Abstract
Stochastic expression is a hallmark of the Ly49 family that encode the main MHC class-I-recognizing receptors of mouse natural killer (NK) cells. This highly polygenic and polymorphic family includes both activating and inhibitory receptor genes and is one of genome's fastest evolving loci. The inhibitory Ly49 genes are expressed in a stochastic mono-allelic manner, possibly under the control of an upstream bi-directional early promoter and show mono-allelic DNA methylation patterns. To date, no studies have directly addressed the transcriptional regulation of the activating Ly49 receptors. Our study shows differences in DNA methylation pattern between activating and inhibitory genes in C57BL/6 and F1 hybrid mouse strains. We also show a bias towards bi-allelic expression of the activating receptors based on allele-specific single-cell RT–PCR in F1 hybrid NK cells for Ly49d and Ly49H expression in Ly49h+/− mice. Furthermore, we have identified a region of high sequence identity with possible transcriptional regulatory capacity for the activating Ly49 genes. Our results also point to a likely difference between NK and T-cells in their ability to transcribe the activating Ly49 genes. These studies highlight the complex regulation of this rapidly evolving gene family of central importance in mouse NK cell function.
INTRODUCTION
Natural killer (NK) cells constitute an important part of the body's defence both as centurions of innate immunity and as communicators and collaborators of the adaptive immune system. They exert their function via the interpretation of signals received from their surface receptors. Normal cells display major histocompatibility complex (MHC) class I molecules that are recognized by a number of inhibitory receptors on the surface of NK cells. The inhibitory receptors prevent activation of NK cells and the destruction of normal, MHC class I-possessing cells (1). NK cells also possess stimulatory (activating) receptors that recognize other molecules on the surface of potential target cells which are mostly pathogen-encoded or stress-induced self proteins (2–4).
NK activation and the killing of target cells therefore depend on the balance between stimulatory and inhibitory signals received from these surface receptors. In primates and some other mammals, the killer cell immunoglobulin-like (KIR) family of genes code for the main MHC class-I recognizing receptors (5). However, in rodents, the structurally unrelated Ly49 family provides this function. Both KIR and Ly49 families are polygenic and polymorphic among different individuals and mouse strains (6,7).
The KIR gene cluster of human is ∼150 kb containing approximately 14 genes and pseudogenes with very high coding and regulatory sequence similarity (8) indicating that they arose through recent gene duplication events (9). There is great diversity in both gene number and sequence polymorphisms for the KIRs among different people (6). The Ly49 gene cluster has also arisen from recent duplications and gene conversions of ancestral genes (10–12). Hence, there is high sequence similarity both in the coding and non-coding regions among the majority of the genes. The Ly49 cluster includes 16 genes and pseudogenes spanning over 600 kb in the C57BL/6 (B6) mouse strain and is located in the natural killer gene complex (NKC) region of chromosome 6 (8,13). There are two functional Ly49 activating receptors, Ly49D and H, coded by the B6 genome. Ly49D binds to the MHC-class-I allele H2-Dd (14,15) and Ly49H binds to the m157 protein of mouse cytomegalovirus (MCMV) conferring resistance towards virus infection in the strains of mice that express Ly49H (2,3,16,17). Unlike the inhibitory Ly49 receptors that are expressed on T and NKT cells as well as NK cells, the stimulatory Ly49 receptors are only expressed on NK cells (18). Lack of a well-defined upstream promoter-1 (Pro-1) region, which likely acts as a stochastic switch for inhibitory receptor genes in immature NK cells (19,20) and reports of the higher co-expression of Ly49D and H and deviation from the product rule (21), suggest that the activating Ly49 genes, despite being surrounded by inhibitory genes in the Ly49 gene cluster, are subject to distinct regulatory mechanisms.
DNA methylation of the 5′-region of the inhibitory Ly49 genes correlates with their expression. In the case of Ly49a and Ly49c, where stochastic mono-allelic expression has been demonstrated, we have previously shown that DNA methylation and mono-allelic receptor expression correlate (22). The existence of stochastic mono-allelic expression and/or correlation with DNA methylation is unknown for the activating receptors. Here, we have investigated the link between DNA methylation of the 5′-region of activating receptors and the maintenance of their expression. Furthermore, we show evidence for a mode of transcriptional regulation for the activating Ly49 genes that differs from that of the inhibitory genes.
MATERIALS AND METHODS
Mice
All C57BL/6 mice were bred and maintained in the animal facility of the British Columbia Cancer Research Centre (Vancouver, British Columbia, Canada). 129SvEvTac and 129SvEvTac/C57BL6 F1 hybrids were ordered from Taconic farms. NOD/ShiLtJ and C57BL6/NOD-ShiLtJ F1 hybrids were ordered from the Jackson Laboratory (Bar Harbor, Maine, USA). All mice used in this study were more than 6-weeks old. The B6.BXD8/B6 (23) and age-matched B6 mice were 6.5-weeks old. All experiments were according to a protocol approved by the Committee on Animal Care of the University of British Columbia.
Antibodies, cell separation and flow cytometry
The monoclonal antibody (mAb) anti-FcRγ (2.4G2) (24,25) and 3D10-FITC (21) have been described before. Anti- CD3ε-PerCP-Cy5.5, anti- NK1.1-PE, anti-NK1.1-APC, anti-DX5-PE (alpha-2 integrin, CD49b), 1F8-FITC (anti-Ly49C/I/H), 5E6-biotin (anti-Ly49C/I), 4E5-FITC (anti-Ly49D), A1-biotin (anti-Ly49AB6) and fluorochrome-conjugated streptavidin were purchased from BD Biosciences (Mississauga, Ontario, Canada). The anti-mouse NKp46-PE antibody was purchased from eBioscience (San Diego, CA, USA). The 12A8 purified antibody was generously provided by Dr Stephen Anderson (NCI, Frederick, MD, USA) and was conjugated to FITC via Thermo Scientific Pierce EZ-label fluorescein isothiocyanate protein labelling kit (Rockford, IL, USA) per manufacturer's protocol. Flow cytometry for cell sorting was performed on Cytopeia Influx cell sorter and Becton Dickinson FACSAria Cell Sorting System (Mississauga, Ontario, Canada). All sorted samples were >95% pure or resorted for high purity. For the single-cell sorting, NK cells were sorted into wells containing JEG-3 (human choriocarcinoma) as carrier cells. Doublet discrimination was applied to avoid more than one cell per well.
Primary cell and tissue genomic DNA extraction
Genomic DNA (gDNA) was obtained from FACS sorted cells as described before (22). gDNA was extracted from fresh B6 mouse liver using DNAzol reagent (Invitrogen) per manufacturer's instructions. Further proteinase K digestion and phenol–chloroform extraction was performed.
Sodium bisulfite conversion and PCR
Bisulfite conversion was performed as described previously (22). The conversion rate was >98%. First-round PCR amplification of Ly49h 5′ region was performed using Ly49h forward and reverse flanking bisulfite primers. Converted DNA was used as template in a 45 µl reaction volume, containing 30 pmol of each primer, 1 mM dNTPs, 3 mM MgCl2 and 0.5 U Taq Platinum DNA polymerase (Qiagen). After initial denaturation for 7 min at 95°C, 30–40 cycles were performed, each consisting of 90 s at 95°C, 55 s at 50°C and 40 s at 72°C with a final extension of 7 min at 72°C. Two microliters of the first PCR was used for nested amplification using Ly49h forward and reverse nested bisulfite primers. The same amplification conditions were chosen as for first-round PCR with the exception that the annealing temperature was changed to 48 + 0.1°C/cycle. All bisulfite primer sequences are shown in Table 1.
Table 1.
Oligonucleotide sequences of primers and probes
| Ly49h forward flanking bisulfite primer | 5′-ATA GGG GAA TGT TAG GGT TAA AAA G-3′ |
| Ly49h reverse flanking bisulfite primer | 5′-ATT TAA CCT AAT ATA ACA CAA CCA A-3′ |
| Ly49h forward nested bisulfite primer | 5′-GGA TAT ATG TTT TGT TTT TTT TGG T-3′ |
| Ly49h reverse nested bisulfite primer | 5′-TAA CAC AAC CAA AAA AAC TCT CAA C-3′ |
| Ly49d forward flanking bisulfite primer | 5′-TAT TAA GAT GTA ATT AGT ATG ATT TAA T-3′ |
| Ly49d reverse flanking bisulfite primer | 5′-ACA ATA CAT TTA TAC ACT TCA CCT AA-3′ |
| Ly49d reverse nested bisulfite primer | 5′-CCA AAT ACT ACA AAA AAA ATA ACT ATA T-3′ |
| Ly49d forward nested bisulfite primer | 5′-AGG TAG AGT TAT AGG TAA TAA TAG T-3′ |
| Ly49d/r reverse nested bisulfite primer | 5′-TTC CTC TAC CTT AAT TTC TTA AC-3′ |
| Ly49d Exon 2 forward primer | 5′-CGG AAG CCT GAA AAA GCT CG-3′ |
| Ly49d Exon 4 reverse primer | 5′-TCA CAC AGT ATG TTT TGA TCC C-3′ |
| Ly49h Exon 2 forward primer | 5′-GAA CAG CCA GGT GAG ACT T-3′ |
| Ly49h Exon 3-4 reverse primer | 5′-TGT TTG TGA CAA AGT TTT TTC AGT-3′ |
| Nkg2d Exon 2 forward primer | 5′-ACT ACC AGT CAA CCT GGA GAA-3′ |
| Nkg2d Exon 6 reverse primer | 5′-GAC ATA TCC AGT TGT TAG GGC AT-3′ |
| Ly49dB6/NOD forward Exon 2 primer | 5′-GCT GTG AGA TTC CAT AAG TCT TC-3′ |
| Ly49dB6/NOD reverse Exon 4 primer | 5′-GAT GCT GCA GTT ATT GTG GTG-3′ |
| Ly49dB6-specific oligonucleotide probe | 5′-CGG AAG CCT GAA AAA GCT CG-3′ |
| Ly49dNOD-specific oligonucleotide probe | 5′-AGC CTC GAA AAG CTG GCC TCA-3′ |
| Nkg2d-specific oligonucleotide probe | 5′-CAA TTC GAT TCA CCC TTA ACA CAT T-3′ |
| Ly49h Exon 2 inner reverse primer | 5′-AAA GTG ACC TCC TGC TCA CT-3′ |
| Ly49d forward EMSA probe | 5′-AGA AAA GGC CCA CAT TAC CCC AAC AGG GAC ATC CAT TCC TTC TAC-3′ |
| Ly49h forward EMSA probe | 5′-AGT AAA GGC CCA CAT TAC CCC AAT TGA GGC ATC CAT TCT TTC TAC-3′ |
| Ly49a forward EMSA probe (Plus two T-10-mers) | 5′-tttttttttt AGA AAA AGC CAA CTT TTT CCT CCA C tttttttttt-3′ |
| Ly49c forward EMSA probe (Plus two T-4-mers) | 5′-tttt AGA AAA CGC CAA CGT TTC AGA CAA ATT TTC CCT CCA C tttt-3′ |
For the Ly49d 5′ region of the B6 strain the following primers were used to amplify two regions for COBRA analysis of two CpG dinucleotides: first round (flanking) PCR was performed with Ly49d forward and reverse flanking bisulfite primers. After initial denaturation for 7 min at 95°C, 35 cycles were performed, each consisting of 90 s at 95°C, 55 s at 51°C and 50 s at 72°C with a final extension of 7 min at 72°C. Two separate nested PCRs were performed on 3 µl of the product of the flanking PCR to amplify two regions containing an upstream and a downstream (in relation to transcription start site) CpG dinucleotide. For the upstream CpG-containing fragment the Ly49d forward flanking and reverse nested bisulfite primers were used. After initial denaturation for 7 min at 95°C, 35 cycles were performed, each consisting of 90 s at 95°C, 55 s at 51°C and 17 s at 72°C with a final extension of 7 min at 72°C. The downstream CpG-containing fragment was amplified via the Ly49d forward nested and reverse flanking bisulfite primers. After initial denaturation for 7 min at 95°C, 35 cycles were performed, each consisting of 90 s at 95°C, 55 s at 53°C and 40 s at 72°C with a final extension of 7 min at 72°C. The resulting products were subjected to combined bisulfite and restriction enzyme analysis (COBRA).
For Ly49d and Ly49r 5′ region amplification from the 129SvEvTac/C57BL6 F1 hybrid cells, first round PCR was performed with the same flanking primers as Ly49d as their sequence is identical to Ly49r with the same PCR conditions as that used for Ly49d of B6. Nested PCR was performed on 2 µl of the flanking PCR product with the forward nested Ly49d primer (which is identical to Ly49r sequence) and Ly49d/r reverse nested bisulfite primer. After initial denaturation for 7 min at 95°C, 35 cycles were performed, each consisting of 90 s at 95°C, 55 s at 50 + 0.1°C/cycle and 45 s at 72°C with a final extension of 7 min at 72°C. The PCR products were electrophoresed on 1% agarose gels and correct size bands were extracted using the MinElute gel extraction kit (Qiagen). The purified products were cloned into the T-vector using the pGEMT-vector kit (Promega). Sequencing was performed using the SP6 primer by McGill University and Genome Québec Innovation Centre sequencing facility. All clones included in the figures are unique as per criteria stated before (22).
Combined bisulfite and restriction enzyme analysis
For COBRA, gel purified fragments of Ly49d upstream and downstream regions were digested with TaqαI and BmgBI restriction enzyme (NEB), respectively to distinguish between methylated (CpG) and unmethylated (TpG). Only fragments that were originally methylated in the gDNA and therefore not converted by sodium bisulfite treatment are cut. Gel purified Ly49h fragments were digested with TaqαI (NEB) and BsaAI (NEB) restriction enzymes.
RNA extraction, RT–PCR and 5′ amplification of cDNA ends
cDNA was generated from total B6 and NOD/ShiLtJ spleens and FACS sorted B6 T-cell RNA per SuperScript III protocol. PCR was performed on the generated cDNAs using primers specific for Ly49d, g, h, Nkg2d and actin. Ly49g and actin primers have been described before (22). All RT–PCR primer sequences are shown in Table 1.
5′-Rapid amplification of cDNA end (5′-RACE) was performed on B6 spleen RNA using the FirstChoice RML-RACE kit (Ambion) per manufacturer's protocol with Ly49h-specific primers. For the outer PCR, the Ly49h Exon 3–4 reverse primer used for RT–PCR was used as the outer 5′-RACE primer in combination with the kit's outer forward primer. For the inner PCR, Ly49h Exon 2 inner reverse primer in combination with the kit's inner forward primer. The products were cloned into the T-vector using the pGEMT-vector kit (Promega). Sequencing was performed using the T7 primer by McGill University and Genome Québec Innovation Centre sequencing facility.
Electrophoretic mobility shift assay
Nuclear extract from FACS sorted B6 T-cells and IL-2-expanded FACS-sorted B6 NK cells were prepared as described before (26). Electrophoretic mobility shift assay (EMSA) was performed with the following double-stranded probes located downstream of the transcriptional start site within exon1 of various Ly49 genes (only the forward strand is shown in Table 1): to control for length differences among the probes, poly-T tails were added to the shorter Ly49a and c probes. The YY1 antibody (sc-281) was purchased from Santa Cruz Biotechnology and supershift experiments were performed as described before (26).
Single-cell RT–PCR
Single-cell RT–PCR and Southern blotting was performed as described before (27) with minor modifications. RNA from JEG-3, a human choriocarcinoma cell line (28), was used as carrier. Ly49d forward exon 2 and reverse exon 4 primers were used to specifically amplify Ly49d cDNA of both B6 and NOD/ShiLtJ. Ly49dB6-specific oligo probe and Ly49dNOD-specific oligo probe were used to specifically detect cDNA from Ly49dB6 or Ly49dNOD in Southern blot analysis. For Nkg2d, the same primers used for RT–PCR (previous section) were used and the products were identified after Southern blotting with an Nkg2d-specific oligo probe. Up to five sets of independent PCRs were performed on each single cell.
RESULTS
Lack of detectable activating Ly49 transcripts in T-cells
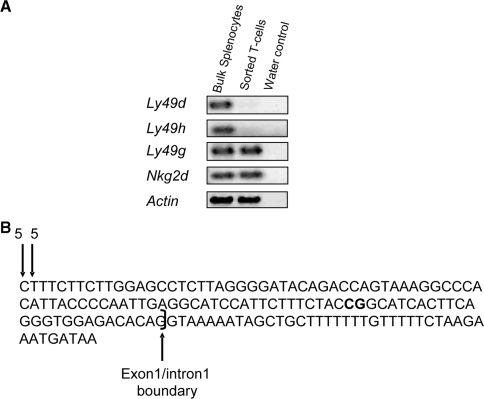
Unlike the inhibitory Ly49 receptors, the activating Ly49 receptors of the B6 strain (Ly49D and H) are not expressed on the surface of T and NKT cells (29,30). This absence of surface expression on T and NKT is most likely due to the negligible amounts of the activating adaptor protein DAP12 that seems to be required for cell surface expression of the B6 activating Ly49 receptors (31,32). However, there are reports of the association of Ly49D with the activating adaptor protein CD3ζ and DAP10 (33,34). We also did not detect Ly49R (an activating receptor in 129 mice) or Ly49D and R surface expression on 129SvEvTac or 129SvS6/B6 F1 hybrid fresh ex vivo splenic T-cells (DX5−, CD3ε+) respectively by flow cytometry (data not shown). To determine if transcripts for the activating receptors exist in T-cells, fresh ex vivo splenic T-cells were sorted with high purity and RT–PCR with Ly49d and Ly49h-specific primers was performed on the isolated RNA (Figure 1A). Transcripts from both Ly49d and Ly49h were detected in whole spleen but very little to none in splenic T-cells, which correlates with the lack of surface expression of these receptors on T-cells. We detected similar amounts of the inhibitory Ly49g and Nkg2d cDNA in both splenic lymphocytes and sorted T-cells.
Figure 1.
Transcription of activating Ly49 genes. (A) RT–PCR on whole spleen and sorted splenic T-cells was performed for actin (25 cycles), the inhibitory Ly49g and activating Ly49d and h as well as Nkg2d (30 cycles) with gene-specific primers. (B) 5′RACE of Ly49h on cDNA from whole spleen. Vertical downward-pointing arrows show the transcription start sites assayed by 5′RACE. The numbers on top of the arrows show the number of sequenced clones beginning at a given nucleotide position. The exon 1/intron 1 boundary is also indicated.
5′-region DNA methylation of Ly49d and Ly49h correlates with state of expression
Ly49 genes have multiple promoters but most are transcribed from a region called Pro-2, which is thought to be the main promoter in mature NK cells (19,35). The transcriptional start site of Ly49d has been mapped to the Pro-2 region (35). To determine the promoter of origin for Ly49h, 5′RACE was performed on whole spleen RNA with gene-specific primers. The Ly49h transcripts detected originated from Pro-2. However, unlike other Ly49 genes (35) we did not detect transcriptional start site variability for Ly49h (Figure 1B).
Based on the 5′RACE results, the region equivalent to the Pro-2 of the inhibitory Ly49 genes is the main area of transcriptional start for Ly49h. We examined the DNA methylation status of the CpG dinucleotides of this region that we shall refer to as the 5′-region of Ly49h from here onwards. There is a cluster of six CpGs within the proximal 5′-region of Ly49h with three CpGs located upstream and three located downstream of the transcription start site (Figure 2A).
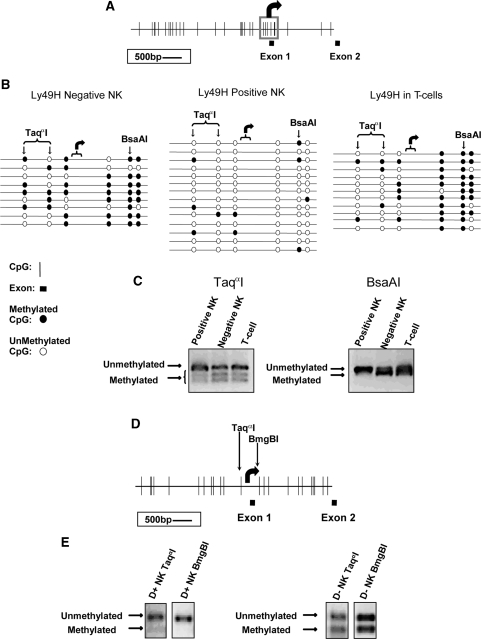
Figure 2.
DNA methylation patterns of the 5′ regions of Ly49d and h in the B6 strain. (A) Location of all CpG dinucleotides in the 5′ region of Ly49h is shown. CpGs are represented by vertical lines, black boxes represent exons and the bent arrow indicates the transcriptional start and the direction of transcription. The CpGs within the boxed region (∼430 bp) were assayed for methylation in primary C57BL/6 splenic NK cells via sodium bisulfite sequencing (B) and COBRA (C). For bisulfite sequencing, each line represents the sequence of an independent clone. The location of the CpG dinucleotides assayed by COBRA are indicated by arrows. Fragments that contain a CpG dinucleotide at these locations are digested by restriction endonuclease indicating methylation in the original genomic DNA. Fragments that remain uncut contain a TpG instead of a CpG, which indicates that in the original genomic DNA template this CpG was unmethylated. (D) CpG dinucleotide distribution in the 5′ region of Ly49d is shown. CpGs are represented by vertical lines, black boxes represent exons and the bent arrow indicates the transcriptional start (35) and the direction of transcription. (E) Two CpGs indicated with vertical arrows were assayed for methylation in primary B6 splenic NK cells by COBRA.
Sodium bisulfite sequencing revealed heavy methylation of this region in FACS-sorted Ly49H-negative (1F8−) splenic NK and T-cells but not in Ly49H-positive (1F8+, 5E6−) NK cells (Figure 2B). As with the inhibitory Ly49a gene (22), the CpG sites downstream of the transcriptional start site are most heavily methylated in Ly49H-negative cells. However, unlike for Ly49a and Ly49c which display a ‘half-and-half’ mono-allelic methylation pattern, the Pro2 region was hypo-methylated in all clones sequenced in the Ly49H+ population. The bisulfite sequencing results were also confirmed by COBRA (Figure 2C).
There are very few CpG dinucleotides in the 5′-region of Ly49d. Hence, we only used COBRA to analyze two CpG dinucletides, one upstream and the other downstream of the transcription start site (Figure 2D). TaqαI and BmgBI restriction endonucleases were used to assay the upstream and downstream CpGs, respectively. Ly49D expressing and non-expressing NK cells of fresh ex vivo spleen were FACS sorted and analyzed for DNA methylation at these two CpGs. The results are the combination of four individual PCRs per each sorted population. Ly49D negative NK cells show moderate amounts of DNA methylation at both assayed CpGs where as the Ly49D positive NK cells show almost no DNA methylation at either site (Figure 2E) reflecting a DNA methylation pattern similar to that observed for the Ly49h 5′-region in Ly49H positive NK cells (Figure 2B and C).
5′-region DNA methylation of Ly49d and Ly49r does not correlate with stochastic mono-allelic receptor expression in 129S6/B6 F1 hybrid
Mono-allelic gene expression has not been shown for the activating Ly49 genes and we did not observe the bimodal DNA methylation pattern typical of the mono-allelically expressed inhibitory Ly49a and c (22) for Ly49d and h in receptor-expressing NK cells. In order to verify the DNA methylation status of both alleles of an activating receptor in receptor-expressing NK cells, we investigated the DNA methylation status of Ly49d in the 129S6/B6 F1 hybrid. Antibody binding and specificities have been examined for the 129S6 strain Ly49 receptors allowing for sorting and analysis of specific Ly49-expressing subpoulations (36). Ly49dB6 and Ly49r129 are considered alleles based on the criteria presented by Makrigiannis et al. (36,37) such as coding region homology, intron homology and gene order. We therefore chose to assay DNA methylation of the 5′-regions of Ly49d and r in the F1 hybrid.
There are no antibodies available that bind Ly49D but not Ly49R or vice-versa, making receptor/allele-specific sorting impossible because the 4E5 and 12A8 antibodies (14) detect both receptors. However, the 4E5 antibody also detects Ly49O129 and Ly49V129 where as 12A8 binds to Ly49AB6 as well (36). The percentage of NK cells expressing Ly49D (stained with 4E5) and R (stained with 12A8 antibody) in B6 and 129S6 strains respectively is 50-60% (data not shown). In order to specifically sort Ly49D and R expressing cells in 129S6/B6 F1 hybrid, splenic lymphocytes were co-stained with 12A8 (anti-Ly49D, R and AB6) and A1 (anti-Ly49AB6) antibodies. The A1 antibody also detects Ly49P129 and V129 with low binding affinity (36).
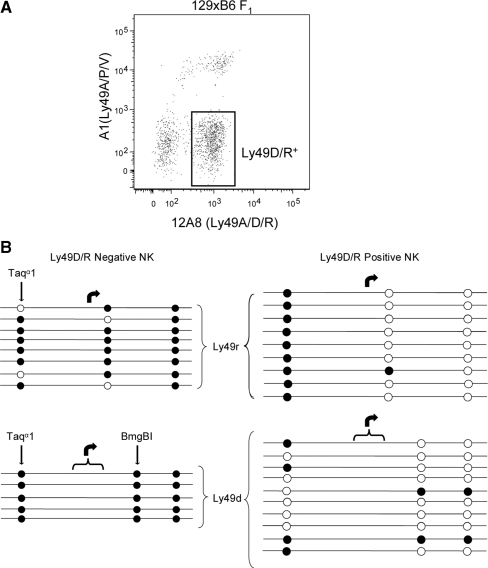
Assuming Ly49d and r are allelic, we hypothesized that, based on the lack of a bimodal DNA methylation pattern observed for Ly49D and H expressing NK cells in B6, both Ly49d and r 5′-regions should be hypo-methylated. We FACS sorted 12A8-positive/A1-negative NK cells (Figure 3A). This population should include Ly49D-single positive, Ly49R single-positive and Ly49D and R double positive cells. A few polymorphisms in the 5′-region of Ly49d and r, assayed by sodium bisulfite sequencing, allow for their distinction (Figure 3B).
Figure 3.
DNA methylation of Ly49d and Ly49r 5′-region in the F1 hybrid of 129/S6 and B6. (A) Ly49D/R expression profile of 129S6xC57Bl/6 F1 hybrid fresh spleen NK cells (DX5+, CD3ε−). The Ly49D/R-expressing (12A8+, A1−) population (gated) and non-expressing (12A8−, A1−) were sorted and analysed for DNA methylation. (B) Sodium bisulfite sequencing of 12A8-positive/A1-negative NK cells (Ly49D/R positive) and 12A8-negative NK cells (Ly49D/R negative). Ly49d and r differ in the position of one CpG dinucleotide but also have other polymorphisms in this region (∼700 bp). All clones presented here are unique.
We also sorted 12A8 negative (Ly49D/R non-expressing) NK cells and analyzed the 5′-region of Ly49d and r for this population as well (Figure 3B). As expected, the 5′-region of Ly49d and r in the Ly49D/R-negative NK cells was hyper-methylated for all the sequenced clones. With the exception of two clones for Ly49d and one clone for Ly49r, all other clones (16 clones) are hypo-methylated in Ly49D/R-positive NK cells. The most 5′ CpG of all of the nine independent Ly49R-positive clones is methylated in Ly49D/R-positive NK cells. However, the rest of the methylation pattern does not resemble that of the Ly49D/R-negative cells. We also sequenced the Ly49r Pro2 region from Ly49R-postive NK cells of the 129/SvEvTac mouse (the parent of the F1 hybrid) to gauge the methylation of the most 5′ CpG. We observed the same methylation pattern as in the F1 (Supplementary Figure 1) possibly indicating that this pattern of methylation is specific to the Ly49r locus. The bimodal DNA methylation observed for the inhibitory Ly49a and c is not observed for Ly49d and r, rather the pattern of DNA methylation resembles that observed for Ly49h.
Detection of bi-allelic expression of Ly49d
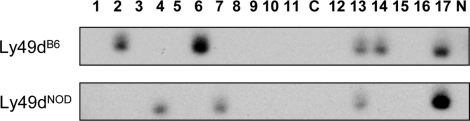
The recent sequencing and assembly of the NOD Ly49 cluster (7) provided an opportunity to test allelic expression of activating Ly49 receptors. While there may be some question as to the allelic relationship of Ly49dB6 and Ly49r129, the NOD strain has a definite Ly49d allele with ∼98.5% identity to the B6 Ly49d sequence at the level of cDNA (7,38). In order to directly test the quality and quantity of allelic expression of Ly49D, we single-cell FACS sorted NK cells (NKp46+, DX5+, NK1.1+, CD3ε−) from B6/NOD F1 hybrid spleen and performed single-cell RT–PCR (27) followed by Southern blot and hybridization with allele-specific probes (see Figure 4 for example).
Figure 4.
Detection of Ly49dB6 and Ly49dNOD cDNA by single-cell RT–PCR and Southern blot. Ly49dB6-specific and Ly49dNOD-specific probes were hybridized to identical blots of amplified cDNA generated from single-cell RT–PCR on FACS sorted F1 hybrid splenic NK cells. This figure shows a representative experiment (one set of PCRs) where seven individual cells show one or more products. Three cells contain Ly49dB6 only, two cells contain Ly49dNOD only and two cells contain both products. C indicates a lane with cDNA from JEG-3 carrier cells only (no sorted NK cell) and N indicates the no-template control lane. After five sets of independent PCRs 4 of 9 single cells in this combination of F1 cells showed bi-allelic expression for Ly49d (data not shown).
We assayed 88 NK cells and detected cDNA for Ly49dB6 and/or Ly49dNOD in 40 cells where cDNA from both Ly49dB6 and Ly49dNOD was detected in 22 of these 40 cells indicating 55% bi-allelic expression (Table 2). We never detected any products from wells that only contained carrier cells. The single-cell RT–PCR technique will underestimate bi-allelic expression due to the fact that one allele may not be detected through inefficient cDNA generation during the RT reaction, multiple sample purification steps and random sampling error (27,39).
Table 2.
Single-cell RT–PCR results
| cDNA type | Ly49DB6 | Ly49DNOD | Ly49DB6 and NOD | Total Ly49D+ | Ly49D− NKG2D+ | Ly49D+ NKG2D+ | Total NKG2D+ | Ly49D+ NKG2D− | Total cells with cDNA |
|---|---|---|---|---|---|---|---|---|---|
| Number of transcribing single cells (n) | 5 | 13 | 22 | 40 | 40 | 33 | 73 | 7 | 80 |
| Percentage positive | 5/40=12.5% | 13/40=32.5% | 22/40=55% | 40/73=55% | 33/73=45% | 7/80=8.8% |
As a control for the efficiency of our method, we used Nkg2d, an activating receptor gene that is transcribed in all NK cells (40) most probably at levels similar to the Ly49 genes, to gauge the efficiency of cell sorting, cDNA generation and detection. We only detected 73 Nkg2d+ cells from the total cells assayed after multiple sets of independent PCRs (Table 2). Nkg2d transcripts went undetected in seven NK cells indicating that possibly 9% of the time the transcript of a gene and/or allele is not detected by our method. In total, we detected transcript for at least one of Ly49dB6, Ly49dNOD and Nkg2d in 80 cells from the total of 88 NK cells assayed.
Interestingly, the difference between the fraction of cells positive for Ly49dNOD versus Ly49dB6 is nearly 3-fold. The allelic expression of Ly49G in various F1 hybrid mice using allele-specific antibodies shows that different alleles of the same gene can be expressed at different levels in various mouse strains (41). This might be due to the MHC class-I background of the mice, the Ly49 cluster gene composition as well as polymorphisms in the promoter regions.
Expression of Ly49H in a single genomic allele mouse model
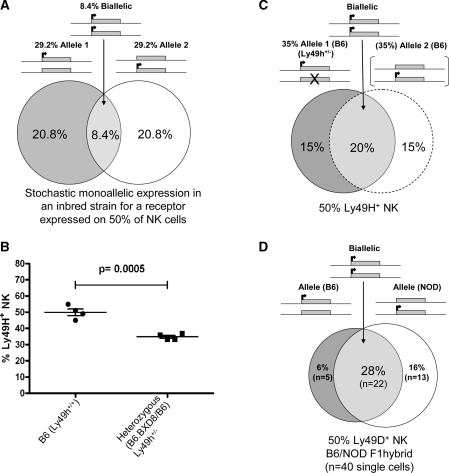
Ly49H is expressed on ∼50% of splenic NK cells in the B6 strain. For a completely bi-allelically transcribed gene expressed in 50% of NK cells, each allele would be expressed in all expressing cells and would contribute half of the total transcript in each cell. In a situation with only one genomic allele present, the fraction of cells expressing the gene should, however, remain the same at 50% but the amount of total transcript should be halved. For a gene expressed mono-allelically through allelic exclusion, one allele is chosen for expression and the other allele is repressed. If this gene is expressed in 50% of cells, each allele is transcribed in 25% of cells only. In contrast, for a stochastic mono-allelic expression pattern, as described for the inhibitory Ly49 receptors, every allele is equally, randomly and independently expressed (42). Based on this model, if 50% of NK cells are positive for a receptor, each allele should be expressed in 29.2% of NK cells with 8.4% of all NK cells expressing this receptor bi-allelically (see model of this situation in Figure 5A).
Figure 5.
Expression of Ly49H in B6 Ly49h+/− NK cells and comparison to the stochastic mono-allelic expression model. (A) Expression of a theoretical Ly49 receptor expressed on 50% of NK cells based on the stochastic mono-allelic model in an inbred mouse strain. (B) The percentage of peripheral blood NK cells expressing Ly49H in B6.BXD8/B6 (Ly49h+/−) mice and age-matched B6 (Ly49h+/+) were determined by FACS. P-value was calculated according to the two-tailed unpaired t-test. (C) Allelic expression of Ly49H in a theoretical B6 mouse based on the expression of this receptor in B6.BXD8/B6 (Ly49h+/−) mice. The dashed circle and brackets indicated predicted expression patterns. (D) Allelic expression of Ly49D on NK cells based on the single-cell RT–PCR results (Table 2) in B6/NOD F1 hybrid. Each chromosome is shown with a line, the alleles are shown with rectangle, arrows indicate transcription and cross shows the deleted allele.
We attempted to distinguish between the bi-allelic versus stochastic mono-allelic model using the B6.BXD8 (B6.Ly49h−/−) mouse, generated recently by two independent groups (23,43), which carries the B6 Ly49 cluster with a ∼26 kb deletion spanning the length of the Ly49h gene. Both groups have independently shown that the expression of other Ly49 receptors does not change in the B6.BXD8 mouse compared to the wild-type B6 mouse indicating that the lack of Ly49H does not affect the expression of other Ly49 receptors (23,43). Unfortunately, an inhibitory Ly49 gene deletion mouse model does not exist for comparison. The B6.BXD8 is unique because the deleted Ly49 genomic allele is in the same Ly49 gene cluster as the other (non-deleted) chromosome. The variable Ly49 clusters as well as possible differences in MHC class-I background can produce confounding effects on expression of Ly49 receptors in F1 hybrids (41).
To investigate the expression pattern of Ly49H on NK cells carrying only one genomic allele of Ly49h on the B6 background, we FACS analyzed peripheral blood NK cells from 6-week old Ly49 h+/− heterozygous (B6.BXD8/B6) mice by staining with the 3D10 antibody. The percentage of NK cells expressing Ly49H in the heterozygous mice (Ly49h+/−) was reduced to 35% from that of age-matched B6 (Ly49h+/+) that have 50% Ly49H+ NK cells (P= 0.0005) (Figure 5B). This fraction of Ly49H expressing cells in the heterozygotes is lower than expected for a completely bi-allelically expressed gene but higher than the expected 29.2% based on the stochastic mono-allelic model (Figure 5A). Thus, based on these data, the situation for Ly49h likely lies between these two extremes. Although we cannot predict the possible effects of the deletion of one Ly49h allele on the expression of the other allele, we can assume that in the wild-type B6 background both alleles should have the same probability of expression. Since Ly49H is expressed on 50% of NK cells and assuming that in the Ly49h+/+ B6 mouse the second allele would also be expressed on 35% of NK cells, the bi-allelic expression of Ly49H would be ∼20% (Figure 5C). This value is strikingly similar to the percentage of Ly49dB6/NOD (bi-allelic) NK cells (assuming 50% Ly49D+ NK) of the B6/NOD F1 hybrid, as assayed with single-cell RT–PCR (Figure 5D).
Low sequence identity in the 5′-UTR of inhibitory and activating Ly49 genes
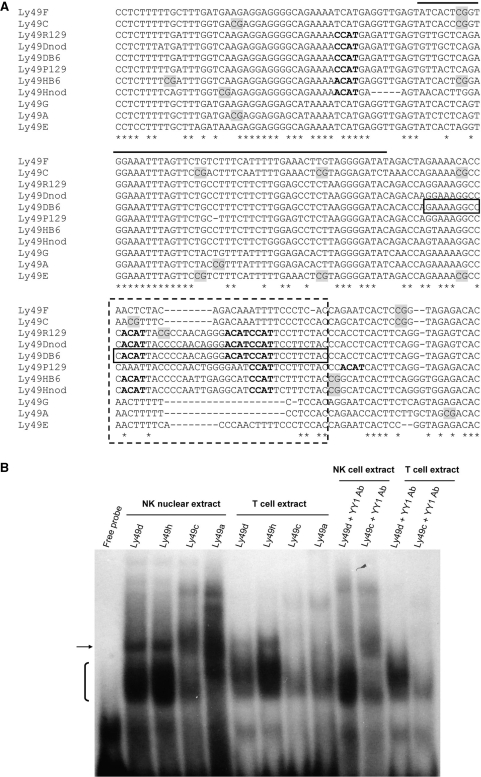
The regulatory elements governing the transcription of activating Ly49 genes have not been analyzed to date. It is unknown if inhibitory and activating genes share common transcription factors. Multiple alignment of a number of inhibitory and activating Ly49 genes from various sequenced mouse strains (performed with Clustal W) shows a region in exon 1 that is present for the activating Ly49 genes but is either absent or different for the inhibitory ones (Figure 6A). This region contains a number of potential binding sites for the transcription factor Yin Yang-1 (YY1) in the activating but not the inhibitory genes.
Figure 6.
Identification of a region of high homology among activating Ly49 genes. (A) Alignment of 5′ regions of inhibitory and activating Ly49 genes. A region, located in exon 1, with high homology among activating receptors only, is boxed (dashed line). The region of transcriptional start site is indicated by the solid black line above the alignment. Promoter region sequences of Ly49d and h of NOD, Ly49p and r of 129/S6 as well as Ly49a, c, d, e, f, g and h of the B6 strain are shown. Ly49a, c, e, f and g are inhibitory genes and all other sequences are from activating genes. CpG dinucleotides are also highlighted. The sequence used as probe in the supershift experiment is indicated for Ly49d with box (solid line). Potential YY1 binding sites (CCAT and ACAT) are shown in bold font. (B) Double-stranded probes harbouring the region of interest in exon 1 for the activating Ly49d and h of the B6 strain and the corresponding region for the inhibitory Ly49a and Ly49c were used in gel-shift experiments with nuclear extract from FACS-sorted T-cells and IL-2 cultured NK cells. A YY1 antibody was used for supershift assays. The bracket indicates the protein complexes bound most significantly by the activating Ly49d and h probes but not the inhibitory Ly49a and c in both NK and T-cells. The arrow indicates another protein complex specific to the activating Ly49 probes that disappears (supershifts) in the presence of anti-YY1 in both NK and T-cells.
Since more than half of the assayed CpG dinucleotides of the 5′-region of Ly49 genes fall downstream of the transcriptional start site, it is possible that differential methylation of this region might affect transcription of the Ly49 genes. Furthermore, investigation of the possible differential regulatory role of this region might explain some of the differences between inhibitory and activating receptor expression. We therefore probed the possible existence of activating gene specific transcription factor binding sites by gel-shift assay with nuclear extracts from FACS sorted IL-2 cultured NK cells and T-cells (Figure 6B and Supplementary Figure 2).
Fragments spanning the region of interest from Ly49a, c, d and h were used as probe. In both NK and T-cells a differential pattern of protein binding was observed between the activating and inhibitory Ly49 probes. A large band (indicated by a bracket in Figure 6B and Supplementary Figure 2), likely consisting of multiple protein complexes, was most prominent for the activating genes. In addition, a protein complex only bound to the activating Ly49 probes (indicated by arrow in Figure 6B and Supplementary Figure 2) disappeared (supershifted) in the presence of YY1 antibody.
DISCUSSION
Here, we have analyzed the DNA methylation of the 5′-region of B6 activating NK receptor genes, Ly49d and h. We have shown that DNA methylation of this region correlates with expression patterns of these receptors as with the inhibitory Ly49A, Ly49C (22) and NKG2A (44). Furthermore, as in the case of Ly49a, the CpG dinucleotides downstream of the transcriptional start site of Ly49h seem to have higher levels of methylation compared to those located upstream in Ly49H-negative cells. This find again raises the possibility of the existence of a proximal downstream regulatory element for the Ly49 genes whose function might be affected by DNA methylation. Indeed for the KIR genes, a region spanning the transcriptional start site and the 5′-UTR provides the core promoter activity in T-cells but not NK cells (45). This region also binds different protein complexes in NK and T-cells as shown with gel shift experiments (45). The multiple alignment of the 5′-region of inhibitory and activating Ly49 genes revealed a region of high homology in exon 1 among the activating genes that is not present in the inhibitory Ly49 sequences (Figure 6A). Further investigation of this region, for the activating Ly49d and h versus the corresponding region for the inhibitory Ly49a and c, in gel shift experiments, showed different patterns of protein complex binding between inhibitory and activating gene regions (Figure 6B). YY1 was also confirmed as a candidate transcription factor binding to this region. YY1 is known to have both transcriptional activating and inhibiting properties (46). In addition, the tested region showed a different protein-binding pattern with nuclear extract from NK and T-cells (Figure 6B). These results combined with the lack of detectable activating Ly49 transcripts in T-cells support a varied mode of transcriptional regulation for the two types of Ly49 genes. Further analysis of this region is needed to confirm the extent of its role in the transcriptional regulation of the activating Ly49 genes.
In contrast to the inhibitory receptors, the DNA methylation pattern of the 5′-region in the Ly49D and H expressing NK populations does not follow the bimodal (half-and-half) methylation pattern. The half-and-half pattern of DNA methylation correlates with the mono-allelic expression of Ly49A, C and NKG2A as was subsequently shown using F1 hybrids for Ly49a and Nkg2a (22,44). If DNA methylation does not correlate with stochastic mono-allelic expression; either stochastic gene expression is maintained at the level of histone modifications only or this pattern is possibly an indication of high, if not complete, bi-allelic expression and even the lack of stochastic expression of the activating receptors.
We have previously shown that the transcription of some Ly49 genes and Nkg2a correlates with histone acetylation levels (22,44,47). Ly49g transcription in the EL4 cell line is activated mostly in response to histone deacetylase inhibitors but is mostly unaffected by DNA methyltransferase inhibitors. Histone acetylation levels of the Pro-2 region as assayed via chromatin immunoprecipitation (ChIP) also correlate with state of expression in EL4-derived subclones (47). It is possible that the stochastic mono-alleleic expression of Ly49G (42) is controlled at the level of histones only. Based on these results, it is also possible that the maintenance of the activating receptor expression patterns is through differential histone modifications.
However, an alternative hypothesis to explain the lack of a half-and-half DNA methylation pattern could be that the expression of the activating receptors is predominantly bi-allelic. Based on this hypothesis, in the Ly49D/R expressing NK cells of the 129/B6 F1 hybrid, the 5′-regions of both Ly49d and r should always be hypomethylated to reflect absolute bi-allelic expression. Our results show that most Ly49d and r clones are hypo-methylated (16/19) in the receptor-expressing NK cells indicating a strong deviation away from the mono-allelic DNA methylation pattern observed for the inhibitory genes (Figure 3B). If Ly49d and r are not alleles but are independent loci, based on the product rule, assuming no bias for co-expression given 50% surface expression of each receptor by NK cells, we would expect 25% of all NK cells to be co-expressing the two receptors. Since the total of NK cells expressing one or both of Ly49D and R would be 75% (assuming independent loci), we would expect that 1/3 of all clones sequenced for each gene (when sorting for D and/or R expressing NK cells) to show hyper-methylation patterns similar to the negative population. Our data indicates a much lower methylation frequency than would be expected with stochastic mono-allelic expression and hence we can conclude that neither Ly49d nor r show bimodal DNA methylation patterns. Our experiment does not prove or disprove the allelic relationship of Ly49d and r.
The bias towards bi-allelic expression of the activating Ly49 receptors is further supported by the results of the single-cell Ly49d allele-specific RT–PCR performed on NK cells of the B6/NOD F1 hybrid (Table 2) as well as the higher than expected expression of Ly49H from a single genomic allele (Figure 5). We detected bi-allelic transcription of Ly49d in 55% (22/40) of Ly49d-transcribing NK cells. If Ly49D is expressed on 50% of NK cells, the percentage of NK cells expressing this receptor bi-allelically would be ∼28% (Figure 5D). The single-cell RT–PCR method tends to be somewhat inefficient in detecting bi-allelic gene expression. The transcription factor, Pax-5, was statistically deemed to be bi-allelically expressed by this method even though only close to 65% of the cells analysed showed bi-allelic expression (39). Transcripts for our control gene Nkg2d, that is expressed by all NK cells (40), went undetected in ∼9% (7/80) of NK cells (Table 2), indicating a likely underestimation of the bi-allelic expression percentage calculated for Ly49d. However, based on the few hyper-methylated clones detected from the Ly49D/R-expressing NK cells as well as the detection of only one Ly49d allele in a few single cells (from the B6/NOD F1 hybrid) even after five sets of single-cell RT–PCR, we believe that a real albeit very small population of NK cells express Ly49D mono-allelically.
It is not inconceivable that the activating receptors are controlled and expressed differently from the inhibitory receptors. The lack of a well-defined Pro-1 element and its transcripts (19,20) in addition to deviation of co-expression percentages from the product rule (21) suggests a different mode of transcriptional regulation from that of the inhibitory genes. Also, in the Ly49h genomic transgenic mouse, Ly49H expression was restricted to NK cells as is the case with the endogenous receptor (48). This might be due to the lack of the adapter protein DAP12 in T and B cells. However, in mice carrying only an Ly49d cDNA transgene (driven by H-2Kb promoter and IgH enhancer) (32) some surface expression of Ly49D was observed on T cells (49) indicating possible pairing of Ly49D with other adaptor proteins in these cells. The ability of T-cells to express some Ly49D on their surface in the absence of DAP12 in this transgenic mouse (49) supports the notion that the endogenous Ly49d promoter might be inactive in T-cells due to lack of necessary transcription factors and/or the presence of repressive epigenetic marks at regulatory sequences. Our results support both possibilities. The Ly49h 5′ region is hypermethylated in T-cells (Figure 2B) indicating a possible epigenetic hindrance to transcription of this gene in these cells. The lack of detectable Ly49d and h transcripts in T-cells (Figure 1A) also supports this notion. Furthermore, the 5′-UTRs of both Ly49d and h show different protein complex binding pattern compared to that of the inhibitory Ly49 genes (Figure 6B).
In contrast to the endogenous receptor, in the Ly49a genomic transgenic mouse, expression of Ly49A was seen on the majority of splenic B-cells for all transgenic lines regardless of copy number of the transgene (50). The 30Kb Ly49a genomic transgene contained the Pro-1 region and it was subsequently shown that the deletion of this region in the same transgene abrogated the expression of Ly49A in NK, T and B cells (50). Mice carrying a 79kb Ly49h genomic transgene spanning the complete length of the gene displayed Ly49H expression patterns similar to the B6 Ly49h gene indicating that the transcriptional regulatory sequences necessary for the ‘wild-type’ expression of this gene are contained within the 79 kb region (48). Ly49h transcript and percentage of NK cells expressing Ly49H correlated in the transgenic mice and increased according to the transgene copy number, but only up to a certain threshold (48). This phenomenon indicates a possible control mechanism for an upper limit of expression likely at the level of transcription and/or at the level of NK selection.
The activating Ly49 genes evolve faster than their inhibitory family members (51). This is evident from the large variation in the number and sequence of the activating Ly49 genes and the high number of activating psuedogenes in nearly all the sequenced mouse strains (7,11). It is also possible that the regulatory sequences of the activating receptors evolved differently from that of the inhibitory receptors in order to allow a tighter control in NK cells and as a side effect of this evolution, mouse T-cells lost the ability to express these receptors. Deviation from stochastic expression might also allow more control on the number of activating receptors on the surface of NK cells because mono-allelic expression might lead to a lower number of receptors on the cell surface compared to bi-alleleic expression (41). Hence, in a non-stochastic system, the expression level of these receptors is homogeneous among different NK cells. In conclusion, our results suggest that distinct modes of transcriptional regulation govern the expression of the activating and inhibitory Ly49 genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Canadian Institutes of Health Research with core support from the British Columbia Cancer Agency. Funding for open access charge: Canadian Institutes of Health Research.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We sincerely thank Liane Gagnier, Hyun-Jung Goo, Vivian Lam and Dr Maura Gasparetto for technical assistance; Dr Florian Kuchenbauer, Dr Sally Rogers and Dr Nooshin Tabatabaei for sharing expertise; Dr Claudia Luther, Evette Haddad and Tim Halim for sharing reagents. We are very grateful to the staff of the TFL cell sorting facility and the BCCRC animal facility. We are thankful to Dr Stephen K. Anderson for the gift of 12A8 antibody.
REFERENCES
- 1.Yokoyama WM, Kim S. How do natural killer cells find self to achieve tolerance? Immunity. 2006;24:249–257. doi: 10.1016/j.immuni.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 2.Smith HR, Heusel JW, Mehta IK, Kim S, Dorner BG, Naidenko OV, Iizuka K, Furukawa H, Beckman DL, Pingel JT, et al. Recognition of a virus-encoded ligand by a natural killer cell activation receptor. Proc. Natl Acad. Sci. USA. 2002;99:8826–8831. doi: 10.1073/pnas.092258599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 4.Eagle RA, Trowsdale J. Promiscuity and the single receptor: NKG2D. Nat. Rev. Immunol. 2007;7:737–744. doi: 10.1038/nri2144. [DOI] [PubMed] [Google Scholar]
- 5.Parham P. The genetic and evolutionary balances in human NK cell receptor diversity. Semin. Immunol. 2008;20:311–316. doi: 10.1016/j.smim.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nat. Rev. Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 7.Belanger S, Tai LH, Anderson SK, Makrigiannis AP. Ly49 cluster sequence analysis in a mouse model of diabetes: an expanded repertoire of activating receptors in the NOD genome. Genes Immun. 2008;9:509–521. doi: 10.1038/gene.2008.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1:e27. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stulberg MJ, Wright PW, Dang H, Hanson RJ, Miller JS, Anderson SK. Identification of distal KIR promoters and transcripts. Genes Immun. 2007;8:124–130. doi: 10.1038/sj.gene.6364363. [DOI] [PubMed] [Google Scholar]
- 10.Anderson SK, Dewar K, Goulet ML, Leveque G, Makrigiannis AP. Complete elucidation of a minimal class I MHC natural killer cell receptor haplotype. Genes Immun. 2005;6:481–492. doi: 10.1038/sj.gene.6364232. [DOI] [PubMed] [Google Scholar]
- 11.Makrigiannis AP, Patel D, Goulet ML, Dewar K, Anderson SK. Direct sequence comparison of two divergent class I MHC natural killer cell receptor haplotypes. Genes Immun. 2005;6:71–83. doi: 10.1038/sj.gene.6364154. [DOI] [PubMed] [Google Scholar]
- 12.Wilhelm BT, Gagnier L, Mager DL. Sequence analysis of the ly49 cluster in C57BL/6 mice: a rapidly evolving multigene family in the immune system. Genomics. 2002;80:646–661. doi: 10.1006/geno.2002.7004. [DOI] [PubMed] [Google Scholar]
- 13.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat. Rev. Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 14.Mason LH, Anderson SK, Yokoyama WM, Smith HR, Winkler-Pickett R, Ortaldo JR. The Ly-49D receptor activates murine natural killer cells. J. Exp. Med. 1996;184:2119–2128. doi: 10.1084/jem.184.6.2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.George TC, Mason LH, Ortaldo JR, Kumar V, Bennett M. Positive recognition of MHC class I molecules by the Ly49D receptor of murine NK cells. J. Immunol. 1999;162:2035–2043. [PubMed] [Google Scholar]
- 16.Lee SH, Girard S, Macina D, Busa M, Zafer A, Belouchi A, Gros P, Vidal SM. Susceptibility to mouse cytomegalovirus is associated with deletion of an activating natural killer cell receptor of the C-type lectin superfamily. Nat. Genet. 2001;28:42–45. doi: 10.1038/ng0501-42. [DOI] [PubMed] [Google Scholar]
- 17.Daniels KA, Devora G, Lai WC, O'Donnell CL, Bennett M, Welsh RM. Murine cytomegalovirus is regulated by a discrete subset of natural killer cells reactive with monoclonal antibody to Ly49H. J. Exp. Med. 2001;194:29–44. doi: 10.1084/jem.194.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Takei F, McQueen KL, Maeda M, Wilhelm BT, Lohwasser S, Lian RH, Mager DL. Ly49 and CD94/NKG2: developmentally regulated expression and evolution. Immunol. Rev. 2001;181:90–103. doi: 10.1034/j.1600-065x.2001.1810107.x. [DOI] [PubMed] [Google Scholar]
- 19.Saleh A, Makrigiannis AP, Hodge DL, Anderson SK. Identification of a novel Ly49 promoter that is active in bone marrow and fetal thymus. J. Immunol. 2002;168:5163–5169. doi: 10.4049/jimmunol.168.10.5163. [DOI] [PubMed] [Google Scholar]
- 20.Saleh A, Davies GE, Pascal V, Wright PW, Hodge DL, Cho EH, Lockett SJ, Abshari M, Anderson SK. Identification of probabilistic transcriptional switches in the Ly49 gene cluster: a eukaryotic mechanism for selective gene activation. Immunity. 2004;21:55–66. doi: 10.1016/j.immuni.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 21.Smith HR, Chuang HH, Wang LL, Salcedo M, Heusel JW, Yokoyama WM. Nonstochastic coexpression of activation receptors on murine natural killer cells. J. Exp. Med. 2000;191:1341–1354. doi: 10.1084/jem.191.8.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rouhi A, Gagnier L, Takei F, Mager DL. Evidence for epigenetic maintenance of Ly49A monoallelic gene expression. J. Immunol. 2006;176:2991–2999. doi: 10.4049/jimmunol.176.5.2991. [DOI] [PubMed] [Google Scholar]
- 23.Cheng TP, French AR, Plougastel BF, Pingel JT, Orihuela MM, Buller ML, Yokoyama WM. Ly49h is necessary for genetic resistance to murine cytomegalovirus. Immunogenetics. 2008;60:565–573. doi: 10.1007/s00251-008-0313-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takei F, Brennan J, Mager DL. The Ly-49 family: genes, proteins and recognition of class I MHC. Immunol. Rev. 1997;155:67–77. doi: 10.1111/j.1600-065x.1997.tb00940.x. [DOI] [PubMed] [Google Scholar]
- 25.Brennan J, Lemieux S, Freeman JD, Mager DL, Takei F. Heterogeneity among Ly-49C natural killer (NK) cells: characterization of highly related receptors with differing functions and expression patterns. J. Exp. Med. 1996;184:2085–2090. doi: 10.1084/jem.184.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maksakova IA, Mager DL. Transcriptional regulation of early transposon elements, an active family of mouse long terminal repeat retrotransposons. J. Virol. 2005;79:13865–13874. doi: 10.1128/JVI.79.22.13865-13874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kubota A, Kubota S, Lohwasser S, Mager DL, Takei F. Diversity of NK cell receptor repertoire in adult and neonatal mice. J. Immunol. 1999;163:212–216. [PubMed] [Google Scholar]
- 28.Landry JR, Rouhi A, Medstrand P, Mager DL. The Opitz syndrome gene Mid1 is transcribed from a human endogenous retroviral promoter. Mol. Biol. Evol. 2002;19:1934–1942. doi: 10.1093/oxfordjournals.molbev.a004017. [DOI] [PubMed] [Google Scholar]
- 29.Coles MC, McMahon CW, Takizawa H, Raulet DH. Memory CD8 T lymphocytes express inhibitory MHC-specific Ly49 receptors. Eur. J. Immunol. 2000;30:236–244. doi: 10.1002/1521-4141(200001)30:1<236::AID-IMMU236>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 30.Assarsson E, Kambayashi T, Sandberg JK, Hong S, Taniguchi M, Van Kaer L, Ljunggren HG, Chambers BJ. CD8+ T cells rapidly acquire NK1.1 and NK cell-associated molecules upon stimulation in vitro and in vivo. J. Immunol. 2000;165:3673–3679. doi: 10.4049/jimmunol.165.7.3673. [DOI] [PubMed] [Google Scholar]
- 31.Bakker AB, Hoek RM, Cerwenka A, Blom B, Lucian L, McNeil T, Murray R, Phillips LH, Sedgwick JD, Lanier LL. DAP12-deficient mice fail to develop autoimmunity due to impaired antigen priming. Immunity. 2000;13:345–353. doi: 10.1016/s1074-7613(00)00034-0. [DOI] [PubMed] [Google Scholar]
- 32.Voyle RB, Beermann F, Lees RK, Schumann J, Zimmer J, Held W, MacDonald HR. Ligand-dependent inhibition of CD1d-restricted NKT cell development in mice transgenic for the activating receptor Ly49D. J. Exp. Med. 2003;197:919–925. doi: 10.1084/jem.20021615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ortaldo JR, Winkler-Pickett R, Willette-Brown J, Wange RL, Anderson SK, Palumbo GJ, Mason LH, McVicar DW. Structure/function relationship of activating Ly-49D and inhibitory Ly-49G2 NK receptors. J. Immunol. 1999;163:5269–5277. [PubMed] [Google Scholar]
- 34.Tassi I, Le Friec G, Gilfillan S, Takai T, Yokoyama WM, Colonna M. DAP10 associates with Ly49 receptors but contributes minimally to their expression and function in vivo. Eur. J. Immunol. 2009;39:1129–1135. doi: 10.1002/eji.200838972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wilhelm BT, McQueen KL, Freeman JD, Takei F, Mager DL. Comparative analysis of the promoter regions and transcriptional start sites of mouse Ly49 genes. Immunogenetics. 2001;53:215–224. doi: 10.1007/s002510100313. [DOI] [PubMed] [Google Scholar]
- 36.Makrigiannis AP, Pau AT, Saleh A, Winkler-Pickett R, Ortaldo JR, Anderson SK. Class I MHC-binding characteristics of the 129/J Ly49 repertoire. J. Immunol. 2001;166:5034–5043. doi: 10.4049/jimmunol.166.8.5034. [DOI] [PubMed] [Google Scholar]
- 37.Makrigiannis AP, Pau AT, Schwartzberg PL, McVicar DW, Beck TW, Anderson SK. A BAC contig map of the Ly49 gene cluster in 129 mice reveals extensive differences in gene content relative to C57BL/6 mice. Genomics. 2002;79:437–444. doi: 10.1006/geno.2002.6724. [DOI] [PubMed] [Google Scholar]
- 38.Silver ET, Gong DE, Chang CS, Amrani A, Santamaria P, Kane KP. Ly-49P activates NK-mediated lysis by recognizing H-2Dd. J. Immunol. 2000;165:1771–1781. doi: 10.4049/jimmunol.165.4.1771. [DOI] [PubMed] [Google Scholar]
- 39.Rhoades KL, Singh N, Simon I, Glidden B, Cedar H, Chess A. Allele-specific expression patterns of interleukin-2 and Pax-5 revealed by a sensitive single-cell RT-PCR analysis. Curr. Biol. 2000;10:789–792. doi: 10.1016/s0960-9822(00)00565-0. [DOI] [PubMed] [Google Scholar]
- 40.Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH. The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity. 2002;17:19–29. doi: 10.1016/s1074-7613(02)00333-3. [DOI] [PubMed] [Google Scholar]
- 41.Makrigiannis AP, Rousselle E, Anderson SK. Independent control of Ly49g alleles: implications for NK cell repertoire selection and tumor cell killing. J. Immunol. 2004;172:1414–1425. doi: 10.4049/jimmunol.172.3.1414. [DOI] [PubMed] [Google Scholar]
- 42.Held W, Kunz B. An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire. Eur. J. Immunol. 1998;28:2407–2416. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 43.Fodil-Cornu N, Lee SH, Belanger S, Makrigiannis AP, Biron CA, Buller RM, Vidal SM. Ly49h-deficient C57BL/6 mice: a new mouse cytomegalovirus-susceptible model remains resistant to unrelated pathogens controlled by the NK gene complex. J. Immunol. 2008;181:6394–6405. doi: 10.4049/jimmunol.181.9.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rogers SL, Rouhi A, Takei F, Mager DL. A role for DNA hypomethylation and histone acetylation in maintaining allele-specific expression of mouse NKG2A in developing and mature NK cells. J. Immunol. 2006;177:414–421. doi: 10.4049/jimmunol.177.1.414. [DOI] [PubMed] [Google Scholar]
- 45.Xu J, Vallejo AN, Jiang Y, Weyand CM, Goronzy JJ. Distinct transcriptional control mechanisms of killer immunoglobulin-like receptors in natural killer (NK) and in T cells. J. Biol. Chem. 2005;280:24277–24285. doi: 10.1074/jbc.M500727200. [DOI] [PubMed] [Google Scholar]
- 46.Shi Y, Lee JS, Galvin KM. Everything you have ever wanted to know about Yin Yang 1. Biochim. Biophys. Acta. 1997;1332:F49–F66. doi: 10.1016/s0304-419x(96)00044-3. [DOI] [PubMed] [Google Scholar]
- 47.Rouhi A, Brooks CG, Takei F, Mager DL. Plasticity of Ly49g expression is due to epigenetics. Mol. Immunol. 2007;44:821–826. doi: 10.1016/j.molimm.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 48.Lee SH, Zafer A, de Repentigny Y, Kothary R, Tremblay ML, Gros P, Duplay P, Webb JR, Vidal SM. Transgenic expression of the activating natural killer receptor Ly49H confers resistance to cytomegalovirus in genetically susceptible mice. J. Exp. Med. 2003;197:515–526. doi: 10.1084/jem.20021713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Merck E, Voyle RB, MacDonald HR. Ly49D engagement on T lymphocytes induces TCR-independent activation and CD8 effector functions that control tumor growth. J. Immunol. 2009;182:183–192. doi: 10.4049/jimmunol.182.1.183. [DOI] [PubMed] [Google Scholar]
- 50.Tanamachi DM, Moniot DC, Cado D, Liu SD, Hsia JK, Raulet DH. Genomic Ly49A transgenes: basis of variegated Ly49A gene expression and identification of a critical regulatory element. J. Immunol. 2004;172:1074–1082. doi: 10.4049/jimmunol.172.2.1074. [DOI] [PubMed] [Google Scholar]
- 51.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J. Exp. Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.