Abstract
The translational activity of the hepatitis C virus (HCV) internal ribosome entry site (IRES) and other HCV-like IRES RNAs depends on structured RNA elements in domains II and III, which serve to recruit the ribosomal 40S subunit, eukaryotic initiation factor (eIF) 3 and the ternary eIF2/Met-tRNAiMet/GTP complex and subsequently domain II assists subunit joining. Porcine teschovirus-1 talfan (PTV-1) is a member of the Picornaviridae family, with a predicted HCV-like secondary structure, but only stem-loops IIId and IIIe in the 40S-binding domain display significant sequence conservation with the HCV IRES. Here, we use chemical probing to show that interaction sites with the 40S subunit and eIF3 are conserved between HCV and HCV-like IRESs. In addition, we reveal the functional role of a strictly conserved co-variation between a purine–purine mismatch near the pseudoknot (A–A/G) and the loop sequence of domain IIIe (GAU/CA). These nucleotides are involved in a tertiary interaction, which serves to stabilize the pseudoknot structure and correlates with translational efficiency in both the PTV-1 and HCV IRES. Our data demonstrate conservation of functional domains in HCV and HCV-like IRESs including a more complex structure surrounding the pseudoknot than previously assumed.
INTRODUCTION
Translation initiation in eukaryotes requires the full complement of canonical eukaryotic initiation factors (eIFs) and a 5′-capped mRNA (1–4). In the first step of 80S ribosome assembly, the 5′-cap structure is recognized by the eIF4E cap-binding protein of the eIF4F complex, which also contains eIF4A (RNA helicase) and the large scaffold protein, eIF4G. This leads to recruitment of the 43S preinitiation complex consisting of the 40S small ribosomal subunit, eIF1, 1A, 3 and the eIF2/GTP/Met-tRNAiMet ternary complex to the 5′end of the mRNA. The ribosomal assembly then scans the 5′ UTR of the mRNA with the help of the helicase eIF4A and its co-factor eIF4B until recognition of the AUG start codon in the ribosomal P-site. This in turn triggers eIF5-mediated hydrolysis of eIF2-bound GTP (5,6), and finally all factors dissociate from the 40S subunit during eIF5B-assisted joining of the large ribosomal 60S subunit to form initiated 80S ribosomes (5–8).
An alternative route to 80S ribosome formation is mediated by IRES RNAs, which are structured RNA elements located in the 5′UTR of many viral and several cellular RNAs (9). In the family of HCV and HCV-like IRES RNAs, translation initiation does not depend on a 5′-cap structure or scanning, and 48S complex formation requires only a subset of the canonical eIFs (10,11). IRES-mediated ribosome assembly is believed to be driven by the formation of a binary IRES–40S subunit complex (12,13), which in turn recruits eIF3 and the ternary complex to form a 48S complex at the AUG start codon (12,14,15). Subunit joining and factor release still not only require eIF5, eIF5B and GTP (16), but also depend on interactions of the IRES with the 40S subunit (17,18). In addition to the general eIF2-dependent pathway of 80S assembly, both the HCV IRES and the classical swine fever virus (CSFV) IRES can also use a bacterial-like pathway, which requires only eIF5B and eIF3 (18,19).
The PTV-1 IRES is a member of the Picornaviridae family, with the same eIF requirement for ribosome assembly as the HCV IRES (16,20). The HCV and PTV-1 IRES elements display 55% sequence identity with a strong conservation of the basal pseudoknot domains IIIef (20). On the basis of this sequence similarity, the structural organization of the PTV-1 IRES is predicted to be very similar to HCV and other HCV-like IRESs, especially in the basal domain III (Figure 1) (21,22). The 5′-boundaries of the PTV-1 IRES have been mapped between nucleotides 125 and 150 of the PTV-1 5′ UTR (20) and the secondary structure prediction displays an organization into two major domains, namely II and III as found in the HCV IRES (Figure 1) (21,22). In the HCV IRES, the basal domains IIIdef contain a four-way junction including a pseudoknot motif (IIIef) and the IIId three-way junction with a branching G-rich stem-loop IIId (23,24). This part of the HCV IRES provides the major 40S binding site (15,25), and the pseudoknot and stem-loops IIId and IIIe are essential for IRES function (26,27). The predicted secondary structure of the PTV-1 IRES preserves these main structural motifs suggesting a conservation of their role in 40S recruitment. Especially, the small stem loop IIIe (GAU/CA) and its adjacent helix, which invariably contain a purine–purine mismatch base pair (A–A/G) in all HCV-like IRESs, display high primary sequence conservation (22). Surprisingly, these sequences display co-variation (GAU/CA with A–A/G) in all HCV-like IRESs, but their functional role is unknown (22).
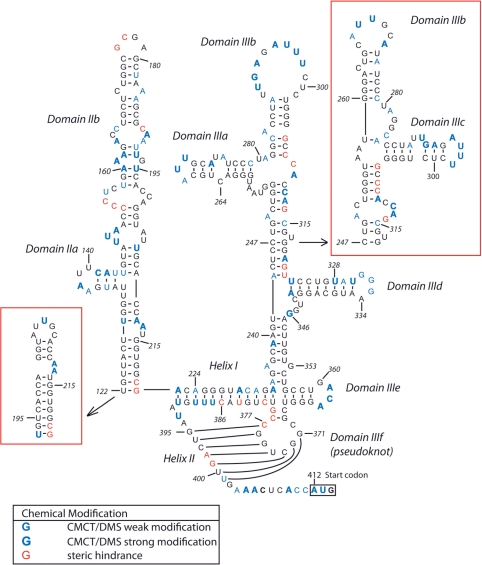
Figure 1.
Schematic representation of the secondary structure of the PTV-1 IRES (21,22) showing bases that are accessible to modification by CMCT and DMS or that caused reverse transcription arrest in untreated RNA as indicated in the lower left. The initiation codon (AUG412–414) is boxed.
The apical domains IIIabc of the HCV IRES contain a four-way junction, which comprises eIF3 binding site (IIIab) and a small stem-loop for 40S interaction (IIIc) (28). The secondary structure of the PTV-1 IRES, on the other hand, predicts an apical three-way junction either lacking stem-loop IIIa or IIIc (Figure 1) (21,22) suggesting either a reduced eIF3 or 40S-binding site compared to the HCV IRES. The smaller domain II displays little sequence and structure conservation between the HCV and PTV-1 IRESs (20,21). Key functional features of HCV IRES domain II, such as the IIb hairpin loop (UAGCCAU) and an apical loop E motif, which are required for 80S formation (17,18), are absent in the predicted PTV-1 structure (21). Moreover, a recent comparative sequence analysis of multiple picornaviral IRESs predicted a much smaller domain II stem-loop structure in the PTV-1 IRES (22). Since there are significant differences in secondary structure within domain II and the apical domain III between the HCV and PTV-1 IRES and since these IRES domains play distinct roles during the 80S ribosome assembly, it is crucial to further define the secondary structure of the PTV-1 IRES RNA and map interactions with the 40S ribosomal subunit and eIF3 (28).
Chemical and enzymatic probing are powerful techniques to study RNA secondary structure and to map regions that are involved in interactions with ligands (29). This technique has been used to establish the magnesium dependence of the HCV IRES fold (24) and to determine interaction sites of the HCV and CSFV IRES on the 40S subunit and the eIF3 complex (8,13,26). On the other hand, little is known about IRES interactions within a 48S complex, and with exception of the encephalomyocarditis virus (EMCV) IRES 48S complex, no other 48S–IRES complex has been analyzed by chemical probing (30).
To investigate the secondary structure of the PTV-1 IRES and interactions with the translational machinery up to 48S complex assembly, we performed comparative chemical probing of the PTV-1 IRES in the free form, in the binary 40S–IRES and in the 48S complex. We show that domain III of the free IRES RNA adopts a fold with exposed stem-loops IIIa, IIIb, IIId and IIIe, and that large parts of domain II are unstructured in the free form. Strong protection from chemical modification in domain II and the basal domain III occurs upon 40S binding. The apical eIF3 binding site in domain III, in contrast, displays enhanced modifications in the binary 40S–IRES complex suggesting an exposed conformation for eIF3 recruitment, and binding of eIF3 and the ternary complex to the binary IRES–40S particle protects the predicted eIF3 binding site from chemical modifications. Mutational analysis of a conserved purine–purine mismatch base pair (A–A in HCV or A–G in PTV-1) and a co-varying nucleotide in hairpin loop IIIe (U or C) in both HCV and PTV-1 IRES reveals a tertiary interaction near the pseudoknot, which helps to stabilize this local structure and is required for efficient translation initiation. Our data propose conservation of functional domains in HCV and HCV-like IRESs including a complex tertiary structure surrounding the pseudoknot.
MATERIALS AND METHODS
Preparation of mRNAs
DNA constructs containing the wild-type PTV-1 and HCV IRESs for in vitro transcription were prepared by standard PCR and cloning techniques as described elsewhere (31). The DNA constructs containing PTV-1 double C362U/G353A and single C362U or G353A and the HCV double A288G/U297C and single A288G or U297C mutants were generated by PCR mutagenesis of the PTV-1 or HCV wild-type plasmids, respectively. PCR products were digested with HindIII and BamHI or EcoRI (HCV) and ligated into pUC18 digested with the same enzymes. The plasmids were linearized with BamHI or EcoRI and in vitro transcription was performed following standard protocols. The transcripts were purified by size-exclusion chromatography as described elsewhere (32). The purified sample was equilibrated with 10 mM Tris pH 7.4 and concentrated to 0.5–1.0 A260 units per microliter using Centriprep concentrators. The samples were stored at –20°C.
Chemical probing on free RNA
Cyclo-3(2-morpholinoethyl) carbodiimide metho-p-toluene sulfate (CMCT) probing on free RNA was performed in 20 µl reactions containing 50 nM free RNA in 50 mM borate NaOH pH 8, 5 mM magnesium acetate, 50 mM KOAc and 2 mM β-mercaphthoethanol. The samples were denatured at 90°C for 1 min and re-natured on ice for a further minute. A 1 µl tRNA (2 mg/ml) and a range of CMCT from 3 to 24 mg/ml were added before incubation for 15 min at 37°C. The samples were precipitated by the addition of 30 mM sodium acetate and 150 µl, 100% ethanol and incubated in dry ice for 20 min, the pellet was washed in 150 µl, 80% ethanol and the resulting pellet was resuspended in nuclease-free water.
Dimethyl sulfate (DMS) probing on free RNA was performed in 20 µl reactions containing 50 nM free RNA in 50 mM HEPES pH 7.5, 5 mM MgCl2, 50 mM KOAc and 2 mM β-mercaphthoethanol. The samples were denatured at 90°C for 1 min and re-natured on ice for further 1 min. A 1 µl tRNA (2 mg/ml) and 1 µl DMS ranging from 1:0, 1:1, 1:4 and 1:9 DMS in ethanol were added before incubation for 5 min at room temperature. The samples were precipitated by the addition of 30 mM sodium acetate and 60 µl, 100% ethanol and incubated in dry ice for 20 min, the pellet was washed in 150 µl, 80% ethanol and the resulting pellet was resuspended in nuclease-free water.
Primer extension
For primer extension, six oligonucleotides complementing nucleotides 186–202, 244–262, 341–358, 402–416 and 453–470 of the RNA were 5′-end labeled with [γ32P]–ATP. The labeled primers were annealed to 2 µg PTV-1 IRES RNA at 90°C for 1 min followed by 1 min incubation on ice. Primer extension was performed at 37°C for 30 min in a 15 µl reaction containing 1U Super RT reverse transcriptase (H T Biotechnology Ltd). The products were precipitated by the addition of 30 mM sodium acetate and 200 µl, 100% ethanol and incubated on dry ice for 20 min, the pellet was washed in 150 µl, 80% ethanol and the resulting pellet resuspended in 8 µl formamide dye (10 ml formamide, 10 mg xylene cyanol, 10 mg bromophenol blue). The cDNA synthesized were analyzed in denaturing 6% acrylamide, 8 M urea gels.
In vitro translational assays
DNA constructs containing the wild-type PTV-1 and HCV IRESs preceded by a SpeI restriction site were prepared from overlapping primers; the PCR products were digested with HindIII and BamHI or EcoRI (HCV) and ligated into pUC18 digested with the same enzymes. DNA templates for PTV-1 and HCV mutations (pRptvF 1–8 and pRhcvF 1–4 as described in the text) were prepared by PCR using the wild-type PTV-1 or HCV plasmids mentioned above and appropriate primers. For coupled transcription/translation, the resulting PCR products were digested with SpeI and NcoI or EcoRI (HCV) and ligated into pRF digested with the same enzymes. Translational assays were performed in Promega TnT quick coupled transcription/translation system supplemented with 0.5 mCi/ml 35S-labeled Met according to manufacturer's instructions. Translation products were analyzed by 4–12% bis–tris SDS gel electrophoresis. Gels were quantified by Molecular Dynamics PhosphorImager analysis.
Preparation and analysis of 40S and 48S initiation complexes
40S and 48S initiation complexes were formed on the PTV-1 IRES, prepared above, and purified as previously described (31). Binary 40S–IRES complexes and 48S complexes were resolved by fractionation of the sucrose density gradient while monitoring the absorbance at 260 nm. Peak fractions were analyzed on 1% agarose gels using a Tris-Borate running buffer at 4°C. The pure fractions were pooled and concentrated in YM-50 centricons and centrifuged at 65 000 rpm in a Beckman TLX ultracentrifuge TLA-100.3 rotor at 4°C for 4 h. The resulting pellets were stored at –20°C.
Toe-printing analysis
Toe printing of binary 40S–IRES and 48S complexes was performed on affinity purified complexes (as described above) by reverse transcription using a primer complimentary to nucleotides 453–470 of the RNA. Toe-printing analysis of 48S and 80S complexes formed on the wild-type and mutant HCV IRES in buffer A (2 mM Tris pH 7.6, 100 mM KOAc, 2.5 mM MgCl2 and 2 mM DTT) as described elsewehere (33) and analyzed by primer extension by AMV reverse transcriptase and a 5′end [γ32P]-labeled primer complementary to the NS’-coding sequence, as previously described (16). The cDNAs synthesized were analyzed in denaturing 6% acrylamide, 8 M urea gels.
Chemical probing of binary 40S–IRES and 48S complexes
DMS probing was performed in 100 µl containing 1 µM free RNA, 40S binary or 48S complex in 20 mM HEPES pH 7.5, 100 mM KCl, 2.5 mM MgCl2, 2 mM DTT, 1% sucrose; 2 µl DMS (1:3 in ethanol) was added and samples were incubated for 10 min at 37°C. The reaction was arrested by the addition of 50 µl stop buffer (1 M Tris pH 7.4, 1M β-mercaphthoethanol, 1 mM EDTA pH 8) and the RNA complexes precipitated with 450 µl ice cold 100% ethanol. The resulting pellets were resuspended in 300 µl of 300 mM sodium acetate, 5 mM EDTA pH 8, 0.5% SDS and phenol–chloroform extracted three times and chloroform extracted twice. The extracted aqueous phase was precipitated with 900 µl of ice cold 100% ethanol, the pellet was washed in 150 µl ice cold 80% ethanol and the resulting pellet was resuspended in nuclease-free water.
RESULTS
Establishing the secondary structure of PTV-1 IRES RNA by chemical probing
To further characterize functional domain boundaries and the secondary structure of the PTV-1 IRES, we employed chemical probing techniques and in vitro translation assays using wild-type and domain-deletion mutant of IRES RNAs. Initially we sought to investigate nucleotide accessibility to the chemical probes, DMS and 1-cyclohexyl-3-(2-morpholinoethyl) carbodiimide metho-p-toluene sulfonate (CMCT), which modify adenine (N-1) and cytosine (N-3), and guanine (N-1) and uridine (N-3), respectively. Such accessibility would thereby indicate nucleotides in single stranded or unstructured regions. Since the HCV IRES had been shown to adopt a magnesium-dependent tertiary fold (24), probing experiments were performed in the presence or absence of 5 mM Mg2+. There was very little variation between conditions (data not shown), and here we present only the chemical probing results of PTV-1 IRES RNA obtained in the presence of 5 mM Mg2+. The modified RNAs were analyzed by end-labeled primer extension using reverse transcription. The position of the chemical modification prevents base pairing with the complementary deoxynucleotide resulting in an arrest in primer extension one nucleotide prior to the modified nucleotide that is not observed in untreated samples. Therefore, increased band intensity can be observed directly before the modified nucleotide. Strong stops in treated and untreated samples, which could not be attributed to chemical modification, were presumed to be the result of steric hindrance in regions displaying strong structural features. The cDNA of the reverse transcribed RNAs were then compared to dideoxy sequencing of the PTV-1 IRES RNA prepared in the same way (Supplementary Figures S1 and S2).
The CMCT and DMS probing results show clusters of modification (C141–U144, U148–A150, A160–A162, A190–U193, A229–A231, A268–U270, U291–A293, A295–U298, U328–G333, A361–A363, U387–U389, A403–405, A409–U413) (Supplementary Figures S1 and S2). These modifications, and the additional single modifications, were mapped onto the predicted structures previously reported (Figure 1) (21,22). The positioning of modifications A403–A405 and A409–U413 matches the prediction of a single-stranded region following the proposed pseudoknot. Interestingly, there are no bases accessible to chemical modification in helix II of the proposed pseudoknot (C371–C377 and G395–U401). However, steric hindrance in an unmodified RNA can be observed at nucleotides A398 and G399 within helix II and at C377–G379 between helix I and helix II (Figure 1, Supplementary Figures S1 and S2, lane 5). The steric hindrance at C377–G379 could be as a result of the interactions required to form a pseudoknot or a strong secondary structure appearing at the four-way junction. The steric hindrance at A398 and G399, however, do not occur at any known or predicted structures other than pseudoknot helix II, suggesting that helix II is at least partially formed in the free form of the PTV-1 IRES RNA. In contrast, modifications at nucleotides A229–A231 and U387–U389 occur within helix I of the proposed pseudoknot indicating that this region is flexible in the free form of the RNA.
Modification mapping also showed that the clustered modifications at A268–U270, U291–A293, A295–U298, U328–A334, A361–A363 (Supplementary Figures S1 and S2) correlated well with the stem loops of domain III subdomains (Figure 1). The basal subdomains (IIId and IIIe) in addition to modification within the stem loops (U328–A334 and A361–A363, respectively) display little or no accessibility to chemical modification within the stem helices suggesting that these domains are also formed within the free form of the RNA. The apical subdomains unlike the structured basal domains showed additional modifications (A250, A273, A275, C278, A281, C284, A285, C312, A313 and C315) (Supplementary Figures S1 and S2). These modifications occur within the stem helices of both predicted structures (Figure 1). To further understand the domain organization of this region, we also looked at the steric hindrance observed in the unmodified samples. Mapping these steric hindrances shows distinct differences on the predicted apical subdomains. The predicted secondary structure with domains IIIb and IIIc (21) positions the steric hindrance within a helical region preceding the subdomains. The IIIa and IIIb predictions, however (22), would place the steric hindrance at the apical three-way junction, where it would be possible for the RNA to form strong secondary and tertiary structural features resulting in the steric hindrance as shown in Figure 1.
Unlike domain III, the modifications mapped onto the predicted domain II structures (21,22) were widespread and did not match with any of the stem loops or internal bulges predicted in either of the proposed models (U129, U135, A137, A138, C141-U144, U148-A150, U156, U158, A160-163, A165, C167, U182, A184, A185, A190-U193, U206, A212 and A213) (Figure 1). This suggests that in the free form of the RNA domain II remains flexible; however, the 5′ boundary of domain II is yet to be determined and the question of domain II length and organization remains unanswered.
Domains II, IIIab and IIId are crucial for PTV-1 IRES-mediated translation
Our chemical probing results suggest a similar secondary structure and domain organization as found in HCV and HCV-like IRESs (28). To test whether these domains are also important for IRES function, we investigated the translation efficiency of several truncation and deletion mutants (Figure 2). In addition, we also wanted to test the 5′ boundaries of the PTV-1 IRES, previously mapped between nucleotides 125 and 150 (20), since a recent study proposed a much shorter stem-loop structure for domain II (22) and the chemical probing results did not allow this shortened domain II to be discounted (Figure 1). The wild-type and mutant PTV-1 IRESs were cloned into a discistronic pRF vector that codes for the Renilla luciferase (RLuc) through the canonical, cap-dependent pathway and for the firefly luciferase (FLuc), which relies on a translationally active 5′IRES for expression. The resulting plasmids were analyzed in rabbit reticulocyte lysate (RRL) using standard coupled transcription/translation assays (TnT Promega) and the RLuc and FLuc expression measured.
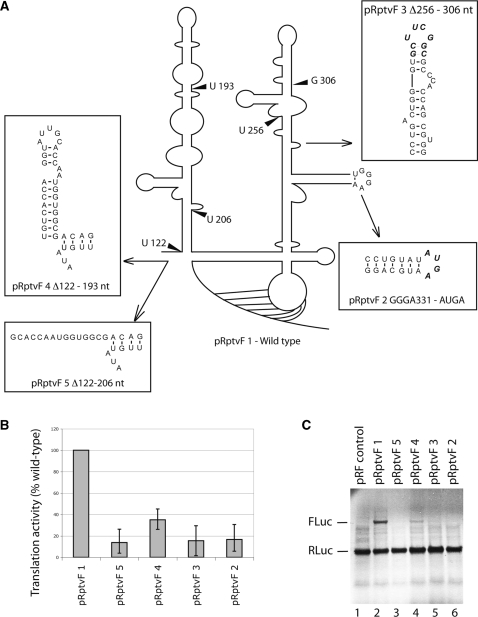
Figure 2.
Comparison of IRES efficiency of PTV-1 IRES wild-type and mutant RNAs in dicistronic translation assays. (A) Schematic representation of the secondary structure of the PTV-1 IRES showing the position of mutations. (B) Quantification of firefly luciferase (FLuc) expression from wild-type and mutant PTV-1 IRES RNA. Data obtained after the subtraction of background and presented as a percentage of a wild-type value and are the mean of three experiments. (C) Autoradiograph of RNAs translated in rabbit reticulocyte lysate with 35S labeled met-tRNAimet and separated by electrophoresis on a 4–12% bis–tris SDS gel. Expression of Renilla luciferase (RLuc) from the upstream cistron had a similar efficiency for all plasmids as expected.
In HCV-like IRESs, the basal domain III constitutes the core 40S binding region (28), which contains a G-rich hairpin loop in domain IIId (G331–G333), that is highly conserved among HCV-like IRESs (22) and essential for translational activity in the HCV and CSFV IRESs (8,26,27). Mutating the domain IIId stem-loop sequence in the PTV-1 IRES (Figure 2A, GGGA to AUGA–pRptvF 2) reduced translational activity more than 5-fold as compared to wild type suggesting conservation of the functional importance of domain IIId among HCV-like IRESs (Figure 2B and C—pRpvtF 2).
The apical domain III constitutes the major eIF3 binding site of HCV-like IRESs and deletion or truncation strongly reduces eIF3 binding and therefore IRES activity (13). When the apical subdomains IIIa and IIIb of the PTV-1 IRES were deleted, Δ256–306 nt (Figure 2A, B and C—pRptvF 3), translation was reduced to 14% of wild-type suggesting that these subdomains are also essential for PTV-1 IRES translation.
Finally, we could also confirm the previously suggested 5′ boundary of the PTV-1 IRES. Truncation of domain II Δ122–193 nt (22) (Figure 2A—pRptvF 4) maintained relatively high levels of IRES activity (34% of wild-type levels, Figure 2B and C), but when domain II was shortened further, Δ122–206 nt, (Figure 2A—pRptvF 5) translation efficiency dropped below 13% (Figure 2B and C). These results show that the shorter stem loop domain II supports lower levels of translation, but that the longer domain II is required for efficient translation as previously shown for the HCV IRES (17). Notwithstanding, our chemical probing results of the free PTV-1 IRES RNA that show both domain II and the apical domain III are flexible and probably less structured as compared to the HCV IRES (13); these domains still share their importance for IRES function.
Purification of binary 40S-IRES and 48S initiation complexes
To map interactions with the translational machinery and possible structural rearrangements of the PTV-1 IRES up to 48S assembly, an RNA construct containing the PTV-1 IRES element, 89 nt of the open reading frame and a streptomycin aptamer [StreptoTag (34)] was designed and this hybrid RNA was then used for affinity purification of binary 40S-IRES and 48S initiation complexes from rabbit reticulocyte lysate (RRL) as previously described (31). Formation of stable 48S initiation complexes on the PTV-1 IRES requires eIF2 and eIF3 (20), and we have previously shown by western and northern blot analyses that 48S initiation complexes purified by this technique contain eIF3, eIF2 and Met-tRNAiMet in a 1:1 stoichiometry with the PTV-1 IRES RNA (31).
The correct positioning of the AUG codon in the mRNA binding cleft of the ribosome in the purified binary 40S–IRES and 48S complexes was analyzed by toe printing (Figure 3A). Inhibition of reverse transcription is observed at positions 16–19 nt downstream of the adenine base of the AUG start codon. This specific pattern of reverse transcription arrest is a characteristic for the AUG codon positioned in the ribosomal P-site and indicates that the isolated complexes are properly assembled.
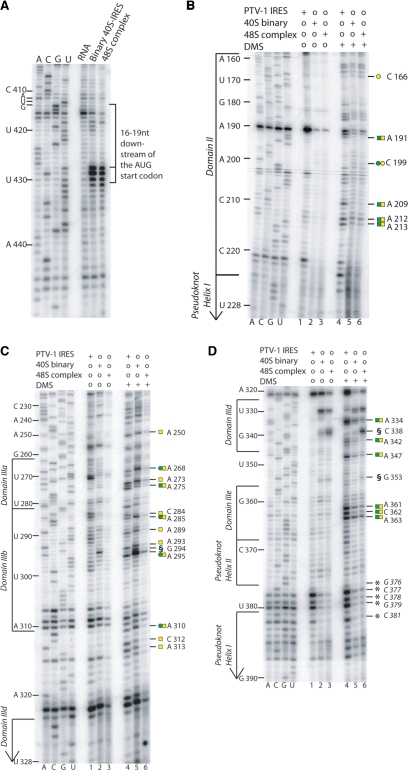
Figure 3.
Toe print and DMS probing of binary 40S–IRES and 48S complexes. (A) Toe-printing analysis of binary 40S–IRES and 48S complexes previously purified as reported (31) and reverse transcribed from a 5′ end-labeled primer complimentary to nucleotides 453–470 of the PTV-1 IRES. The toe print at position 16–19 nt downstream from A412 of the initiation codon represents binding of the 40S subunit. Lanes A, C, G and U show cDNA sequences corresponding to wild-type PTV-1 IRES. The position of the AUG initiation codon (AUG412–414) is indicated on the left. (B, C and D) DMS probing of PTV-1 IRES in the free form, binary 40S–IRES and 48S complexes (lanes 4, 5 and 6) reverse transcribed from 5′ end-labeled primers complimentary to nucleotides 186–202 (B), 244–262 (C) and 402–416 (D). Domain organization shown on the left. Binary 40S–IRES and 48S complexes, shown in A, B, C and D were phenol extracted prior to reverse transcription to remove bound proteins, whereas the free RNA samples did not undergo this procedure. This difference in the experimental procedure may account for the changes observed between the free RNA sample and the complex samples (binary 40S–IRES and 48S complexes) at nucleotides G376–G379 and C381 [represented by asterisk] (C and D), such differences are observed in both DMS treated (compare lane 4 to lanes 5 and 6) and untreated samples (compare lane 1 to lanes 2 and 3), therefore no conclusions can be drawn in this region. The strong arrest observed at G294 and C338 and G353 in the DMS treated and untreated samples [represented by section symbol (C and D respectively)] are also believed to be a result of this difference in the experimental procedure, where hydrolysis may have occurred as a result of the phenol extraction procedure.
Chemical probing of the binary 40S–PTV-1 IRES complex
First we analyzed changes in chemical reactivity to DMS between the free PTV-1 IRES and the binary 40S–IRES complex (Figure 3B). As expected, the majority of reactivity changes occur within the major 40S binding domain comprising the basal subdomains of domain III (Figure 3B–D, compare lanes 4 and 5). A334 in domain IIId, A347 in the IIId three-way junction of domain IIId, A361–A363 in domain IIIe are all protected from chemical attack upon 40S subunit binding (Figure 3D compare lanes 4 and 5, protection to DMS attack represented by green squares). Changes can also be seen at positions G376–379 and C381 located within the pseudoknot (Figure 3D, compare lanes 4 and 5); however, the primer arrest at these positions are thought to be due to a strong structure in this region as it can also be observed in the unmodified samples (Figure 3D, compare lane 1 and 4). Since the binary 40S–IRES and 48S complexes were subjected to phenol/chloroform extraction while the free RNA was not, changes in primer arrest are most likely due to difference in the experimental procedure between these samples (Figure 3D compare lanes 1 and 4, lanes 2 and 5, and lanes 3 and 6). However, we cannot conclusively confirm that 40S binding causes structural rearrangements within the pseudoknot due to a difference in the experimental procedure between the free RNA and the binary 40S–IRES and 48S complexes that could also account for this difference.
The changes in chemical reactivity suggest either direct interactions of the respective nucleotides with the 40S subunit (e.g. hairpin loops IIId and IIIe), but protection around the IIId three-way junction could also be indicative of structural rearrangements in the IRES RNA upon 40S binding (Figure 4). Protection against chemical attack also occurs in domain II (Figure 3B), which might not only reflect an interaction with the 40S subunit, but also that domain II is more structured in the binary complex (Figure 4). The apical subdomains of domain III, in contrast, display an enhanced reactivity to DMS upon 40S binding (Figure 3C compare lanes 4 and 5, enhanced DMS reactivity represented by green circles). The predicted stem loops (A268 in IIIa and A293–A295 in IIIb) specifically show increased modifications in the binary complex (Figure 4). In the HCV IRES RNA, this region comprises the eIF3 binding site (13) and the cryo-EM structure of the binary 40S–HCV IRES complex showed that the apical parts of domain III are not in contact with the 40S subunit (35). Our probing data are consistent with the apical domain III of the PTV-1 IRES also being exposed in the binary 40S–IRES complex and undergoing conformational changes leading to enhanced reactivity to DMS.
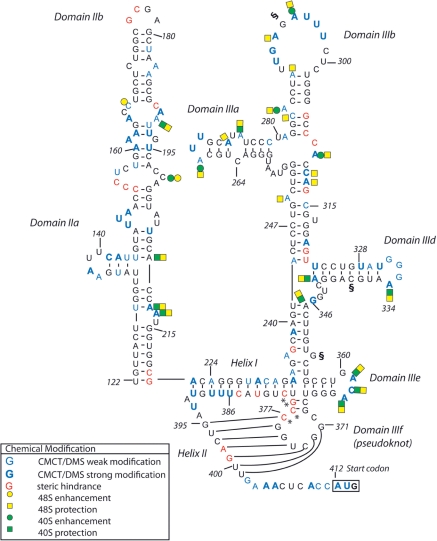
Figure 4.
Schematic representation of the changes in DMS reactivity between free RNA, binary 40S–IRES and 48S complexes mapped on the secondary structure of the PTV-1 IRES RNA as indicated in the lower left. The initiation codon (AUG412-414) is boxed. Asterisk and section symbol annotations are as described in Figure 3.
Chemical probing of the PTV-1 IRES–48S complex
Next we monitored changes in the chemical reactivity of the PTV-1 IRES RNA in the 48S initiation complex. The pattern of DMS modification in the 48S complex shows widespread protection throughout the IRES RNA (Figure 3B–D, lanes 4–6, DMS protection represented by yellow squares). The basal region of domain III and domain II, which displayed major changes in reactivity upon 40S binding, show a very similar pattern of chemical modification between binary 40S–IRES and 48S complexes (Figure 3D, compare lanes 5 and 6). The apical subdomains of domain III (IIIa and IIIb), however, exhibit distinct differences (Figure 3C, compare lanes 5 and 6). Upon binding of eIF3 and the ternary complex within a 48S complex, nucleotides in the hairpin loops and surrounding the apical three-way junction are protected from modification (Figure 4). Protection of the apical domains IIIa and IIIb in the 48S complex might reflect a direct interaction with eIF3 as seen for the analogous domains in the HCV IRES RNA (13), but changes around the apical three-way junction point towards additional structural rearrangement in this part of the IRES upon recruitment of eIF3 and the ternary complex.
A tertiary interaction in domain IIIe enhances translation efficiency of HCV-like IRESs
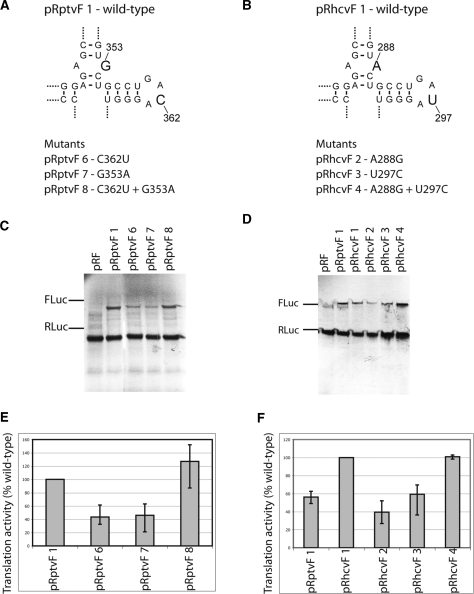
Domain IIIe and the preceding stem including a mismatched A–G or A–A base pair display strong sequence conservation among HCV-like IRESs (Figure 5A and B) (22). Interestingly, in the PTV-1 IRES, the mismatch A–G base pair occurs with a GACA loop sequence in domain IIIe as opposed to the HCV or CSFV IRES, where a GAUA loop in IIIe occurs with a mismatch A–A base pair (Figure 5B) (22). This conserved co-variation maintains the possibility to form a potential Watson–Crick base pair between these two elements and prompted us to investigate whether this covariant substitution is functionally relevant in HCV-like IRESs. Mutations in the purine–purine base pair or the IIIe loop were made in both PTV-1 and HCV IRES (Figure 5A and B), cloned into a pRF vector and the resulting plasmids assayed using standard coupled transcription/translation assays. The results show that single mutation in either of these positions in the PTV-1 IRES (G353A or C362U, Figure 5A), which would disrupt a potential Watson–Crick base pair, reduces translation to ∼40–45% (Figure 5C and E), while a double mutation restoring a potential base pair at these positions (G353A and C362U, Figure 5A) in the PTV-1 IRES showed wild-type translation levels (Figure 5C and E). The same result is obtained for the HCV IRES where single mutation at either A288G or U297C cause translation to decrease to ∼40% and ∼60% of wild-type levels and a double mutation restores activity (Figure 5B, D and F). Together these results suggest that the potential to form a canonical base pair between the mismatched base pair and the hairpin loop in domain IIIe plays a role in enhancing translation efficiency of these IRESs, possibly by stabilizing tertiary structure in this region.
Figure 5.
Influence of the co-variation substitution between the mismatch purine–purine and loop IIIe on PTV-1 IRES activity. (A and B) Schematic representation of the wild-type four-way junction at domain IIIe of the PTV-1 and HCV IRESs. Single and double mutants are listed below. (C and D) Autoradiographs of RNAs translated in rabbit reticulocyte lysate with 35S labeled met-tRNAimet and separated by electrophoresis on a 4–12% bis–tris SDS gel. Expression of Renilla luciferase (RLuc) from the upstream cistron had a similar efficiency for all plasmids as expected. (E and F) Quantification of firefly luciferase (FLuc) expression from wild-type and mutant PTV-1 and HCV IRES RNA. Data obtained after the subtraction of background and presented as a percentage of a wild-type value and are the mean of three experiments.
The conserved tertiary interaction stabilizes the pseudoknot structure
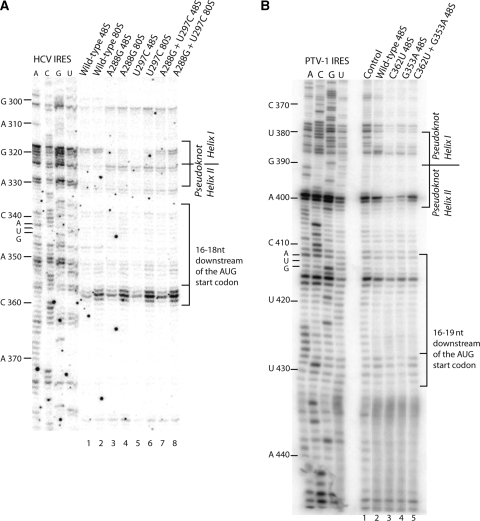
To test the effect of the tertiary interaction between the purine–purine mismatch and loop IIIe on the local structure, we performed toe-print assays in RRL using wild type, single mutant (A288G or U297C) and double mutant (A288G–U297C) HCV IRESs. This assay not only yields the characteristic toe print 16–18 nt downstream of the AUG start codon indicative of its base pairing with Met-tRNAiMet in the ribosomal P-site, but also upstream stops due to the strong secondary structure or the leading edge of eIF3 (16). Assembly of ribosomal complexes in RRL onto the wild-type and mutant HCV IRES RNAs was performed either in the presence of the non-hydrolysable GTP analog, GMPPNP, to inhibit eIF2 release and accumulate fully assembled 48S complexes (36) or in the presence of cycloheximide, to inhibit the elongation process, and accumulate fully assembled 80S ribosomes (37). Both wild-type and mutant complexes displayed very similar toe prints at positions +16 to +18 from the adenine (+1) in the AUG start codon indicative of 48S complex formation and an intensified middle band corresponding to the position +17 upon 80S assembly (Figure 6A). In addition, both wild-type and the compensatory double mutant 48S and 80S complexes display stops at positions G318 and G319 in helix I of the pseudoknot, while the single mutants do not (Figure 6A). This suggests that in the presence of the tertiary interaction near the pseudoknot, the reverse transcriptase encounters a stronger secondary structure in helix I as compared to the single mutants which disrupt the tertiary interaction.
Figure 6.
Toe-print analysis of wild-type and mutant HCV and PTV-1 IRESs. (A) Toe-print analysis of 48S and 80S wild-type and mutant HCV IRES formed in rabbit reticulocyte lysate in the presence of GMPPNP or cycloheximide, respectively. Toe-print positions, 16–18 nt downstream from A432 of the AUG initiation codon (indicated on the left), represent binding of the 40S subunit. Lanes A, C, G and U show cDNA sequences corresponding to wild-type HCV IRES. Pseudoknot helix positions at nucleotides C317 and G318 are indicated on right. (B) Toe-printing analysis of purified wild-type and mutant 48S complexes, reverse transcribed from a 5′ end-labeled primer complimentary to nucleotides 453–470 of the PTV-1 IRES. The toe print at position 16–19 nt downstream from A412 of the initiation codon represents binding of the 40S subunit. Lanes A, C, G and U show cDNA sequences corresponding to wild-type PTV-1 IRES. The position of the AUG initiation codon (AUG412–414) is indicated on left and pseudoknot helices and toeprints are indicated on right.
In the same assay performed on purified wild type, the single mutants (G353A or C362U) and double mutant (G353A–C362U) PTV-1 IRES 48S complexes, we observed a similar effect on the local pseudoknot structure (Figure 6B). In the latter, strong stops were observed at positions A398 and G399 in helix II of the PTV-1 pseudoknot, which were absent in the single mutants, which would disrupt the G353–C362 base pairing. Therefore, our data suggest that the conserved tertiary interaction enhances local stability of the pseudoknot structure to increase translational efficiency in HCV and HCV-like IRESs.
DISCUSSION
All HCV-like IRESs differ in terms of size and number of branching stem-loop structures in domain III, but preserve a structurally conserved core in domain III with a pseudoknot, domains IIId and IIIe and the helix connecting these stem-loop structures (22). This suggests a functional importance of this core structure in all HCV-like IRESs. Chemical probing of binary HCV or CSFV IRES 40S complexes revealed that these structural elements comprise the major 40S binding site and that the conserved G-rich loop in domain IIId is crucial for 40S binding and IRES activity (8,13,26,27). We show that this functional role is also conserved in the PTV-1 IRES. Major protections from chemical modifications occur in domains IIId and IIIe upon 40S binding of the PTV-1 IRES, and mutation of the G-rich loop in domain IIId abolishes IRES activity (Figure 2B and C—pRptvF 2).
Other functionally important elements are more variable among HCV-like IRESs (22). Domain II, which adopts an L-shaped conformation in both CSFV and HCV IRES (17,38) and contains a hairpin loop (5′-UAGCCAU-3′) and a loop E motif required for IRES function, displays a very different predicted secondary structure in the PTV-1 IRES lacking these elements. The HCV IRES domain II is not required for 40S binding, but the interaction of domain IIb with the 40S subunit near the E-site mediates opening of the mRNA channel and aids to stably accommodate the HCV ORF (8,35). The main role of the HCV domain II-dependent conformational changes of the 40S subunit lies in stimulating efficient hydrolysis of eIF2-bound GTP, eIF2-release and subunit joining (17,18). Although the PTV-1 IRES domain II does not preserve the key elements and appears highly unstructured in the free form according to our probing results, it also interacts with the 40S subunit in binary 40S–IRES and 48S complexes. Deletion of PTV-1 IRES domain II renders the IRES inactive, while shortening the sequence by 69 nt, which would still allow formation of a small hairpin stem-loop structure, still retains 34% IRES activity compared to wild type (Figure 2B and C—pRptvF 4 and 5). These data as well as a previous study (21) show that the PTV-1 IRES domain II, while being comprised of different structural elements, still interacts with the 40S subunit and is also required for efficient IRES-mediated translation.
The other domain displaying little sequence conservation among HCV-like IRESs comprises the eIF3 binding site in the apical domain III. Both the HCV and CSFV IRES contain a four-way junction with branching stem-loops IIIabc, which are crucial for IRES activity (13), while the predicted PTV-1 IRES secondary structure lacks either stem-loop IIIa or IIIc. Nevertheless, we show that also this reduced apical domain III in the PTV-1 IRES that is essential for activity (Figure 2B and C—pRptvF 3). Chemical and enzymatic probing as well as modification interference experiments on the HCV IRES identified stem-loops IIIa and IIIb as the major eIF3 interaction sites, while IIIc binds the 40S subunit (13,39). Our chemical probing experiments confirm that PTV-1 IRES lacks an equivalent stem-loop IIIc and therefore displays a reduced 40S binding domain as compared to the HCV IRES, but conserves stem-loops IIIa and IIIb, which are incorporated into an apical three-way junction (Figure 4). This domain seems to undergo restructuring upon 40S binding of the PTV-1 IRES, and strong modifications in the corresponding loop sequences propose their exposure for subsequent eIF3 binding (Figures 3C and 4). Upon recruitment of eIF3 and the ternary complex to the binary 40S–IRES particle, this region displays strong protections suggesting direct interaction with eIF3 and functional conservation of this domain (Figures 3C and 4). Interestingly, all other domains of the PTV-1 IRES display almost an identical chemical modification pattern in the binary 40S–IRES and 48S complex indicating that no significant additional changes in the IRES–40S interaction occur upon 48S complex formation (Figure 4).
The pseudoknot structure in the core 40S binding site is a highly conserved structural feature found in all HCV-like IRESs (22), but its precise role during IRES-mediated translation initiation is unknown. The maintenance of this structural element is critical for HCV IRES function (40). Disruption of CSFV IRES pseudoknot helix I abrogated stable binding of 40S subunits to the IRES and thereby abolished IRES activity (8). Abrogation of HCV IRES pseudoknot helix II, on the other hand, results only in a 2- to 3-fold reduction in 40S binding affinity, but again IRES activity is completely lost (13) suggesting a functional role of the pseudoknot beyond 40S subunit binding.
This role might also depend on a more complex tertiary structure surrounding the pseudoknot. We show that preserving the possibility to form long-range Watson–Crick base pairing between two nucleotides in hairpin loop IIIe and the conserved purine–purine mismatch in the adjacent helix connecting to domain IIId correlates with efficient translational activity in both PTV-1 and HCV IRESs (Figure 5). Comparative sequence analysis of HCV-like IRESs reveals strong sequence conservation and covariant nucleotide substitutions, suggesting that this tertiary interaction near the pseudoknot domain is conserved in all HCV-like IRESs (22). Previous evidence for a more complex pseudoknot structure came from enzymatic probing of the HCV IRES in the presence of magnesium ions, which revealed strong protections from cleavage not only in the pseudoknot, but also in the adjacent hairpin loop IIIe. Mutational studies of domain IIIe and the adjacent helix connecting to domain IIId also showed abolishment of translational activity or affected toe prints in the pseudoknot helices (8,12,24). Our toe print experiments of PTV-1 and HCV IRES mutants with disrupted long-range base pairing also show weakened toe prints in the pseudoknot helices indicative of the local destabilization of the pseudoknot fold (Figure 6). In the free form of the PTV-1 IRES, the pseudoknot might be at least partially formed as judged from strong steric hindrance in helices I and II in our chemical probing experiments, but strong modifications in loop IIIe do not support formation of the tertiary interaction in the unbound IRES (Figure 1). The interaction with the 40S subunit stabilizes the pseudoknot, and the conserved long-range base pair might provide additional stabilization for efficient recruitment of initiation factors or events downstream of 48S complex assembly.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Medical Research Council. Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank members of the K. Nagai, D. Neuhaus and P.J. Lukavsky groups for helpful discussion and comments on the manuscript.
REFERENCES
- 1.Kapp LD, Lorsch JR. The molecular mechanics of eukaryotic translation. Annu. Rev. Biochem. 2004;73:657–704. doi: 10.1146/annurev.biochem.73.030403.080419. [DOI] [PubMed] [Google Scholar]
- 2.Maag D, Fekete CA, Gryczynski Z, Lorsch JR. A conformational change in the eukaryotic translation preinitiation complex and release of eIF1 signal recognition of the start codon. Mol. Cell. 2005;17:265–275. doi: 10.1016/j.molcel.2004.11.051. [DOI] [PubMed] [Google Scholar]
- 3.Pestova TV, Borukhov SI, Hellen CU. Eukaryotic ribosomes require initiation factors 1 and 1A to locate initiation codons. Nature. 1998;394:854–859. doi: 10.1038/29703. [DOI] [PubMed] [Google Scholar]
- 4.Pestova TV, Lorsch JR, Hellen CUT. Translational Control in Biology and Medicine. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 87–128. [Google Scholar]
- 5.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon-anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Algire MA, Maag D, Lorsch JR. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 7.Pisarev AV, Kolupaeva VG, Pisareva VP, Merrick WC, Hellen CU, Pestova TV. Specific functional interactions of nucleotides at key -3 and +4 positions flanking the initiation codon with components of the mammalian 48S translation initiation complex. Genes Dev. 2006;20:624–636. doi: 10.1101/gad.1397906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kolupaeva VG, Pestova TV, Hellen CU. Ribosomal binding to the internal ribosomal entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belsham GJ, Sonenberg N. RNA-protein interactions in regulation of picornavirus RNA translation. Microbiol. Rev. 1996;60:499–511. doi: 10.1128/mr.60.3.499-511.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hellen CU, Sarnow P. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes Dev. 2001;15:1593–1612. doi: 10.1101/gad.891101. [DOI] [PubMed] [Google Scholar]
- 11.Sachs AB, Sarnow P, Hentze MW. Starting at the beginning, middle, and end: translation initiation in eukaryotes. Cell. 1997;89:831–838. doi: 10.1016/s0092-8674(00)80268-8. [DOI] [PubMed] [Google Scholar]
- 12.Otto GA, Puglisi JD. The pathway of HCV IRES-mediated translation initiation. Cell. 2004;119:369–380. doi: 10.1016/j.cell.2004.09.038. [DOI] [PubMed] [Google Scholar]
- 13.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hellen CU, Pestova TV. Translation of hepatitis C virus RNA. J. Viral Hepat. 1999;6:79–87. doi: 10.1046/j.1365-2893.1999.00150.x. [DOI] [PubMed] [Google Scholar]
- 15.Ji H, Fraser CS, Yu Y, Leary J, Doudna JA. Coordinated assembly of human translation initiation complexes by the hepatitis C virus internal ribosome entry site RNA. Proc. Natl Acad. Sci. USA. 2004;101:16990–16995. doi: 10.1073/pnas.0407402101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pestova TV, Shatsky IN, Fletcher SP, Jackson RJ, Hellen CU. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Locker N, Easton LE, Lukavsky PJ. HCV and CSFV IRES domain II mediate eIF2 release during 80S ribosome assembly. EMBO J. 2007;26:795–805. doi: 10.1038/sj.emboj.7601549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova TV, de Breyne S, Pisarev AV, Abaeva IS, Hellen CU. eIF2-dependent and eIF2-independent modes of initiation on the CSFV IRES: a common role of domain II. EMBO J. 2008;27:1060–1072. doi: 10.1038/emboj.2008.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Terenin IM, Dmitriev SE, Andreev DE, Shatsky IN. Eukaryotic translation initiation machinery can operate in a bacterial-like mode without eIF2. Nat. Struct. Mol. Biol. 2008;15:836–841. doi: 10.1038/nsmb.1445. [DOI] [PubMed] [Google Scholar]
- 20.Pisarev AV, Chard LS, Kaku Y, Johns HL, Shatsky IN, Belsham GJ. Functional and structural similarities between the internal ribosome entry sites of hepatitis C virus and porcine teschovirus, a picornavirus. J. Virol. 2004;78:4487–4497. doi: 10.1128/JVI.78.9.4487-4497.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chard LS, Kaku Y, Jones B, Nayak A, Belsham GJ. Functional analyses of RNA structures shared between the internal ribosome entry sites of hepatitis C virus and the picornavirus porcine teschovirus 1 Talfan. J. Virol. 2006;80:1271–1279. doi: 10.1128/JVI.80.3.1271-1279.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hellen CU, de Breyne S. A distinct group of hepacivirus/pestivirus-like internal ribosomal entry sites in members of diverse picornavirus genera: evidence for modular exchange of functional noncoding RNA elements by recombination. J. Virol. 2007;81:5850–5863. doi: 10.1128/JVI.02403-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown EA, Zhang H, Ping LH, Lemon SM. Secondary structure of the 5′ nontranslated regions of hepatitis C virus and pestivirus genomic RNAs. Nucleic Acids Res. 1992;20:5041–5045. doi: 10.1093/nar/20.19.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kieft JS, Zhou K, Jubin R, Murray MG, Lau JY, Doudna JA. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 1999;292:513–529. doi: 10.1006/jmbi.1999.3095. [DOI] [PubMed] [Google Scholar]
- 25.Otto GA, Lukavsky PJ, Lancaster AM, Sarnow P, Puglisi JD. Ribosomal proteins mediate the hepatitis C virus IRES-HeLa 40S interaction. RNA. 2002;8:913–923. doi: 10.1017/s1355838202022057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lukavsky PJ, Otto GA, Lancaster AM, Sarnow P, Puglisi JD. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 2000;7:1105–1110. doi: 10.1038/81951. [DOI] [PubMed] [Google Scholar]
- 27.Jubin R, Vantuno NE, Kieft JS, Murray MG, Doudna JA, Lau JY, Baroudy BM. Hepatitis C virus internal ribosome entry site (IRES) stem loop IIId contains a phylogenetically conserved GGG triplet essential for translation and IRES folding. J. Virol. 2000;74:10430–10437. doi: 10.1128/jvi.74.22.10430-10437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lukavsky PJ. Structure and function of HCV IRES domains. Virus Res. 2009;139:166–171. doi: 10.1016/j.virusres.2008.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel JP, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pestova TV, Shatsky IN, Hellen CU. Functional dissection of eukaryotic initiation factor 4F: the 4A subunit and the central domain of the 4G subunit are sufficient to mediate internal entry of 43S preinitiation complexes. Mol. Cell Biol. 1996;16:6870–6878. doi: 10.1128/mcb.16.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Locker N, Easton LE, Lukavsky PJ. Affinity purification of eukaryotic 48S initiation complexes. RNA. 2006;12:683–690. doi: 10.1261/rna.2227906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lukavsky PJ, Puglisi JD. Large-scale preparation and purification of polyacrylamide-free RNA oligonucleotides. RNA. 2004;10:889–893. doi: 10.1261/rna.5264804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 34.Bachler M, Schroeder R, von Ahsen U. StreptoTag: a novel method for the isolation of RNA-binding proteins. RNA. 1999;5:1509–1516. doi: 10.1017/s1355838299991574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spahn CM, Kieft JS, Grassucci RA, Penczek PA, Zhou K, Doudna JA, Frank J. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 36.Merrick WC. Evidence that a single GTP is used in the formation of 80 S initiation complexes. J. Biol. Chem. 1979;254:3708–3711. [PubMed] [Google Scholar]
- 37.Godchaux W, III, Adamson SD, Herbert E. Effects of cycloheximide on polyribosome function in reticulocytes. J. Mol. Biol. 1967;27:57–72. doi: 10.1016/0022-2836(67)90351-8. [DOI] [PubMed] [Google Scholar]
- 38.Lukavsky PJ, Kim I, Otto GA, Puglisi JD. Structure of HCV IRES domain II determined by NMR. Nat. Struct. Biol. 2003;10:1033–1038. doi: 10.1038/nsb1004. [DOI] [PubMed] [Google Scholar]
- 39.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J. Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang C, Le SY, Ali N, Siddiqui A. An RNA pseudoknot is an essential structural element of the internal ribosome entry site located within the hepatitis C virus 5′ noncoding region. RNA. 1995;1:526–537. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.