Abstract
In vivo nucleosomes often occupy well-defined preferred positions on genomic DNA. An important question is to what extent these preferred positions are directly encoded by the DNA sequence itself. We derive here from in vivo positions, accurately mapped by partial micrococcal nuclease digestion, a translational positioning signal that identifies the approximate midpoint of DNA bound by a histone octamer. This midpoint is, on average, highly A/T rich (∼73%) and, in particular, the dinucleotide TpA occurs preferentially at this and other outward-facing minor grooves. We conclude that in this set of sequences the sequence code for DNA bending and nucleosome positioning differs from the other described sets and we suggest that the enrichment of AT-containing dinucleotides at the centre is required for local untwisting. We show that this signature is preferentially associated with nucleosomes flanking promoter regions and suggest that it contributes to the establishment of gene-specific nucleosome arrays.
INTRODUCTION
In vivo nucleosomes often occupy well-defined preferred positions on genomic DNA (1–5). It is well established that binding of DNA to the histone octamer is strongly favoured both by DNA sequences with a preferred bending trajectory (6–12) and by DNA flexibility (13). However the nature of the signal(s) that defines precise translational positioning relative to the DNA sequence has remained elusive.
A variety of techniques, including partial micrococcal nuclease (MNase) digestion utilizing either indirect end-labelling or primer extension protocols (1–5), parallel sequencing (14,15) and tiled microarrays (16,17) have been used to define nucleosome positions in vivo while the sequence organization of the octamer-binding site has been characterized by analysis of nucleosome core DNA sequences. A current view of the sequence pattern of nucleosomal DNA so derived is that DNA bending around the octamer is facilitated by the periodic occurrence of A/T and G/C containing dinucleotides such that the A/T containing dinucleotides ApA, ApT, TpA and TpT are preferentially situated where the minor groove points in towards the histone octamer and the G/C containing dinucleotides CpC, CpG, GpC and GpG are preferentially situated where the minor groove points out (7,18,19). This experimentally derived pattern is, however, at variance with both theoretical and other experimental studies that suggest that TpA, in particular, could be preferentially located at an outward-facing minor groove (10,20–26). An additional complication is that different studies of the sequence organization of nucleosomal DNA derived from isolated core particles have arrived at disparate conclusions. Whereas Satchwell et al. (7) found that in chicken erythrocyte core particle DNA the midpoint, equating to the presumed dyad, was marginally A/T rich, Field et al. (19) showed that in their collection of yeast core nucleosomal DNA sequences the DNA at the midpoint was enriched in G/C containing dinucleotides and correspondingly depleted in A/T containing dinucleotides.
With the exception of partial MNase digestion all the techniques used to define both nucleosome positions (14–19,27–29) and sequence organization (9,18,27) depend on a limit MNase digestion to core nucleosomes. Programs that predict nucleosome positions, apart from two that depend only on the calculated physicochemical properties of DNA (26,30), largely use as their primary database DNA sequences derived from limit MNase digests (18,19,27,31–33) or sequences of mixed origin (34). These predictive programs thus depend on the implicit, but so far untested assumption, that the population of core particles so isolated is wholly representative of the actual occupancy in chromatin in vivo. The preferred cleavage specificity of MNase for certain A/T rich sequences (35), or more particularly for the TpA dinucleotide (18), is well established. If, during digestion of chromatin to nucleosome core particles, MNase cleavage occurred only outside the DNA wrapped by the octamer, the resulting core particles should in principle be representative of the in vivo situation. If, however, MNase cleaved within the wrapped region, even at low frequency, the resulting isolated core particle population would likely be biased by the selective MNase cleavage.
To address this possibility we have analysed the DNA sequences of nucleosome positions mapped by partial MNase digestion and find that, in contrast to octamer-bound sequences derived from limit MNase digestion, the former sequences are enriched in preferred MNase cleavage sites. Since these sites are most apparent at the midpoint we further conclude that the preference for A/T rich sequences at the midpoint could constitute a sequence signal for the translational positioning of a histone octamer at a specified site on genomic DNA.
MATERIALS AND METHODS
Sequence analysis
The periodicity signature of DNA sequences was calculated using the parameters derived by Drew and Calladine (36) from the original sequence data of Satchwell et al. (7). These parameters are the calculated likelihood of each of 10 dinucleotides occurring at each position in a single turn of bent DNA. The average periodicity value imparted by dinucleotides was determined over a defined sequence window starting from nucleotide n, then from n + 1, and so on. Each value was plotted with reference to the midpoint of the window. A scanning window of m bp is equivalent to a window of m −1 base-steps. This technique determines the likelihood of a coherent periodic sequence pattern over any given window. The computed value of the periodicity index is dependent on window size so that for larger windows the average value is higher. The average stacking energy was calculated in a similar way using the values defined experimentally by Yavokchuk et al. (37). Average dinucleotide or mononucleotide composition in the set of in vivo mapped sequences was determined by aligning these sequences on the midpoints of the mapped positions. The amplitudes and phases of the sequences were computed as previously described (7). The compiled list comprised the sequences of mapped positions from ADH2 (5), ADY2 (this paper), BAR1 (3), CHA1 (38), GAL1-10 (39), HMR (40), MET16 (41), MET17 (41), PHO5 (1), recombination enhancer (42), RNR3 (43), STE6 (3), SUC2 (44) and URA3 (4) from Saccharomyces cerevisiae. This set is similar to that used by Segal et al. (27) for illustrative purposes. ‘Promoter’ nucleosomal DNA sequences are defined as those in the immediate vicinity of the transcription startpoint. Where the TATA box is occluded by a nucleosome as in, for example, the ADH2, ADY2, CHA1, GAL1 and GAL10 loci, that nucleosome was also included. For example, in the CHA1/VAC17 locus the CHA1 TATA box, but not that of VAC17 is occluded so that CHA1 −1 and +1 nucleosomes and VAC17 + 1 nucleosome (corresponding to CHA1 −2) were included. Where multiple possible overlapping positions have been reported, as in the CUP1-2 locus, none were included. To allow direct comparisons with other compilations of Saccharomyces cerevisiae nucleosome associated DNA sequences mapped positions in MOX from Pichia augusta (45) with a different A/T content were added only for the periodicity profile analysis and promoter/non-promoter nucleosomal DNA comparison. Chicken erythrocyte core nucleosome DNA sequences were as compiled by Satchwell et al. (7), while yeast core nucleosome DNA sequences and in vitro selected octamer-binding sequences were obtained from: http://genie.weizmann.ac.il/software/nucleo_prediction.html.
Nucleosome mapping
Mapping of nucleosome positions in the ADY2 locus was performed as previously described (5). Mapping of the H2A.Z containing nucleosomes in the ADY2 and ADH2 locus was performed by ChIP using a yeast strain containing TAP-tagged H2A.Z (obtained from Open Biosystems).
RESULTS
Positioned nucleosomes possess a distinct sequence periodicity signature
Can positioning information be derived from the sequences bound by nucleosomes mapped by partial MNase digestion in vivo and to what extent the sequence organization of these positions reflected that derived from sequences obtained from isolated core particles? Partial MNase digestion identifies a preferred nucleosome array at a given locus but does not directly identify nucleosomes as such. Typically the bounds of the mapped positions encompass >145 bp and thus do not necessarily correspond to core particles.
To probe for positioning information we used as an analytical tool the sequence periodicity profile, as originally described by Satchwell et al. (7) and quantitated by Calladine and Drew (36). This parameter measures the extent to which a DNA sequence periodicity of ∼10 bp with A/T containing and G/C containing dinucleotides in opposite phases is apparent in a chosen length of DNA and is normally taken as a measure of anisotropic bendability (36). Importantly the phase of the periodic modulations of both ApA/TpT and GpC is reversed in the centre of the chicken erythrocyte octamer-bound sequence (7). We therefore asked whether a similar phase reversal, or more simply a loss of periodicity without reversal could be found in the DNA of nucleosomes mapped in vivo. With a scanning window of 20–50 bp, such reversal or loss would appear as a minimum in the periodicity plot.
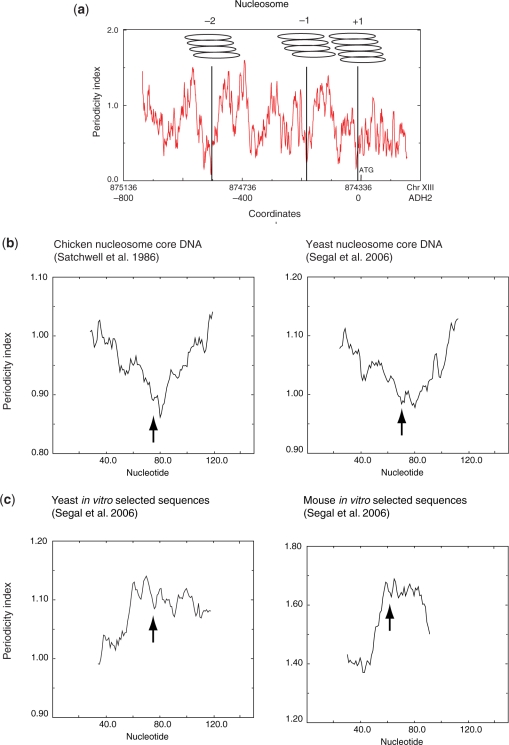
To validate this approach we first analysed nucleosomes +1, −1 and −2 from the yeast ADH2 promoter because their positions have been mapped to base-pair resolution (5). In this example, each ‘position’ was shown to consist of a family of four to five closely overlapping positions (5). A periodicity plot for the whole promoter region (Figure 1a) showed that, with an averaging window of 51 bp, the midpoint of each family of overlapping positions corresponded with a minimum value of the periodicity index. In two cases (nucleosomes −2 and −1) the minimum value was flanked by substantially higher values of the index resulting in a similar overall profile to the cloned sequences. The positions mapped by partial MNase digestion agreed to within ±10 bp of those mapped with parallel sequencing and tiled microarray analyses (15,17) (Supplementary Figure S1.1). Extending this analysis to nucleosomes −4, −3 and +2 to +4, mapped to a lower resolution of ±20 bp (5), we observed the same correspondence between midpoints and periodicity minima (Supplementary Figure S1.1) However, within the periodicity profile there were other prominent dips which did not correspond to the midpoint of a mapped nucleosome.
Figure 1.
(a) In vivo nucleosome positions on the ADH2 promoter have reduced periodicity index at their centres. The figure shows the multiple setting for each of the –2, –1 and +1 nucleosomes mapped by Verdone et al. (5). Coordinates are shown for chromosome XIII and to the ATG of ADH2. The periodicity profile was calculated using a 51-bp scanning window. (b) Compilations of ‘in vivo’ core nucleosome DNA sequences from chicken and yeast have a similar periodicity profile. (c) Compilation of octamer-binding sequences selected in vitro have a profile with a periodicity maximum at the centre. For both (b) and (c) the periodicity profile was calculated using a 51-bp window. The average sequence lengths for the chicken in vivo, yeast in vivo and yeast in vitro sequences are 145 bp and that of the mouse in vitro 119 bp. Arrows indicate the midpoints of the sequences.
We next asked whether this signature was also apparent in compilations of histone octamer-binding sites. We chose two compilations: 177 and 199 nucleosome core DNA sequences from chicken erythrocyte chromatin (7) and yeast chromatin (27), respectively. The average periodicity signature of these sets, containing in total 376 sequences, was similar, although attenuated by at least 5-fold, to that derived from the ADH2 promoter nucleosomes, with a minimum at or close to the midpoint (Figure 1b). In contrast, as expected from the high G/C content at the midpoint the periodicity signature for octamer binding sequences isolated from nucleosomes reconstituted in vitro by salt dilution (19,46) showed a maximum at the midpoint—essentially the opposite of the pattern of DNA isolated from nucleosomes in chromatin (Figure 1c). We conclude that the periodicity signature for the ADH2 promoter nucleosomes is similar to the average signal of sequenced nucleosomal DNA in chromatin and infer that the point of minimum average periodicity may correspond to a potential translational positioning signal. Second, we conclude that the sequences selected by the histone octamer depend strongly on the assembly conditions such that different selection conditions select DNA molecules with different sequence organizations.
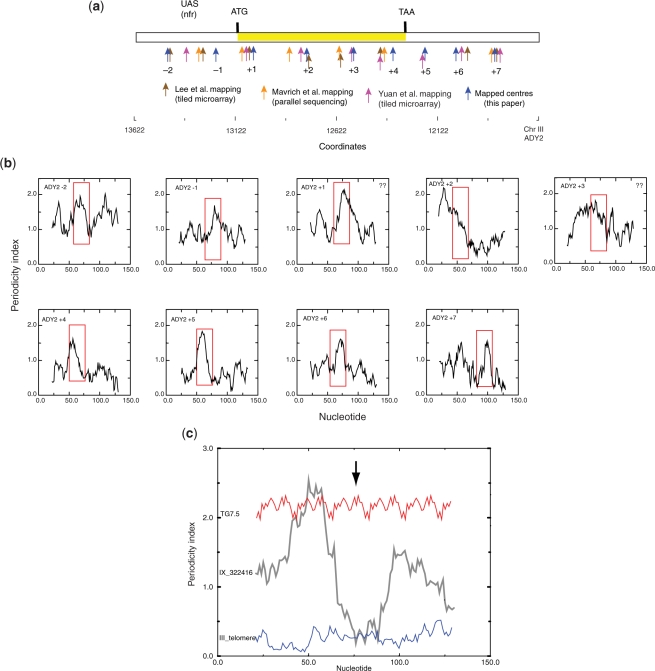
To ascertain whether the periodicity signature we had identified correlated with other positioned nucleosomes in vivo we first determined positions in the yeast ADY2 gene de novo by partial MNase digestion as a control for our previous mapping procedures (Figure 2a and Supplementary Figure S1.2). In all we identified nine nucleosomes spanning the gene, including two in the upstream region. With the exception of nucleosome −1 these positions agreed well but not precisely with the positions determined both by microarray mapping (17) and by parallel sequencing (14,15). Nevertheless, the agreement between the positions mapped by partial MNase digestion and either those mapped by parallel sequencing or by tiled microarray individually were comparable to that between positions mapped by the latter two methods. We determined the periodicity signature for each positioned nucleosome and found that seven out of the nine mapped nucleosomes exhibited a region in which there is a change from a high to low periodicity index within a length of up to 26 bp with the minimum being located within 10 bp of the centre of the mapped position (Figure 2b). In several cases, this pattern was asymmetric about the midpoint.
Figure 2.
(a) Comparison of nucleosome positions mapped by partial MNase digestion (this work), tiled-microarray (17) and parallel sequencing (15). In all cases, the midpoint of the mapped positions is shown but this does not take into account either the resolution of the different methods, which can vary depending on the nucleosome, or the ‘fuzziness’ of the mapping. The positions shown for the parallel sequencing data (15) are the averages of the positions determined on the two DNA strands. Nucleosome positions (–1, +1, etc.) apply to the set mapped by partial MNase digestion. The periodicity profile was calculated using a 51-bp window. (b) High-resolution periodicity profiles (21-bp scanning window) of nucleosomes mapped by partial MNase digestion in the ADY2 gene; 151 bp centred on the midpoint (75.5 bp) of each mapped position were analysed. The boxed regions indicate transitions from low to high periodicity close to the midpoint. With the exception of nucleosomes +1 and +3 all nucleosomes have such a signal. A low-resolution profile (window = 51 bp) is presented in Supplementary Figure S1.2. (c) Comparison of high-resolution periodicity profiles (21-bp scanning window) of an in vivo mapped nucleosome position (from the MET17 locus; see Figure 3) with that of an artificial DNA sequence (8) and a yeast telomeric repeat from the left telomere of chromosome III. An arrow indicates the sequence midpoints.
Comparison of the conserved periodicity pattern revealed by this analysis with other DNA sequences used for positioning studies revealed that an artificial positioning sequence consisting only of tandem 20-bp repeats (9) that fails to position nucleosomes translationally in vivo (47,48) lacked a significant periodicity minimum at the midpoint (Figure 2c). The average periodicity index for this sequence was of comparable magnitude to regions of high periodicity index in in vivo mapped nucleosomes. Furthermore, telomeric DNA sequences which, in vitro, have a low affinity for the histone octamer and poorly position octamers (49) possess a consistently low periodicity index. From these findings we identify a putative positioning signal as a region in which there is a change from a high to low periodicity index within a length of up to ∼40 bp. The low value of the index would correspond to the central part of the bound DNA and the high value to a region of tight bending around the octamer. Since DNA bends coherently around the octamer (50) this signature implies that the sequence preferences for DNA bending are context dependent, differing at the dyad and the immediately flanking regions.
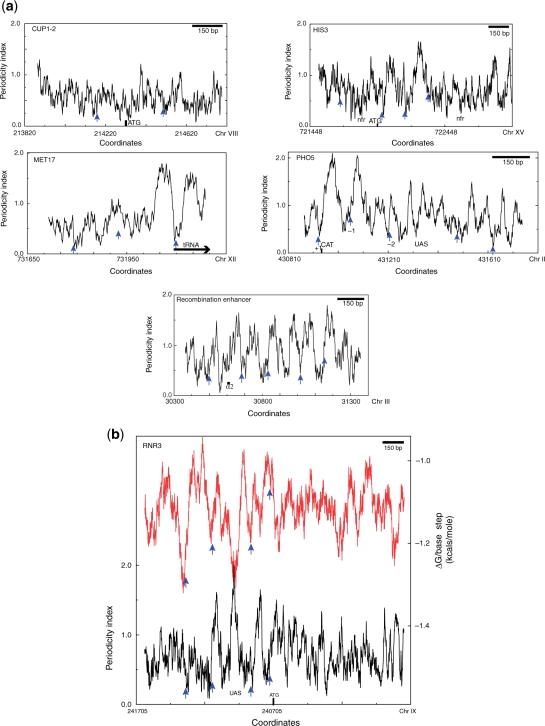
To ascertain whether this periodicity signal is also associated with partial MNase mapped positions in other genes we analysed the periodicity profiles of several genes reported in the literature. A representative selection is shown in Figure 3 and Supplementary Figure S1. The observed patterns are quite variable. In some cases, for example CUP1-2 and HIS3, the periodicity index is on average low relative to other genes. In other examples, as shown for MET17, PHO5 and RNR3, like ADY2, prominent peaks with a maximal value of ∼2.0 occur in the vicinity of transcription startpoint but, unlike ADY2, the maximal values then decrease away from the transcription startpoint. A third type of pattern is a more regular array of strong maxima as exemplified by the recombination enhancer (Figure 3) and as also seen in the SAC7 gene (not mapped by partial MNase digestion) (Supplementary Figure S1.10). When these periodicity profiles were related to 109 positions mapped by partial MNase digestion we found only a single example in the HMRα locus, where the midpoint of the mapped position corresponds to a prominent periodicity maximum. In most other cases the midpoint is located at a position where the periodicity index is <1.0. In regions of prominent periodicity maxima, the midpoint is located at or close to a periodicity minimum. This result is opposite to that predicted by models for nucleosome positioning in which the phase of the periodic modulations of dinucleotides is maintained throughout the octamer-binding site (18,19).
Figure 3.
(a) Low-resolution periodicity profiles (51-bp scanning window) for CUP1-2, HIS3, MET17, PHO5 and the recombination enhancer. (b) Low-resolution periodicity and stacking energy profiles (51-bp scanning window) for the RNR3 gene. One region of the recombination enhancer exhibits a pattern of strong periodicity variation correlating with positioned nucleosomes throughout the region analysed. RNR3, PHO5 and MET17 exhibit strong periodicity variation in the region bounding the transcription start site but the signal becomes less apparent in flanking regions. Note that for RNR3 the strong periodicity signal centred at coordinate 240975 (Chr IX) is also a region of high stacking energy which would be expected to increase the bending energy required for wrapping. In PHO5 nucleosomes ±1 correspond well with periodicity signals and are well positioned in vivo (1). The periodicity signals in HIS3 and CUP1-2 are generally lower that in the Adr1-regulated genes, especially for the latter two, but periodicity minima are still observed. In the RNR3 profile the periodicity is indicated by a black line and the stacking energy by a red line.
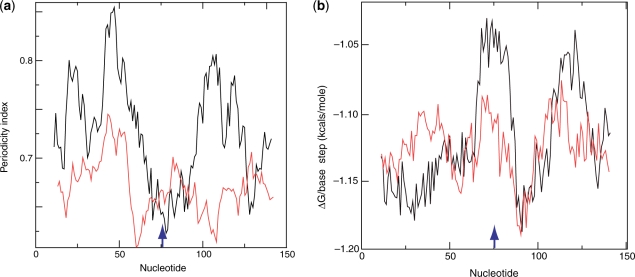
Because of the greater apparent prominence of the periodicity signal around the startpoint, we separated the compilation of positions into two sets, the first, 31 examples, comprising DNA sequences from nucleosomes in the immediate vicinity of the transcription startpoint and the remainder comprising DNA sequences from non-promoter nucleosomes. The A/T content of these two sets was similar (Supplementary Figure S5.1) but whereas the averaged periodicity signal for the promoter nucleosomes contained a very prominent dip at the midpoint that for the non-promoter nucleosomes lacked such a feature (Figure 4a and Supplementary Figure S3). We conclude the sequence organization of promoter nucleosomes is markedly different from non-promoter nucleosomes.
Figure 4.
(a) Averaged periodicity profiles of the set of ‘promoter’ nucleosomes (31 examples) (black line) and non-promoter nucleosomes (78 examples) (red line). (b) Averaged stacking-energy profiles of the set of ‘promoter’ nucleosomes and non-promoter nucleosomes. For both (a) and (b) the scanning window is 21 bp. All sequences in the compilations contained 151 bp. Sequence midpoints are arrowed in (a) and (b).
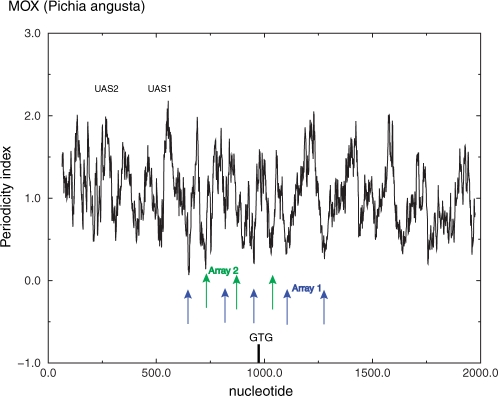
The form and length of the translational signature are such that more than one such signal could be accommodated within a 145-bp octamer binding site. In addition to mobilization correlated with histone acetylation (51) there is a well-documented example of the phenomenon of alternative exclusive overlapping nucleosome arrays under different environmental conditions at the Pichia angusta MOX promoter (45). In the MOX promoter region, we can identify periodicity minima within the promoter region that correspond to both possible arrays (Figure 5).
Figure 5.
The DNA signature is consistent with overlapping arrays observed experimentally in the Pichia angusta methanol oxidase (MOX) gene (45). The figure shows a low-resolution periodicity profile plot (51-bp window) compared with the mapped positions.
DNA flexibility may also have a role for nucleosome positioning
Stacking energy (52) is a sequence-dependent parameter that influences DNA stiffness or persistence length, which in turn is a major determinant of the affinity of a DNA sequence for the histone octamer (13). With the same scanning window that we used to probe sequence periodicity, we found that the stacking-energy profiles of yeast genes contained conserved patterns. Notably stacking energy is often low (<1.1 kcal/mole/base-step) in the immediate 5′ and 3′ flanking regions and high in the UAS transcription factor binding sites. This pattern is apparent in both the ADH2 and ADY2 genes (Supplementary Figure S4.1). In the 5′ flanking regions, low stacking energy is coincident with the centres of nucleosomes −2 (−1 of UBP15) −1 and +I of ADH2 and -1 of ADY2 (Supplementary Figure S4.1). This pattern is also observed for CHA1, GAL10 and GAL1 (Supplementary Figure S4.2). Analysis of the ‘promoter’ and’non-promoter’ sets of mapped nucleosomes sequences with a high-resolution 21-bp window confirmed that sequences in the region of the midpoint of promoter nucleosomes are enriched in dinucleotides with low stacking energy relative to non-promoter nucleosomes (Figure 4b). We conclude that the DNA signature associated in particular with promoter nucleosomes has two components—a strong coherent periodicity signal 20–25 bp from the midpoint and a region of low stacking energy at the midpoint itself.
In contrast, high stacking energy is associated with the UAS regions of ADH2, and ADY2 (Supplementary Figures S4.1), RNR3 (Figure 3b) and also CHA1 and GAL1-10 (Supplementary Figure S4.2), although this is not a universal feature of UAS regions. The high stacking energy and the correlated decreased flexibility could in principle decrease the affinity for histone octamers (13), thereby, consistent with the calculations of others (30), contribute to the formation of a nucleosome-free region. In several examples, e.g. RNR3, CHA1 and GAL1-10, the UAS is situated in a region that also contains a high periodicity signal that does not correspond to a mapped positioned nucleosome.
Nucleosomes mapped by partial MNase digestion are enriched in TpA at the midpoint
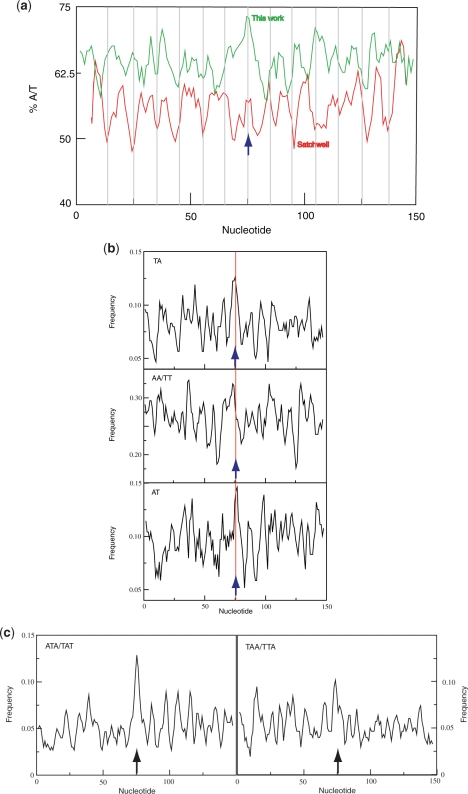
A sequence periodicity minimum does not distinguish between a simple loss of periodicity and a reversal of phase. To ascertain the sequence characteristic responsible for the minimum we collated 109 positions for which the coordinates of the nucleosome limits are reported (also including positions mapped de novo as described above) by alignment about the midpoint of the protected sequence. Although this method might be expected to lack the precision of aligning the sequences of DNA of core particles, we observed a robust signal, especially at the midpoint. In contrast to some other published yeast data sets (18,19) the average A/T content at the midpoint is ∼73% (Figure 6a) while the total average A/T composition (62%) is essentially identical to that of yeast genomic DNA (61.7%). The separated promoter and non-promoter sets of nucleosomal DNA sequences had similar compositions (Supplementary Figure S5.1a). For comparison the midpoint A/T content of the set of core nucleosome DNA sequences isolated from yeast by H2A.Z tagging is 61% (14,53) and that of the yeast set characterized by Segal et al. (27) is ∼59%. In sharper contrast high-affinity octamer-binding sequences selected in vitro are G/C-rich at the midpoint (18,19,27). Sets of 500 random yeast sequences and 500 randomized sequences of the same base composition lacked this signal (data not shown). In the vicinity of the midpoint the ApA/TpT, ApT and TpA dinucleotides are preferentially represented and the trinucleotides TAT/ATA and TAA/TTA are also over-represented at the same position (Figures 6b and c). The enrichment of TpA at the midpoint is observed for both the ‘promoter’ and ‘non-promoter’ sets of sequences (Supplementary Figure S5.1b). The preferred periodic occurrences of the dinucleotide TpA itself are, on average, in the opposite phase to that determined for the chicken erythrocyte nucleosomal core DNA sequences (Table 1) (7). This distribution represents a preference, not necessarily exclusive, for TpA to be located at outward-facing minor grooves on nucleosomal DNA from arrays characterized by partial MNase cleavage. The ApA/TpT maximum at the midpoint of in vivo mapped yeast nucleosomal DNA is similar to that observed for chicken nucleosomal DNA sequences but TpA has opposite profiles at the midpoint in these two sets (Supplementary Figure S5.2) (7). We conclude that the marked dip in the periodicity pattern in the vicinity of the midpoint of the yeast sequences results from a phase change in the ApA/TpT periodicity and the low stacking energy at the same position from the high frequency of occurrence of TpA.
Figure 6.
(a) Average base composition of collated set of DNA sequences mapped by partial MNase digestion (green line) compared with the set of chicken erythrocyte (7) (red line). Bars are placed at 10-bp intervals from the dyad. The chicken erythrocyte plot reveals minima at or close to many of the bars consistent with the conclusion that A/T containing dinucleotides occur at a lower frequency where the DNA minor groove points away from the surface of the histone octamer. The lower average A/T content of the chicken erythrocyte nucleosomal DNA reflects the lower A/T content (∼57%) of chicken DNA relative to yeast DNA (∼62%). (b) Frequency of occurrence of TpA and ApA/TpT dinucleotides in collated set of nucleosome positions mapped by partial MNase digestion. (c) Frequency of occurrence of TAT/ATA and TAA/TTA trinucleotides. Sequence midpoints are arrowed in all panels. In all panels the distribution of nucleotides was calculated using a 3-bp scanning window.
Table 1.
Principal dinucleotide periodicities in chicken erythrocyte nucleosome core DNA sequences (7) and yeast nucleosome positions mapped by partial MNase digestion
| Dinucleotide | Chicken |
Yeast |
||||
|---|---|---|---|---|---|---|
| Phase | Amplitude | Period (bp) | Phase | Amplitude | Period (bp) | |
| TpA | 176° | 0.12 | 10.26 | 5° | 0.17 | 10.11 |
| ApA/TpT | ±180° | 0.16 | 10.26 | 162° | 0.09 | 9.91 |
| GpC | 13° | 0.20 | 10.15 | 40° | 0.11 | 9.83 |
In each case the midpoint of the sequences is assigned a phase of 0°. For the chicken sequences the amplitude values (fractional variation in occurrence) were higher than at all other frequencies; for the yeast sequences only the amplitude for TpA met this condition.
DISCUSSION
Provenance of analysed sequences
The provenance of the nucleosomal DNA positions used for analytical programmes is of crucial importance in assessing their reliability. Our analysis depends on the accuracy of mapping nucleosome positions in vivo by partial MNase digestion. A major caveat is that this method relies on a regular pattern of MNase cleavage and does not positively identify nucleosomes. Nevertheless, while it is likely that the accuracy of published positions varies over a wide range this variation reflects more probably the variation between different studies than within a single study. Two observations indicate that the mapped positions primarily delineate nucleosomes. First, periodicity of the occurrences of TpA is ∼10.1 bp, a similar value to the dominant periodicities for ApA/TpT and GpC observed in chicken erythrocyte core DNA (7). Second, especially for the set of promoter nucleosomal DNA sequences, the distributions of stacking energy and periodicity profiles are symmetric about the midpoint (Figure 4). A second potential problem, raised by Segal et al. (27), is that because the lengths of nucleosomal DNA delimited by partial MNase digestion are >147 bp and the identified cleavage sites are averaged alignment of such sequences about their midpoints could result in insufficient resolution. Both the observation of distinctive signals at the midpoint (for example, the distribution of ATA/TAT) and the apparent symmetry of the DNA signature (Figure 4) indicate that this is not a valid concern. The corollary is that the initial MNase cleavage sites occur at positions that are approximately equidistant from the midpoint of the nucleosome and must therefore likely reflect some intrinsic structural feature of the yeast chromatin.
Novel sequence code for DNA bending and nucleosome positioning in yeast
Our results depend strongly on the provenance of the positioning information. In yeast, many accurately mapped in vivo nucleosome positions are derived from measurements derived from partial MNase digestion on repressed transcription units, for example, those regulated by the Adr1 activator and by the α2 repressor (3,54) and thus may be particularly well defined. Although the periodic sequence organization of our compilation of these sequences corresponds reasonably well with that reported by Satchwell et al. (7), it also exhibits unique features. In particular, the preferred rotational location of TpA at the midpoint is opposite to that reported for high affinity octamer-binding sequences selected in vitro (27) and also to recent compilations of yeast nucleosome core DNA sequences obtained by parallel sequencing (18,19). For example, the relative frequency of occurrence of the A/T and G/C containing dinucleotides are respectively 0.33 and 0.17 for the set reported here and 0.23 and 0.27 for that reported by Field et al. (18) Nonetheless, the rotational orientation of TpA in our compilation is consistent with theoretical predictions (20,26,49), with DNA structural data from both crystal and NMR studies (10,23,24) and also with DNA circularization data (25). In all the structural studies, TpA preferentially adopts, on average, a significant positive roll angle (12,24), compatible with an outward-facing minor groove in bent DNA, while the circularization studies indicate that the sequence TATAT is located where the minor groove is on the outside of a small DNA circle (25).
These observations provide an exception to the long-held view that A/T-rich sequences always preferentially occur at inward-facing minor grooves in tightly bent DNA (9,21). Our sequence data are derived from nucleosomal arrays identified by partial MNase digestion and are therefore not necessarily representative of yeast nucleosomal DNA sequences in general. Nevertheless, it is also possible that mapping procedures that depend on MNase digestion to core nucleosomes may bias the resulting data sets with the degree of difference depending on the extent of digestion. Since partial MNase digestion also delimits particles with more DNA the difference between our compilation and others for nucleosomal DNA isolated from chromatin may reflect either a different type of nucleosomal particle or a selective loss of nucleosomal DNA species with TpA, ApT and/or ApA/TpT dinucleotides and TAT/ATA and TAA/TTA trinucleotides at outward-facing minor grooves during limit digestion by MNase to mononucleosomes. TpA is a highly preferred substrate for MNase cleavage (18,35) and, indeed, overall depletion of the TpA dinucleotide relative to genomic occurrence was noted for the set of chicken core particle sequences (7). The DNA at outward-facing minor grooves is more mobile than that at inward-facing minor grooves (55) and could thus be more accessible to cleavage by a nuclease. In particular, the dyad DNA is more extensively cleaved by hydroxyl radicals than any other position (56,57). Any such preferential cleavage by MNase could bias a recovered set of nucleosomal DNA sequences and result in selection of a different overlapping set of sequences. Nevertheless, the average sequence organization of nucleosomal DNA sequences recovered from nucleosomes isolated by limit MNase digestion is entirely consistent with the structure of the core nucleosome (12,55) and therefore these sequences must represent bona fide octamer-binding sites. Given that both in vivo and in vitro the octamer can bind to a large number of overlapping sites with varying affinities (29,58), we suggest that where the positioning sequences identified by partial and limit MNase digestion or by different analyses of limit MNase digestions [for example, Supplementary Figures S1.4 (CHA1), S1.8 (recombination enhancer)], disagree all identifications may be valid. They may simply reflect overlapping sets of positions, some of which may be sub-optimal and possibly analogous to those demonstrated in the CUP1-2 and HIS3 genes (42,59). Since measurements of nucleosome occupancy depend on the isolation of unbiased populations this caveat also applies to such measurements that utilize limit MNase digestion (19)
The strong preference for A/T rich DNA at the centre of the mapped sequences is in accordance with other findings. Notably, Satchwell et al. (7) showed that in an average half-site not only the amplitude of the periodic oscillations of AA/TT frequency correlated with DNA bending increase from the midpoint to the periphery but also that at the midpoint the phase of the AA/TT frequency was reversed. Both these phenomena would, when averaged over a window of 51 bp, create a periodicity minimum at the midpoint. Similarly, Fitzgerald et al. (60) identified a periodicity pattern for nucleosomes reconstituted in vitro that is qualitatively identical to that reported here. Again, however, our result is at variance with other data showing the preferred occurrence of G/C rich sequences at the midpoint (18,19). Internal cleavage by MNase could again account for the different pattern of nucleosomal DNA sequences isolated from chromatin. However, the selection of sequences with a G/C rich dyad in vitro must however be due to other causes. These high-affinity sequences are selected at high salt concentrations and low temperature with limiting octamer. Yet, under only slightly different in vitro reconstitution conditions, the CHA1 -1 octamer-binding sequence (with an A/T rich midpoint) outcompetes an in vitro selected sequence (61). We suggest that under the selection conditions normally used stable binding to the octamer requires sequences with the low flexibility imparted by G/C-rich sequences rather than more flexible TpA-rich sequences.
The data reported here imply that for at least a set of yeast nucleosomes the sequence code for DNA bending and nucleosome positioning differs from that reported previously. This difference would strongly affect predictive accuracy. For example, whereas the midpoint of >95% of the nucleosome locations analysed here falls at a position of low periodicity index (for example, see Figure 3), a dinucleotide pattern in which periodic phases of G/C and A/T containing dinucleotides is maintained without interruption would predict that the midpoint should fall at a position of high periodicity index in a similar manner to the octamer binding sequences selected in vitro (Figure 1c).
We note that although −1 nucleosomes often contain the variant histone H2A-Z (14,53) this is not the case for ADY2 [ref. (14) and our own observations]. Instead, H2A-Z is preferentially associated with nucleosomes within the transcription unit. We conclude that preferred DNA sequence organization of promoter nucleosomes does not mark H2A-Z containing nucleosomes.
Structural consequences on an A/T rich dyad
What is the physical significance of the two novel features of the DNA sequences associated with natural positioned nucleosomes in yeast—the A/T-rich region of low stacking energy around the midpoint and a conserved periodicity signal of ∼40–50 bp with a minimum value close to the midpoint and a maximum value at 25–30 bp from the midpoint? The latter feature indicates that the minimal signal for nucleosome positioning can be short relative to the length of DNA bound in a core particle. We suggest that the periodicity signal corresponds to a half-site for H3–H4 tetramer binding such that the region of high coherent periodicity corresponds to the most tightly bent DNA on the surface of the tetramer (12,54). The short extent of the signal would be consistent with the finding that preferred ‘positions’ mapped by either partial MNase digestion or parallel sequencing may represent tight distributions of rotationally related positions (5,15) because the binding of an anisotropically bendable sequence of about two double-helical turns would not necessarily be uniquely defined. TpA preferentially adopts a positive roll angle and thus the preferred occurrence of TpA at the midpoint with an outward-facing minor groove is consistent with the coherent wrapping of DNA all around the octamer and is thus consistent with the crystal structure (12). The occurrence of sequences of low stacking energy at the midpoint is also consistent with the observation that the surface helical periodicity of nucleosomal DNA is 10.7 bp around the dyad but only 10 bp in the flanking region (22). Although a 10.7-bp surface helical periodicity is not necessarily indicative of a higher intrinsic helical periodicity (implying a low twist in the DNA), the enrichment of TpA at the midpoint would impart a torsional flexibility that would facilitate a local limited untwisting and so provide a physical characteristic restricting the positioning of the octamer on DNA. A requirement for a local low twist value could also be met by other sequences, e.g. TpG or CpG, and consequently could be relatively independent of overall base composition. In yeast, the high A/T content of genomic DNA would favour TpA and other A/T containing dinucleotides.
Genomic nucleosome organization
We have identified a DNA periodicity signature in yeast DNA that is strongly correlated with translational nucleosome positioning in vivo. This signature has two features: a preference for A/T rich DNA at the midpoint and as a consequence, a distinct sequence periodicity profile that is low at the midpoint and high on one or both sides. The nature of the positioning signal required to define an octamer-binding site implies a minimum length substantially shorter than the entire interaction region. Importantly, this frequency also implies that octamers can potentially occupy alternative positions at a given locus. Indeed, evidence from overlapping arrays, alternative nucleosome positions and shifts in positions in the absence of a chromatin remodelling complex indicates that this is the case (51,62–64). In the CUP1-2 and HIS3 genes, clusters of overlapping positioned nucleosomes of high to moderate occurrence have average separations between nucleosome centres varying from 35 to 90 bp, implying that these genes can accommodate multiple array settings (51,59,65). Both the HIS3 and the CUP1-2 gene DNA lack multiple strong periodicity signals in contrast to the mating type recombination enhancer (Figure 3) while in, for example, MET17, PHO5 and RNR3 strong periodicity maxima are associated with nucleosomes bordering the transcription start site. It remains to be established whether the affinity of the octamer for a given site correlates with the strength of the associated periodic signal but, were it to do so, a strong signal at the 5′-end of an array and weaker signals downstream would be consistent with the observations of Mavrich et al. (15) and Lee et al. (17) that for many nucleosome arrays the definition of nucleosome positions becomes increasingly ‘fuzzy’ with increasing distance from the 5′-end of an array. It is important to note that not all prominent periodicity signals are necessarily associated with nucleosomes since other DNA-binding proteins and protein complexes wrap DNA. For example, the signal in the UAS region of RNR3 may be one such case (Figure 3b).
From the potential multiplicity of octamer-binding sites and the variation in the sequence signals associated with positioned nucleosomes it follows that the documented strongly positioned nucleosome arrays are likely aligned by an ‘organizer’ acting in concert with nucleosome remodelling complexes, a principle similar to that proposed by Mavrich et al. (15). Such an organizer could be a strong intrinsic positioning signal specified by the DNA sequence or alternatively could be a sequence-specific DNA-binding protein, or indeed both. Potential examples of both mechanisms have been described. A strong intrinsically positioned nucleosome, also identified by our algorithm, is found in the MMTV LTR promoter (2,66), while the binding of the yeast α2 repressor correlates with aligned nucleosome arrays occluding the TATA boxes of repressed genes (3). Our data, which shows that the signature for translational positioning is most prominent in ‘promoter’ nucleosomes implies that DNA sequence can define a nucleosome position and in turn a strong positioning signal may contribute to the establishment of nucleosome arrays in vivo. This proposed mode of organizing eukaryotic chromatin is paralleled in bacterial chromatin by the H-NS protein which nucleates silencing at high-affinity binding sites and then spreads along the DNA by binding to low affinity, less precise, sites interspersed with occasional other high-affinity sites (67).
Kornberg (68) originally posed the question as to whether the location of nucleosomes in chromatin is specific or statistical. This proposition was subsequently refined to conclude that the observed distribution of nucleosomes in gene-specific arrays could be explained by statistical positioning associated with a boundary constraint for each array (69). Neglecting end effects statistical positioning implies that the potential number of histone octamer binding sites in a DNA sequence is essentially equivalent to the number of base pairs in that sequence, whereas specific positioning implies that, at one extreme, the observed pattern of nucleosome positions is uniquely defined by the DNA sequence. This latter possibility is countered by the observation that the repeat length of nucleosomal DNA in higher eukaryotes is tissue dependent (70,71). Our analysis indicates that the number of potential nucleosome positions within a gene is on average significantly greater than the number of nucleosomes and that for several genes the periodicity signals at the 5′-end of an array are stronger than internal signals towards the 3′-end (e.g. RNR3). In addition, the amplitude of the positioning signals varies between genes. These observations argue that observed preferred nucleosome arrays may indeed require a boundary constraint but, in contrast to a recent conclusion (15), that the associated positioning is neither strictly statistical nor uniquely defined. This conclusion for yeast agrees well with the deduced genomic nucleosome organization of the nematode Caenorhabditis elegans (28,29). In this case, at most loci a diversity of nucleosome positions was observed. In yeast the frequent identification of overlapping sets of positions by different techniques points to the same conclusion. Our results support a model in which the overall nucleosome organization in Saccharomyces cerevisiae would provide a basis for both a flexible chromatin organization and a concomitant ability to maintain DNA compaction.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Medical Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We especially thank Dr Graeme Mitchison of the Department of Applied Mathematics and Theoretical Physics, University of Cambridge for writing the computer programs. E.H. thanks EMBO for a fellowship. E.A., M.C., E.D.M. and L.V. thank Ministero Ambiente - Progetto Polveri for support. A.A.T. thanks Dr M. Buckle for stimulating discussions and l’Agence Nationale de Recherche for the award of a ‘chaire d’excellence’.
REFERENCES
- 1.Almer A, Rudolph H, Hinnen A, Hörz W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 1986;5:2689–2696. doi: 10.1002/j.1460-2075.1986.tb04552.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richard-Foy H, Hager GL. Sequence-specific positioning of nucleosomes over the steroid-inducible MMTV promoter. EMBO J. 1987;6:2321–2328. doi: 10.1002/j.1460-2075.1987.tb02507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shimizu M, Roth SY, Szent-Gyorgyi C, Simpson RT. Nucleosomes are positioned with base pair precision adjacent to the α2 operator in Saccharomyces cerevisiae. EMBO J. 1991;10:3033–3041. doi: 10.1002/j.1460-2075.1991.tb07854.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanaka S, Livingstone-Zatchej M, Thoma F. Chromatin structure of the yeast URA3 gene at high resolution provides insight into structure and positioning of nucleosomes in the chromosomal context. J. Mol. Biol. 1996;257:919–934. doi: 10.1006/jmbi.1996.0212. [DOI] [PubMed] [Google Scholar]
- 5.Verdone L, Camilloni G, Di Mauro E, Caserta M. Chromatin remodeling during Saccharomyces cerevisiae ADH2 gene activation. Mol. Cell Biol. 1996;16:1978–1988. doi: 10.1128/mcb.16.5.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drew HR, Travers AA. DNA bending and its relation to nucleosome positioning. J. Mol. Biol. 1985;186:773–790. doi: 10.1016/0022-2836(85)90396-1. [DOI] [PubMed] [Google Scholar]
- 7.Satchwell SC, Drew HR, Travers AA. Sequence periodicities in chicken nucleosome core DNA. J. Mol. Biol. 1986;191:659–675. doi: 10.1016/0022-2836(86)90452-3. [DOI] [PubMed] [Google Scholar]
- 8.Shrader TE, Crothers DM. Artificial nucleosome positioning sequences. Proc. Natl Acad. Sci. USA. 1989;86:7418–7422. doi: 10.1073/pnas.86.19.7418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Piña B, Truss M, Ohlenbusch H, Postma J, Beato M. DNA rotational positioning in a regulatory nucleosome is determined by base sequence. An algorithm to model the preferred superhelix. Nucleic Acids Res. 1990;18:6981–6987. doi: 10.1093/nar/18.23.6981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olson WK, Gorin AA, Lu XJ, Hock LM, Zhurkin VB. DNA sequence-dependent deformability deduced from protein-DNA crystal complexes. Proc. Natl Acad. Sci. USA. 1998;95:11163–11168. doi: 10.1073/pnas.95.19.11163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Widom J. Role of DNA sequence in nucleosome stability and dynamics. Q. Rev. Biophys. 2001;34:269–324. doi: 10.1017/s0033583501003699. [DOI] [PubMed] [Google Scholar]
- 12.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 13.Virstedt J, Berge T, Henderson RM, Waring MJ, Travers AA. The influence of DNA stiffness upon nucleosome formation. J. Struct. Biol. 2004;148:66–85. doi: 10.1016/j.jsb.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 14.Albert I, Mavrich TN, Tomsho LP, Qi J, Zanton SJ, Schuster SC, Pugh BF. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature. 2007;446:572–576. doi: 10.1038/nature05632. [DOI] [PubMed] [Google Scholar]
- 15.Mavrich TN, Ioshikhes IP, Venters BJ, Jiang C, Tomsho LP, Qi J, Schuster SC, Albert I, Pugh BF. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 2008;18:1073–1083. doi: 10.1101/gr.078261.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan GC, Liu YJ, Dion MF, Slack MD, Wu LF, Altschuler SJ, Rando OJ. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 17.Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat. Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- 18.Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhurkin VB. Specific alignment of nucleosomes on DNA correlates with periodic distribution of purine-pyrimidine and pyrimidine-purine dimers. FEBS Lett. 1983;158:293–297. doi: 10.1016/0014-5793(83)80598-5. [DOI] [PubMed] [Google Scholar]
- 21.Hagerman PJ. Sequence-directed curvature of DNA. Nature. 1986;321:449–450. doi: 10.1038/321449a0. [DOI] [PubMed] [Google Scholar]
- 22.Travers AA, Klug A. The bending of DNA in nucleosomes and its wider implications. Phil. Trans. Roy. Soc. Lond. B Biol. Sci. 1987;317:537–561. doi: 10.1098/rstb.1987.0080. [DOI] [PubMed] [Google Scholar]
- 23.Stefl R, Wu H, Ravindranathan S, Sklénar V, Feigon J. DNA A-tract bending in three dimensions: solving the dA4T4 vs. dT4A4 conundrum. Proc. Natl. Acad. Sci. USA. 2004;101:1177–1182. doi: 10.1073/pnas.0308143100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.El Hassan MA, Calladine CR. Conformational characteristics of DNA: empirical classifications and a hypothesis for the conformational behaviour of dinucleotide steps. Phil. Trans. Roy. Soc. Lond. A Phys. Sci. 1997;355:43–100. [Google Scholar]
- 25.Ulanovsky L, Bodner M, Trifonov EN, Choder M. Curved DNA: design, synthesis, and circularization. Proc. Natl. Acad. Sci. USA. 1986;83:862–866. doi: 10.1073/pnas.83.4.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Anselmi C, Bocchinfuso G, De Santis P, Savino M, Scipioni A. A theoretical model for the prediction of sequence-dependent nucleosome thermodynamic stability. Biophys. J. 2000;79:601–613. doi: 10.1016/S0006-3495(00)76319-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal E, Fondufe-Mittendorf Y, Chen L, Thastrom A, Field Y, Moore IK, Wang JP, Widom J. A genomic code for nucleosome positioning. Nature. 2006;442:772–778. doi: 10.1038/nature04979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Johnson SM, Tan FJ, McCullough HL, Riordan DP, Fire AZ. Flexibility and constraint in the nucleosome core landscape of Caenorhabditis elegans chromatin. Genome Res. 2006;16:1505–1516. doi: 10.1101/gr.5560806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valouev A, Ichikawa J, Tonthat T, Stuart J, Ranade S, Peckham H, Zeng K, Malek JA, Costa G, McKernan K, et al. A high-resolution, nucleosome position map of C. elegans reveals a lack of universal sequence-dictated positioning. Genome Res. 2008;18:1051–1063. doi: 10.1101/gr.076463.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miele C, Vaillant Y, d'Aubenton-Carafa C, Thermes C, Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan GC, Liu JS. Genomic sequence is highly predictive of local nucleosome depletion. PLoS Comput. Biol. 2008;4:e13. doi: 10.1371/journal.pcbi.0040013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peckham HE, Thurman RE, Fu Y, Stamatoyannopoulos JA, Noble WS, Struhl K, Weng Z. Nucleosome positioning signals in genomic DNA. Genome Res. 2007;17:1170–1177. doi: 10.1101/gr.6101007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chung HR, Vingron M. Sequence-dependent nucleosome positioning. J. Mol. Biol. 2009;386:1411–1422. doi: 10.1016/j.jmb.2008.11.049. [DOI] [PubMed] [Google Scholar]
- 34.Ioshikhes I, Bolshoy A, Derenshteyn K, Borodovsky M, Trifonov EN. Nucleosome DNA sequence pattern revealed by multiple alignment of experimentally mapped sequences. J. Mol. Biol. 1996;262:129–139. doi: 10.1006/jmbi.1996.0503. [DOI] [PubMed] [Google Scholar]
- 35.Hörz W, Altenburger W. Sequence specific cleavage of DNA by micrococcal nuclease. Nucleic Acids Res. 1981;9:2643–2658. doi: 10.1093/nar/9.12.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Drew HR, Calladine CR. Sequence-specific positioning of core histones on an 860 base-pair DNA. Experiment and theory. J. Mol. Biol. 1987;195:143–173. doi: 10.1016/0022-2836(87)90333-0. [DOI] [PubMed] [Google Scholar]
- 37.Yakovchuk P, Protozanova E, Frank-Kamenetskii MD. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 2006;34:564–574. doi: 10.1093/nar/gkj454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moreira JM, Holmberg S. Nucleosome structure of the yeast CHA1 promoter: analysis of activation-dependent chromatin remodeling of an RNA-polymerase-II-transcribed gene in TBP and RNA pol II mutants defective in vivo in response to acidic activators. EMBO J. 1998;17:6028–6038. doi: 10.1093/emboj/17.20.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li S, Smerdon MJ. Nucleosome structure and repair of N-methylpurines in the GAL1-10 genes of Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:44651–44659. doi: 10.1074/jbc.M206623200. [DOI] [PubMed] [Google Scholar]
- 40.Ravindra A, Weiss K, Simpson RT. High-resolution structural analysis of chromatin at specific loci: Saccharomyces cerevisiae silent mating-type locus HMRa. Mol. Cell Biol. 1999;19:7944–7950. doi: 10.1128/mcb.19.12.7944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kent NA, Tsang JS, Crowther DJ, Mellor J. Chromatin structure modulation in Saccharomyces cerevisiae by centromere and promoter factor 1. Mol. Cell Biol. 1994;14:5229–5241. doi: 10.1128/mcb.14.8.5229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weiss K, Simpson RT. Cell type-specific chromatin organization of the region that governs directionality of yeast mating type switching. EMBO J. 1997;16:4352–4360. doi: 10.1093/emboj/16.14.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li B, Reese JC. Ssn6-Tup1 regulates RNR3 by positioning nucleosomes and affecting the chromatin structure at the upstream repression sequence. J. Biol. Chem. 2001;276:33788–33797. doi: 10.1074/jbc.M104220200. [DOI] [PubMed] [Google Scholar]
- 44.Gavin IM, Simpson RT. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–6271. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Costanzo G, Di Mauro E, Negri R, Pereira G, Hollenberg C. Multiple overlapping positions of nucleosomes with single in vivo rotational setting in the Hansenula polymorpha RNA polymerase II MOX promoter. J. Biol. Chem. 1995;270:11091–11097. doi: 10.1074/jbc.270.19.11091. [DOI] [PubMed] [Google Scholar]
- 46.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 47.Tanaka S, Zatchej M, Thoma F. Artificial nucleosome positioning sequences tested in yeast minichromosomes: a strong rotational setting is not sufficient to position nucleosomes in vivo. EMBO J. 1992;11:1187–1193. doi: 10.1002/j.1460-2075.1992.tb05159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wallrath LL, Lu Q, Granok H, Elgin SCR. Architectural variations of inducible eukaryotic promoters: preset and remodeling chromatin structures. BioEssays. 1994;16:165–170. doi: 10.1002/bies.950160306. [DOI] [PubMed] [Google Scholar]
- 49.Pisano S, Pascucci E, Cacchione S, De Santis P, Savino M. AFM imaging and theoretical modeling studies of sequence-dependent nucleosome positioning. Biophys. Chem. 2006;124:81–89. doi: 10.1016/j.bpc.2006.05.012. [DOI] [PubMed] [Google Scholar]
- 50.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–150. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 51.Kim Y, McLaughlin N, Lindstrom K, Tsukiyama T, Clark DJ. Activation of Saccharomyces cerevisiae HIS3 results in Gcn4p-dependent, SWI/SNF-dependent mobilization of nucleosomes over the entire gene. Mol. Cell Biol. 2006;26:8607–8622. doi: 10.1128/MCB.00678-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Protozanova E, Yakovchuk P, Frank-Kamenetskii MD. Stacked-unstacked equilibrium at the nick site of DNA. J. Mol. Biol. 2004;342:775–785. doi: 10.1016/j.jmb.2004.07.075. [DOI] [PubMed] [Google Scholar]
- 53.Guillemette B, Bataille AR, Gévry N, Adam M, Blanchette M, Robert F, Gaudreau L. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 2005;3:e384. doi: 10.1371/journal.pbio.0030384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Agricola E, Verdone L, Xella B, Di Mauro E, Caserta M. Common chromatin architecture, common chromatin remodeling, and common transcription kinetics of Adr1-dependent genes in Saccharomyces cerevisiae. Biochemistry. 2004;43:8878–8884. doi: 10.1021/bi049577+. [DOI] [PubMed] [Google Scholar]
- 55.Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 56.Gottesfeld JM, Melander C, Suto RK, Raviol H, Luger K, Dervan PB. Sequence-specific recognition of DNA in the nucleosome by pyrrole-imidazole polyamides. J. Mol. Biol. 2001;309:615–629. doi: 10.1006/jmbi.2001.4694. [DOI] [PubMed] [Google Scholar]
- 57.Ragab A, Travers A. HMG-D and histone H1 alter the local accessibility of nucleosomal DNA. Nucleic Acids Res. 2003;31:7083–7089. doi: 10.1093/nar/gkg923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gencheva M, Boa S, Fraser R, Simmen MW, Whitelaw CBA, Allan J. In vitro and in vivo nucleosome positioning on the ovine β-lactoglobulin gene are related. J. Mol. Biol. 2006;361:216–230. doi: 10.1016/j.jmb.2006.06.039. [DOI] [PubMed] [Google Scholar]
- 59.Shen CH, Clark DJ. DNA sequence plays a major role in determining nucleosome positions in yeast CUP1 chromatin. J. Biol. Chem. 2001;276:35209–35216. doi: 10.1074/jbc.M104733200. [DOI] [PubMed] [Google Scholar]
- 60.Fitzgerald DJ, Dryden GL, Bronson EC, Williams JS, Anderson JN. Conserved patterns of bending in satellite and nucleosome positioning DNA. J. Biol. Chem. 1994;239:21303–21314. [PubMed] [Google Scholar]
- 61.Wu C, Travers A. Relative affinities of DNA sequences for the histone octamer depend strongly upon both the temperature and octamer concentration. Biochemistry. 2005;44:14329–14334. doi: 10.1021/bi050915w. [DOI] [PubMed] [Google Scholar]
- 62.Fazzio TG, Tsukiyama T. Chromatin remodeling in vivo: evidence for a nucleosome sliding mechanism. Mol. Cell. 2003;12:1333–1340. doi: 10.1016/s1097-2765(03)00436-2. [DOI] [PubMed] [Google Scholar]
- 63.Whitehouse I, Tsukiyama T. Antagonistic forces that position nucleosomes in vivo. Nat. Struct. Mol. Biol. 2006;13:633–640. doi: 10.1038/nsmb1111. [DOI] [PubMed] [Google Scholar]
- 64.Xella B, Goding C, Agricola E, Di Mauro E, Caserta M. The ISWI and CHD1 chromatin remodelling activities influence ADH2 expression and chromatin organization. Mol. Microbiol. 2006;59:1531–1541. doi: 10.1111/j.1365-2958.2005.05031.x. [DOI] [PubMed] [Google Scholar]
- 65.Sekinger EA, Moqtaderi Z, Struhl K. Intrinsic histone-DNA interactions and low nucleosome density are important for preferential accessibility of promoter regions in yeast. Mol Cell. 2005;18:735–748. doi: 10.1016/j.molcel.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 66.Piña B, Bruggemeier U, Beato M. Nucleosome positioning modulates accessibility of regulatory proteins to the mouse mammary tumor virus promoter. Cell. 1990;60:719–731. doi: 10.1016/0092-8674(90)90087-u. [DOI] [PubMed] [Google Scholar]
- 67.Bouffartigues E, Buckle M, Badaut C, Travers A, Rimsky S. H-NS cooperative binding to high-affinity sites in a regulatory element results in transcriptional silencing. Nat. Struct. Mol. Biol. 2007;14:441–448. doi: 10.1038/nsmb1233. [DOI] [PubMed] [Google Scholar]
- 68.Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981;292:579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- 69.Kornberg RD, Stryer L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 1988;16:6677–6690. doi: 10.1093/nar/16.14.6677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Compton JL, Bellard M, Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc. Natl Acad. Sci. USA. 1976;73:4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Thomas JO, Thompson RJ. Variation in chromatin structure in two cell types from the same tissue: a short DNA repeat length in cerebral cortex neurons. Cell. 1977;10:633–640. doi: 10.1016/0092-8674(77)90096-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.