while the sixth domain is dispensable for its activity in the phage replication (19). We demonstrated that the S1 fragment formed by the third, fourth and fifth domains enhances the activity of the T4 phage ribonuclease RegB as efficiently as the whole protein (8). Accordingly, S1 seems formed of three main regions: the N-terminal region, formed of the first and second domains and involved in the interaction with S1 other partners in the cell (ribosome, Qβ replicase), the intermediate region, formed of the third, fourth and fifth domains and involved in the interactions with the RNAs (translation or replication initiation region, RegB substrates) and, finally, the sixth domain, whose role remains to be elucidated.
We hoped that the comparison of the structures and RNA-binding properties of the fourth and sixth domains of S1 could help us to understand the origin and the functional differences observed between the intermediate region and the sixth domain. In fact, the two structures are very similar to each other and very close to those of many other S1 domains (as revealed by a VAST search, not shown). In addition, our RNA titration experiments ((7) and this study) indicate that the four C-terminal domains (third, fourth, fifth and sixth) interact with RNAs and that their RNA-binding areas are located on the same side of the β-barrel. We observed some differences: the number of residues involved in the RNA binding at the surface of the β-barrel is smaller in the case of the sixth domain and several residues of the unstructured fragment following this domain could play a role, while the corresponding positions in the other domains are not affected. However, these differences are difficult to interpret: we do not know the real target of the sixth domain and we studied its interaction with RNAs by using a protein fragment containing only the domain (while our previous study was performed using the F3–5 fragment containing the associated third, fourth and fifth domains).
However, the possibility to define a consensus sequence for each of the β-strands of the β-barrels of the proteins S1 domains gave us the opportunity to progress in the analysis of their specialization. Indeed, this allowed us to precisely align the sequences of the domains of very different protein S1 belonging to different Gram-negative bacteria and consequently to analyze the relationship between these domains. The Phylogenic trees built using this sequence alignment not only confirmed the functional specialization already observed but also extended it (Figure 3A). Domains 1 and 2 are found in two different branches indicating that they do not play the same role in ribosome binding. Neither are domains 3, 4 and 5 equivalent. Domains 4 and 5 are found in the same branche, while domains 3 are well segregated. It is tempting to relate this to the functional and structural differences observed between the E. coli domains 3, 4 and 5 in the F3–5 fragment. Indeed, in this fragment, the domains 4 and 5 are associated while the domain 3 is free to move in the absence of RNAs (7). In addition, the subfragment formed of the domains 4 and 5 has an activity by itself in RegB activition while the domain 3 is only active when associated to the two others (8). The other trees of the Figure 3, built by using weighted alignments of the sequences, reveal that the differences observed between the domains 3, 4, 5 and 6 are mainly due to differences in the RNA-binding surface, in the extremities and in the long loop likely involved in the domain/domain interactions, while the β-barrel hydrophobic core seems conserved. On the opposite, domains 1 and 2 present differences, between them and with the others, at the level of their hydrophobic cores, suggesting that the specialization between RNA- and ribosome- (presumably protein) binding domains could induce repercussions on the structure of their β-barrels. In view of this hypothesis, the determination of the structures of the domains 1 and 2 becomes an important objective.
Yet another open question is the relationship between the various forms of the proteins S1 (noted as S1 in the data banks). Using hmm profiles built from the alignment of the sequences of the domains of the Gram-negative proteins, we were able to identify the type of the domains of the Gram-positive proteins. This first show that most of the retrieved proteins are likely related to the Gram-negative forms. They are mainly composed of S1 domains clearly related to those of the Gram-negative proteins and, more significantly, disposed in the same order. Nevertheless, a few are questionable, in particular the ‘S1’ proteins of S. kunkelii and M. pulmonis (tenericutes) that are formed of a unique domain and that of F. nucleatum, which possesses four S1 domains (two domains 4/5 and two domains 6) but in fusion with another, unrelated, large protein. Second, in 1991, Farwell and Rabinowitz (20) proposed that both the Gram-negative and the high G+C content Gram-positive bacteria possess a functional ribosomal protein S1, but not the low G+C Gram-positive bacteria. They related the presence of S1 to the absence of translational specificity in the two first groups. However, their study was based on limited data concerning the presence or absence of S1 and on contradictory results concerning the functionality of the putative S1 proteins. In particular, even now the statement that the S1 protein of the low G+C content Gram-positive bacteria is not a ribosomal protein seems to lay on only two studies, the first realized by Isono and Isono in 1976 and showing that there is no equivalent of the S1 protein in the Bacillus stearothermophilus ribosome (21), the second realized in 1982 by Higo and co-workers (22) on the B. subtilis ribosome (22) and leading to the same result. Similarly, the statement that high G+C content Gram-positive bacteria contain an active S1 ribosomal protein mainly relies on the report that the protein S1 of M. luteus increases the translation of poly(U) and of several natural mRNAs by both the E. coli and M. luteus ribosome (23, 24). But, at the same time, the protein S1 of Streptomices aurefasciens (another high G+C content Gram-positive bacteria) was shown to have no activity on the ribosome (25). In the case of the high G+C content Gram-positive bacteria, our results suggest that their S1 proteins are indeed ribosomal proteins. They possess the ribosome-binding domains 1 and 2 at their N-terminal end. These proteins also possess a domain 3 and a domain 4/5 at the correct positions, but the second domain 4/5 is replaced by an unknown domain of 86–100 amino acids, which seems specific of the actinobacteria (as shown by a BLAST search against all bacteria genomes). Accordingly, it is likely that these proteins are not functionally equivalent to the Gram-negative S1. In the case of the low G+C Gram-positive bacteria, the situation seems more complicated. As previously shown, it is not clear whether the tenericutes S. kunkelii and M. pulmonis possess a protein related to the Gram-negative S1. The other firmicutes and tenericutes we looked at (L. reuterii, B. subtilis, Lactococcus lactis, A. laidlawii) and the clostridia C. perfringens possess a protein related to S1. In the case of the firmicutes and tenericutes, the first domain of the protein is never a domain 1 (the score against any profile is very low and the ‘best’ value never corresponds to a domain 1). In the case of C. perfringens, the first domain is identified as a domain 1, but with a lower score than that obtained by the first domain of the high G+C proteins (22 instead of 70–75). This suggests a more or less pronounced loss of the first domain ‘identity’ that supports the idea that these proteins loss the ability to bind the ribosome (but the case of C. perfringens would deserve a deeper investigation).
Besides the Gram-negative, we identified two other groups of bacteria that potentially have a functional protein S1, the thermotogae and the deinococcus-thermus. The protein S1 of T. maritima is formed of six domains, the only difference with the proteins of the Gram-negative bacteria being the fact that the third domain is slightly closer of a domain 6 than of a domain 3 or 4/5 (score of 59 instead of 58 for a domain 4/5 and 50 for a domain 3). The proteins S1 of T. thermophilus and D. radiodurans seem formed of five domains corresponding to the first five domains of the Gram-negative S1. The protein S1 of T. thermophilus was indeed shown to be bound to the ribosome (26). It was described to be formed of six domains, but the alignment reported by Shiryaev et al. for the presumed sixth domain is very poor and this domain lacks the residues characteristic of the β-barrel. Finally, it seems that the S1 protein of the chloroflexi (that are found next to the deinococcus-thermus in the classification) is similar to that of the clostridiae and that the cyanobacteria, as the firmicutes and tenericutes, possess a short form (three domains) with a degenerated domain 1.
From our study, it seems thus possible to form four groups of protein S1. The Gram-negative bacteria (including the aquificae), the thermotogae and the deinococcus-thermus possess a protein S1 formed of, at least, the five first domains in the correct order. They very likely correspond to functional ribosomal proteins. The actinobacteria (high G+C content Gram-positive bacteria) possess a shorter form that have conserved the two first domains, but have an unknown domain instead of the fifth. They are likely ribosomal proteins, but their function in translation initiation is questionable. The firmicutes, tenericutes and cyanobacteria possess shorter forms of the protein in which the first domains seem no more a domain 1, suggesting that they all lost the ability to bind the ribosome. Finally, the chloroflexi and the clostridiae seem intermediate between the second (actinobacteria) and the third groups. They lost the fifth domain and possess a first domain identifiable as a domain 1 but with a very low score. Considering the respective positions of these groups and their interweaving, this strongly suggests that the protein S1 found in the Gram-negative bacteria correspond to an ancient function conserved in some branches (the aquificae, thermotogae, deinococcus-thermus, proteobacterie, chlamidiae, spirochetes and bacteroides) and lost in the others through the loss of functional domains (the fifth in most case, the fourth and the fifth in others) and/or the loss of their ability to bind ribosome. To test these hypotheses, experimental evidences are of course needed. It would be interesting to verify if the identification of a domain 1 and a domain 2 by our method in the sequence of an S1 protein indeed correlates with the ability of this protein to bind the ribosome. It would also be interesting to verify if the S1 proteins of T. maritima and D. radiodurans are necessary to the translation initiation.
ACCESSION NUMBERS
2KHI and 2KHJ.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Centre National pour la Recherche Scientifique; the Universités Paris VI et Paris VII; the Institut de Chimie des Substances Naturelles (UPR2301 du CNRS) (to P.S. and P.A.); and the Fondation pour la Recherche Medicale (to M.B. and P.A.). Funding for open access charge: Institut de Chimie des Substances Naturelles.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
We are grateful to Bernard Gilquin for the time he spent helping us running INCA and to Joel Pothier and Mathilde Carpentier for their advice concerning the bioinformatics aspect of this study. We also are grateful to Eric Jacquet for the technical support we received in his laboratory, and to Eric Guittet's team members for many interesting and helpful discussions.
REFERENCES
- 1.Schmitt E, Guillon JM, Meinnel T, Mechulam Y, Dardel F, Blanquet S. Molecular recognition governing the initiation of translation in Escherichia coli. A review. Biochimie. 1996;78:543–554. doi: 10.1016/s0300-9084(96)80001-0. [DOI] [PubMed] [Google Scholar]
- 2.Komarova AV, Tchufistova LS, Supina EV, Boni IV. Protein S1 counteracts the inhibitory effect of the extended Shine-Dalgarno sequence on translation. RNA. 2002;8:1137–1147. doi: 10.1017/s1355838202029990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sorensen MA, Fricke J, Pedersen S. Ribosomal protein S1 is required for translation of most, if not all, natural mRNAs in Escherichia coli in vivo. J. Mol. Biol. 1998;280:561–569. doi: 10.1006/jmbi.1998.1909. [DOI] [PubMed] [Google Scholar]
- 4.Boni IV, Isaeva DM, Musychenko ML, Tzareva NV. Ribosome-messenger recognition: mRNA target sites for ribosomal protein S1. Nucleic Acids Res. 1991;19:155–162. doi: 10.1093/nar/19.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subramanian AR. Structure and functions of ribosomal protein S1. Prog. Nucleic Acid Res. Mol. Biol. 1983;28:101–142. doi: 10.1016/s0079-6603(08)60085-9. [DOI] [PubMed] [Google Scholar]
- 6.Boni IV, Artamonova VS, Dreyfus M. The last RNA-binding repeat of the Escherichia coli ribosomal protein S1 is specifically involved in autogenous control. J. Bacteriol. 2000;182:5872–5879. doi: 10.1128/jb.182.20.5872-5879.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aliprandi P, Sizun C, Perez J, Mareuil F, Caputo S, Leroy JL, Odaert B, Laalami S, Uzan M, Bontems F. S1 ribosomal protein functions in translation initiation and ribonuclease RegB activation are mediated by similar RNA-Protein interactions: an NMR and SAXS analysis. J. Biol. Chem. 2008;283:13289–13301. doi: 10.1074/jbc.M707111200. [DOI] [PubMed] [Google Scholar]
- 8.Bisaglia M, Laalami S, Uzan M, Bontems F. Activation of the RegB endoribonuclease by the S1 ribosomal protein is due to cooperation between the S1 four C-terminal modules in a substrate-dependant manner. J. Biol. Chem. 2003;278:15261–15271. doi: 10.1074/jbc.M212731200. [DOI] [PubMed] [Google Scholar]
- 9.Bycroft M, Hubbard TJ, Proctor M, Freund SM, Murzin AG. The solution structure of the S1 RNA binding domain: a member of an ancient nucleic acid-binding fold. Cell. 1997;88:235–242. doi: 10.1016/s0092-8674(00)81844-9. [DOI] [PubMed] [Google Scholar]
- 10.Malliavin TE, Pons JL, Delsuc MA. An NMR assignment module implemented in the Gifa NMR processing program. Bioinformatics. 1998;14:624–631. doi: 10.1093/bioinformatics/14.7.624. [DOI] [PubMed] [Google Scholar]
- 11.Bartels C, Xia T-Hh, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 12.Savarin P, Zinn-Justin S, Gilquin B. Variability in automated assignment of NOESY spectra and three-dimensional structure determination: a test case on three small disulfide-bonded proteins. J. Biomol. NMR. 2001;19:49–62. doi: 10.1023/a:1008311110174. [DOI] [PubMed] [Google Scholar]
- 13.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J. Biomol. NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 14.Olsen GJ, Woese CR, Overbeek R. The winds of (evolutionary) change: breathing new life into microbiology. J. Bacteriol. 1994;176:1–6. doi: 10.1128/jb.176.1.1-6.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 16.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 17.Lopez-Mendez B, Guntert P. Automated protein structure determination from NMR spectra. J. Am. Chem. Soc. 2006;128:13112–13122. doi: 10.1021/ja061136l. [DOI] [PubMed] [Google Scholar]
- 18.Bernstein JR, Bulter T, Shen CR, Liao JC. Directed evolution of ribosomal protein S1 for enhanced translational efficiency of high GC Rhodopseudomonas palustris DNA in Escherichia coli. J. Biol. Chem. 2007;282:18929–18936. doi: 10.1074/jbc.M701395200. [DOI] [PubMed] [Google Scholar]
- 19.Guerrier-Takada C, Subramanian AR, Cole PE. The activity of discrete fragments of ribosomal protein S1 in Qbeta replicase function. J. Biol. Chem. 1983;258:13649–13652. [PubMed] [Google Scholar]
- 20.Farwell MA, Rabinowitz JC. Protein synthesis in vitro by Micrococcus luteus. J. Bacteriol. 1991;173:3514–3522. doi: 10.1128/jb.173.11.3514-3522.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isono K, Isono S. Lack of ribosomal protein S1 in Bacillus stearothermophilus. Proc. Natl Acad. Sci. USA. 1976;73:767–770. doi: 10.1073/pnas.73.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Higo K, Otaka E, Osawa S. Purification and characterization of 30S ribosomal proteins from Bacillus subtilis: correlation to Escherichia coli 30S proteins. Mol. Gen. Genet. 1982;185:239–244. doi: 10.1007/BF00330792. [DOI] [PubMed] [Google Scholar]
- 23.Muralikrishna P, Suryanarayana T. Structural and immunochemical characterization of a ribosomal protein from gram-positive Micrococcus luteus which is functionally homologous to Escherichia coli ribosomal protein S1. Eur. J. Biochem. 1987;167:299–305. doi: 10.1111/j.1432-1033.1987.tb13336.x. [DOI] [PubMed] [Google Scholar]
- 24.Farwell MA, Roberts MW, Rabinowitz JC. The effect of ribosomal protein S1 from Escherichia coli and Micrococcus luteus on protein synthesis in vitro by E. coli and Bacillus subtilis. Mol. Microbiol. 1992;6:3375–3383. doi: 10.1111/j.1365-2958.1992.tb02205.x. [DOI] [PubMed] [Google Scholar]
- 25.Mikulik K, Smardova J, Jiranova A, Branny P. Molecular and functional properties of protein SS1 from small ribosomal subunits of Streptomyces aureofaciens. Eur. J. Biochem. 1986;155:557–563. doi: 10.1111/j.1432-1033.1986.tb09524.x. [DOI] [PubMed] [Google Scholar]
- 26.Shiryaev VM, Selivanova OM, Hartsch T, Nazimov IV, Spirin AS. Ribosomal protein S1 from Thermus thermophilus: its detection, identification and overproduction. FEBS Lett. 2002;525:88. doi: 10.1016/s0014-5793(02)03092-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.