Abstract
Quantitative reverse transcription polymerase chain reaction (qRT-PCR) is a valuable tool for measuring gene expression in biological samples. However, unique challenges are encountered when studies are performed on cells microdissected from tissues derived from animal models or the clinic, including specimen related issues, variability of RNA template quality and quantity, and normalization. qRT-PCR using small amounts of mRNA derived from dissected cell populations requires adaptation of standard methods to allow meaningful comparisons across sample sets. The protocol described here presents the rationale, technical steps, normalization strategy, and data analysis necessary to generate reliable gene expression measurements of transcripts from dissected samples. The entire protocol from tissue microdissection through qRT-PCR analysis requires approximately 16 hours.
Keywords: quantitative measurements, microdissected tissues, qRT-PCR, validation, gene expression analysis, protocol, normalization strategy
INTRODUCTION
There is significant interest in the biomedical community in quantitative mRNA expression analysis of microdissected cells from animal models or clinical specimens. These measurements can be performed as either a primary investigative tool or as a method to independently validate results from expression microarray experiments1–15. The present report provides a comprehensive protocol for analyzing microdissected samples5;16. To date, this is the only available protocol detailing the use of qRT-PCR for the comparative analysis of diseased and normal frozen microdissected tissues using an appropriate normalization strategy. The protocol is generally accessible to researchers with the potential application to all cell type from snap-frozen specimens, but as with any scientific methodology the user may need to modify conditions to meet their individual experimental requirements.
In our experience, qRT-PCR can be applied successfully to microdissected samples, though there are several challenges and caveats that need to be considered. Clinical specimens in particular require special diligence and care as upstream processing steps can significantly influence downstream molecular results17–21. Due to the potential importance of these caveats in influencing expression results, a brief overview of each issue is first provided followed by the detailed protocol for measuring mRNA.
TISSUE CONSIDERATIONS
Investigators need to be aware of upstream influences on data derived from tissue specimens, especially those from the clinic. Potentially confounding patient-related issues include demographics, disease status, and past and present therapies. In addition, one needs to be cognizant of changes that can occur in the clinic during surgery, tissue acquisition, and in the pathology laboratory, including; time to freezing, freezing method, tissue type, storage, and the effects of endogenous proteases and RNases. More information on these topics and a general review can be found at the National Cancer Institute Office of Biorepositories and Biospecimen Research (http://biospecimens.cancer.gov/) and is addressed by other researchers22–34.
TISSUE HANDLING, PROCESSING, AND EMBEDDING
It is important to consider the route the specimen takes from the time it leaves the patient30 or animal model to the purification of analytes, including how the tissue is handled up until it is processed. Tissue handling steps are dependent on tissue sample size35. Biopsies are typically small and are snap-frozen or placed in fixative immediately after removal from the body. Alternatively, surgical specimens (e.g. entire prostate) are large and need gross processing into smaller pieces prior to snap-freezing or fixing. Snap-frozen tissue is embedded in Optimum Cutting Temperature Compound (OCT) and stored at −80°C, and fixed tissue is almost always formalin-fixed and embedded in paraffin to form a tissue block and stored at room temperature (20 – 25°C). Fixation and embedding provides for optimal histopathological review under light microscopy, although the biomolecules are compromised due to the cross-linking effects of formalin, as well as the embedding process. In contrast, snap-frozen samples provide the highest quality DNA, mRNA, and protein for analysis, although the histological detail of the sections is inferior to that of fixed and embedded samples. The majority of microdissection-based mRNA expression studies are performed on frozen samples. If the tissue is adequately preserved, it is feasible to recover sufficient RNA quantities (100 to 200 ng) to make accurate measurements without pre-amplification of the transcriptome16. If it is not possible to immediately freeze tissue during collection then RNase inhibitors such as RNA Later can be used. While this approach is useful for preserving RNA in bulk tissue, it may not be suitable for microdissection due to the effect on tissue histology. If microdissection is required, it is recommended that the effect of RNA Later on tissue histology be investigated before using it on samples.
TISSUE SECTIONING AND STAINING
Once the tissue sample has been located in the database and retrieved from the freezer or archive, it is ready for sectioning onto glass slides (see Step 3 and Fig. 3a–e). The thickness that tissue sections are cut at varies depending upon the experimental design, with 3–5 μm being typical26;36–38 for ethanol fixed paraffin embedded (EFPE) and formalin fixed paraffin embedded (FFPE) tissues. However, we have found that cutting OCT-embedded frozen tissues at 8 μm gives excellent results for downstream RNA analysis, without increasing the tissue opacity or chance of dissecting contaminating cells. Frozen tissue sections are generally stored for no more than one month36 and microdissection is performed within a few weeks of cutting. However, improved RNA recovery has been observed from frozen tissue sections stored for ≤ two weeks. When cryostat sectioning, investigators should be aware of contamination issues that may be due to tissue carryover from other tissue blocks. Thus, clean uncharged slides, disposable cryostat blades, clean brushes, and RNA clean protocols are recommended. To reduce tissue waste, it is recommended that the tissue be aligned as closely as possible with the cryostat blade prior to cutting tissue sections. In addition, investigators often prepare 10 recuts (or more) at a time, with sections one, five, and 10 hematoxylin and eosin (H&E) stained and coverslipped for review of the histopathology.
Figure 3.
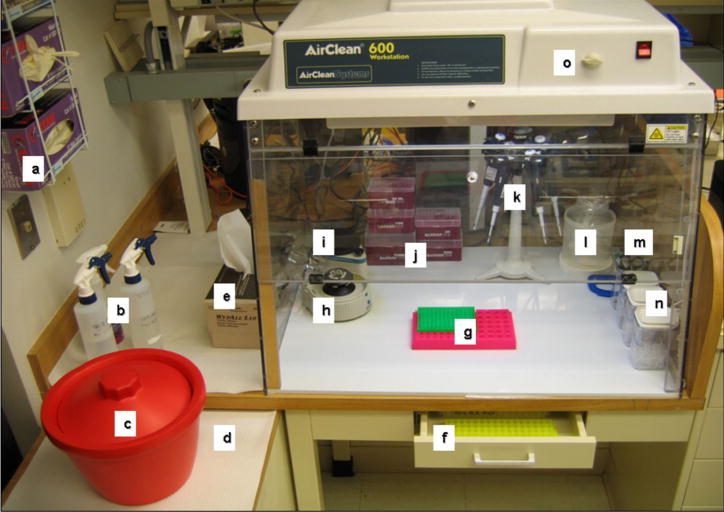
Dedicated RT-PCR Hood Setup. The clean dead air hood with UV sterilization capability used for setting up RT and PCR reactions is imperative in preventing contamination. All necessary equipment should be dedicated and remain beside or in the hood at all times, depending on the specific use. Dedicated equipment and consumables presented in this figure outside the hood remains outside the hood. This includes: (a) gloves, (b), 10% bleach, 70% ethanol, and RNase Away solutions, (c) ice bucket, (d) blotter paper, (e) paper towels, and (f) tube racks in the drawer which provides a dark environment for thawing out light sensitive reagents. The dedicated equipment and consumables presented inside the hood remain inside the hood at all times except when adding sterile pipette tip boxes, tubes, and clean waste bags or removing empty pipette tip boxes or full waste bags. This includes: (g) tube and plate racks, (h) mini-centrifuge, (i) vortexer, (j) pipette tips, (k), pipettes, (l) waste receptacle with bag inside, (m) pens, scissors, tube cap sealing tool, and (n) tube containers with sterile tubes. The UV bulb is located in the top of the dead air hood and the timer is located outside the hood (o). cDNA tubes (after RT) should be quenched on ice in dedicated ice bucket. The tubes containing reagents or cDNA may be brought into the hood for use and removed after use. After each use and between RT and PCR, the hood should be cleaned and UV sterilized to prevent contamination.
Prior to microdissection, it is necessary to stain the tissue section to allow visualization of the cells of interest. H&E is a standard approach for visualization and is used widely. However, more recently methyl green was shown to have the least fluorescent interference for qRT-PCR39. Therefore, to investigate this more thoroughly, we performed analysis of three different staining methods (H&E, hematoxylin, and methyl green) on replicate histological sections prior to microdissection to determine if any of the stains caused a detectable fluorescent interference of subsequent qRT-PCR reactions. No statistically significant CT value difference for the three stains was found using an F-test16. Therefore, H&E continues to be an excellent choice for staining tissue sections prior to LCM (see Step 4, Box 1 and Fig. 3f).
BOX 1. FROZEN TISSUE SECTION STAINING FOR HISTOPATHOLOGY CONSIDERATION AND ARCHIVE.
Frozen tissue sections numbers 1, 5, and 10 should be H&E stained and cover slipped for histopathology consideration (Box 2) and archival reference. The fixation step (Step 1, 70% ethanol) should be at least one minute for tissue morphology preservation. It is recommended to use the same fixative as the one used for tissue sections intended for microdissection (i.e. 70% ethanol). This assures the generation of similar tissue morphology for histologic review. In general, 70% ethanol fixation is an acceptable fixative for tissue morphology preservation. However, other fixatives, such as formalin, can be used to improve tissue morphology preservation for histopathologic assessment. Dependent upon the tissue, times in hematoxylin and eosin should be modified to obtain the desired staining tone.
Prepare individual staining dishes with the following solutions and treat slides for the described durations. All solution preparation and staining are conducted at room temperature. After each incubation, slides should be briefly drained and then moved to the next solution:
70% ethanol, 1 – 2 min
DI H2O, 30 s
Mayer's Hematoxylin, 1 min
DI H2O, 30 s
Scott’s Bluing, 30 s
70% ethanol, 30 s
Eosin Y, 30 s
95% ethanol, 1 min
95% ethanol, 1 min
100% ethanol, 1 min
100% ethanol, 1 min
Xylene, 1 min
Xylene, 1 min
Xylene, 1 min
Coverslip using xylene-based mounting media.
Allow slide to dry, 10 – 15 min. Be sure that the mounting media is dry before storing the slide sideways in a slide box.
Stained tissue section is now ready for histopathology review.
PATHOLOGY EVALUATION OF TISSUE FOR MICRODISSECTION
Tissue sections need to be reviewed and annotated prior to microdissection to histologically identify the desired cells for microdissection. Therefore, an evaluation of the tissue samples by a pathologist or a scientist trained in histologic cell identification of frozen tissues is needed36 before, during and after microdissection (see Step 5, Box 2 and Fig. 2). Also of importance is the orientation of the specimen in the tissue block. This is particularly important so that the cells of interest are adequately represented on the slide. Pathology slide review includes the evaluation of the tissue integrity, histopathology, determination of the adequacy of the sample for microdissection based on the amount of the target cell population, and annotation of the target cells on the slide. The pathologist also can give advice on the staining procedure that will help to better identify the cells of interest under the microscope during dissection.
BOX 2. HISTOPATHOLOGICAL CONSIDERATIONS FOR TISSUE MICRODISSECTION.
The goals of histopathology consideration before proceeding with tissue microdissection are: 1) to evaluate the total amount of tissue and the amount of the target cells in the tissue section present in the block, 2) to study the histopathology of the tissue specimen and identify the target cells, and 3) to plan the microdissection for each specimen, taking in account the heterogeneity of tissue samples.
Recommendations
Always make a regular H&E slide for histopathologic analysis (Box 1) before proceeding with tissue microdissection. Traditional H&E staining (using longer times in each solution than H&E for LCM) and cover slipping of sections 1, 5, and 10 for histopathologic analysis are recommended prior to beginning the study. These sections will serve as a permanent record of tissue specimen status and show histologic changes that occur in the deeper sections.
Always label the slides with the tissue block identification label, the number of the recut (e.g. 1, 2, 3, etc.), and the date the section was cut from the tissue block.
If you are unsure of the tissue histology represented in the traditional H&Es, consult with a pathologist to review the slides to accomplish the general goals listed above.
Keep the traditional H&Es used for histopathologic assessment with you when performing the tissue microdissection. These slides will help in identifying the cells of interest in the tissue section being used for microdissection. Both slides can be compared side by side.
Only dissect cells that can be clearly identified. If there is any doubt, do not dissect it without consulting with a pathologist.
Figure 2.
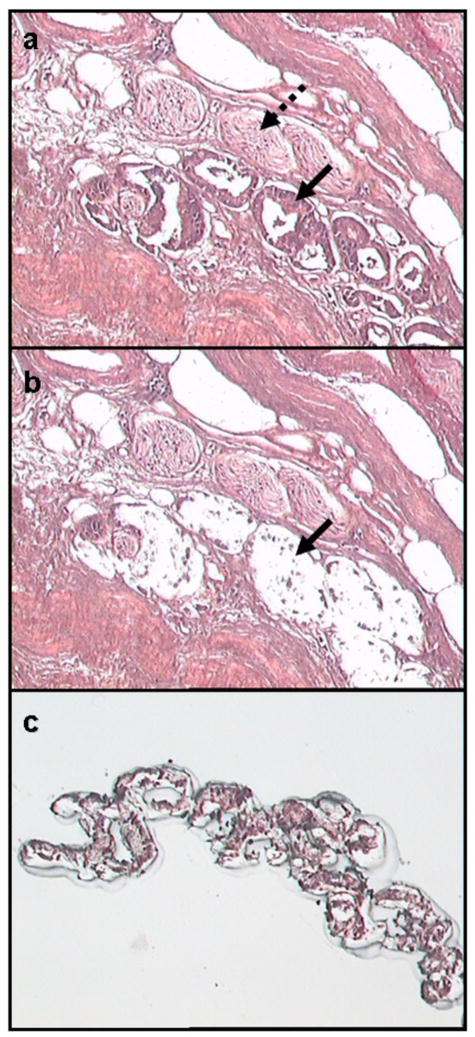
Frozen prostate epithelium laser capture microdissection (LCM) procurement sequence. (a) Roadmap prior to microdissection (solid arrow indicates epithelium; dashed arrow indicates nerve). (b) Post microdissection (solid arrow indicates lifted epithelial area). (c) Microdissected epithelial cells on LCM cap (Hematoxylin and eosin (H&E) stained. Original magnification 100X).
TISSUE MICRODISSECTION
Laser based techniques have now made tissue microdissection a routine step in obtaining precise quantitative gene expression measurements from dissected tissue samples. Laser capture microdissection (LCM) was invented at the National Institutes of Health (NIH) and developed through a cooperative research and development agreement with Arcturus Engineering, Inc. (www.arctur.com), to facilitate fast, simple, and reliable tissue microdissection37;40. Several laser based microdissection platforms are now available for procurement of pure populations of cells. A thorough review of the various microdissection platforms and protocols for LCM has been detailed previously in Nature Protocols36.
RNA RECOVERY AND ASSESSMENT
Tissue samples may contain qPCR inhibitory agents41–46. In our experience OCT is a major factor in qPCR inhibition, therefore it is important to extract and isolate RNA from the microdissected sample prior to qRT-PCR analysis. Many methods for RNA extraction and isolation are available on the market. Choice of the appropriate method depends on the type of starting tissue sample, whether it is derived from cells in culture, bulk tissue, tissue scrapes, or small cell quantities (e.g. microdissection). In addition, it is recommended that an RNA only workspace is available for RNA extraction.
RNA purification methods for downstream qRT-PCR analysis have been discussed in detail previously47. For a microdissection-based approach, samples are placed in lysis buffer and stored at −80°C. To be rigorous in ensuring the total RNA isolated is free of large DNA fragments, the sample is subjected to DNase during the extraction process because DNA is often a contaminant when using glass filter RNA extraction methods2;16. To reduce bias, total RNA from all microdissected samples in a given study should be extracted using the exact same method and aliquotted prior to analysis and storage at −80°C.
RNA quantitation methods are numerous and each technique has its own strengths and weaknesses47;48. However, it is imperative that the quantitation method chosen is appropriate for the range of RNA obtained during dissection which is generally on the order of 5–20 pg per cell. Since microdissection yields low quantities of RNA (Table 1), NanoDrop total RNA quantitation technique (NanoDrop Technologies, Wilmington, DE, USA) is an excellent choice due to the small quantity of sample that is required for the assay.
Table 1.
Representative total RNA quantity and quality from microdissected frozen tissue samples a.
| Tissue | Neoplastic Status | # LCM Shots | Total RNA Quantity (ng/ul) | Total RNA Quality (RINb) |
|---|---|---|---|---|
| Epithelium: | Range = 8.4 – 20.42 | Range = 4.2 – 7.8 | ||
| Breast | Normal | 2000 | 12.29 | 7.6 |
| Tumor | 3000 | 13.51 | 7.8 | |
| Colon | Normal | 3000 | 13.2 | 4.2 |
| Tumor | 3000 | 12.57 | 6.5 | |
| Esophagus | Normal | 4000 | 16.59 | 5.1 |
| Tumor | 3700 | 20.42 | 4.5 | |
| Prostate | Normal | 3000 | 8.4 | 6.2 |
| Tumor | 3000 | 10.8 | 5.6 | |
| Urethra | Normal | 2000 | 13.87 | 6.1 |
| Stroma: | Range = 4.81 – 8.86 | Range = 4.8 – 6.9 | ||
| Breast | Normal | 3000 | 6.53 | 6.8 |
| Tumor | 2400 | 8.86 | 6.5 | |
| Prostate | Normal | 20,000 | 6.65 | 6.9 |
| Tumor | 20,000 | 6.37 | 5.7 | |
| Urethra | Normal | 3000 | 4.81 | 4.8 |
This data was generated at the Pathogenetics Unit, NCI and is representative of frozen microdissected total RNA from these tissues.
RIN = RNA Integrity Number. Algorithm assigning value to RNA quality, as calculated by Bioanalyzer 2100 (Agilent, Inc.).
The RNA quality in frozen tissue samples can vary due to the upstream effects of tissue handling, but are generally higher than EFPE and FFPE processed specimens. There are several schools of thought regarding RNA quality related to qRT-PCR that are reviewed elsewhere48. For assessing total RNA from frozen microdissected tissues, the Bioanalyzer system (Agilent Technologies, Inc., Santa Clara, CA, USA) and RNA integrity number (RIN) provides an adequate assessment of total RNA quality (Table 1). This is best utilized in concert with NanoDrop quantitation to select the appropriate Bioanalyzer kit (either Pico- or Nano-Chip) since the recovered total RNA quantity varies per microdissection (see Box 5).
BOX 5. CONSIDERATIONS FOR QUALITATION OF TOTAL RNA FROM MICRODISSECTED TISSUES.
When generating total RNA from microdissected tissues, the quantity of total RNA recovered per sample varies depending on many variables, including, but not limited to, the subject, tissue type, tissue processing and storage. As such, total RNA concentrations per μl of elution buffer often fall in range of one of the detection capabilities of the Bioanalyzer quantitation Pico- and Nano-Assays. The Agilent RNA 6000 Pico Kit has a qualitative range of 50 – 5000 pg/μl and is not suitable for quantitation. The Agilent RNA 6000 Nano Kit has a qualitative range of 5 – 500 ng/μl and quantitative range of 25–500 ng/μl. Therefore it is imperative to perform NanoDrop quantitation prior to Bioanalyzer qualitation analysis to determine which kit (i.e. Pico or Nano) is appropriate. In addition, one method of quantitation and one method of qualitation should be used throughout the study for all tissue samples. Therefore, since RNA quantities vary and are predominantly < 25 ng/μl (the minimum for Bioanalyzer quantitation analysis is ≥ 25ng/μl), NanoDrop is the preferred technique for RNA quantitation.
Perform total RNA quantitation of samples using NanoDrop quantitation technique for RNA.
-
Choose appropriate Bioanalyzer qualitation method.
Use Agilent 6000 Pico Kit for samples with total RNA quantities < 5ng/μl.
Use Agilent 6000 Nano Kit for samples with total RNA quantities >5ng/μl.
Perform total RNA qualitation of samples using appropriate Agilent RNA 6000 kit.
qRT-PCR ASSAY
Since limited quantities of total RNA from clinical or animal model tissue samples are typically used as the starting template, it is recommended to perform two-step qRT-PCR, with one RT tube per sample producing enough cDNA to perform 12 qPCR reactions, allowing for triplicate analysis of three genes of interest and one endogenous control reference gene. The present protocol is optimized for bias reduction; therefore, RNA pre-amplification is not used. However, if an investigator chooses to use an RNA pre-amplification step (see below), the protocol should be adjusted accordingly.
qPCR assays use one of two fluorescent detection chemistries. The first, SybrGreen, employs one pair of specific primers and the fluorescent dye incorporates into the DNA during amplification49. The second utilizes one pair of specific primers and a gene specific fluorescent probe that increases the specificity of the assay. The most common probe detection chemistry is TaqMan (Applied Biosystems, Foster City, CA, USA) in which the signal is generated by cleavage of the fluorescent quencher during the reaction cycle. This is the detection chemistry described in the protocol below; however, fluorescent probe expense may be an issue for some users.
For downstream qRT-PCR application, RNA quality of RIN >5 is considered good and RIN >8 is considered excellent50. In general, highly fragmented RNA from FFPE samples is not optimal for qRT-PCR amplification; however, informative qRT-PCR results can be obtained from frozen samples that demonstrate some RNA degradation (RIN 2<x<5). The use of random hexamers as primers for RT diminishes the effects of degradation by generating more complete cDNA coverage of genes than with oligo dT priming. A more complete review of issues related to RT and qPCR has been discussed elsewhere47;48;51–58. In addition, our experience suggests that single-plexing of the qPCR reaction (i.e. separate reactions from the same cDNA sample for the gene(s) of interest and the endogenous control reference gene) reduces the potential for competitive inhibition by the two primer and probe sets and subsequently improves detection capabilities by 2–3 CT values, which is important when analyzing transcripts that may be expressed at a lower level. Spiking the master mix with additional Taq polymerase (1 μl/qPCR reaction) increases the detection capabilities by another 2–3 CT values. In addition, we have found that by using larger qPCR reaction volumes (50μl) CT values were further improved. Smaller reaction volumes (e.g. 20 μl) without additional Taq polymerase spiking may be used successfully to obtain quantitative gene expression measurements; however, investigators must weigh the cost of larger reaction volumes and additional Taq polymerase to the potential decrease in detection capabilities for the specific experiment and transcripts to be detected.
The number of gene transcripts that can be analyzed from a typical microdissection from frozen tissue samples using qRT-PCR is limited. Moreover, the ability to analyze qPCR efficiency and standard curves is not possible due to the small sample quantity generated by microdissected tissues48. Therefore, the use of a comparable positive control total RNA sample (i.e. if using microdissected frozen human esophageal tissue the positive control would be purchased total human esophagus RNA) in the qRT-PCR is necessary.
As an example, a typical prostate epithelium microdissection of 3000 laser shots (~10,000 cells), allows for the procurement of ±160 ng of total RNA, which is enough to analyze the transcripts of nine genes of interest and one endogenous control reference gene. For prostate stroma, microdissection of 3000 laser shots typically yields ±24 ng total RNA, which is only enough template to analyze one gene of interest and one endogenous control reference gene. To procure ~10,000 cells from stroma the microdissection laser shots are generally increased four times, depending on the cellular density.
NORMALIZATION STRATEGY
A normalization control is critical when attempting to quantitatively compare gene expression levels between two biologic samples (e.g. diseased cells versus normal cells). Housekeeping genes such as ACTB and GAPDH have historically been used for normalization because they were presumed to be stably expressed in all cell types. In addition, cell analyte content (i.e. amounts of RNA, DNA, or protein), numbers of cells, and averages of multiple genes from expression arrays are sometimes used as normalization approaches. The majority of the qRT-PCR studies in the literature use a common set of housekeeping genes as endogenous controls for normalization. These mRNAs are generally effective, but their expression levels have been shown to vary among different cell types, samples, and environments59–70. Therefore, universally stable endogenous control genes have not been found71. For example, ACTB has consistently stable levels of gene expression in cultured prostate cancer DU-145 and PC-3 cells 67. But our experience and the experience of others72 have demonstrated that the same in vitro expression levels are not always observed in ex vivo tissue samples. Therefore, a housekeeping gene such as ACTB may not be the most stable endogenous control gene for use in qRT-PCR studies using tissue samples. In addition to issues related to tissue type, disease states (e.g. cancer or other proliferative processes) and procurement methods may introduce variability in housekeeping gene expression60;73–77. Therefore, for each tissue type and experimental system a reliable and accurate normalization strategy must be determined and validated.
To address this in prostate, our group analyzed three gene expression normalization strategies (LCM cell count; total RNA measurement; and endogenous housekeeping genes) using microdissected frozen samples16. Briefly, the data demonstrated that microdissection cell counting was not a reliable normalization method. Within replicate dissections, a sizeable variation of up to 1.84 CT values was seen which could induce a bias of up to 3.6 fold between samples. Since gene expression changes of approximately three to four fold (1.5 – 2 CT values) are regarded as potentially biologically important78, the use of microdissected cell count is not recommended as a normalization strategy.
The second normalization method analyzed was the use of total RNA quantity as an internal control for qRT-PCR from microdissected tissues. Total RNA was recovered and quantified by the NanoDrop method from microdissected cells from triplicate serial sections. The data demonstrated that RNA quantitation alone produces a large variance which prohibits using this approach for normalization.
Even though the data showed that neither microdissection cell count nor total RNA measurement are precise enough to serve as a normalization strategy for microdissected tissue, they are practical first and second steps in calibrating ‘ballpark’ RNA input levels that can successfully be examined by qRT-PCR.
The third normalization method analyzed was the use of endogenous housekeeping genes as an internal reference control for qRT-PCR from microdissected tissues. Endogenous housekeeping genes were found to offer an excellent representation of the cellular transcriptome and the use of only one or two housekeeping genes (i.e. PGK1 or 18s and HPRT for paired samples) was precise enough to serve as an internal reference control for quantitative gene expression analysis. The need to normalize with only one or two housekeeping genes is important, since limited amounts of RNA are typically procured from microdissected tissue samples.
Several methods for relative quantitation exist, including the most commonly used 2−ΔΔCT method79;80. This approach is recommended for dissected cells as it allows for the maximal number of genes that can be analyzed from one sample. Assuming comparable amplification efficiencies between the gene(s) of interest and the calibrator, this method eliminates the need for generating standard curves since the gene of interest data is normalized to the calibrator (endogenous housekeeping reference control gene)81.
EXPERIMENTAL DESIGN
General considerations
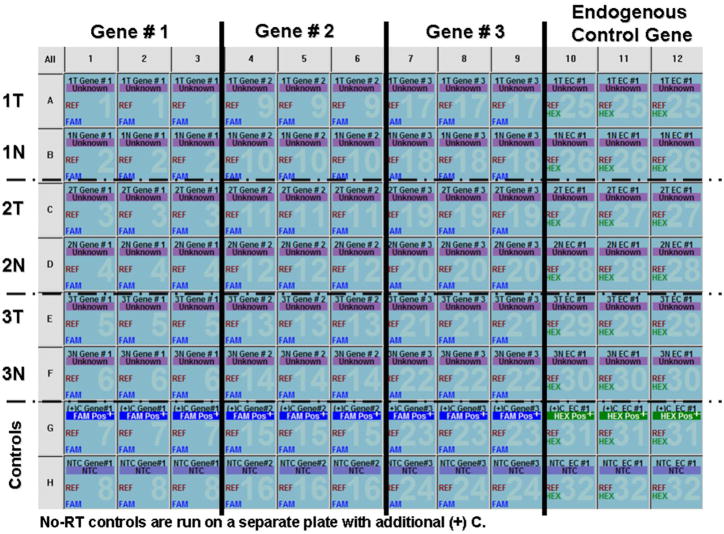
Before beginning the protocol, the following general guidelines should be considered: 1) Use a clean dead air hood that is dedicated for setting up RT and PCR reactions in preparing all qRT-PCR reactions to avoid contamination. All pipettes, pipette tips, tubes, pens, min-centrifuge, and vortex should be contained inside the hood. 2) Keep all reagents on ice. 3) Protect all TaqMan probes and master mixes from light (i.e. keep in dark). 4) Mix (vortex or swirl, depending) and spin down all source vials prior to opening. 5) Always run a no template control (NTC), a no RT, and a positive control ((+) C) for each RT and primer/probe set for qPCR. 6) In loading reaction tubes, load in this order: 1) NTC; 2) no RT; 3) unknowns; and 4) (+) C. This will minimize the potential for contamination carryover. 7) Aliquot source vials to minimize freeze-thaw cycles. 8) To prevent pipetting errors, prepare master mixes of reagents wherever possible to minimize the number of pipette steps made. 9) Use powder-free gloves to prevent optical reader interference. 10) Use a common positive control commercial total RNA for all qRT-PCR reactions for the same tissue type. This will allow a common reference from plate to plate, allowing data to be analyzed across batches.
Tissues
To obtain total RNA from microdissected tissues, the following considerations must be taken into account: 1) Sample has been snap-frozen properly prior to storage. 2) Tissue block is maintained at optimal temperature during sectioning. 3) Cryostat and all sectioning and staining consumables are free of contaminating tissues. 4) Once the tissue section is cut from the block, mount the section on an uncharged glass slide at room temperature. An uncharged slide will allow the tissue to more easily be procured/lifted off the glass slide during the microdissection process. The glass slide with the tissue section on it should then be placed immediately on dry ice. Do not let the tissue section defrost. 5) Prior to choosing the tissue block for use in the study, do a scrape of the tissue section and do RNA extraction and isolation followed by quantitation and qualitation analysis to assess RNA integrity. 6) Frozen tissue sections should not be stored for more than 2 weeks prior to use. 7) Once the tissue section is out of the freezer, no more than 30 min should elapse before putting the microdissected tissue into lysis buffer.
RNA Isolation and Measurement
RNase-free techniques need to be followed stringently to prevent experiment failure due to contamination. This can be accomplished by: (1) wearing powder-free disposable gloves, (2) avoiding touching RNase contaminated surfaces, (3) changing gloves often, (4) cleaning bench tops and hood surfaces with an RNase decontaminant solution prior to use, (5) using a dedicated RNA bench and equipment, and (6) using reagents and consumables that are free of contaminants (e.g. RNases, other RNA or DNA, PCR products, thawed tissue fragments, or chemical contaminants).
Extract and isolate RNA using a total RNA extraction kit that is appropriate for the amount of microdissected tissue procured. In our experience the PicoPure RNA extraction kit (Arcturus Engineering, Inc.), a glass filter-based method, provides the largest and most reproducible total RNA yields from microdissected tissue samples.
After RNA purification, DNase treatment for 15 min is recommended2;16. For most genes a DNase step is not necessary. It is best to design and use cDNA-specific primers and probes to reduce the need for a DNase treatment. But for some pseudogenes a DNase step is necessary. Therefore, it is generally recommended to add a DNase treatment to the RNA extraction if the kit chosen does not include one. In this protocol, DNase treatment is recommended to prevent interference by the PicoPure RNA Isolation Kit (Agilent, Inc.) components that may cause artifacts during downstream Agilent 2100 Bioanalyzer assessment of quality and gDNA contamination. However, if investigators choose not to use a DNase step, downstream quality assessment using the Agilent 2100 Bioanalyzer may be confounded.
Immediately after total RNA extraction and before storage at −80°C, be sure to aliquot samples into volumes appropriate for quantitation (1 μl), qualitation (2 μl), and multiple RT reactions (4 μl each). It is recommended to use an elution buffer volume of 20 μl to recover enough total RNA to make the previous aliquots. Keep total RNA aliquots frozen at −80°C until ready for use in RT reaction.
This RNA extraction and isolation protocol eliminates the presence of qRT-PCR inhibitors in the total RNA sample; however, qRT-PCR inhibition due to template overload is still possible.
Following total RNA purification, quality and quantity measurement is recommended16. Because of the very small total RNA quantities obtained from microdissected tissues (Table 1), it is difficult to impossible to adjust or equalize total RNA quantities across samples prior to qRT-PCR. However, it is important to know the starting quantities and qualities of total RNA in order to “ballpark” starting template input16 and it is recommended to quantify total RNA using the NanoDrop ND-1000 spectrophotometer (260/280 nm; NanoDrop Technologies, Inc.). This method only requires 1 μl of sample and can be used with concentrations as low as 2 ng/μl. Since total RNA concentrations per μl from microdissected tissue samples (Table 1) generally fall below the quantity detection limits of the Bioanalyzer (>25 ng/μl; Agilent Technologies, Inc.), this method of total RNA quantitation is not sensitive enough for routine use. RNA qualitation of total RNA from microdissected tissue samples yields quantities too small to routinely use conventional gel electrophoresis to determine RNA integrity. Therefore, it is recommended to use the Agilent 2100 Bioanalyzer (Agilent Technologies, Inc.) because it only requires 1 μl of sample for analysis of 18S and 28S rRNA and provides an RNA integrity number (RIN).
Controls
An LCM negative control is prepared for each tissue section by making 3000 laser shots in a tissue free portion of the tissue section. This sample is processed throughout the entire experiment in parallel with microdissected tissue samples to control for contaminating RNA.
The choice of positive control total RNA origin (i.e. organism, organ, and tissue type) is dependent on the tissue samples being analyzed, but is typically from the same organism and tissue type, e.g. positive control of total human prostate RNA for experimental microdissected frozen human prostate tumor and normal tissue samples. However, if the desired tissue type is not commercially available, a good alternative is to use organism specific universal RNA.
Once initial positive control concentrations are determined, the investigator should choose the three positive controls that are closest in concentration to the total RNA amount from the microdissected sample.
An endogenous control reference gene(s) that shows stable expression in the samples being analyzed is used in the third step of the normalization strategy16. It is recommended that if a single endogenous control reference gene can be used for the study, that just one is used, see Erickson et al for a discussion of this topic. Primer and probe design has been discussed extensively in a previous issue of Nature Protocols48. TaqMan® chemistry based primer/probe sets (e.g. AB Assays-on-demand, AB Assays-by-Design, or other TaqMan chemistry primer/probe sets) are recommended for use with RNA from microdissected samples because of improved specificity over primer only based detection chemistries (e.g. SYBR). For microarray gene expression profile validation, it is recommended to design all primer/probe sets to amplify the same region of the cDNA sequence as the gene specific microarray probe set used to derive the data.
Using a single endogenous control housekeeping gene, it is possible to analyze up to 9 genes of interest from a single microdissection (Fig. 4). For both the genes of interest and the endogenous control reference gene(s), non-primer-limited primer/probe sets can be used in single-plex reactions. If performing multi-plex reactions, it is recommended to use primer-limited primer/probe sets for amplification of the endogenous control gene, as this limits amplification of the gene once the signal threshold is reached. In multi-plex reactions, choose different reporter dyes for the genes of interest (e.g. FAM) and the endogenous control reference gene (e.g. VIC/HEX). For single-plex reactions, it is not necessary to choose different reporter dyes or use primer-limited primer/probe sets. However, if using the endogenous control reference gene(s) for other types qRT-PCR experiments, it may be more cost effective to use VIC labeled primer-limited primer/probe sets.
Figure 4.
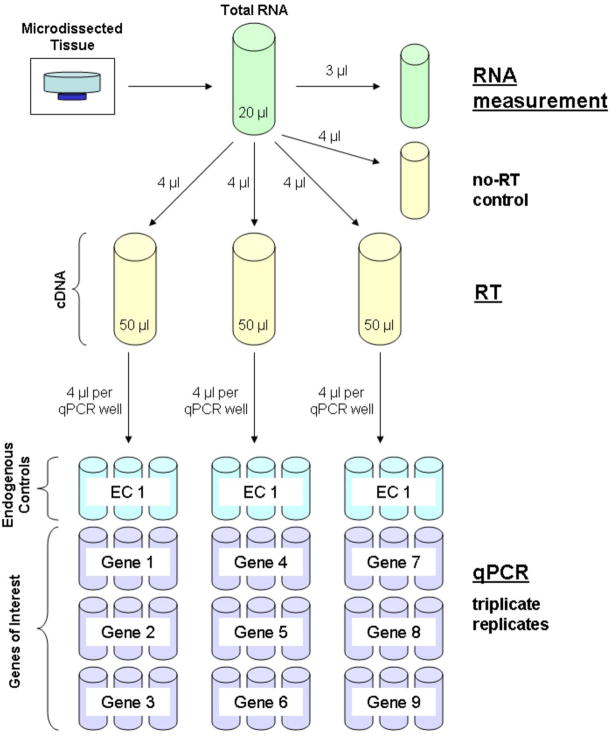
qRT-PCR from microdissected tissue process flowchart. Beginning with a typical dissection of ~10,000 cells (3,000 laser shots), total RNA is extracted and isolated. This process yields ~160 ng of total RNA in a volume of 20 μl. The total RNA is aliquotted into four 4 μl volumes and one 3 μl volume. The 3 μl volume is used for RNA measurement. The remaining aliquots are used for qRT-PCR with one aliquot used as the no-RT control and the remaining three aliquots are used in RT to generate three 50 μl volumes of cDNA. Each of these 50 μl volumes is used for qPCR triplicate replicate analysis of three genes of interest and one endogenous control housekeeping gene. The genes of interest may be different between the three sub-samples, but the same endogenous control housekeeping gene must be analyzed from each of the RT tubes.
qRT-PCR
Total RNA yield from microdissected tissue determines the number of genes of that can be analyzed by qRT-PCR. Each qPCR reaction requires 4 ng of total RNA starting template that was used for RT. For example, if the microdissection of epithelium yielded ~160 ng of total RNA, this would allow for the analysis of 9 genes of interest and one endogenous control housekeeping gene. However, if the microdissection of stroma yielded ~24 ng of total RNA, only one gene of interest and one housekeeping gene can be analyzed. Stroma should be analyzed for cellular density and the number of laser shots adjusted to increase the total RNA yield.
Total RNA samples from microdissected tissues are limited and precious. Therefore, it is imperative that primers and probe sets are validated to ensure that they specifically amplify their intended target sequence linearly and reproducibly prior to using the microdissected RNA samples. This can be accomplished by using a commercially available tissue specific total RNA to perform serial dilutions. The serially diluted RNA is then applied to the qRT-PCR protocol using the specific primers and probe sets. The CT values, PCR efficiencies, and standard curves are evaluated. Following primer and probe set specificity and sensitivity, cDNA generated from the RT portion of the protocol can be used for qPCR assay. qPCR assays should be performed in triplicate as technical replicates to assess precision of assay.
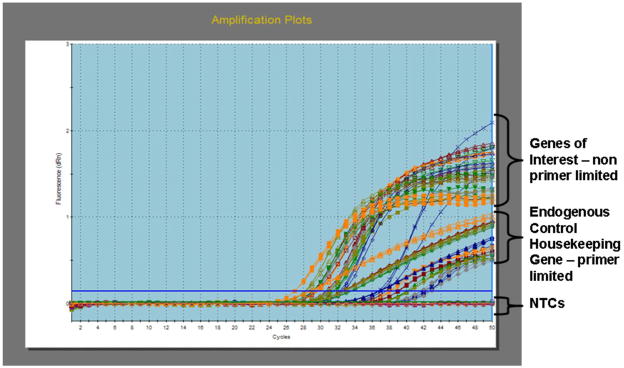
qRT-PCR of microdissected tissues is done in two steps (RT and qPCR) to maximize the number of genes that can be analyzed from one sample (Fig. 4). During RT, random hexamers should be used as reverse primers to counter the effects of degradation of mRNA. Perform RT and qPCR of matched disease and normal tissue sample total RNA at the same time. Three technical replicates of qPCR are used to assess precision of the assay, and endogenous control gene primer/probe sets are analyzed for each sample and control. This qPCR protocol described here employs 50 PCR cycles instead of the usual 40, although with microdissected samples we find that more than 90% of genes do not have CT values > 40. 50 cycles are used for qPCR to ensure that the NTCs for a gene of interest have 10 CT values later than the sample gene of interest expression in cases where low level of gene expression (e.g. 37 CT values) are encountered. This ensures that if the NTCs demonstrate ‘no CT’ at 50 cycles, the qPCR reagents are free of contamination. In addition, the observation of 10 CT difference comparing no-RT with RT-positive samples means there is negligible contamination (0.1%) of genomic DNA92;93. However, because PCR efficiency changes with the length of the experiment, it is recommended to test the in-run PCR efficiency of gene specific primer/probe sets at the Ct range of the gene of interest using a standard cDNA dilution series. In our experience adding additional Taq polymerase is valuable in lowering CT values and improving technical replicate standard deviations16. However, investigators should be aware that changes such as Taq concentrations can change PCR efficiencies.
Analysis
To ensure standardization of analysis techniques, one qPCR method and software analysis method should be used throughout an entire study. Although beyond the scope of this protocol, a detailed review of qRT-PCR data analysis for amplification plots, primer/probe efficiencies, standard curve generation, and general qRT-PCR analysis has been described in a previous issue of Nature Protocols48.
The normalization strategy dictates the analysis method that can be used. Based upon the optimal normalization strategy for qRT-PCR from microdissected tissues16 when comparing diseased and normal tissues, data is analyzed via relative quantitation analysis using the 2−ΔDelta;CT method 94. This is not the only analysis method; however, investigators should assess if their study requires absolute quantitation using a standard curve. It is always recommended to assay the PCR efficiencies of each gene primer/probe set to be used in relative or absolute quantitation studies. In summary, all samples are analyzed relative to an endogenous control housekeeping gene from the same sample total RNA to generate the normalized ΔCT value. And subsequently, normalized disease tissue qPCR data is analyzed relative to normalized matched normal tissue qPCR data to generate the Δ ΔCT value.
CONCLUSION
The protocol described in this report allows for quantitative analysis of mRNA dissected from animal model tissues or clinical specimens. However, it is important that investigators factor tissue-related variables, especially from clinical samples, into their experimental design and data analysis. Precise expression measurements from phenotypicaly- or molecularly-defined cell populations in tissue samples likely will have significant value for both laboratory researchers and clinical investigators.
MATERIALS
REAGENTS
Dry ice. ! CAUTION Dry ice can burn skin on contact and vapors can cause asphyxiation. Take appropriate precautions when using dry ice.
Optimal Cutting Temperature (OCT) compound (Sakura Finetek Corp., Tissue-Tek, cat. no. 4583) for cryo-preservation.
Frozen tissue specimens sectioned at 8 μm. ▲CRITICAL Do not store tissue sections for more than one month (≤ two weeks is preferred) prior to microdissection. ! CAUTION Follow established protocols for safely working with bloodborne pathogens for all human and animal biologic samples. For animal samples, follow all Institutional Animal Care and Use Committee protocol regulations. For human samples, follow all established Institutional Review Board clinical protocol regulations, including patient consent and sample anonymization.
Mayer’s Hematoxylin solution (Sigma, cat. no. MHS128). ! CAUTION Contact hazard.
Scott’s Blueing Solution (Fisher, cat. no. CS410-4D) or alkaline water. ▲CRITICAL The reddish color of Mayer’s Hematoxylin will turn blue with mild alkaline treatment, such as that of Scott’s Blueing Solution. Omitting the alkaline treatment step will make histopathological analysis of tissue and cell morphology difficult.
Eosin Y solution (Sigma, cat. no. HT110116). ! CAUTION Contact hazard. Flammable.
Absolute (100%) ethanol (molecular grade, Sigma, cat. no. E7023). ! CAUTION Contact hazard. Flammable. ▲CRITICAL In humid climates (> 40%), monitor the water content of the ethanol, and change the solution as often as necessary. It is generally recommended to change all solutions in staining process every week or more frequently if staining > 20 slides.
95% and 70% ethanol. Prepare with Milli-Q-filtered water (Millipore). ▲CRITICAL If making solutions in batches do not store for more than one week at 4 °C.
Xylene (Sigma Aldrich, cat. no. 247642) ! CAUTION Vapor and contact hazard. Vapor is harmful or fatal. Use appropriate safety measures for working with and disposing of hazardous materials. ▲CRITICAL Xylene substitutes (e.g. Citra-Solv) may diminish microdissection efficiency of some tissue types.
RNA extraction buffers (PicoPure RNA extraction kit, Arcturus Molecular Devices, cat. no. KIT0204).
RNase-free DNase Set (50) (Qiagen, cat. no. 79254).
RNA qualitation (see Reagent Setup): RNA 6000 Pico Series II Assay (Agilent Technologies, 5067-1513) and/or RNA 6000 Nano Series II Assay (Agilent Technologies, 5067-1511).
RNase/DNase-free water (e.g. Ambion Applied Biosystems, cat. no. AM9937).
Tissue specific total RNA (e.g. Ambion Applied Biosystems, cat. nos. vary depending on the species and organ) to use as positive control template.
RT-PCR grade water (e.g. Ambion Applied Biosystems, cat. no. AM9935).
RT assay: Reverse transcription reagents (e.g. Applied Biosystems, High Capacity cDNA Reverse Transcription Kit, cat. no. 4368813, TaqMan® Reverse Transcription Reagents, cat. no. N8080234), Random hexamers (e.g. Applied Biosystems, cat. no. N8080127). Additional information for other reverse transcription reagents has been covered in a previous issue of Nature Protocols48.
Primer/probe sets for genes of interest and endogenous controls: Assays-on-Demand TaqMan® MGB Probe (Applied Biosystems, Inventoried or Made to order, cat. no. 4331182 or 4351372), Assays-by-Design: Primers (Applied Biosystems, cat. no. 4304970) and TaqMan® MGB Probe (Applied Biosystems, cat. no. 4316034). Primer and probe design considerations have been covered in a previous issue of Nature Protocols48.
TE Buffer pH 7.0–8.0 (e.g. 1x TE Buffer pH 8.0, Quality Biological, Inc. cat. no. 351-011-131).
qPCR assay: 2x TaqMan Universal Master Mix (Applied Biosystems, cat. no. 4304437), AmpliTaq Gold® DNA Polymerase, LD (Applied Biosystems, cat. no. 4338857). Other companies such as Stratagene offer qPCR reagents. Additional information for non-TaqMan chemistry based qPCR assay reagents has been covered in a previous issue of Nature Protocols48.
EQUIPMENT
Protective personal wear, including lab coats, powder free latex and/or nitrile gloves, and safety glasses.
Biohazard or medical pathological waste container.
−80 °C freezer.
−20 °C freezer.
4 °C refrigerator.
Cryomolds (Sakura Finetek Corp., Tissue-Tek, cat. no. 4557).
Cryostat (e.g. Leica CM 1900 UV, Leica Microsystems).
Uncharged slides and coverslips of the size appropriate for the tissue specimen to be sectioned. There are many distributors for these consumables (e.g. Fisher Scientific, cat. no. NC9744786, cat. no. 22-037-169).
LCM system (e.g. PixCell II, Arcturus Molecular Devices). Considerations for the choice of LCM systems have been discussed in a previous issue of Nature Protocols36.
Adhesive pads (e.g. Post-it Note®, 3M)
-
CapSure Macro LCM Caps (Arcturus Molecular Devices, cat. no. LCM0211).
▲CRITICAL Because tissue is in direct contact with the cap, it is important to use the adhesive pad to remove any tissue debris. If extraneous tissue is not carefully removed, it will be available for RNA extraction and may contaminate the sample.
Microcentrifuge tubes: MicroAmp 500 μl Thin-walled PCR Reaction Tubes (Applied Biosystems, cat. no. 9N801-0611) or Safe-Lock 500 μl Eppendorf Tubes (Brinkmann Instruments, cat. no. 2236361-1). ▲CRITICAL To prevent lysis buffer leakage during pre-extraction heat incubation, the use of either of these tubes are recommended.
Oven (e.g. Hybaid Mini MK-II hybridization oven, Hybaid, cat. no. Mini Oven MK II)
Agilent 2100 Bioanalyzer (Agilent Technologies, Inc., cat. no. G2940CA).
NanoDrop® ND-1000 UV-Vis Spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
Dedicated PCR Hood (e.g. AirClean 600 PCR Workstation, AirClean Systems, cat. no. AC632DB Dead Air Box).
Microfuge tubes: 1.7 ml DNase/RNase free microfuge tubes (Costar, cat. no. 3560), 0.65 ml DNase/RNase free microfuge tubes (Costar, cat. no. 32090, 0.65 ml DNase/RNase free PCR thin-walled tubes (e.g. .PGC, cat. no. 502-075). Considerations for the choice of plastic ware have been discussed in a previous issue of Nature Protocols48.
Barrier filter pipette tips (e.g. CLP Direct Barrier Tips cat. nos. BT10XL, BT20, BT100, BT200, BT1000).
PCR thermocycler (e.g. MJ Research PTC-200 thermocylcer, MJ Research).
qPCR thermocycler: (e.g. Stratagene Mx3000P™ Real-Time PCR machine, Stratagene, Inc.; Applied Biosystems 7500 Real-Time PCR System, Applied Biosystems). Considerations for the choice of qPCR thermocycler have been discussed in a previous issue of Nature Protocols48.
REAGENT SETUP
RNA qualitation reagents for RNA 6000 Pico Series II Assay (Agilent Technologies, 5067-1513) and/or RNA 6000 Nano Series II Assay (Agilent Technologies, 5067-1511): For primer/probe sets delivered as individual components, prepare a 100 μl 20x working solution in TE for TaqMan Gene Expression qPCR assays according to calculations detailed in the table below.
20x Primer/Probe Gene Expression Assay Working Solution (100 μl).
| Component | Stock Concentration | Volume | Final Working Concentration |
|---|---|---|---|
| Forward Primer | 100 μM | 18 μl | 18 μM |
| Reverse Primer | 100 μM | 18 μl | 18 μM |
| MGB Probe | 100 μM | 5 μl | 5 μM |
| 1x TE Buffer | 59 μl |
Store all primer/probe stocks and working solutions at −20 °C until ready for use. For primer/probe sets that are delivered in pre-prepared working solution, aliquot into 60 μl volumes and store at −20 °C until ready for use.
PROCEDURE
Frozen Tissue Sectioning and Staining for Histopathological Consideration • TIMING 1.5 to 3 h
-
1| Select frozen tissue sample to be used in study from database of available tissue specimens (Fig. 1a).
▲CRITICAL STEP Assess that the tissue specimen has been promptly snap-frozen and embedded in OCT. Rapid processing of the tissue specimen limits RNase activity and subsequent RNA degradation. If the frozen tissue specimen has not been previously embedded in OCT do so at this time. The tissue block is now ready to have tissue sections cut onto glass slides.
2| Remove selected tissue block(s) to be sectioned from freezer and put on dry ice (Fig. 1b).
-
3| Make 10 tissue sections by sectioning the tissue block onto standard histology glass slide using a cryostat and place on dry ice (Figs. 1c,d).
▲CRITICAL STEP It is recommended to cut OCT-embedded tissue blocks at eight microns. This thickness allows for excellent histology visualization and is technically easy to cut. Decreased tissue thicknesses are technically difficult to cut an intact tissue section, while increased tissue thicknesses increase tissue opacity and the possibility of microdissecting contaminating cells. Frozen tissues most often can be sectioned onto non-charged slides; however, small tissue samples tend to fall off the slides while undergoing staining or microdissection; therefore, it is recommended that small tissue be sectioned onto charged slides
! CAUTION Tissue blocks preserve biomolecules better than tissue sections. Do not store tissue sections for more than two weeks after cutting from tissue block prior to microdissection (Fig. 1e). Longer storage increases biomolecule deterioration and makes the tissue unsuitable for downstream RNA analysis.
4| H&E stain and coverslip tissue sections 1,5, and 10 (Fig. 1f and Box 1)
5| Obtain histopathology consult to review tissues and select the cells of interest for microdissection procurement (Box 2).
Figure 1.
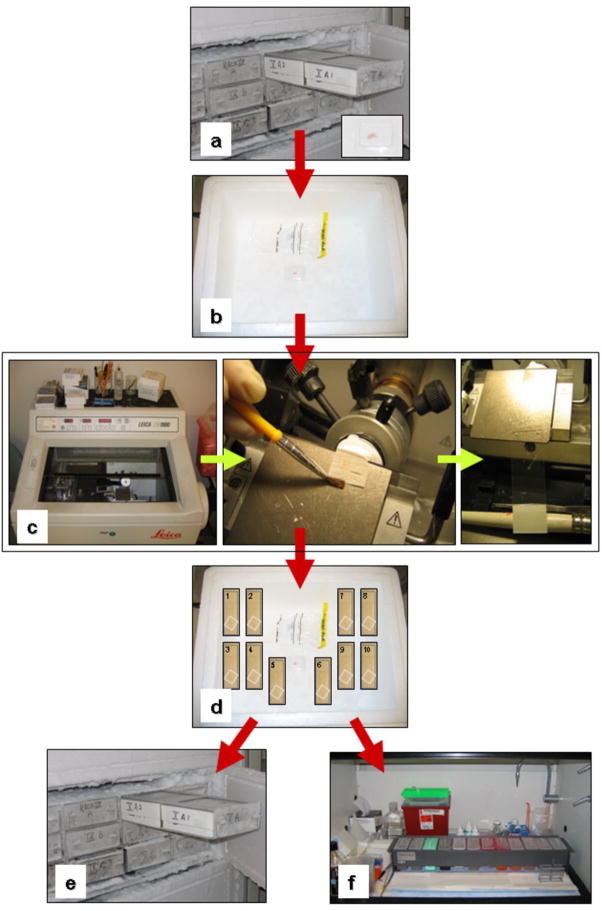
Tissue sectioning and staining flowchart. (a) Tissue block is removed from the −80 °C biorepository and (b) placed immediately on dry ice. (c) The OCT embedded tissue block is placed in the cryostat and adhered to a chuck using OCT. It is important to level the block to reduce facing off tissue loss. Make 8 μm thick sections and place onto labeled glass slide. Repeat 9 times for a total of 10 tissue sections. (d) Place the tissue sections immediately on dry ice. (e) Replace tissue block back into the −80 °C biorepository. If tissue sections are not going to be used immediately, store at −80 °C until use. (f) If tissue sections are going to be used immediately, stain tissue sections 1, 5, and 10 by traditional H&E staining method (Box 1). Follow with histopathologic analysis to assess the cells of interest to be microdissected (Box 2). Keep the other sections on dry ice or store at −80 °C until ready to perform microdissection (Box 4). Immediately prior to microdissection, use the same tissue staining station to stain sections for microdissection (Box 3).
Tissue section staining and microdissection • TIMING 45 to 55 min
Timing reflects the maximum time for microdissection of a given slide. The number of slides that can be microdissected in this time is dependent on the ease of recognition of cells to be dissected and the number of cells present on the tissue section.
6| Stain tissue section for microdissection (Box 3).
-
7| Microdissect cells of interest (Box 4, Fig. 2).
▲CRITICAL STEP The first step in normalization is to ensure the same number of shots (assuming similar cellular density) are microdissected for both the cells of interest (e.g. diseased epithelium) and the calibrator cells (e.g. normal epithelium)16. It is recommended to aim for about 10,000 cells / 3,000 shots for each of the tissue samples to be procured by microdissection if there is no previous experience with RNA quantity and quality of this particular tissue type / cell type to be microdissected. If there is no previous experience, perform a test microdissection followed by RNA quantity and quality assessment to determine the number of cells / number of laser shots that will be required for the study tissues.
BOX 3. FROZEN TISSUE SECTION STAINING FOR MICRODISSECTION.
To improve cell adhesion to the LCM cap, create a “rougher” tissue surface prior to staining by melting the frozen tissue section. This is achieved by placing the underside of the slide on the back of the hand for ~30 s after the slide containing the tissue section is removed from the freezer. Times in parentheses should be used in laboratories located in humid environments. In addition, the longer times in absolute ethanol and xylene are recommended, even in semi-arid environments, to achieve strong dehydration to facilitate lifting cells from the glass slide.
! CAUTION In our experience, the times in ethanol and xylenes baths does not affect the quality of RNA. But, always use RNase free water for the staining baths and always use fresh solutions and cleaned jars.
Prepare individual staining dishes with the following solutions and treat slides for the described durations. All solution preparation and staining are conducted at room temperature. After each incubation, slides should be briefly drained and then moved to the next solution:
70% ethanol, 15 s (30 s)
DI H2O, 15 s
Mayer's Hematoxylin, 30 s
DI H2O, 15 s
Scott’s Bluing, 15 s
70% ethanol, 15 s
Eosin Y – 2–5 s
95% ethanol, 15 s (30 – 60 s)
95% ethanol, 15 s (30 – 60 s)
100% ethanol, 15 s (30 s – 2 min)
100% ethanol, 15 s (30 s – 2 min)
Xylene, 60 s (1 min)
Xylene, 60 s (1 min)
Xylene, 60 s (1 min)
Completely remove xylene from tissue by air-drying for approximately 2 min (5 min). ! CAUTION The use of an air gun is not recommended as it may remove tissue.
Stained tissue section is now ready for microdissection. ! CAUTION No more than 30 minutes should elapse from staining until tissue is microdissected and placed into lysis buffer.
BOX 4. FROZEN TISSUE MICRODISSECTION METHOD.
Appropriate frozen tissue processing, sectioning (Box 1) and staining for microdissection (Box 3) should be observed. Histopathology consultation should be used to ensure that the cells of interest are the only cells procured (Box 2). If in doubt, do not procure those cells.
Microdissection
Put the glass slide with the stained tissue section on the laser microscope stage.
Put a LCM cap (cap) onto the tissue. Use microscopic visualization (lowest magnification) to guide cap placement onto the tissue.
Take road map image. This is done initially while visualizing the tissue section under the microscope.
Prior to beginning microdissection, focus the laser (infrared or UV) and set up the appropriate laser parameters (power, duration of the pulse). Test the laser in a free space within the cap area, with no tissue, to evaluate the laser parameters.
Fire the laser over the cells to be dissected until the desired number of laser shots has been taken.
Take a pre-dissection image. This is done prior to lifting the tissue.
Lift the cap up from the tissue to remove the laser targeted cells of interest.
Take a post-dissection image. This is done after lifting the cells of interest from the remaining tissue section.
Take a cap image. This is done to visualize the dissected cells.
Check efficiency of tissue lifting onto cap under a light microscope. Record the percentage of lifting by noting the overall average percentage of the laser spots occupied by dissected cells. In addition, the presence of any contaminating cells should be noted.
If contaminating cells are present on the cap, clean the cap with a sterile adhesive tape or similar note paper. This is accomplished by gently placing the cap on top of the sticky border of the paper a couple of times.
Re-examine the cap by light microscopy. If contaminating cells are still present, repeat step 10 and 11 until no contaminating cells are present.
Place the cap onto a microcentrifuge tube containing lysis buffer. Lysis buffer is provided in the RNA extraction kit.
? TROUBLESHOOTING
-
8| Place LCM cap on dry ice and proceed directly to pre-RNA extraction, prepare LCM negative control, or dissection of another area of the tissue with another cap.
▲CRITICAL STEP If more than one cap needs to be used to procure the necessary number of cells for a single cell type sample (e.g. 20,000 shots for prostate stroma), pre-RNA extraction should begin for the first cap (cap A) before moving to microdissection with the second cap (cap B). Begin microdissection using cap B while cap A is undergoing heat incubation (see step 12.). An LCM negative control is prepared for each tissue section by making 3000 laser shots in a tissue free portion of the tissue section. This sample is processed throughout the rest of the Protocol in parallel with microdissected tissue samples to control for contaminating RNA.
! CAUTION If time is running out (i.e. time elapsed from beginning of staining to end of microdissection for a single slide is quickly approaching the 30 min time limit), it is best to put cap A on dry ice while proceeding to microdissection with cap B. In this instance, pre-RNA extraction batch processing can be done once all caps containing microdissected cells for a single tissue section have been collected.
Pre-RNA extraction • TIMING 1 hr 40 min
9| Add 50 μl extraction buffer into labeled microfuge tube.
-
10| Place cap containing microdissected cells onto microcentrifuge tube containing lysis buffer.
▲CRITICAL STEP If multiple caps (e.g. cap A, cap B, cap C, etc.) containing microdissected cells were generated for one tissue cell type sample, the caps can be processed sequentially in the single volume of extraction buffer36. For cap A, follow steps 10 through 14. Discard cap A and transfer the lysate to a clean labeled microfuge tube. Repeat steps 10 through 15 for additional caps.
11| Invert tube and flick it to cause the buffer to cover the cap.
12| Heat at 42 oC for 30 min.
13| Mix by pulse vortex.
14| Centrifuge for 2 min at 800 x g in room temperature.
-
15| Discard cap and close microfuge tube containing cell extract.
▲CRITICAL STEP If multiple pre-RNA extraction cell extracts for one tissue cell type sample were batch processed from multiple caps (e.g. cap A, cap B, cap C, etc.), instead of sequential processing as described in step 10, pool all cell extracts into a clean labeled microfuge tube and record total volume of pooled cell extracts. This volume will be in multiples of 50 μl depending on the number of single cap derived cell extracts that were pooled (e.g. 3 caps = 150 μl total cell extract volume).
-
16| Place in −80 oC freezer for 1 hr or until ready to extract RNA. It is recommended to incorporate this step for all microdissected tissue samples for RNA extraction and subsequent transcript analysis. This ensures equal treatment of all tissue samples to be analyzed and reduces bias that may be incorporated by not all microdissected tissues being subjected to −80 oC freezing.
■ PAUSE POINT Samples may be stored at −80 oC at this point until ready to extract RNA.
RNA Extraction • TIMING 45 min
17| Remove microfuge tubes containing cell extract from pre-RNA extraction step from the −80oC freezer.
18| Place in wet ice to transfer to RNA clean bench for RNA extraction.
19| Pre-condition the RNA purification columns: add 250 μl of conditioning buffer to the RNA purification columns, incubate at room temperature for 5 min, then centrifuge the columns at 16,000 x g for 1 min at room temperature.
-
20| Add 50 μl of 70% ethanol to the microfuge tube containing the cell extract from Pre-RNA extraction. This results in a 1:1 ratio of 70% ethanol:cell extract.
▲CRITICAL STEP If multiple pre-RNA extraction cell extracts for one tissue cell type sample were batch processed from multiple caps (e.g. cap A, cap B, cap C, etc.), as described in step 17, use the recorded pooled cell extract volume to determine the necessary volume of 70% ethanol (e.g. 150 μl pooled cell extract requires 150 μl 70% ethanol).
-
21| Add cell extract and ethanol mixture to the RNA purification columns.
▲CRITICAL STEP There is no limit to the amount of RNA the column can handle. However, the column can only handle 200 μl of ethanol:cell extract at one time. Therefore, for volumes > 200 μl, repeat step 21 and 22 using sequential aliquots of the ethanol:cell extract that are 200 μl until all ethanol:cell extract has been loaded to the column and spun down.
22| Centrifuge at 100 x g K for 2 min at room temperature. Flow-through collection tube is large enough to collect all flow-through between steps 22 to 33 and steps 35 to 38. Therefore it is not necessary to discard flow-through between these steps. If processing an ethanol:cell extract volume > 200 μl, discard flow through after the final time repeating step 23.
23| Centrifuge at 16,000 x g for 30 s at room temperature.
24| Add 100 μl of wash buffer 1 (W1) to the columns.
25| Centrifuge at 8,000 x g for 1 min at room temperature.
26| Add 40 μl of DNAse solution (35 μl RDD buffer and 5 μl DNAse) to the columns.
27| Incubate at room temperature for 15 min.
28| Add 40 μl of W1 to the columns.
29| Centrifuge at 8,000 x g for 15 s at room temperature.
30| Add 100 μl wash buffer 2 (W2) to the columns.
31| Centrifuge at 8,000 x g for 1 min at room temperature..
32| Add another 100 μl of W2 to the columns.
33| Centrifuge at 16,000 x g for 2 min at room temperature.
34| Transfer each of the columns to a labeled microfuge tube.
-
35| Add 10 μl of elution buffer to the columns.
▲CRITICAL STEP To improve RNA elution, it is recommended to elute the RNA in two 10 μl volumes instead of using one 20 μl volume. Depending upon experimental tissues being used, an investigator may choose to elute in one 20 μl volume, and still recover adequate quantities of total RNA.
36| Centrifuge at 1,000 x g for 1 min at room temperature.
37| Centrifuge at 16,000 x g for 1 min at room temperature to elute the RNA.
38| Repeat steps 35 through 37.
39| Remove column from the labeled microfuge tube. The elution buffer containing the total RNA sample will be collected at the bottom of the microfuge tube.
40| Save column and flow through until after RNA quantitation analysis.
41| Aliquot each total RNA sample as follows for quantitation, qualitation, and qRT-PCR analysis (Fig. 4). Pipette 3 μl of total RNA sample into a microfuge tube for quantitation and qualitation analysis. Aliquot the remaining total RNA into 4 μl volumes by pipetting into labeled microfuge tubes for qRT-PCR analysis.
-
42| As total RNA samples are aliquoted, place on either dry or wet ice depending on downstream analysis. If time permits, it is recommended to immediately proceed to RNA quantitation and qualitation at this point using the 3 μl aliquot intended for quantitation and qualitation analysis; therefore, put these samples on wet ice. If total RNA measurement cannot be conducted promptly, place samples on dry ice and move to −80 oC freezer until ready to use.
▲CRITICAL STEP Place total RNA aliquots intended for qRT-PCR analysis in −80 oC freezer for 1 hr or until ready to run qRT-PCR. It is recommended to incorporate this step for all total RNA samples for qRT-PCR analysis. This ensures equal treatment of all tissue samples to be analyzed and reduces bias that may be incorporated by not subjecting all RNA samples to −80 oC freezing.
■ PAUSE POINT Samples may be stored at −80 oC at this point until ready for qRT-PCR analysis.
Preparation of Positive Control Total RNA Dilutions • TIMING 20 min
Prepare dilution series of commercially available total RNA sample covering the expected ranges of total RNA concentrations recovered from microdissected tissue samples, as follows. These will be used as positive controls throughout total RNA measurement, primer/probe amplification efficiency assay, and qRT-PCR analysis of samples from microdissected tissues. All dilutions, other than 1:1, are made in RNase/DNase free microcentrifuge tubes using RNase/DNase free molecular grade water (H2O).
| Dilution | Total RNA concentration | Recipe |
|---|---|---|
| 1:1 | 1000.0 ng/μl | 100 μl of 1000.0 ng/μl total RNA |
| 1:5 | 200.0 ng/μl | 20 μl of 1:1 in 80 μl of H2O |
| 1:10 | 100.0 ng/μl | 10 μl of 1:1 in 90 μl of H2O |
| 1:20 | 50.0 ng/μl | 5 μl of 1:1 in 95 μl of H2O |
| 1:50 | 20.0 ng/μl | 10 μl of 1:5 in 90 μl of H2O |
| 1:100 | 10.0 ng/μl | 10 μl of 1:10 in 90 μl of H2O |
| 1:200 | 5.0 ng/μl | 10 μl of 1:20 in 90 μl of H2O |
| 1:500 | 2.0 ng/μl | 10 μl of 1:50 in 90 μl of H2O |
43| Pulse vortex and pulse centrifuge all dilutions at room temperature and place on wet ice.
44| Aliquot 3 μl of diluted samples to use as positive control total RNA measurement and place on wet ice. As samples are aliquoted, place on either dry or wet ice depending on time constraints. If time permits, it is recommended to immediately proceed to RNA quantitation and qualitation at this point using the 3 μl aliquots intended for quantitation and qualitation analysis; therefore, put these positive control dilution samples on wet ice. If total RNA measurement cannot be conducted promptly, place positive control dilution samples on dry ice and move to −80 oC freezer until ready to use.
-
45| Prepare 20 μl aliquots of diluted samples for future use in total RNA measurements and qRT-PCR analysis. Place positive control dilution samples on dry ice and move to −80 oC freezer until ready to use.
■ PAUSE POINT Samples may be stored at −80 oC at this point until ready for downstream analysis.
-
46| Aliquot 4 μl of commercially available RNase/DNase free molecular grade water to use as negative control and for blanking the system.
▲CRITICAL STEP Use water that does not have DEPC or any other chemical compound in it. The quantitation method is spectrophotometry based; therefore, any additional compound that may be in the water used as a blank or negative control will confound the data and not give accurate representation of the total RNA concentration found within the sample aliquots.
Total RNA Quantitation Analysis • TIMING 2 min per sample
47| Place 3 μl aliquots of all controls (see step 45) and total RNA samples from microdissected tissue samples (from step 42) on wet ice. Mix the controls and samples by vortexing and spin down by centrifugation. Place tubes back on wet ice.
-
48| Follow NanoDrop manufacturer’s protocol for RNA analysis.
▲CRITICAL STEP To prevent contamination carryover and ensure accurate data analysis, analyze samples and controls in the following order: (1) negative control water, (2) all experimental total RNA samples from microdissected tissues, (3) positive control RNA samples, and (4) negative control water. The use of the negative control water at the end of quantitation analysis used to determine whether residual total RNA may be present in the NanoDrop equipment. If negative control water analysis demonstrates the presence of total RNA, clean the analysis equipment and re-measure total RNA quantities of all samples and controls.
? TROUBLESHOOTING
49| Place remaining 2μl of RNA sample back on wet ice after quantitation analysis.
Total RNA Integrity Qualitation Analysis • TIMING 50 min per chip plus an initial 30 min to bring kit reagents to room temperature
50| Analyze sample total RNA concentrations (ng/μl) to determine which total RNA qualitation assay to use (i.e. BioAnalyzer RNA 6000 Pico Assay or RNA 6000 Nano Assay) for each individual sample (Box 5).
51| Use the remaining 2μl from the above 3 μl aliquot used for quantitation analysis that is on wet ice for qualitation analysis.
52| Bring appropriate kit reagents to room temperature in the dark for 30 min.
-
53| Follow manufacturer’s protocol with the following modification: After preparing the gel-dye mix, incubate all samples, controls, and ladder at 70°C for 2 minutes. Then quench on ice for 2 min, vortex, and spin down. Load sample within 10 min. Continue with manufacturer’s protocol until completion of quantitation analysis
▲CRITICAL STEP Due to potential evaporation during the sample denaturation, it is recommended to use the remaining 2 μl of total RNA aliquot for heat incubation. This ensures that a full 1 μl is available to meet sample requirement volume for qualitation analysis. Accurate pipetting is critical since only 1 μl of the samples are loaded into the chip wells. During chip and sample loading process, ensure that no air bubbles are formed as this may confound qualitation analysis.
54| Review data for each sample and controls. Assess ladder, peaks, electropherograms, and RINs. RNA quality of RIN >5 is considered good quality total RNA and RIN >8 is considered excellent total RNA quality 50.
? TROUBLESHOOTING
■ PAUSE POINT All sample aliquots (see steps 42 and 44) are stored at −80 oC and do not need to be removed until ready for downstream analysis.
Primer/Probe Specific qPCR Assay Efficiency Determination • TIMING 3.5 hr
-
55| Test assay efficiency, sensitivity, and reproducibility of all primer/probe sets using the positive control total RNA dilution series prepared in step 12. The qRT-PCR assay will be carried out in a two-step reaction; therefore, specific primer/probe qPCR test assays will be conducted using the same parameters as the experimental assay parameters for template, RT, and qPCR reagent concentrations. See step 25 below for experimental protocol for RT and qPCR. For one RT tube of each positive control dilution series sample, 12 qPCR assays will be able to be performed. Using three qPCR technical replicates per cDNA sample and primer/probe set, four primer/probe sets can be analyzed. Usual convention is to test three genes of interest and one endogenous control primer/probe set per RT tube generated cDNA sample.
▲CRITICAL All qRT-PCR set up should be done in a dedicated PCR hood providing a clean controlled environment (Fig. 3; Box 6).
56| Analyze each qPCR assay individually by evaluating efficiencies and standard curves. The detection limits will be determined by the lowest dilution sample concentration that generates repeatable CT values within the three technical replicates.
BOX 6. CLEANING PROCEDURES FOR MAINTENANCE OF THE RT-PCR PREPARATION HOOD.
To maintain a clean environment for qRT-PCR setup, it is imperative to use a dedicated dead air hood and dedicated equipment such as pipettes, pipette tips, tubes, racks, vortexer, and mini-centrifuge. PCR hood and/or reagent/template contamination may adversely affect the results of the qRT-PCR experiment. This contamination potential can be negated by optimal hood and equipment cleaning as described below.
Regular cleaning should take place before and after each experiment is completed.
Weekly cleaning and maintenance should also take place.
Additional barrier tips, caps, tubes, extra trash bags bottles of 10% bleach, 70% ethyl alcohol, RNase Away, and paper towels and gloves should be located within reach of the hood.
Any time one goes in and out of the PCR hood, gloves must be changed.
While working in the PCR hood, it is necessary to wear the designated clean PCR hood lab coat with elastic cuffs.
For UV light use, make sure all pipette tips are unstacked and the centrifuge lid is open. This allows UV to reach all areas of potential contamination during the UV sterilization process.
Fresh 10% bleach solution should be made for each weekly cleaning
As supplies dwindle in the PCR hood, they should be replaced as soon as possible.
Regular Cleaning
-
Use in succession: 10% bleach followed by 70% ethanol on the inside of the hood to:
Wipe all sides and back of the hood.
Wipe bottom of hood.
Wipe outside of all containers.
Wipe around both centrifuge and vortex.
Wipe outsides of pipette tip boxes.
Wipe down pipettes.
Wipe sides of the hood.
Wipe all parts of the pull up access panel on both front and back.
Wipe and dry PCR-only ice bucket.
Turn UV light switch all the way to start.
Replace plastic backed paper mats (“diapers”) on outside counter of PCR hood as needed.
Weekly Maintenance
In addition to regular single use cleaning, perform the following each week:
Follow all bleach and ethanol steps by RNase-Away.
Replace plastic backed paper mats on outside counter for PCR hood. Tape down mats to counter area to prevent it from sliding.
Mark mats with “FOR PCR ONLY” to remind others to respect the integrity of the PCR clean area.
Compile list of consumables that are running low/need to be ordered (tips, tubes, caps, plates, and plastic bags). Keep two boxes minimum of the consumables at all times. When the second to last box of consumables gets opened, an order should be placed.
Record date weekly maintenance was completed by writing on tape and placing on top of hood.
UV light
The UV integrity is good for 1000 hrs per bulb. One hour is clocked/used every time the light is turned on, even if the light is only on for 5 minutes.
Change UV light every six months to a year, depending on use. If heavy use of UV, record each time UV is turned on and change when 1000 uses has occurred.
Update tape on hood with date UV light changed.
A review of qPCR efficiencies and standard curves has been discussed in a previous issue of Nature Protocols48.
qRT-PCR Assay
▲CRITICAL All qRT-PCR set up should be done in a dedicated PCR hood providing a clean controlled environment (Fig. 3; Box 6).
Reverse Transcription (RT) • TIMING 1.5 to 2 hr
-
57| Prepare cDNA by reverse transcription (RT) of all individual total RNA samples and controls (NTC, no-RT, and (+) C) using TaqMan RT Reagents (Applied Biosystems, Inc. (ABI), Foster City, CA, USA; Cat # N808-0234), with random hexamers as the RT primers. Random hexamers have been shown to minimize potential effects of starting template degradation and improve qPCR analysis of short amplicons (<200 bp).
▲CRITICAL STEP Prior to planning and running the RT, plan the qPCR plate setup and determine the number of qPCR reactions (Fig. 5) that will be run from each of the samples and controls to determine the number of RT reactions that will be needed.
-
58| In a 1.7 or 2 ml microfuge tube, prepare a master mix of the following RT reaction components multiplied by the number of reactions plus one (for pipetting loss) as outlined in the table below. Vortex and spin down all RT master mix components and templates prior to use.
▲ CRITICAL STEP Prepare no-RT mastermix for each of the samples. Do not add reverse transcriptase to the RT master mix, instead add 1.25 μl additional H2O to the mastermix. Prepare one additional RT reaction with (+) C to be analyzed with the no-RT controls.Reagent Volume to add per reaction 10x Buffer 5.00 μl MgCl2 11.00 μl 10mM dNTP 10.00 μl RNase Inhibitor 1.00 μl Random Hexamers 2.50 μl Multiscribe RT Enzyme 1.25 μl H2O 15.25 μl 59| Vortex and spin down master mix.
60| Add 46 μl of RT master mix to labeled PCR tubes.
61| Add 4 μl H2O to the NTC RT reaction tube. Add 4 μl of template total RNA per RT reaction tube and no-RT control reaction tube. Add 4 μl (+) C total RNA to the positive control RT reaction tube. The final reaction volume will be 50 μl. Vortex and spin down, avoiding bubbles.
- 62| Run all RT reactions in PCR thermocycler using the following protocol as suggested by manufacturer (ABI):
Cycles Temperature Time 1 cycle 25 oC 10 min 48 oC 30 min 95 oC 5 min 63| Immediately place samples on ice and spin down after 2 min.
-
64| Use RT product cDNA immediately in qPCR reactions or store at −20 oC.
▲CRITICAL STEP If analyzing nine genes of interest from one microdissected sample, prepare three RT reactions from each of the microdissected RNA samples (Fig. 4). The first of three cDNA samples will be used in the first qPCR run. Place the other two cDNA samples on wet ice until use. All three qPCR runs should be done consecutively on the same day as RT reaction. After running all three qPCR plates, analyze all no-RT controls and the additional (+) C RT cDNA in an additional qPCR plate.
■ PAUSE POINT If this cannot be done, store remaining cDNA samples at −80 oC until use.
Figure 5.
Example qPCR 96-well setup. Using this layout, three genes of interest and one endogenous control housekeeping gene can be analyzed by qPCR from six samples and appropriate control cDNAs (one RT tube per template). This setup may be used to analyze additional genes of interest from the microdissected sample using the same endogenous control housekeeping gene. Example cDNA samples (e.g. 1N, 1T, 2N, 2T, 3N, 3T) are from three cases of matched tumor (T) and normal (N) tissues. To prevent the introduction of bias, it is important to process both diseased and normal tissue samples at the same time. Plate two would include genes of interest four through 6. Plate three would include genes of interest seven through nine. All no-RT controls are run on a separate plate using (+) C and NTCs.
qPCR • TIMING 3 hr per 96-well plate (1 hr for setup, 2 hr for qPCR)
Conduct single-plex qPCR analysis of all individual cDNA samples and controls (NTC, no-RT, and (+) C) generated from above RT using 2x TaqMan Universal Master Mix and AmpliTaq Gold, and specific primer/probe sets (Applied Biosystems, Inc. (ABI), Foster City, CA, USA).
-
65| For each of the primer/probe sets, prepare a master mix in a 1.7 or 2 ml microfuge tube of the following qPCR reaction components multiplied by the number of reactions plus one (for pipetting loss) as outlined in the table below. Vortex and spin down all cDNA templates and qPCR master mix components, except for 2x TaqMan Universal PCR Master Mix, prior to use.
! Caution The 2x TaqMan Universal Master Mix is viscous and will bubble upon vortexing. Mix gently instead of vortexing.Reagent Volume to add per reaction 2x TaqMan Universal PCR MM 25.00 μl Amplitaq Gold 1.00 μl H2O 17.50 μl FAM or VIC primer/probe set 2.50 μl -
66| Mix gently and spin down master mix.
! Caution Do not vortex qPCR master mix as micro bubbles will form, interfering with qPCR optical detection analysis.
67| Add 46 μl of qPCR master mix to designated wells in 96-well plate (Fig. 5).
-
68| Add 4 μl H2O to the NTC qPCR wells. Add 4 μl of template cDNA per RT reaction tube and no-RT control reaction tube. Add 4 μl (+) C cDNA to the (+) C wells. Pipette up and down, avoiding bubbles, when adding template to master mix to mix thoroughly.
! Caution Do not depress the pipette all the way down as this may cause bubbles to occur in the well.
69| Place optical caps or film over the wells of the plate. Mix gently and spin down
-
70| Run all qPCR reactions in qPCR thermocycler using the following protocol:
Cycles Temperature Time 1 cycle 50 oC 2 min 50 cycles 95 oC 15 s 60 oC 1 min ▲CRITICAL STEP 50 cycles are used for qPCR to ensure that the NTCs for a gene of interest have 10 CT values later than the sample gene of interest expression in cases where low level of gene expression (e.g. 37 CT values) are encountered. This ensures that if the NTCs demonstrate ‘no CT’ at 50 cycles, the qPCR reagents are free of contamination. In addition, the observation of 10 CT difference comparing no-RT with RT-positive samples, negligible contamination (0.1%) of genomic DNA contamination is confirmed92;93.
? TROUBLESHOOTING
■ PAUSE POINT Upon qPCR data collection, data is saved at this point, and can be analyzed later.
Data Analysis • TIMING 2.5 h
71| Data from qRT-PCR of microdissected tissue samples is normalized using endogenous control housekeeping gene expression. Therefore, relative quantitation is the analysis method that must be used. The recommended method for relative quantitation is the 2−Δ ΔCt method94 summarized in the following table. A brief discussion of biologic fold change expression classifications can be found in Box 7.
BOX 7. BIOLOGIC FOLD CHANGE EXPRESSION CLASSIFICATIONS.
Biologic fold change (relative quantitation) of a diseased tissue sample compared to a normal tissue sample can be calculated using the relative quantitation 2−ΔΔCT method 94. Expression classifications (adapted from Xu et al. 77) can be used to define general thresholds of overexpression, underexpression, or no change for a gene of interest.
-
Overexpression in disease tissue (D) compared to normal tissue (N) (D>N), defined as
1+ (1.5–5-fold),
2+ (5.1–10-fold),
3+ (10.1–20-fold) and
4+ (>20-fold);
-
Underexpression in disease tissue (D<N), defined as
1– (1.5–5-fold),
2– (5.1–10-fold),
3– (10.1–20-fold) and
4– (>20-fold);
No change (D=N), refers to the difference of gene expression between D and N as <1.5-fold.
? TROUBLESHOOTING
| Relative Quantitation: | ||
|---|---|---|
| Steps | Formula | Definitions |
| 1. Gene expression normalization within a sample | Δ CT = X – EC | For a given sample (diseased or normal), X is the gene of interest CT value and EC is the endogenous control CT value. |
| 2. Gene expression comparison | Δ Δ CT = CTx - CTcb | For a given case (comparing diseased (D) and normal (N) tissues from the same organism) x is the sample of interest and cb is the calibrator. The calibrator can be D or N sample if we are asking different questions. |
| 3. Biologic fold change | 2−Δ ΔCT | Translates the CT value to assign biologic significance in terms of biologic fold change. If CT is a positive number after calculating 2- CT transform by 1/x to obtain the underexpression value. |
TIMING
Steps 1–5 , 1.5 to 3 h
Steps 6–8, 45 to 55 min
Steps 9–16, 1 hr 40 min
Steps 17–42, 45 min
Steps 43–46, 20 min
Steps 47–49, 2 min per sample
Steps 50 – 54, 50 min per chip plus an initial 30 min to bring kit reagents to room temperature
Steps 55 – 56, 3.5 h
Steps 57 – 64, 1.5 to 2 h
Steps 65 –70, 3 h per 96-well plate
Step 71, 2.5 h
ANTICIPATED RESULTS
Fig. 4 depicts the specific capture of the cells of interest from a heterogeneous cellular environment via laser capture microdissection using the PixCell II (Arcturus Molecular Devices).
Table 1 describes the quantity and quality of RNA yields from typical microdissections from various tissues. Attention to cellular density per laser shot is important to estimate in advance the total RNA recovery (e.g. prostate stroma compared to breast stroma number of laser shots to obtain similar quantities of total RNA).
Fig. 6 presents an example of the amplification plot results from qRT-PCR analysis of epithelial microdissected prostate tumor and matched normal tissue samples.
Figure 6.
Typical TaqMan qPCR amplification plots for 96-well setup. ΔRn versus cycles linear view. Primer/probe concentrations are optimized for highest ΔRn and lowest CT values for microdissected tissue sample templates. All samples endogenous control housekeeping gene analysis used VIC/HEX labeled primer-limited primer/probe sets. Genes of interest were FAM labeled non-primer limited primer/probe sets. NTCs were all negative as evidenced by no amplification crossing the threshold resulting in no CT values. Triplicate technical replicates are good with similar amplification efficiencies and standard of pipetting.
A review of example CT value ranges that can be anticipated for endogenous control housekeeping genes and the corresponding RNA quantity/quality measurements can be found in Erickson et al.’s Laboratory Investigation paper16. Other examples of CT value ranges have previously been published4;16 and can be reviewed for further information.
? TROUBLESHOOTING
Troubleshooting qRT-PCR data is a complex process when working with tissue samples. Standard total RNA controls, such as commercially available, high-quality total RNA from tissue, should be used for qRT-PCR, including primer/probe optimization and efficiency assessment, as well as standard curve generation. It is recommended that the total RNA chosen for this purpose be from the same organ as the one being analyzed in the qRT-PCR study. However, when working with animal model or clinical tissue samples, RT and qPCR only represent the last two steps of a series of protocols that have been applied sequentially to generate the quantitative gene expression data. Incorporating appropriate controls during each step, these steps leading to qRT-PCR include: tissue collection, tissue processing, tissue sectioning and staining, microdissection, and RNA extraction and isolation. Assuming proper techniques have been used for collecting the samples (in particular, reasonable reduction of the time from tissue removal to freezing), as well as adequate snap freezing techniques, the issue of little or no RNA recovery can still be faced.
In general, sets of tissues from different animal subjects or patients are analyzed in each experiment. Therefore, the situation varies if the negative results are observed in all or just in one or a few samples. In the former case general explanations are found, such as an improper RNA extraction technique. Investigators can even use high quality RNA to “re-extract” and make sure its integrity is intact throughout the extraction. It is ideal to use a RNA quality and quantity “proven” tissue control side by side with the experimental samples when perform the extraction. This “proven” tissue serves as a “tissue extraction positive control”. DNase treatment is one common source for RNA degradation. Another general problem could be improper storing such as freezer malfunctions, or storing the tissues too long in the freezer. It has been observed that after storage for a year, RNA can be compromised even at -80C.
If only a few individual samples demonstrate poor RNA recovery or higher than expected CT values are seen, general experimental protocol issues can be ruled out. In this case, the most typical explanation is RNA integrity in the individual sample. This can often be attributed to RNA degradation prior to freezing or poor tissue processing. In addition, certain organ types may also contain higher level of RNases. Brain tissue inherently contains less RNases than other tissues, with pancreas, spleen, and intestinal mucosa having the most34. Another step in the protocol to examine is the dissection process. Issues of cell procurement (lifting on the cap) during the laser process as well as cellular density of the dissection area of interest need to be taken into account. In the case of variable cell density, it has been observed that five to six times more laser shots are needed to obtain equivalent total RNA yields in stroma compared to epithelium.
Finally, troubleshooting issues can also arise at the phase of the analysis of the data. This includes, but is not limited to, the use of inappropriate normalization strategies, high variability (i.e. large standard deviations) between qPCR replicates that is sometimes observed in low expressed transcripts, and the appropriate data analysis strategy (e.g. relative quantitation for data obtained from qRT-PCR of microdissected tissues). Also, a key point in the analysis is to define a threshold of what fold change (2−ΔΔCT) is meaningful between samples (e.g. paired diseased and normal tissue samples). The natural trend is to consider a larger fold change as biologically more relevant. However, smaller differences in expression can still be important in disease progression. For instance, the loss of function of just one allele (haploinsufficiency) may be critical in several neoplastic and non-neoplastic processes96;97.
General qRT-PCR and LCM troubleshooting issues have been addressed previously in a previous issues of Nature Protocols36;48. Additional troubleshooting advice specifically related to qRT-PCR of microdissected tissue samples can be found in Table 2.
Table 2.
Troubleshooting.
| Problems | Possible reason | Solution |
|---|---|---|
| LCM (Step 7) | ||
| No cells captured in LCM cap. | Tissue section mounted on a charged (plus) slide. | Mount the tissue section on uncharged slides for LCM. |
| Tissue section on slide not sufficiently dry. | Change solutions including ethanol and xylenes. Now increase the times of dehydration in ethanol 95%, 100% and xylene at 2 minutes each bath. | |
| Contamination of LCM cap with non-dissected extra tissue. | Tissue section could be too dry. | Check the dissected cells in the cap under the microscope. If there is extra tissue, remove it with a sterile adhesive tape or similar note paper. Gently place the cap with the film on the sticky border of the sterile adhesive tape or similar note paper a couple of times. Eliminate the used sterile adhesive tape or similar note paper. Now, check the cap again under the microscope. The extra tissue will be removed from the cap (attached to the sticky paper) while the dissected cells will remain embedded in the film and will be not removed by the sticky paper. |
| RNA Quantity and Quality (Steps 49 and 53) | ||
| No quantity or quality data reported by NanoDrop or Bioanalyzer analysis, respectively. | Tissue section stored in freezer too long. | Use tissue section within 2 weeks of sectioning and freezing. |
| RNase rich tissue (e.g. pancreas, duodenum, colon, etc.). | Stain and dissect as quickly as possible. Add RNase inhibitors to staining solutions. | |
| Cell not captured by LCM in the cap. | Always check lifting of captured cells on the cap under the microscope after LCM before proceeding to RNA extraction. | |
| Nanogram quantity of sample appropriate for the Bioanalyzer chip chosen (Pico v. Nano) | Use the appropriate chip for the quantity of total RNA determined by NanoDrop. | |
| Washed RNA through the column during RNA extraction | Pool all flow-through and re-extract all flow through and recover the RNA. | |
| Total RNA not aliquotted appropriately leading to multiple free-thaw degradation. | Aliquot total RNA prior to initial storage at −80°C | |
| Tissue block assessment by scraping tissue section and performing RNA extraction, quantity, and quality assessments prior to microdissection was not done. | Perform tissue block assessment prior to microdissection. | |
| Low RIN number. | Tissue or RNA degradation | Continue with qRT-PCR analysis. Use random hexamers in RT step to minimize effects of degradation. Determine acceptable RIN range for the tissue type being used in the study. For prostate, esophagus and urethra, we have found that even low RINs (RIN >2) produce specific repeatable qRT- PCR results. |
| qRT-PCR (Step 67) | ||
| No CT values reported. | Poor quality and quantity or total RNA template used for RT. | See above RNA Quantity and Quality section. |
| RT failed to generate cDNA. | Check positive control. If positive control for a particular primer/probe set did generate CT values and the endogenous control gene analyzed for the sample generated CT values, then it can be concluded that cDNA was generated and the sample may not be expressing the transcript of interest within qRT-PCR detectable quantities. If not, repeat RT. | |
| Only ran qPCR for 40 cycles. | Use RT generated cDNA to run qPCR for 50 cycles. | |
| No CT values reported in one paired sample and not the other. | Identical number of LCM shots/number of cells not collected for RNA extraction. | Use comparable number of cells/laser shots for RNA extraction and downstream qRT-PCR. If comparable numbers of cells and total RNA were used for downstream qRT-PCR, transcript in one sample (e.g. diseased sample or normal sample) may not be expressed within detectable levels. At this point it is imperative to ascertain that the endogenous control qRT-PCR for that sample did generate a CT value. If not, see No CT values reported section above. |
| Analysis (Step 68) | ||
| Under expression or down regulation of gene expression unable to be determined. | ΔΔCT value is positive. | Multiply positive ΔΔCT value by 1/x. Then perform 2−ΔΔCT fold change calculation using this number. This fold change value will represent the under expression compared to the calibrator. |
Acknowledgments
The authors appreciatively thank S. Gonzalez and A. Velasco (The Catholic University, Santiago, Chile) for their collaboration in providing the frozen prostate whole-mount tissue blocks used to assess normalization strategies. RFC and MRE-B are Federal employee inventors on NIH patents covering LCM and xMD technologies and are entitled to receive royalty payments through the NIH Technology Transfer program. This research was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
Footnotes
Research was performed at the Pathogenetics Unit, Laboratory of Pathology and Urologic Oncology Branch, National Cancer Institute, National Institutes of Health, Bethesda, Maryland 20892
COMPETING INTERESTS STATEMENT
The authors declare no competing financial interests.
References
- 1.Best CJ, et al. Molecular differentiation of high- and moderate-grade human prostate cancer by cDNA microarray analysis. Diagn Mol Pathol. 2003;12:63–70. doi: 10.1097/00019606-200306000-00001. [DOI] [PubMed] [Google Scholar]
- 2.Best CJ, et al. Molecular alterations in primary prostate cancer after androgen ablation therapy. Clin Cancer Res. 2005;11:6823–34. doi: 10.1158/1078-0432.CCR-05-0585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Best CJ, Emmert-Buck MR. Molecular profiling of tissue samples using laser capture microdissection. Expert Rev Mol Diagn. 2001;1:53–60. doi: 10.1586/14737159.1.1.53. [DOI] [PubMed] [Google Scholar]
- 4.Richardson AM, et al. Global expression analysis of prostate cancer-associated stroma and epithelia. Diagn Mol Pathol. 2007;16:189–97. doi: 10.1097/PDM.0b013e3180de20ac. [DOI] [PubMed] [Google Scholar]
- 5.Wiese AH, et al. Identification of gene signatures for invasive colorectal tumor cells. Cancer Detect Prev. 2007;31:282–95. doi: 10.1016/j.cdp.2007.07.003. [DOI] [PubMed] [Google Scholar]
- 6.Lee S, et al. Alterations of gene expression in the development of early hyperplastic precursors of breast cancer. Am J Pathol. 2007;171:252–62. doi: 10.2353/ajpath.2007.061010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turashvili G, et al. Novel markers for differentiation of lobular and ductal invasive breast carcinomas by laser microdissection and microarray analysis. BMC Cancer. 2007;7:55. doi: 10.1186/1471-2407-7-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jaeger J, et al. Gene expression signatures for tumor progression, tumor subtype, and tumor thickness in laser-microdissected melanoma tissues. Clin Cancer Res. 2007;13:806–15. doi: 10.1158/1078-0432.CCR-06-1820. [DOI] [PubMed] [Google Scholar]
- 9.Sunde JS, et al. Expression profiling identifies altered expression of genes that contribute to the inhibition of transforming growth factor-beta signaling in ovarian cancer. Cancer Res. 2006;66:8404–12. doi: 10.1158/0008-5472.CAN-06-0683. [DOI] [PubMed] [Google Scholar]
- 10.Dahl E, et al. Molecular profiling of laser-microdissected matched tumor and normal breast tissue identifies karyopherin alpha2 as a potential novel prognostic marker in breast cancer. Clin Cancer Res. 2006;12:3950–60. doi: 10.1158/1078-0432.CCR-05-2090. [DOI] [PubMed] [Google Scholar]
- 11.Tsai MF, et al. A new tumor suppressor DnaJ-like heat shock protein, HLJ1, and survival of patients with non-small-cell lung carcinoma. J Natl Cancer Inst. 2006;98:825–38. doi: 10.1093/jnci/djj229. [DOI] [PubMed] [Google Scholar]
- 12.Scott M, et al. Altered patterns of transcription of the septin gene, SEPT9, in ovarian tumorigenesis. Int J Cancer. 2006;118:1325–9. doi: 10.1002/ijc.21486. [DOI] [PubMed] [Google Scholar]
- 13.Luzzi VI, Holtschlag V, Watson MA. Gene expression profiling of primary tumor cell populations using laser capture microdissection, RNA transcript amplification, and GeneChip microarrays. Methods Mol Biol. 2005;293:187–207. doi: 10.1385/1-59259-853-6:187. [DOI] [PubMed] [Google Scholar]
- 14.Yao F, et al. Microarray analysis of fluoro-gold labeled rat dopamine neurons harvested by laser capture microdissection. J Neurosci Methods. 2005;143:95–106. doi: 10.1016/j.jneumeth.2004.09.023. [DOI] [PubMed] [Google Scholar]
- 15.Chan S, et al. The use of laser capture microdissection (LCM) and quantitative polymerase chain reaction to define thyroid hormone receptor expression in human 'term' placenta. Placenta. 2004;25:758–62. doi: 10.1016/j.placenta.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Erickson HS, et al. Assessment of normalization strategies for quantitative RT-PCR using microdissected tissue samples. Lab Invest. 2007;87:951–62. doi: 10.1038/labinvest.3700659. [DOI] [PubMed] [Google Scholar]
- 17.Ransohoff DF. Lessons from controversy: ovarian cancer screening and serum proteomics. J Natl Cancer Inst. 2005;97:315–9. doi: 10.1093/jnci/dji054. [DOI] [PubMed] [Google Scholar]
- 18.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–9. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 19.Ransohoff DF. Rules of evidence for cancer molecular-marker discovery and validation. Nat Rev Cancer. 2004;4:309–14. doi: 10.1038/nrc1322. [DOI] [PubMed] [Google Scholar]
- 20.Ransohoff DF, McNaughton Collins M, Fowler FJ. Why is prostate cancer screening so common when the evidence is so uncertain? A system without negative feedback. Am J Med. 2002;113:663–7. doi: 10.1016/s0002-9343(02)01235-4. [DOI] [PubMed] [Google Scholar]
- 21.Twombly R. Identity crisis: finding, defining, and integrating biomarkers still a challenge. J Natl Cancer Inst. 2006;98:11–2. doi: 10.1093/jnci/djj029. [DOI] [PubMed] [Google Scholar]
- 22.Compton C. Getting to personalized cancer medicine: taking out the garbage. Cancer. 2007;110:1641–3. doi: 10.1002/cncr.22966. [DOI] [PubMed] [Google Scholar]
- 23.Vaught JB. Biorepository and biospecimen science: a new focus for CEBP. Cancer Epidemiol Biomarkers Prev. 2006;15:1572–3. doi: 10.1158/1055-9965.EPI-06-0632. [DOI] [PubMed] [Google Scholar]
- 24.Lin DW, et al. Influence of surgical manipulation on prostate gene expression: implications for molecular correlates of treatment effects and disease prognosis. J Clin Oncol. 2006;24:3763–70. doi: 10.1200/JCO.2005.05.1458. [DOI] [PubMed] [Google Scholar]
- 25.Micke P, et al. Biobanking of fresh frozen tissue: RNA is stable in nonfixed surgical specimens. Lab Invest. 2006;86:202–11. doi: 10.1038/labinvest.3700372. [DOI] [PubMed] [Google Scholar]
- 26.Bova GS, et al. Optimal molecular profiling of tissue and tissue components: defining the best processing and microdissection methods for biomedical applications. Mol Biotechnol. 2005;29:119–52. doi: 10.1385/MB:29:2:119. [DOI] [PubMed] [Google Scholar]
- 27.Gillespie JW, et al. Molecular profiling of cancer. Toxicol Pathol. 2004;32 (Suppl 1):67–71. doi: 10.1080/01926230490430728. [DOI] [PubMed] [Google Scholar]
- 28.Ahram M, et al. Evaluation of ethanol-fixed, paraffin-embedded tissues for proteomic applications. Proteomics. 2003;3:413–21. doi: 10.1002/pmic.200390056. [DOI] [PubMed] [Google Scholar]
- 29.Perlmutter MA, et al. Comparison of snap freezing versus ethanol fixation for gene expression profiling of tissue specimens. J Mol Diagn. 2004;6:371–7. doi: 10.1016/S1525-1578(10)60534-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Leiva IM, Emmert-Buck MR, Gillespie JW. Handling of clinical tissue specimens for molecular profiling studies. Curr Issues Mol Biol. 2003;5:27–35. [PubMed] [Google Scholar]
- 31.Chuaqui RF, et al. Post-analysis follow-up and validation of microarray experiments. Nat Genet. 2002;32 (Suppl):509–14. doi: 10.1038/ng1034. [DOI] [PubMed] [Google Scholar]
- 32.Gillespie JW, et al. Evaluation of non-formalin tissue fixation for molecular profiling studies. Am J Pathol. 2002;160:449–57. doi: 10.1016/S0002-9440(10)64864-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Emmert-Buck MR, et al. Molecular profiling of clinical tissues specimens: feasibility and applications. J Mol Diagn. 2000;2:60–6. doi: 10.1016/s1525-1578(10)60617-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ellem KA, Colter JS. A consideration of the ribonucleic acid depolymerase-inhibitor systems of mouse tissues. J Cell Comp Physiol. 1961;58:267–76. doi: 10.1002/jcp.1030580308. [DOI] [PubMed] [Google Scholar]
- 35.Erickson HS, Gillespie JW, Emmert-Buck MR. Tissue microdissection. 2008;424:433–48. doi: 10.1007/978-1-60327-064-9_34. [DOI] [PubMed] [Google Scholar]
- 36.Espina V, et al. Laser-capture microdissection. Nat Protoc. 2006;1:586–603. doi: 10.1038/nprot.2006.85. [DOI] [PubMed] [Google Scholar]
- 37.Emmert-Buck MR, et al. Laser capture microdissection. Science. 1996;274:998–1001. doi: 10.1126/science.274.5289.998. [DOI] [PubMed] [Google Scholar]
- 38.Bonner RF, et al. Laser capture microdissection: molecular analysis of tissue. Science. 1997;278:1481–1483. doi: 10.1126/science.278.5342.1481. [DOI] [PubMed] [Google Scholar]
- 39.Okuducu AF, et al. Influence of histochemical stains on quantitative gene expression analysis after laser-assisted microdissection. Int J Mol Med. 2003;11:449–53. [PubMed] [Google Scholar]
- 40.Rubin MA. Tech.Sight. Understanding disease cell by cell. Science. 2002;296:1329–30. doi: 10.1126/science.296.5571.1329. [DOI] [PubMed] [Google Scholar]
- 41.Radstrom P, et al. Pre-PCR processing: strategies to generate PCR-compatible samples. Mol Biotechnol. 2004;26:133–46. doi: 10.1385/MB:26:2:133. [DOI] [PubMed] [Google Scholar]
- 42.Lefevre J, et al. Prevalence of selective inhibition of HPV-16 DNA amplification in cervicovaginal lavages. J Med Virol. 2004;72:132–7. doi: 10.1002/jmv.10539. [DOI] [PubMed] [Google Scholar]
- 43.Sunen E, et al. Comparison of two methods for the detection of hepatitis A virus in clam samples (Tapes spp.) by reverse transcription-nested PCR. Int J Food Microbiol. 2004;91:147–54. doi: 10.1016/S0168-1605(03)00374-X. [DOI] [PubMed] [Google Scholar]
- 44.Perch-Nielsen IR, et al. Removal of PCR inhibitors using dielectrophoresis as a selective filter in a microsystem. Lab Chip. 2003;3:212–6. doi: 10.1039/b304549h. [DOI] [PubMed] [Google Scholar]
- 45.Jiang J, et al. Development of procedures for direct extraction of Cryptosporidium DNA from water concentrates and for relief of PCR inhibitors. Appl Environ Microbiol. 2005;71:1135–41. doi: 10.1128/AEM.71.3.1135-1141.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guy RA, et al. Real-time PCR for quantification of Giardia and Cryptosporidium in environmental water samples and sewage. Appl Environ Microbiol. 2003;69:5178–85. doi: 10.1128/AEM.69.9.5178-5185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bustin SA, Nolan T. Pitfalls of quantitative real-time reverse-transcription polymerase chain reaction. J Biomol Tech. 2004;15:155–66. [PMC free article] [PubMed] [Google Scholar]
- 48.Nolan T, Hands RE, Bustin SA. Quantification of mRNA using real-time RT-PCR. Nat Protoc. 2006;1:1559–82. doi: 10.1038/nprot.2006.236. [DOI] [PubMed] [Google Scholar]
- 49.Morrison TB, Weis JJ, Wittwer CT. Quantification of low-copy transcripts by continuous SYBR Green I monitoring during amplification. Biotechniques. 1998;24:954–8. 960–962. [PubMed] [Google Scholar]
- 50.Fleige S, Pfaffl MW. RNA integrity and the effect on the real-time qRT-PCR performance. Mol Aspects Med. 2006;27:126–39. doi: 10.1016/j.mam.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 51.Hilscher C, Vahrson W, Dittmer DP. Faster quantitative real-time PCR protocols may lose sensitivity and show increased variability. Nucleic Acids Res. 2005;33 :e182. doi: 10.1093/nar/gni181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanley KK, Szewczuk E. Multiplexed tandem PCR: gene profiling from small amounts of RNA using SYBR Green detection. Nucleic Acids Res. 2005;33:e180. doi: 10.1093/nar/gni182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Suslov O, Steindler DA. PCR inhibition by reverse transcriptase leads to an overestimation of amplification efficiency. Nucleic Acids Res. 2005;33:e181. doi: 10.1093/nar/gni176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stahlberg A, et al. Properties of the reverse transcription reaction in mRNA quantification. Clin Chem. 2004;50:509–15. doi: 10.1373/clinchem.2003.026161. [DOI] [PubMed] [Google Scholar]
- 55.Stahlberg A, Kubista M, Pfaffl M. Comparison of reverse transcriptases in gene expression analysis. Clin Chem. 2004;50:1678–80. doi: 10.1373/clinchem.2004.035469. [DOI] [PubMed] [Google Scholar]
- 56.Lewis FMN. Extraction of Total RNa from Formalin-Fixed Paraffin-Embedded Tissue. IUL Press; La Jolla, California: 2004. [Google Scholar]
- 57.Lekanne Deprez RH, et al. Sensitivity and accuracy of quantitative real-time polymerase chain reaction using SYBR green I depends on cDNA synthesis conditions. Anal Biochem. 2002;307:63–9. doi: 10.1016/s0003-2697(02)00021-0. [DOI] [PubMed] [Google Scholar]
- 58.Vandesompele J, De Paepe A, Speleman F. Elimination of primer-dimer artifacts and genomic coamplification using a two-step SYBR green I real-time RT-PCR. Anal Biochem. 2002;303:95–8. doi: 10.1006/abio.2001.5564. [DOI] [PubMed] [Google Scholar]
- 59.Simon R, et al. Design and analysis of DNA microarray investigations. Springer; New York: 2004. [Google Scholar]
- 60.Murphy RM, et al. Effects of creatine supplementation on housekeeping genes in human skeletal muscle using real-time RT-PCR. Physiol Genomics. 2003;12:163–74. doi: 10.1152/physiolgenomics.00060.2002. [DOI] [PubMed] [Google Scholar]
- 61.Khimani AH, et al. Housekeeping genes in cancer: normalization of array data. Biotechniques. 2005;38:739–45. doi: 10.2144/05385ST04. [DOI] [PubMed] [Google Scholar]
- 62.Warrington JA, et al. Comparison of human adult and fetal expression and identification of 535 housekeeping/maintenance genes. Physiol Genomics. 2000;2:143–7. doi: 10.1152/physiolgenomics.2000.2.3.143. [DOI] [PubMed] [Google Scholar]
- 63.Ross DT, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–35. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 64.Thellin O, et al. Housekeeping genes as internal standards: use and limits. J Biotechnol. 1999;75:291–5. doi: 10.1016/s0168-1656(99)00163-7. [DOI] [PubMed] [Google Scholar]
- 65.Suzuki T, Higgins PJ, Crawford DR. Control selection for RNA quantitation. Biotechniques. 2000;29:332–7. doi: 10.2144/00292rv02. [DOI] [PubMed] [Google Scholar]
- 66.Bustin SA. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. J Mol Endocrinol. 2000;25:169–93. doi: 10.1677/jme.0.0250169. [DOI] [PubMed] [Google Scholar]
- 67.Aerts JL, Gonzales MI, Topalian SL. Selection of appropriate control genes to assess expression of tumor antigens using real-time RT-PCR. Biotechniques. 2004;36:84–6. 88, 90–1. doi: 10.2144/04361ST04. [DOI] [PubMed] [Google Scholar]
- 68.Biederman J, Yee J, Cortes P. Validation of internal control genes for gene expression analysis in diabetic glomerulosclerosis. Kidney Int. 2004;66:2308–14. doi: 10.1111/j.1523-1755.2004.66016.x. [DOI] [PubMed] [Google Scholar]
- 69.Tsuji N, et al. Selection of an internal control gene for quantitation of mRNA in colonic tissues. Anticancer Res. 2002;22:4173–8. [PubMed] [Google Scholar]
- 70.Gorzelniak K, et al. Validation of endogenous controls for gene expression studies in human adipocytes and preadipocytes. Horm Metab Res. 2001;33:625–7. doi: 10.1055/s-2001-17911. [DOI] [PubMed] [Google Scholar]
- 71.Vandesompele J, et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:34. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mamo S, et al. Quantitative evaluation and selection of reference genes in mouse oocytes and embryos cultured in vivo and in vitro. BMC Dev Biol. 2007;7:14. doi: 10.1186/1471-213X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mahoney DJ, et al. Real-time RT-PCR analysis of housekeeping genes in human skeletal muscle following acute exercise. Physiol Genomics. 2004;18:226–31. doi: 10.1152/physiolgenomics.00067.2004. [DOI] [PubMed] [Google Scholar]
- 74.Pogue-Geile KL, Greenberger JS. Effect of the irradiated microenvironment on the expression and retrotransposition of intracisternal type A particles in hematopoietic cells. Exp Hematol. 2000;28:680–9. doi: 10.1016/s0301-472x(00)00165-x. [DOI] [PubMed] [Google Scholar]
- 75.Barnard GF, et al. Increased expression of human ribosomal phosphoprotein P0 messenger RNA in hepatocellular carcinoma and colon carcinoma. Cancer Res. 1992;52:3067–72. [PubMed] [Google Scholar]
- 76.Henry JL, Coggin DL, King CR. High-level expression of the ribosomal protein L19 in human breast tumors that overexpress erbB-2. Cancer Res. 1993;53:1403–8. [PubMed] [Google Scholar]
- 77.Vaarala MH, et al. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int J Cancer. 1998;78:27–32. doi: 10.1002/(sici)1097-0215(19980925)78:1<27::aid-ijc6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 78.Xu LL, et al. Quantitative expression profile of PSGR in prostate cancer. Prostate Cancer Prostatic Dis. 2006;9:56–61. doi: 10.1038/sj.pcan.4500836. [DOI] [PubMed] [Google Scholar]
- 79.Morrison T, et al. Nanoliter high throughput quantitative PCR. Nucleic Acids Res. 2006;34 :e123. doi: 10.1093/nar/gkl639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Fink L, et al. Real-time quantitative RT-PCR after laser-assisted cell picking. Nat Med. 1998;4:1329–33. doi: 10.1038/3327. [DOI] [PubMed] [Google Scholar]
- 81.Tsai WJ, et al. Real-Time PCR Quantification Using Cloned Standards and Multiple Housekeeping Genes. Protocol Online. 2007 [Google Scholar]
- 82.Fend F, et al. Immuno-LCM: laser capture microdissection of immunostained frozen sections for mRNA analysis. Am J Pathol. 1999;154:61–6. doi: 10.1016/S0002-9440(10)65251-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tangrea MA, et al. Expression microdissection: operator-independent retrieval of cells for molecular profiling. Diagn Mol Pathol. 2004;13:207–12. doi: 10.1097/01.pdm.0000135964.31459.bb. [DOI] [PubMed] [Google Scholar]
- 84.Grover AC, et al. Tumor-associated endothelial cells display GSTP1 and RARbeta2 promoter methylation in human prostate cancer. J Transl Med. 2006;4:13. doi: 10.1186/1479-5876-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hanson JA, et al. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255–61. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 86.Fink L, et al. Laser-microdissection for cell type- and compartment-specific analyses on genomic and proteomic level. Exp Toxicol Pathol. 2006;57 (Suppl 2):25–9. doi: 10.1016/j.etp.2006.02.010. [DOI] [PubMed] [Google Scholar]
- 87.Wilhelm J, et al. Systematic comparison of the T7-IVT and SMART-based RNA preamplification techniques for DNA microarray experiments. Clin Chem. 2006;52:1161–7. doi: 10.1373/clinchem.2005.062406. [DOI] [PubMed] [Google Scholar]
- 88.Fink L, et al. cDNA array hybridization after laser-assisted microdissection from nonneoplastic tissue. Am J Pathol. 2002;160:81–90. doi: 10.1016/S0002-9440(10)64352-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chenchik A, et al. Gene Cloning and Analysis by RT-PCR. 98 BioTechniques Books; Westborough, MA: Generation and use of high-quality cDNA from small amounts of total RNA byu SMART PCR. [Google Scholar]
- 90.Brenan CJ, Morrison T. High throughput, nanoliter quantitative PCR. Drug Discovery Today: Technologies. 2005;2:247–253. doi: 10.1016/j.ddtec.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 91.Zhang A, et al. Small interfering RNA and gene expression analysis using a multiplex branched DNA assay without RNA purification. J Biomol Screen. 2005;10:549–56. doi: 10.1177/1087057105277414. [DOI] [PubMed] [Google Scholar]
- 92.Canales RD, et al. Evaluation of DNA microarray results with quantitative gene expression platforms. Nat Biotechnol. 2006;24:1115–22. doi: 10.1038/nbt1236. [DOI] [PubMed] [Google Scholar]
- 93.Schmid H, et al. Validation of endogenous controls for gene expression analysis in microdissected human renal biopsies. Kidney Int. 2003;64:356–60. doi: 10.1046/j.1523-1755.2003.00074.x. [DOI] [PubMed] [Google Scholar]
- 94.Cohen CD, et al. Quantitative gene expression analysis in renal biopsies: a novel protocol for a high-throughput multicenter application. Kidney Int. 2002;61:133–40. doi: 10.1046/j.1523-1755.2002.00113.x. [DOI] [PubMed] [Google Scholar]
- 95.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 96.Guidi CJ, et al. Functional interaction of the retinoblastoma and Ini1/Snf5 tumor suppressors in cell growth and pituitary tumorigenesis. Cancer Res. 2006;66:8076–82. doi: 10.1158/0008-5472.CAN-06-1451. [DOI] [PubMed] [Google Scholar]
- 97.King TA, et al. Heterogenic loss of the wild-type BRCA allele in human breast tumorigenesis. Ann Surg Oncol. 2007;14:2510–8. doi: 10.1245/s10434-007-9372-1. [DOI] [PubMed] [Google Scholar]