Abstract
Objective
The cells of the immune system originate from the bone marrow (BM), where many of them also mature. To better understand the aberrant immune response in systemic lupus erythematosus (SLE), we examined the BM in lupus patients using DNA microarrays and compared it to the peripheral blood (PB).
Patients and Methods
Bone marrow mononuclear cells (BMMCs) from 20 SLE patients (11 with active disease and 9 with inactive disease) and peripheral blood mononuclear cells (PBMCs) from 27 patients (16 active/ 11 inactive); BMMCs and PBMCs from 7 healthy individuals and 3 osteoarthritis patients served as controls. Samples were analyzed on genome-scale microarrays with 21,329 genes represented.
Results
We found 102 differentially expressed genes between patients’ and controls’ BMMCs (unpaired student t-test), involved in various biologic processes; 53 of them are involved in major networks including cell death, growth, signaling and proliferation. Comparative analysis between BM and PB of patients identified 88 genes differentially expressed; 61 out of 88 participate in cell growth and differentiation, cellular movement and morphology, immune response and other hematopoietic cell functions. Unsupervised clustering of highly expressed genes revealed two major SLE patient clusters (active and inactive) in BM, but not in PB. The upregulated genes in the bone marrow of active patients included genes involved in cell death and granulopoiesis.
Conclusion
Microarray analysis of the bone marrow differentiates active from inactive lupus patients and provides further evidence for the role of apoptosis and granulocytes in the pathogenesis of the disease.
Systemic lupus erythematosus (SLE) is the prototypic systemic autoimmune disease characterized by the production of autoantibodies to components of the cell nucleus in association with diverse clinical manifestations encompassing almost all organ systems. Although its etiology is not established, much is known about the pathogenic pathways that lead to tissue injury (1, 2)
Bone marrow is a central lymphoid organ consisting of various types of hemopoietic and non- hemopoietic cells and the “stroma” that supports their growth, differentiation and function, collectively called as “the BM micro-environment” (3) In addition to production, maturation and activation of neutrophils, monocytes/macrophages and B cells, BM has a central role in regulating the immune response (4, 5). In lupus patients, bone marrow exhibits a variety of histopathologic findings including necrosis, stromal alterations and abnormal localization of immature precursors (3, 6)
Microarray analysis is a broad-based profiling method that permits the concomitant comparison of gene expression profiles among different study groups revealing active networks of interrelated genes within subpopulations under study (7–11). In SLE these studies have shown interferon-inducible and granulopoiesis signatures that correlate with both disease severity (12) and activity (13). Several IFN related genes were found highly overexpressed in the peripheral blood and kidney glomeruli of lupus patients (14). These data have provided important insights into genetic pathways underlying SLE, as well as the effector cells and molecules involved in its pathogenesis.
In view of the central role of BM in regulating the immune response, we sought to explore bone marrow gene expression profiles using the microarrays in lupus patients and healthy individuals and compare it to the peripheral blood.
PATIENTS AND METHODS
Patients and controls
Twenty seven patients with SLE - followed by the Rheumatology Department of the University Hospital of Crete, a tertiary referral center- were studied following written informed consent. All bone marrow samples were obtained from patients that provided peripheral blood. All patients met the 1982 American College of Rheumatology revised criteria for the classification of SLE (15). In order to capture patients with higher disease activity, we used a SLE Disease Activity Index score cut-off of (SLEDAI) ≥8. Clinical and laboratory characteristics of the patients included in the study are summarized in Table 1. Seven patients had active proliferative and/ or membranous nephritis, while six had active neuropsychiatric lupus with manifestations such as psychosis, major depression, myelitis and polyneuropathy. Patients had not received steroids for at least 24 hours before blood and bone marrow was obtained for study. Controls enrolled in the study included seven healthy individuals and three osteoarthritis patients (5 males and 5 females, age ranging from 35–55 years) from the Department of Transfusion Medicine, Hematology Clinic and Rheumatology Clinic of University Hospital of Crete.
Table 1.
Clinical and demographic characteristics of SLE patients *
| Sex, female/male | 26 /1 |
| Age, mean ± SD | 47.28 ± 17.2 |
| Active / Inactive | 16 (59%) / 11 (41%) |
| SLE duration, mean ± SD | 6.7 ±5.6 |
| SLEDAI (mean ± SD) | |
| Active SLE | 13.86 ± 4.99 |
| Inactive SLE | 4.22 ± 1.56 |
| Nephritis | |
| Active SLE | 7 / 16 |
| Inactive SLE | 0 /11 |
| Total | 7 / 27 |
| CNS | |
| Active SLE | 6 / 16 |
| Inactive SLE | 0 /11 |
| Total | 6 / 27 |
| Cytotoxic therapy | |
| Active SLE | 4 / 16 |
| Inactive SLE | 1 / 11 |
| Total | 5 / 27 |
| Steroids | |
| Active SLE | 9 / 16 |
| Inactive SLE | 3 / 11 |
| Total | 12 / 27 |
| Hydroxychloroquine | |
| Active SLE | 10 / 16 |
| Inactive SLE | 3 / 11 |
| Total | 13 / 27 |
Active SLE was defined as an SLE Disease Activity Index score of ≥8 and inactive as SLEDAI < 8. Controls included 10 healthy bone marrow donors (5 men and 5 women), 35–55 years of age.
Processing of peripheral blood mononuclear cells
(PBMCs) and bone marrow mononuclear cells (BMMCs) and RNA extraction
PBMCs and BMMCs from lupus patients or healthy volunteers were isolated by Ficoll-Histopaque (Sigma-Aldrich, St. Louis, MO) density-gradient centrifugation of heparinized venous blood and bone marrow aspirates immediately after peripheral blood and bone marrow draw. Cells were immediately placed in Trizol (Invitrogen, Carlsbad, CA, USA) and processed for RNA extraction using the RNeasy kit (Qiagen) according to the manufacturer’s instructions. RNA integrity was assessed using capillary gel electrophoresis (Agilent 2100 BioAnalyzer, Agilent Technologies, Santa Clara CA) to determine the ratio of 28s:18s rRNA in each sample. A threshold of 1.0 was used to define samples of sufficient quality and only samples above this limit were used for microarray studies.
cDNA synthesis
cDNA was synthesized using Omniscript reverse transcriptase (Qiagen) with direct incorporation of Cy3-dUTP from 2 ug of RNA. Labeled cDNA was purified using a Montage 96-well vacuum system (Millipore). The cDNA was added to hybridization buffer containing human CoT-1 DNA (0.5 mg/ml final concentration), yeast tRNA (0.2 mg/ml), and poly(dA)40–60 (0.4 mg/ml).
Microarrays
A commercially available genome-scale oligonucleotide library containing gene-specific 70 mer oligonucleotides representing 21,329 human genes was used for microarray production. The library includes 16 replicate spots of 12 random negative controls that are designed to have no significant homology to known human DNA sequences (Qiagen Inc., Valencia, CA, USA). Oligonucleotides were spotted onto Corning UltraGAPS™ amino-silane coated slides, which were then rehydrated with water vapor and then, snap dried at 90°C. Oligonucleotide DNAs were covalently fixed to the surface of the glass using 300 mJ of ultraviolet radiation at a 254nm wavelength. Unbound, free amines on the glass surface were blocked for 15 min with moderate agitation in a solution of 143mM succinic anhydride dissolved in 1-methyl-2-pyrolidinone, 20mM sodium borate, pH 8.0. Slides were rinsed for 2 min in distilled water, immersed for 1 min in 95% ethanol and dried with a stream of nitrogen gas. Hybridization was performed in an automated liquid delivery, air-vortexed, hybridization station for 9 h at 58°C under an oil based cover slip (Ventana Medical Systems Inc., Tucson, AZ, USA). Microarrays were washed at a final stringency of 0.1X SSC. Microarrays were scanned using a simultaneous dual-colour, 48-slide scanner (Agilent Technologies). Background substracted luorescent intensity values were determined using Kaodarray software (Koada Technology, Stirling, UK)
Statistical analysis
a)Normalization: The R/Bioconductor Package “Affy” was used to perform quantile normalization to adjust the marginal distribution of each sample. b) Filtering: Genes that had an average background adjusted fluorescent intensity value > 50 across all arrays were retained in the analysis. Additionally, the variance across all genes was calculated. Genes that have a variance below the median variance are unlikely to be differentially expressed and are therefore removed from further analysis. c) Class Comparison: Genes that are differentially expressed between two classes were identified through an unpaired Student’s t test. A 10% false discovery rate p-value multiplicity adjustment was used. The false discovery rate is the proportion of the list of genes claimed to be differentially expressed that are false positives. Only statistically significant differentially expressed genes with greater than a 2- fold change in expression between groups were retained. d) SLEDAI Modeling: The SLEDAI score was modelled as a continuous variable according to the Generalized Linear Model (GLM) equation: Log2 (Expression) = B1*SLEDAI + B2*Disease_State + B3*SLEDAI * Disease_State + Intercept, where Disease_State is a categorical indicator variable for Active or Inactive Disease, and B# are beta coefficients. A 10% false discovery rate was used on the SLEDAI term to determine statistical significnce. All analysis was performed in JMP Genomics 6.0.3 (Cary, NC). e) SLEDAI Modeling (PBMC): The SLEDAI score was modeled as a continuous variable according to the GLM equation: SLEDAI = B1*Gene X + B2 * Gene Y + … + BN * Gene N, where B1 through BN are beta coefficients and Gene X through Gene N are log base 2 normalized expression values. Multivariate models were created using only genes identified to be significantly associated to SLEDAI in the BM comparison f) Granulopoiesis Score: The score was created by taking genes known to be associated with granulopoiesis from the literature (LYZ, CD63, DEFA4, ELA2, S100A8, S100A12,S100P, CD24, NCF4) and adding their log expression values together. This resulted in a score that was used to correlate this plurality of genes vs SLEDAI through a GLM.
Real-time PCR validation
Nine genes were selected out of the set related to granulopoeisis for confirmation by PCR: S100A8, DEFA4, S100A12, ELA2, S100P, CD24, CD63, LYZ, NCF4. Eight patients were selected, four that represented patients in the "active" group and four that represented patients in the "inactive" group according to SLEDAI.
Reverse Transcription: cDNA was generated from 1.0 ug of total RNA per sample according to the OmniScript Reverse Transcriptase (Qiagen, Valencia, CA) manual, with the replacement of the RT primer mix with for 500ng anchored oligo dT(dT20VN). cDNA was purified with the Montage PCR Cleanup kit (Millipore, Billerica, MA) according to manufacturer's instructions. cDNA was diluted 1:20 in water and stored at −20° C.
Quantitative PCR: Gene-specific primers for the human genes S100A8, DEFA4, S100A12, ELA2, S100P, CD24, CD63, LYZ, NCF4 were designed with a melting temperature close to 60°C length of 19–25 bp for PCR products with a length of 110–150 bp, using Applied Biosystems Inc.(ABI) Primer Express 1.5 software. PCR was run with 2 ul cDNA template in 15ul reactions in triplicate on an ABI SDS 7700 using the ABI SYBR Green I Master Mix and gene specific primers at a concentration of 1uM each. The temperature profile consisted of an initial 95° C step for 10 minutes (for Taq activation), followed by 40 cycles of 95° C for 15 sec, 60° C for 1 min, and then a final melting curve analysis with a ramp from 60° C to 95° C over 20 min. Gene-specific amplification was confirmed by a single peak in the ABI Dissociation Curve software. No template controls were run for each primer pair and no RT controls were run for each sample to detect nonspecific amplification or primer dimers. Average Ct values for B-Actin (run in parallel reactions to the gene of interest) were used to normalize average Ct values of the gene of interest. These values were used to calculate the average group (active vs inactive) and the relative change in Ct was used to calculate fold change between the two groups.
RESULTS
Differentially expressed genes in the bone marrow of SLE patients vs controls
A total of 102 genes were found to have differential levels of expression between the SLE patients and the control subjects using unpaired student t-test. Of the 102 differentially expressed genes, 53 genes are involved in major networks including cell death, differentiation, cell signaling and cellular growth and proliferation (Table 2A). Data mining was performed to identify genes that were expressed in various subpopulations of BMMCs including B and T cells, monocytes and neutrophils. Of the 102 differentially expressed genes, 37 were up-regulated in the bone marrow of patients relative to controls including: TNFR17 (Tumor necrosis factor receptor superfamily, member 17) usually expressed in mature B lymphocytes and may be important for B cell development and autoimmune response. B cell involvement was highlighted by the presence of genes involved in the antigen presentation pathway such as, HLA-F (major histocompatibility complex, class I, F), and IGHG3 (Immunoglobulin heavy constant gamma 3). We found ITPR1, belonging to the family of Inositol 1,4,5-trisphosphate receptors (IP3Rs) which are expressed in most hematopoietic cells, including B cells, upregulated in the bone marrow of patients as well as a transcriptional co-activator BCL3 (B-cell CLL/lymphoma 3) reported to be upregulated in polyclonal plasmablastic cells(16). In mouse bone marrow, transgenic human BCL3 protein increases accumulation of mature B lymphocytes (17).
Table 2.
A. Selected up- and down-regulated genes in the bone marrow of SLE patients relative to controls. B. Selected upregulated genes in BMMCs relative to PBMCs of lupus patients
| A. | |||||
|---|---|---|---|---|---|
| Name | Description upregulated genes in BM |
Genbank | Normalized ratio | Location | Family |
| BCL3 | B-cell CLL/lymphoma 3 | NM_005178 | 4,24 | Nucleus | transcription regulator |
| ITPR1 | inositol 1,4,5-triphosphate receptor, type 1 | NM_002222 | 3,388 | Cytoplasm | ion channel |
| RPL32 | ribosomal protein L32 | NM_000994 | 2,883 | Cytoplasm | other |
| PHACTR1 | phosphatase and actin regulator 1 | AB051520 | 2,311 | Cytoplasm | other |
| RPS2 | ribosomal protein S2 | NM_002952 | 2,258 | Cytoplasm | other |
| RPS13 | ribosomal protein S13 | NM_001017 | 2,094 | Cytoplasm | other |
| ABCG2 | ATP-binding cassette, sub-family G (WHITE), member 2 | NM_004827 | 2,062 | Plasma Membrane | transporter |
| EPS15L1 | epidermal growth factor receptor pathway substrate 15-like 1 | AK023744 | 2,056 | Plasma Membrane | other |
| ABLIM1 | actin binding LIM protein 1 | NM_002313 | 1,987 | Cytoplasm | other |
| HLA-F | major histocompatibility complex, class I, F | NM_018950 | 1,904 | Plasma Membrane | transmembrane receptor |
| NPTX1 | neuronal pentraxin I | NM_002522 | 1,85 | Extracellular Space | other |
| TNFRSF17 | tumor necrosis factor receptor superfamily, member 17 | NM_001192 | 1,837 | Plasma Membrane | other |
| ADD3 | adducin 3 (gamma) | U92992 | 1,781 | Cytoplasm | other |
| TEAD2 | TEA domain family member 2 | BC007556 | 1,735 | Nucleus | transcription regulator |
| BAIAP3 | BAI1-associated protein 3 | NM_003933 | 1,726 | Unknown | other |
| PRKAA2 | protein kinase, AMP-activated, alpha 2 catalytic subunit | NM_006252 | 1,641 | Cytoplasm | kinase |
| GMFG | glia maturation factor, gamma | NM_004877 | 1,613 | Cytoplasm | growth factor |
| BACE1 | beta-site APP-cleaving enzyme 1 | NM_012104 | 1,505 | Cytoplasm | peptidase |
| TMEPAI | transmembrane, prostate androgen induced RNA | AF305616 | 1,481 | Plasma Membrane | other |
| K-ALPHA-1 | alpha tubulin | NM_006082 | 1,165 | Cytoplasm | other |
| downregulated in BM | |||||
| PRKD1 | protein kinase D1 | NM_002742 | 4,724 | Cytoplasm | kinase |
| CCR5 | chemokine (C-C motif) receptor 5 | NM_000579 | 3,877 | Plasma Membrane | G-protein coupled receptor |
| CRHR1 | corticotropin releasing hormone receptor 1 | X72304 | 3,387 | Plasma Membrane | G-protein coupled receptor |
| GJB3 | gap junction protein, beta 3, 31kDa (connexin 31) | NM_024009 | 3,376 | Plasma Membrane | transporter |
| GUCY2D | guanylate cyclase 2D, membrane (retina-specific) | NM_000180 | 3,32 | Plasma Membrane | kinase |
| MPHOSPH1 | M-phase phosphoprotein 1 | NM_016195 | 3,023 | Nucleus | enzyme |
| GAP43 | growth associated protein 43 | NM_002045 | 2,938 | Plasma Membrane | other |
| MBP | myelin basic protein | NM_002385 | 2,901 | Extracellular Space | other |
| PCLO | piccolo (presynaptic cytomatrix protein) | AB011131 | 2,895 | Cytoplasm | transporter |
| MYH10 | myosin, heavy polypeptide 10, non-muscle | AK026977 | 2,786 | Cytoplasm | other |
| AEBP2 | AE binding protein 2 | BC015624 | 2,7 | Nucleus | transcription regulator |
| PAX6 | paired box gene 6 (aniridia, keratitis) | NM_001604 | 2,646 | Nucleus | transcription regulator |
| MAG | myelin associated glycoprotein | NM_002361 | 2,541 | Plasma Membrane | other |
| USP33 | ubiquitin specific peptidase 33 | AB029020 | 2,407 | Cytoplasm | peptidase |
| ACE | angiotensin I converting enzyme (peptidyl-dipeptidase A) 1 | NM_000789 | 2,385 | Plasma Membrane | peptidase |
| CX3CR1 | chemokine (C-X3-C motif) receptor 1 | U20350 | 2,334 | Plasma Membrane | G-protein coupled receptor |
| OTP | orthopedia homolog (Drosophila) | NM_032109 | 2,27 | Nucleus | transcription regulator |
| SRGAP1 | SLIT-ROBO Rho GTPase activating protein 1 | AB037725 | 2,227 | Unknown | other |
| RGS11 | regulator of G-protein signalling 11 | NM_003834 | 2,176 | Plasma Membrane | enzyme |
| HOXB3 | homeobox B3 | U59298 | 2,171 | Nucleus | transcription regulator |
| SLAMF6 | SLAM family member 6 | NM_052931 | 2,164 | Plasma Membrane | transmembrane receptor |
| SSX2IP | synovial sarcoma, X breakpoint 2 interacting protein | NM_014021 | 2,076 | Unknown | other |
| CDH2 | cadherin 2, type 1, N-cadherin (neuronal) | NM_001792 | 2,064 | Plasma Membrane | other |
| DUSP4 | dual specificity phosphatase 4 | NM_057158 | 2,015 | Nucleus | phosphatase |
| GRB2 | growth factor receptor-bound protein 2 | NM_002086 | 1,938 | Plasma Membrane | other |
| EIF4EBP2 | eukaryotic translation initiation factor 4E binding protein 2 | AK057643 | 1,82 | Cytoplasm | other |
| CD276 | CD276 molecule | NM_025240 | 1,813 | Plasma Membrane | other |
| PRKCG | protein kinase C, gamma | NM_002739 | 1,768 | Cytoplasm | kinase |
| C10ORF10 | chromosome 10 open reading frame 10 | NM_007021 | 1,662 | Unknown | other |
| CTNNAL1 | catenin (cadherin-associated protein), alpha-like 1 | NM_003798 | 1,65 | Plasma Membrane | other |
| MAGI1 | membrane associated guanylate kinase, WW and PDZ domain containing 1 | AK023358 | 1,635 | Plasma Membrane | kinase |
| CDKN3 | cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | NM_005192 | 1,426 | Nucleus | phosphatase |
| SNX2 | sorting nexin 2 | NM_003100 | 1,306 | Cytoplasm | transporter |
| B. | ||||
|---|---|---|---|---|
| Symbol | Short Description | Normalized ratio | Location | Family |
| CTSG | Cathepsin G | 6,65 | cytoplasm | peptidase |
| HBD | Hemoglobin, delta | 5,84 | cytoplasm | transporter |
| ELA2 | Elastase 2, neutrophil | 4,49 | extracellular space | peptidase |
| MPO | Myeloperoxidase | 3,3 | cytoplasm | enzyme |
| S100A9 | S100 calcium binding protein A9 (calgranulin B) | 2,83 | cytoplasm | other |
| NR2C2 | Nuclear receptor subfamily 2, group C, member 2 | 2,58 | nucleus | ligand-dependent nuclear receptor |
| PRDX2 | Peroxiredoxin 2 | 2,41 | cytoplasm | enzyme |
| S100A12 | S100 calcium binding protein A12 (calgranulin C) | 2,41 | cytoplasm | other |
| PPIB | Peptidylprolyl isomerase B (cyclophilin B) | 2,34 | cytoplasm | enzyme |
| ITPR1 | Inositol 1,4,5-triphosphate receptor, type 1 | 2,23 | cytoplasm | ion channel |
| STAG3 | Stromal antigen 3 | 2,2 | nucleus | other |
| CRMP1 | Collapsin response mediator protein 1 | 2,18 | cytoplasm | enzyme |
| HP | Haptoglobin | 2,15 | extracellular space | peptidase |
| SDHA | Succinate dehydrogenase complex, subunit A, flavoprotein (Fp) | 2,14 | cytoplasm | enzyme |
| PSCD1 | Pleckstrin homology, Sec7 and coiled/coil domains 1(cytohesin 1) | 2,02 | cytoplasm | other |
| K-ALPHA-1 | Tubulin, alpha, ubiquitous | 2,01 | cytoplasm | other |
| PGLYRP | Peptidoglycan recognition protein | 1,96 | plasma membrane | transmembrane receptor |
| S100P | S100 calcium binding protein P | 1,95 | cytoplasm | other |
| LTF | Lactotransferrin | 1,91 | extracellular space | peptidase |
| ATP7A | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | 1,86 | plasma membrane | transporter |
| LCN2 | Lipocalin 2 (oncogene 24p3) | 1,78 | extracellular space | transporter |
| SNCA | Synuclein, alpha (non A4 component of amyloid precursor) | 1,77 | cytoplasm | other |
| DUSP4 | Dual specificity phosphatase 4 | 1,74 | nucleus | phosphatase |
| CSPG4 | Chondroitin sulfate proteoglycan 4 (melanoma-associated) | 1,7 | plasma membrane | other |
| PHKA1 | Phosphorylase kinase, alpha 1 (muscle) | 1,64 | cytoplasm | kinase |
| DLC1 | Deleted in liver cancer 1 | 1,56 | cytoplasm | other |
| UQCRC2 | Ubiquinol-cytochrome c reductase core protein II | 1,54 | cytoplasm | enzyme |
| CD24 | CD24 antigen (small cell lung carcinoma cluster 4 antigen) | 1,41 | plasma membrane | other |
| CDKN3 | Cyclin-dependent kinase inhibitor 3 (CDK2-associated dual specificity phosphatase) | 1,34 | nucleus | phosphatase |
Sixty five (65 genes) were expressed at lower levels in patients than controls including the chemokine receptors CX3CR1 and CCR5, the latter is normally expressed by T cells and macrophages. Also downregulated were CDH2 (Cadherin 2), CTNNAL1 (Catenin,cadherin-associated protein), CDKN (Cyclin-dependent kinase inhibitor 3) and KALI (Activating NK receptor) whose protein is expressed on Natural Killer, T and B lymphocytes and lung. CD276 (B7-H3) a costimulatory molecule for T cell activation and IFN-gamma production was also found downregulated in the bone marrow of patients.
Bone marrow genes associated with SLE disease activity
By the use of multiple regression analysis, as outlined in Materials and Methods, seven genes were statistically associated with SLEDAI. KIAA1674 (GenBank AB051461) (r2 =0.82), NY-REN-25 antigen, an ankyrin repeat domain (GenBank AF155103) (r2=0.81), cDNA FLJ32586 fis (GenBank AK057148) (r2 = 0.79), the hypothetical protein FLJ10254 (GenBank NM_018041) (r2 = 0.83) and the coiled coil domain CCDC91 (GenBank NM_018318) (r2 = 0.84), CENPH (Centromere protein H, GenBank NM_022909) (r2 =0.79) and EBI3 (Epstein-Barr virus induced gene 3, GenBank NM_005755)) (r2 = 0.77) a subunit of IL-27 which may play an important role in initiation of Th1 responses. The expression of these genes was highly correlated with one another producing Pearson correlation coefficients of 0.89 to 0.97. Due to co-linearity concerns we were not able to combine these terms into a single multivariate model.
In order to test if these 7 SLEDAI-associated bone marrow expressed genes were also associated with SLEDAI in the peripheral blood, we re-analyzed the data and used the genes selected in the BM and refitted the terms to the PBMC data to predict SLEDAI. Only 2 genes, NY-REN-25 antigen, an ankyrin repeat domain (GenBank AF155103) and coiled coil domain CCDC91 (GenBank NM_018318) associated with SLEDAI in the active PBMC patients (r2=0.37, p=0.0108) (Figure 1).
Figure 1. SLEDAI-associated bone marrow expressed genes were also associated with SLEDAI in the periphery.
Two genes, NY-REN-25 antigen, an ankyrin repeat domain (GenBank AF155103) and coiled coil domain CCDC91 (GenBank NM_018318) associated with SLEDAI in active PBMC patients thorugh a GLM (r2=0.37, P=0.0108). The Y Axis represents the predicted SLEDAI from our model of 2 genes and the X axis is the observed SLEDAI from the patient’s medical records.
Differentially expressed genes in BMMCs relative to PBMCs of SLE patients
We next compared bone marrow derived mononuclear cells with peripheral blood mononuclear cells in the lupus cohort. Eighty eight (88) genes were differentially expressed, 41 out of 88 were up-regulated in the bone marrow of lupus patients relative to the peripheral blood (Table 2B) while the remaining 47 were up-regulated in the peripheral blood. Among the lupus bone marrow up-regulated genes, the highest overexpression was found in granulopoiesis- related genes. These genes include major components of neutrophils such as myeloperoxidase MPO, ELA2 (elastase 2) responsible for hydrolyzing proteins within granules, CTSG (cathepsin G), DEFA4 (defensin), LTF (lactotransferrin) and CD24 found on mature granulocytes. Three small abundant proteins found in human neutrophil cytosol S100A9, S100A12 and S100P were upregulated in the bone marrow of SLE patients. Finally, in the peripheral blood of lupus patients we identified a number of chemokines such as CCR5, CXCL3L1, CXCL2 and CXCL3 that are overexpressed relative to the bone marrow. These genes participate in processes such as chemotaxis and migration of leukocytes. Comparison of bone marrow versus peripheral blood in the control cohort revealed that most of the neutrophil related genes found in the previous comparison, were also overexpressed in the bone marrow of control subjects relative to the peripheral blood with minor differences in gene expression level (data not shown). For example, cathepsin CTSG was overexpressed by 7.3-fold in bone marrow of controls relative to peripheral blood and by 6.6-fold in the bone marrow of lupus patients when compared to the peripheral blood. Thus many of these genes are tissue associated differences rather than disease-associated differences.
Differentially expressed genes in the peripheral blood of SLE patients vs controls
Among the SLE upregulated genes in the PBMCs two IFN-inducible genes were overexpressed in lupus patients: IL6R whose expression is regulated by IFNα and PRKCG (protein kinase C, gamma) that is involved in antiviral response of IFNγ and its signaling (data not shown). In total 35 genes were upregulated in SLE peripheral blood including a number of regulatory molecules such as: TCF7 (transcription factor 7, T-cell specific), CYC1 (cytochrome c-1), UBTF (upstream binding transcription factor), HDAC10 (histone deacetylase 10) involved in the acetylation status of histone tails, a ubiquitin-conjugating enzyme UBE2D3 and U2AF1 ( U2(RNU2) small nuclear RNA auxillary factor 1) belonging to the splicing factor SR family of genes. The expression of 18 genes was downregulated in the PBMCs of SLE patients compared to controls including: BACE (beta-site APP-cleaving enzyme), HOXD13 (homeo box D13), K-ALPHA-1 (tubulin), PHKA1 (phosphorylase kinase,alpha) and PRKAA2 (protein kinase AMP-activated).
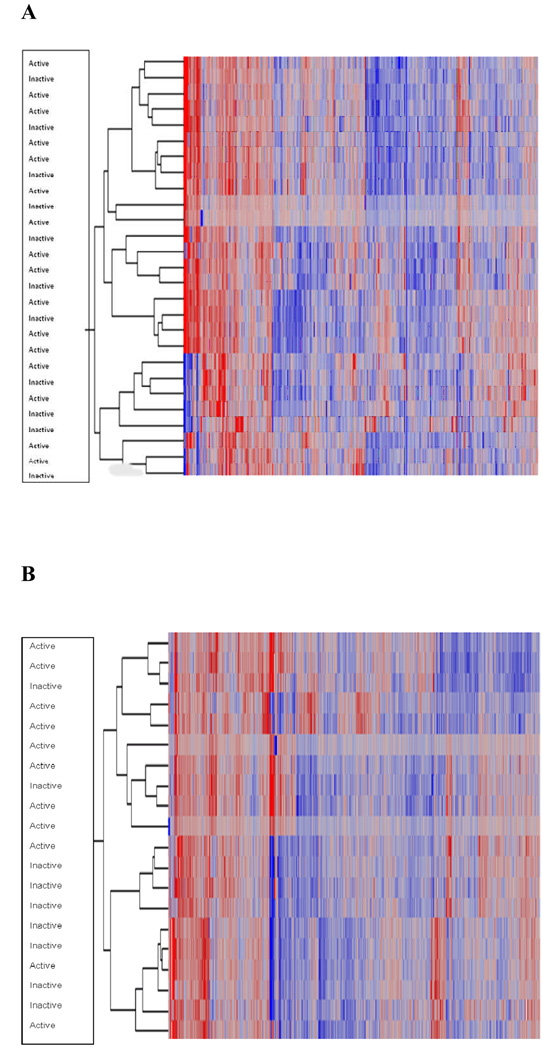
Hierarchical clustering reveals patient subgroups in the bone marrow
In order to group individuals with similar expression profiles in their peripheral blood mononuclear cells (PBMCs) and bone marrow mononuclear cells (BMMCs), we used unsupervised hierarchical clustering of the top 25% of expressed genes (n= 2652) in each group separately. Lupus samples derived from peripheral blood did not form distinct, well-characterized clusters but scattered across the graph regardless of disease activity (Figure 2A). In contrast, hierarchical clustering in the bone marrow demonstrated that lupus patients fell into two groups that displayed different pattern of expression for these 2652 genes (Figure 2B) and the controls formed another distinct cluster. Of interest, these two groups were primarily active patients in one cluster and inactive patients in the other with 20% and 30% misclassification respectively (p=0.07, Fisher’s Exact test). These clusters were not associated with disease manifestations or drug treatment. Taken together, these findings suggest that bone marrow in SLE provides supplemental information to that obtained from peripheral blood in the context of disease activity.
Figure 2. Hierarchical clustering of the top 25% (2652) of highly expressed genes in the peripheral blood and bone marrow of SLE patients.
Each column represents a gene and each row shows the expression for the top 25% genes expressed by each individual. (A). Peripheral blood samples from SLE patients scattered across the graph regardless of disease activity. (B). Hierarchical clustering distinguish SLE active patients from inactive in the bone marrow. The first big cluster corresponds to the active patients and the second one to the inactive
Granulopoiesis and apoptosis signature in the bone marrow of active patients
To explore any gene signatures that characterize the two patient subgroups in the bone marrow, we further analyzed on a whole-genome scale the differentially expressed genes between patients in these two clusters. We found 245 differentially expressed genes that were up-regulated in the active patient cluster as compared to the inactive (Table 3). No genes were identified as significantly repressed in the active relative to inactive group. Genes involved in antigen presentation such as HLA-A, HLA-C and CD74 were expressed in higher levels in the bone marrow of active SLE patients. Among the up-regulated genes we noticed granulopoiesis-related genes such as ELA2 (elastase) which is usually transcribed within the earliest granulocytes, LYZ (lysozyme) a component of azurophil and specific granules which was overexpressed in active patients, DEFA (defensin), CD63 a marker of azurophil granules and CD24 expressed on mature granulocytes. Three family members of S100 calcium-binding proteins, S100A6, S100A8 and S100P were also upregulated in the active group. Among these three genes, S100A8 a leukocyte chemoattractant protein was overexpressed 10-fold compared to the inactive group. All genes that were upregulated on the microarrays were also upregulated when analyzed by quantitative real-time PCR. The expression values of these granulopoiesis genes correlated highly with each other. Additional up-regulated genes included genes involved in processes such as cell death, immune response and cellular movement. The genes implicated in the cell death of leukocytes include ANXA1, CD24, CST3, CXCR4, ELA2, FOS, FOXO3A, IER3, ITGB2, LCN2, LYN, and PCBP2. We also identified the genes involved in the apoptosis of granulocytes which include ANXA1 (annexin 1), chemokine CXCR4, FOXO3A, ITGB2 (integrin) and LYN.
Table 3.
Selected up-regulated genes in the bone marrow of active patients relative to inactive patients after unsupervised hierarchical clustering. Shown highlighted are selected genes involved in granulopoiesis, apoptosis and antigen presentation.
| Name | Description | Genbank | Normalized ratio | Location | Family |
|---|---|---|---|---|---|
| HSPB1 | heat shock 27kDa protein 1 | NM_001540 | 19,969 | Cytoplasm | other |
| RAP1B | RAP1B, member of RAS oncogene family | NM_015646 | 17,784 | Cytoplasm | enzyme |
| ITGB2 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | NM_000211 | 16,509 | Plasma Membrane | other |
| H3F3A | H3 histone, family 3A | M11354 | 14,347 | Nucleus | other |
| G0S2 | G0/G1switch 2 | NM_015714 | 14,192 | Unknown | other |
| MRLC2 | myosin regulatory light chain MRLC2 | NM_033546 | 14,025 | Cytoplasm | other |
| MRCL3 | myosin regulatory light chain MRCL3 | NM_006471 | 11,738 | Unknown | other |
| S100A8 | S100 calcium binding protein A8 (calgranulin A) | NM_002964 | 10,686 | Cytoplasm | other |
| GRN | granulin | NM_002087 | 10,351 | Extracellular Space | growth factor |
| LSP1 | lymphocyte-specific protein 1 | AK056576 | 8,902 | Cytoplasm | other |
| PRG1 (includes EG:55 | proteoglycan 1, secretory granule | NM_002727 | 8,773 | Extracellular Space | other |
| CD74 | CD74 molecule, major histocompatibility complex, class II invariant chain | NM_004355 | 7,637 | Plasma Membrane | transmembrane receptor |
| DDX5 | DEAD (Asp-Glu-Ala-Asp) box polypeptide 5 | NM_004396 | 7,573 | Nucleus | enzyme |
| TMSB10 | thymosin, beta 10 | NM_021103 | 6,974 | Cytoplasm | other |
| AMPH | amphiphysin (Stiff-Man syndrome with breast cancer 128kDa autoantigen) | NM_001635 | 6,745 | Plasma Membrane | other |
| HNRPA1 | heterogeneous nuclear ribonucleoprotein A1 | NM_031157 | 6,67 | Nucleus | other |
| TPT1 | tumor protein, translationally-controlled 1 | NM_003295 | 6,586 | Cytoplasm | other |
| S100A6 | S100 calcium binding protein A6 (calcyclin) | NM_014624 | 6,251 | Cytoplasm | other |
| ATP5G2 | ATP synthase, H+ transporting, mitochondrial F0 complex, subunit C2 (subunit | NM_005176 | 6,065 | Cytoplasm | transporter |
| UBE2D3 | ubiquitin-conjugating enzyme E2D 3 (UBC4/5 homolog, yeast) | NM_003340 | 5,866 | Cytoplasm | enzyme |
| TMC6 | transmembrane channel-like 6 | NM_007267 | 5,324 | Unknown | transporter |
| SF3A3 | splicing factor 3a, subunit 3, 60kDa | NM_006802 | 5,26 | Nucleus | other |
| SFRS2 | splicing factor, arginine/serine-rich 2 | NM_003016 | 5,252 | Nucleus | other |
| PAPOLA | poly(A) polymerase alpha | NM_032632 | 5,208 | Nucleus | enzyme |
| ELA2 | elastase 2, neutrophil | NM_001972 | 5,171 | Extracellular Space | peptidase |
| PFN1 | profilin 1 | NM_005022 | 5,146 | Cytoplasm | other |
| PMPCB | peptidase (mitochondrial processing) beta | NM_004279 | 5,119 | Cytoplasm | peptidase |
| SRP14 | signal recognition particle 14kDa (homologous Alu RNA binding protein) | NM_003134 | 5,034 | Cytoplasm | other |
| COTL1 | coactosin-like 1 (Dictyostelium) | BC010039 | 4,898 | Cytoplasm | other |
| ERAF | erythroid associated factor | NM_016633 | 4,881 | Cytoplasm | other |
| CST7 | cystatin F (leukocystatin) | NM_003650 | 4,701 | Extracellular Space | other |
| HLA-A | major histocompatibility complex, class I, A | NM_002116 | 4,612 | Plasma Membrane | transmembrane receptor |
| PCBP2 | poly(rC) binding protein 2 | AK023529 | 4,557 | Nucleus | other |
| PKM2 | pyruvate kinase, muscle | NM_002654 | 4,47 | Cytoplasm | kinase |
| ATP7A | ATPase, Cu++ transporting, alpha polypeptide (Menkes syndrome) | NM_000052 | 4,343 | Plasma Membrane | transporter |
| ANXA1 | annexin A1 | NM_000700 | 4,273 | Plasma Membrane | other |
| TBCA | tubulin-specific chaperone a | NM_004607 | 4,088 | Cytoplasm | other |
| ALOX5 | arachidonate 5-lipoxygenase | NM_000698 | 4,046 | Cytoplasm | enzyme |
| HP | haptoglobin | AK055872 | 4,03 | Extracellular Space | peptidase |
| LCN2 | lipocalin 2 (oncogene 24p3) | NM_005564 | 4,029 | Extracellular Space | transporter |
| C6ORF115 | chromosome 6 open reading frame 115 | AF116682 | 4,004 | Unknown | other |
| ARD1A | ARD1 homolog A, N-acetyltransferase (S. cerevisiae) | NM_003491 | 3,983 | Nucleus | enzyme |
| TMSB4X | thymosin, beta 4, X-linked | AK055976 | 3,942 | Cytoplasm | other |
| PPAP2B | phosphatidic acid phosphatase type 2B | NM_003713 | 3,872 | Plasma Membrane | phosphatase |
| CD63 | CD63 molecule | NM_001780 | 3,861 | Plasma Membrane | other |
| YWHAB | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, protein, beta p | NM_003404 | 3,841 | Cytoplasm | other |
| EXOSC6 | exosome component 6 | NM_058219 | 3,784 | Nucleus | other |
| HLA-C | major histocompatibility complex, class I, C | M12679 | 3,754 | Plasma Membrane | transmembrane receptor |
| SNCA | synuclein, alpha (non A4 component of amyloid precursor) | NM_000345 | 3,689 | Cytoplasm | other |
| PDHA1 | pyruvate dehydrogenase (lipoamide) alpha 1 | NM_000284 | 3,652 | Cytoplasm | enzyme |
| HSPE1 | heat shock 10kDa protein 1 (chaperonin 10) | NM_002157 | 3,605 | Cytoplasm | other |
| SNRPD2 | small nuclear ribonucleoprotein D2 polypeptide 16.5kDa | NM_004597 | 3,57 | Nucleus | other |
| ACTG1 | actin, gamma 1 | NM_001614 | 3,52 | Cytoplasm | other |
| SYNGR2 | synaptogyrin 2 | NM_004710 | 3,493 | Plasma Membrane | other |
| NDUFA13 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 13 | NM_015965 | 3,449 | Cytoplasm | enzyme |
| CXCR4 | chemokine (C-X-C motif) receptor 4 | NM_003467 | 3,411 | Plasma Membrane | G-protein coupled receptor |
| AURKAIP1 | aurora kinase A interacting protein 1 | NM_017900 | 3,407 | Nucleus | other |
| CYC1 | cytochrome c-1 | NM_001916 | 3,386 | Cytoplasm | enzyme |
| MAFK | v-maf musculoaponeurotic fibrosarcoma oncogene homolog K (avian) | AK056767 | 3,38 | Nucleus | transcription regulator |
| BLOC1S1 | biogenesis of lysosome-related organelles complex-1, subunit 1 | NM_001487 | 3,375 | Cytoplasm | other |
| BPI | bactericidal/permeability-increasing protein | NM_001725 | 3,368 | Plasma Membrane | other |
| FOS | v-fos FBJ murine osteosarcoma viral oncogene homolog | NM_005252 | 3,295 | Nucleus | transcription regulator |
| S100P | S100 calcium binding protein P | NM_005980 | 3,204 | Cytoplasm | other |
| PSMB1 | proteasome (prosome, macropain) subunit, beta type, 1 | AK023290 | 3,174 | Cytoplasm | peptidase |
| APOB | apolipoprotein B (including Ag(x) antigen) | NM_000384 | 3,131 | Extracellular Space | transporter |
| GTF3C5 | general transcription factor IIIC, polypeptide 5, 63kDa | NM_012087 | 3,117 | Nucleus | transcription regulator |
| SF3A2 | splicing factor 3a, subunit 2, 66kDa | NM_007165 | 3,11 | Nucleus | other |
| MAP3K11 | mitogen-activated protein kinase kinase kinase 11 | NM_002419 | 3,078 | Cytoplasm | kinase |
| NRG1 | neuregulin 1 | NM_013957 | 3,069 | Extracellular Space | growth factor |
| CST3 | cystatin C (amyloid angiopathy and cerebral hemorrhage) | NM_000099 | 3,046 | Extracellular Space | other |
| PSCD1 | pleckstrin homology, Sec7 and coiled-coil domains 1(cytohesin 1) | NM_004762 | 3,038 | Cytoplasm | other |
| COPE | coatomer protein complex, subunit epsilon | NM_007263 | 3,014 | Cytoplasm | transporter |
| TALDO1 | transaldolase 1 | NM_006755 | 3,003 | Cytoplasm | enzyme |
| YBX1 | Y box binding protein 1 | NM_004559 | 3 | Nucleus | transcription regulator |
| RNASE3 | ribonuclease, RNase A family, 3 (eosinophil cationic protein) | NM_002935 | 2,889 | Extracellular Space | enzyme |
| SERPINB1 | serpin peptidase inhibitor, clade B (ovalbumin), member 1 | NM_030666 | 2,886 | Cytoplasm | other |
| SAT | spermidine/spermine N1-acetyltransferase 1 | NM_002970 | 2,881 | Cytoplasm | enzyme |
| ARHGAP4 | Rho GTPase activating protein 4 | NM_001666 | 2,863 | Cytoplasm | other |
| LAPTM5 | lysosomal associated multispanning membrane protein 5 | NM_006762 | 2,861 | Plasma Membrane | other |
| ABCA7 | ATP-binding cassette, sub-family A (ABC1), member 7 | NM_019112 | 2,859 | Plasma Membrane | transporter |
| CALCR | calcitonin receptor | NM_001742 | 2,844 | Plasma Membrane | protein coupled receptor |
| HNRPM | heterogeneous nuclear ribonucleoprotein M | AK024911 | 2,794 | Plasma Membrane | transmembrane receptor |
| CD24 | CD24 molecule | NM_013230 | 2,77 | Plasma Membrane | other |
| TM7SF3 | transmembrane 7 superfamily member 3 | NM_016551 | 2,734 | Plasma Membrane | other |
| GYPC | glycophorin C (Gerbich blood group) | NM_002101 | 2,705 | Plasma Membrane | other |
| ATP6V1F | ATPase, H+ transporting, lysosomal 14kDa, V1 subunit F | NM_004231 | 2,702 | Cytoplasm | transporter |
| HMGB2 | high-mobility group box 2 | NM_002129 | 2,661 | Nucleus | transcription regulator |
| EIF4B | eukaryotic translation initiation factor 4B | NM_001417 | 2,569 | Cytoplasm | translation regulator |
| NDUFA1 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 1, 7.5kDa | NM_004541 | 2,568 | Cytoplasm | enzyme |
| WBSCR1 | Williams-Beuren syndrome chromosome region 1 | NM_022170 | 2,562 | Cytoplasm | translation regulator |
| NDUFS5 | NADH dehydrogenase (ubiquinone) Fe-S protein 5, 15kDa (NADH-coenzyme Q red | NM_004552 | 2,498 | Cytoplasm | enzyme |
| BCCIP | BRCA2 and CDKN1A interacting protein | NM_078469 | 2,459 | Nucleus | other |
| SNIP1 | Smad nuclear interacting protein 1 | NM_024700 | 2,456 | Nucleus | other |
| UBA52 | ubiquitin A-52 residue ribosomal protein fusion product 1 | NM_003333 | 2,376 | Cytoplasm | transcription regulator |
| USP7 | ubiquitin specific peptidase 7 (herpes virus-associated) | NM_003470 | 2,374 | Nucleus | peptidase |
| NR6A1 | nuclear receptor subfamily 6, group A, member 1 | NM_033334 | 2,36 | Nucleus | ligand-dependent nuclear receptor |
| ZNF9 | CCHC-type zinc finger, nucleic acid binding protein | NM_003418 | 2,334 | Nucleus | transcription regulator |
| GPSM3 | G-protein signalling modulator 3 (AGS3-like, C. elegans) | NM_022107 | 2,332 | Unknown | other |
| IDI1 | isopentenyl-diphosphate delta isomerase 1 | NM_004508 | 2,324 | Cytoplasm | enzyme |
| KLF6 | Kruppel-like factor 6 | NM_001300 | 2,311 | Nucleus | transcription regulator |
| UQCRC2 | ubiquinol-cytochrome c reductase core protein II | NM_003366 | 2,308 | Cytoplasm | enzyme |
| RAN | RAN, member RAS oncogene family | NM_006325 | 2,289 | Nucleus | enzyme |
| SUB1 | SUB1 homolog (S. cerevisiae) | NM_006713 | 2,286 | Nucleus | transcription regulator |
| TGOLN2 | trans-golgi network protein 2 | AK025557 | 2,282 | Cytoplasm | other |
| BZRP | translocator protein (18kDa) | NM_000714 | 2,273 | Cytoplasm | transmembrane receptor |
| SCPEP1 | serine carboxypeptidase 1 | NM_021626 | 2,249 | Cytoplasm | peptidase |
| FOXO3A | forkhead box O3A | AK024103 | 2,241 | Nucleus | transcription regulator |
| BLVRB | biliverdin reductase B (flavin reductase (NADPH)) | NM_000713 | 2,233 | Cytoplasm | enzyme |
| C1D | nuclear DNA-binding protein | NM_006333 | 2,212 | Nucleus | transcription regulator |
| TAF9 | TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa | NM_003187 | 2,177 | Nucleus | transcription regulator |
| RAD23A | RAD23 homolog A (S. cerevisiae) | NM_005053 | 2,175 | Nucleus | other |
| UGCG | UDP-glucose ceramide glucosyltransferase | AJ420423 | 2,165 | Cytoplasm | enzyme |
| NFKBIZ | nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta | NM_031419 | 2,157 | Nucleus | transcription regulator |
| ATP6AP2 | ATPase, H+ transporting, lysosomal accessory protein 2 | NM_005765 | 2,155 | Cytoplasm | transporter |
| CTSW | cathepsin W (lymphopain) | NM_001335 | 2,153 | Cytoplasm | peptidase |
| CCNI | cyclin I | NM_006835 | 2,138 | Unknown | other |
| UBE1 | ubiquitin-activating enzyme E1 (A1S9T and BN75 temperature sensitivity complem | NM_003334 | 2,136 | Cytoplasm | enzyme |
| ARPC3 | actin related protein 2/3 complex, subunit 3, 21kDa | NM_005719 | 2,13 | Cytoplasm | other |
| DEK | DEK oncogene (DNA binding) | NM_003472 | 2,129 | Nucleus | transcription regulator |
| SSBP3 | single stranded DNA binding protein 3 | NM_018070 | 2,107 | Nucleus | transcription regulator |
| CAMLG | calcium modulating ligand | NM_001745 | 2,102 | Cytoplasm | other |
| CHAF1A | chromatin assembly factor 1, subunit A (p150) | NM_005483 | 2,102 | Nucleus | other |
| F11R | F11 receptor | AF172398 | 2,093 | Plasma Membrane | other |
| NCF4 | neutrophil cytosolic factor 4, 40kDa | NM_013416 | 2,092 | Cytoplasm | enzyme |
| COX7A2 | cytochrome c oxidase subunit VIIa polypeptide 2 (liver) | NM_001865 | 2,089 | Cytoplasm | enzyme |
| H1FX | H1 histone family, member X | NM_006026 | 2,083 | Nucleus | other |
| FOSL1 | FOS-like antigen 1 | NM_005438 | 2,077 | Nucleus | transcription regulator |
| ACTB | actin, beta | NM_001101 | 2,075 | Cytoplasm | other |
| PNN | pinin, desmosome associated protein | NM_002687 | 2,068 | Plasma Membrane | other |
| LYN | v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | NM_002350 | 2,054 | Cytoplasm | kinase |
| TNC | tenascin C (hexabrachion) | AK024586 | 2,049 | Extracellular Space | other |
| SUMO2 | SMT3 suppressor of mif two 3 homolog 2 (S. cerevisiae) | NM_006937 | 2,045 | Nucleus | other |
| CKAP4 | cytoskeleton-associated protein 4 | NM_006825 | 2,043 | Cytoplasm | other |
| SLC44A2 | solute carrier family 44, member 2 | AK027519 | 2,036 | Extracellular Space | other |
Granulopoiesis signature in the bone marrow as a marker for SLE activity
Nine genes selected a priori were used to investigate associations between a granulopoeisis signature and SLEDAI. To facilitate these comparisons we created a granulopoeisis “score” for both PBMCs and BMMCs, as described above in Materials and Methods and found a correlation between granulopoiesis score and SLEDAI (r = 0.33) in the periphery of the SLE patient subset that had also provided bone marrow (Figure 3A). Linear regression analysis showed that granulopoiesis signature significantly correlated with SLEDAI in the bone marrow (r = 0.55, p = 0.013), (Figure 3B) and the granulopoiesis score from bone marrow active group of patients was higher versus the inactive (p =0.004), (Figure 3C)
Figure 3. Granulopoiesis signature in the bone marrow as a marker for SLE activity.
Nine genes selected a priori were used to investigate associations between a granulopoeisis signature and SLEDAI. These genes resulted in a statistically significant model of SLEDAI. (A) A numerical score was calculated by using the normalized expression levels of 9 granulopoiesis-related genes that comprise the granulopoiesis signature. Linear regression analysis demonstrates a correlation between granulopoiesis score and SLEDAI (r = 0.33) in the peripheral blood of patient subset that have also provided BM. (B) Granulopoiesis signature of the patients analyzed on Panel A, correlates stronger with SLEDAI in the bone marrow (r = 0.55, p = 0.013). C. The granulopoiesis score from bone marrow was higher in the active group of SLE patients vs the inactive (p =0.004)
DISCUSSION
This study sought to shed additional light on the pathogenesis of SLE by analyzing gene expression in the bone marrow, an organ with a central role not only in hemopoiesis but also in the immune response. Herein, we report that bone marrow analysis discriminates active from inactive lupus, and displays apoptosis and granulopoiesis signatures
Our microarray analysis has several strong points. First, we only selected the statistically significant genes with a change in expression of at least 2-fold when comparing the means of the two groups; the increased stringency increases the specificity of the results. Second, hierarchical clustering revealed two overlapping groups: active and inactive patients and we determined the differentially expressed genes between these two clusters. By doing so, the analysis was not biased by any arbitrary clustering. Third, our clinical data on organ involvement were obtained at the time on the blood drawn and were not based on historical data on involvement of a particular organ at some time in the course of the disease. Lastly, in addition to the peripheral blood, our analysis included the bone marrow an important organ in the biology of lymphocytes and neutrophils. Using this analysis, we were able to duplicate-only in part- data suggesting an interferon signature in SLE by finding up-regulation of only two IFN regulated genes: IL6R whose expression is regulated by IFNα and PRKCG which is involved in IFNγ signaling. This may be due to differences in the analysis between our study and those of Baechler et al (12) and Bennet et al (13) or-more likely-to genetic differences in the populations studied ( North American vs Mediterranean). The later is supported by data both in murine models (18) and in humans whereby the relative contribution of IFN in the pathogenesis of the disease may depend upon the genetic background (19).
Ficol-Histopaque density gradient preparations of peripheral blood mononuclear cells of patients with lupus contain high numbers of low buoyant density activated neutrophils (20). We found increased expression of early neutrophil genes; most of them encode products of immature granulocytes and their expression is regulated during myeloid cell differentiation. These proteins include components of the neutrophil granules (such as defensin A4, cathepsin, myeloperoxidase, lactoferrin and elastase 2) that may be released by neutrophils and initiate the production of autoantibodies directed against constituents of neutrophil cytoplasm (ANCA) described in patients with systemic vasculitis and autoimmune diseases such as lupus (21–31).
It is of particular interest that we found a significant increase in the levels of β2-integrin and other genes involved in the integrin signaling pathway such as MAP3K11, ACTB, ACTG1, ARPC3, MRCL2 and MRCL3 in the bone marrow of active patients relative to inactive. The integrin beta 2, LFA-1 (leukocyte function-associated antigen-1) and its ligand ICAM-1, has a key role in tissue injury in lupus (32) and other inflammatory diseases (33–36). On the other hand, neutrophils are important effector cells in a variety of acute and chronic inflammatory states including lupus (37). Our data corroborate and expand those from Bennet et al (13). Although we did not observe the granulopoiesis signature in the peripheral blood, we found it in the BM when comparing active vs inactive and BM vs peripheral blood. Common genes overexpressed in our and their study include myeloperoxidase, elastase, cathepsin, CD24, S100P and defensin. These results suggest that the granulopoiesis signatures reported by others in SLE peripheral blood may in fact originate from bone marrow cells and may persist or expand in the blood of certain SLE patients.
In our study we also identified genes involved in the apoptosis of granulocytes in the bone marrow of active SLE, such as FOXO3A which has been described as one of the genes induced after phagocytosis of pathogens (38) as well as annexin 1 reported to have a proapoptotic role in human neutrophils (39) and CXCR4 usually expressed on senescent neutrophils. Accelerated apoptosis of lymphocytes in the peripheral blood of lupus patients and impaired clearance of apoptotic cells due to the decreased phagocytic ability of macrophages, monocytes and neutrophils has a pathogenic role in SLE (40–47). This has been speculated to be the result of unidentified serum factors (48). In addition to the necrotic changes in the bone marrow reported in SLE by Voulgarelis et al. (3) apoptotic bodies were also recently observed in the bone marrow of 8 of 10 SLE patients, many of whom had cytopenias(49). Both alive and apoptotic neutrophils have been implicated in tissue injury in patients with Wegener granulomatosis (50). We speculate that neutrophil apoptosis is probably a result of the excessive activation of polymorphonuclear cells.
In our analysis, we found seven genes correlating with SLEDAI in the bone marrow. Although these genes are highly correlated, they share no known functional relationships. Out of these 7 genes associated with SLEDAI in the bone marrow, only two genes, NY-REN-25 antigen and the coiled-coil domain CCDC91 associated with SLEDAI in active PBMC patients. For both genes there is considerable variation between predicted and actual SLEDAI probably because these genes were selected within the bone marrow comparison.
In summary, these data support the use of microarray analysis to uncover novel immunopathologic pathways in the disease and have shown that the bone marrow, distinguishes active from inactive lupus patients. These data provide additional credence to the role of bone marrow and neutrophils in the pathogenesis of the disease and suggest additional pathways for potential therapeutic modulation targeted at the effector cells to minimize tissue injury.
Acknowledgments
We thank Dr Sanoudou and Dr E. Papadimitraki for helpful discussions and critical review of the manuscript.
This work was supported by FP6 European AUTOCURE program and by NIH grants: P20RR016478, P20RR020143, P20RR017703, P20RR15577, and U19AI062629.
This project has been co-funded in part by the European Social Fund and National Resources (EPEAK ’Pythagoras II), and a grant from the Hellenic Society of Rheumatology. Magda Nakou is a graduate student of the Graduate Program of the “Molecular Basis of Human Diseases”
REFERENCES
- 1.Kyttaris VC, Tsokos GC. T lymphocytes in systemic lupus erythematosus: an update. Curr Opin Rheumatol. 2004;16:548–552. doi: 10.1097/01.bor.0000132646.55056.e0. [DOI] [PubMed] [Google Scholar]
- 2.Papadimitraki ED, Choulaki C, Koutala E, Bertsias G, Tsatsanis C, Gergianaki I, Raptopoulou A, et al. Expansion of toll-like receptor 9-expressing B cells in active systemic lupus erythematosus: implications for the induction and maintenance of the autoimmune process. Arthritis Rheum. 2006;54:3601–3611. doi: 10.1002/art.22197. [DOI] [PubMed] [Google Scholar]
- 3.Voulgarelis M, Giannouli S, Tasidou A, Anagnostou D, Ziakas PD, Tzioufas AG. Bone marrow histological findings in systemic lupus erythematosus with hematologic abnormalities: a clinicopathological study. Am J Hematol. 2006;81:590–597. doi: 10.1002/ajh.20593. [DOI] [PubMed] [Google Scholar]
- 4.Minges Wols HA, Underhill GH, Kansas GS, Witte PL. The role of bone marrow-derived stromal cells in the maintenance of plasma cell longevity. J Immunol. 2002;169:4213–4221. doi: 10.4049/jimmunol.169.8.4213. [DOI] [PubMed] [Google Scholar]
- 5.Kondo M, Weissman IL, Akashi K. Identification of clonogenic common lymphoid progenitors in mouse bone marrow. Cell. 1997;91:661–672. doi: 10.1016/s0092-8674(00)80453-5. [DOI] [PubMed] [Google Scholar]
- 6.Papadaki HA, Boumpas DT, Gibson FM, Jayne DR, Axford JS, Gordon-Smith EC, Marsh JC, et al. Increased apoptosis of bone marrow CD34(+) cells and impaired function of bone marrow stromal cells in patients with systemic lupus erythematosus. Br J Haematol. 2001;115:167–174. doi: 10.1046/j.1365-2141.2001.03076.x. [DOI] [PubMed] [Google Scholar]
- 7.Ramaswamy S, Golub TR. DNA microarrays in clinical oncology. J Clin Oncol. 2002;20:1932–1941. doi: 10.1200/JCO.2002.20.7.1932. [DOI] [PubMed] [Google Scholar]
- 8.Staudt LM. Gene expression profiling of lymphoid malignancies. Annu Rev Med. 2002;53:303–318. doi: 10.1146/annurev.med.53.082901.103941. [DOI] [PubMed] [Google Scholar]
- 9.Goertsches R, Serrano-Fernandez P, Moller S, Koczan D, Zettl UK. Multiple sclerosis therapy monitoring based on gene expression. Curr Pharm Des. 2006;12:3761–3779. doi: 10.2174/138161206778559786. [DOI] [PubMed] [Google Scholar]
- 10.Kappos L, Achtnichts L, Dahlke F, Kuhle J, Naegelin Y, Sandbrink R, Lindberg RL. Genomics and proteomics: role in the management of multiple sclerosis. J Neurol. 2005;252 Suppl 3:iii21–iii27. doi: 10.1007/s00415-005-2013-3. [DOI] [PubMed] [Google Scholar]
- 11.Peterson KS, Huang JF, Zhu J, D'Agati V, Liu X, Miller N, Erlander MG, et al. Characterization of heterogeneity in the molecular pathogenesis of lupus nephritis from transcriptional profiles of laser-captured glomeruli. J Clin Invest. 2004;113:1722–1733. doi: 10.1172/JCI19139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci U S A. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. J Exp Med. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirou KA, Lee C, George S, Louca K, Papagiannis IG, Peterson MG, Ly N, et al. Coordinate overexpression of interferon-alpha-induced genes in systemic lupus erythematosus. Arthritis Rheum. 2004;50:3958–3967. doi: 10.1002/art.20798. [DOI] [PubMed] [Google Scholar]
- 15.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, Schaller JG, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1982;25:1271–1277. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 16.Tarte K, Zhan F, De Vos J, Klein B, Shaughnessy J., Jr Gene expression profiling of plasma cells and plasmablasts: toward a better understanding of the late stages of B-cell differentiation. Blood. 2003;102:592–600. doi: 10.1182/blood-2002-10-3161. [DOI] [PubMed] [Google Scholar]
- 17.Ong ST, Hackbarth ML, Degenstein LC, Baunoch DA, Anastasi J, McKeithan TW. Lymphadenopathy, splenomegaly, and altered immunoglobulin production in BCL3 transgenic mice. Oncogene. 1998;16:2333–2343. doi: 10.1038/sj.onc.1201771. [DOI] [PubMed] [Google Scholar]
- 18.Lauwerys BR, Wakeland EK. Genetics of lupus nephritis. Lupus. 2005;14(2):2–12. doi: 10.1191/0961203305lu2052oa. [DOI] [PubMed] [Google Scholar]
- 19.Pascual V, Farkas L, Banchereau J. Systemic lupus erythematosus: all roads lead to type I interferons. Curr Opin Immunol. 2006;18:676–682. doi: 10.1016/j.coi.2006.09.014. [DOI] [PubMed] [Google Scholar]
- 20.Hacbarth E, Kajdacsy-Balla A. Low density neutrophils in patients with systemic lupus erythematosus, rheumatoid arthritis, and acute rheumatic fever. Arthritis Rheum. 1986;29:1334–1342. doi: 10.1002/art.1780291105. [DOI] [PubMed] [Google Scholar]
- 21.van der Woude FJ, Rasmussen N, Lobatto S, Wiik A, Permin H, van Es LA, van der Giessen M, et al. Autoantibodies against neutrophils and monocytes: tool for diagnosis and marker of disease activity in Wegener's granulomatosis. Lancet. 1985;1:425–429. doi: 10.1016/s0140-6736(85)91147-x. [DOI] [PubMed] [Google Scholar]
- 22.Tamiya H, Tani K, Miyata J, Sato K, Urata T, Lkhagvaa B, Otsuka S, et al. Defensins- and cathepsin G-ANCA in systemic lupus erythematosus. Rheumatol Int. 2006;27:147–152. doi: 10.1007/s00296-006-0173-9. [DOI] [PubMed] [Google Scholar]
- 23.Schnabel A, Csernok E, Isenberg DA, Mrowka C, Gross WL. Antineutrophil cytoplasmic antibodies in systemic lupus erythematosus. Prevalence, specificities, and clinical significance. Arthritis Rheum. 1995;38:633–637. doi: 10.1002/art.1780380509. [DOI] [PubMed] [Google Scholar]
- 24.Molnar K, Kovacs L, Kiss M, Husz S, Dobozy A, Pokorny G. Antineutrophil cytoplasmic antibodies in patients with systemic lupus erythematosus. Clin Exp Dermatol. 2002;27:59–61. doi: 10.1046/j.0307-6938.2001.00964.x. [DOI] [PubMed] [Google Scholar]
- 25.Lash JA, Coates TD, Lafuze J, Baehner RL, Boxer LA. Plasma lactoferrin reflects granulocyte activation in vivo. Blood. 1983;61:885–888. [PubMed] [Google Scholar]
- 26.Kuwana T, Sato Y, Saka M, Kondo Y, Miyata M, Obara K, Nishimaki T, et al. Anti-cathepsin G antibodies in the sera of patients with ulcerative colitis. J Gastroenterol. 2000;35:682–689. doi: 10.1007/s005350070047. [DOI] [PubMed] [Google Scholar]
- 27.Jennette JC, Falk RJ, Andrassy K, Bacon PA, Churg J, Gross WL, Hagen EC, et al. Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum. 1994;37:187–192. doi: 10.1002/art.1780370206. [DOI] [PubMed] [Google Scholar]
- 28.Hoffman GS, Specks U. Antineutrophil cytoplasmic antibodies. Arthritis Rheum. 1998;41:1521–1537. doi: 10.1002/1529-0131(199809)41:9<1521::AID-ART2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 29.Davies DJ, Moran JE, Niall JF, Ryan GB. Segmental necrotising glomerulonephritis with antineutrophil antibody: possible arbovirus aetiology? Br Med J(Clin Res Ed) 1982;285:606. doi: 10.1136/bmj.285.6342.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caccavo D, Rigon A, Picardi A, Galluzzo S, Vadacca M, Ferri GM, Amoroso A, et al. Anti-lactoferrin antibodies in systemic lupus erythematosus: isotypes and clinical correlates. Clin Rheumatol. 2005;24:381–387. doi: 10.1007/s10067-004-1040-2. [DOI] [PubMed] [Google Scholar]
- 31.Caccavo D, Pellegrino NM, Altamura M, Rigon A, Amati L, Amoroso A, Jirillo E. Antimicrobial and immunoregulatory functions of lactoferrin and its potential therapeutic application. J Endotoxin Res. 2002;8:403–417. doi: 10.1179/096805102125001000. [DOI] [PubMed] [Google Scholar]
- 32.Kevil CG, Hicks MJ, He X, Zhang J, Ballantyne CM, Raman C, Schoeb TR, et al. Loss of LFA-1, but not Mac-1, protects MRL/MpJ-Fas(lpr) mice from autoimmune disease. Am J Pathol. 2004;165:609–616. doi: 10.1016/S0002-9440(10)63325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Torsteinsdottir I, Arvidson NG, Hallgren R, Hakansson L. Monocyte activation in rheumatoid arthritis (RA): increased integrin, Fc gamma and complement receptor expression and the effect of glucocorticoids. Clin Exp Immunol. 1999;115:554–560. doi: 10.1046/j.1365-2249.1999.00817.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas RM, Schmedt C, Novelli M, Choi BK, Skok J, Tarakhovsky A, Roes J. C-terminal SRC kinase controls acute inflammation and granulocyte adhesion. Immunity. 2004;20:181–191. doi: 10.1016/s1074-7613(04)00023-8. [DOI] [PubMed] [Google Scholar]
- 35.Gautam N, Herwald H, Hedqvist P, Lindbom L. Signaling via beta(2) integrins triggers neutrophil-dependent alteration in endothelial barrier function. J Exp Med. 2000;191:1829–1839. doi: 10.1084/jem.191.11.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Giblin PA, Lemieux RM. LFA-1 as a key regulator of immune function: approaches toward the development of LFA-1-based therapeutics. Curr Pharm Des. 2006;12:2771–2795. doi: 10.2174/138161206777947731. [DOI] [PubMed] [Google Scholar]
- 37.Niwa Y, Sakane T, Shingu M, Miyachi Y. Role of stimulated neutrophils from patients with systemic lupus erythematosus in tissue injury, with special reference to serum factors and increased active oxygen species generated by neutrophils. Inflammation. 1985;9:163–172. doi: 10.1007/BF00917588. [DOI] [PubMed] [Google Scholar]
- 38.Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Solito E, Kamal A, Russo-Marie F, Buckingham JC, Marullo S, Perretti M. A novel calcium-dependent proapoptotic effect of annexin 1 on human neutrophils. Faseb J. 2003;17:1544–1546. doi: 10.1096/fj.02-0941fje. [DOI] [PubMed] [Google Scholar]
- 40.Courtney PA, Crockard AD, Williamson K, Irvine AE, Kennedy RJ, Bell AL. Increased apoptotic peripheral blood neutrophils in systemic lupus erythematosus: relations with disease activity, antibodies to double stranded DNA, and neutropenia. Ann Rheum Dis. 1999;58:309–314. doi: 10.1136/ard.58.5.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Emlen W, Niebur J, Kadera R. Accelerated in vitro apoptosis of lymphocytes from patients with systemic lupus erythematosus. J Immunol. 1994;152:3685–3692. [PubMed] [Google Scholar]
- 42.Georgescu L, Vakkalanka RK, Elkon KB, Crow MK. Interleukin-10 promotes activation-induced cell death of SLE lymphocytes mediated by Fas ligand. J Clin Invest. 1997;100:2622–2633. doi: 10.1172/JCI119806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lorenz HM, Grunke M, Hieronymus T, Herrmann M, Kuhnel A, Manger B, Kalden JR. In vitro apoptosis and expression of apoptosis-related molecules in lymphocytes from patients with systemic lupus erythematosus and other autoimmune diseases. Arthritis Rheum. 1997;40:306–317. doi: 10.1002/art.1780400216. [DOI] [PubMed] [Google Scholar]
- 44.Perniok A, Wedekind F, Herrmann M, Specker C, Schneider M. High levels of circulating early apoptic peripheral blood mononuclear cells in systemic lupus erythematosus. Lupus. 1998;7:113–118. doi: 10.1191/096120398678919804. [DOI] [PubMed] [Google Scholar]
- 45.Richardson BC, Yung RL, Johnson KJ, Rowse PE, Lalwani ND. Monocyte apoptosis in patients with active lupus. Arthritis Rheum. 1996;39:1432–1434. doi: 10.1002/art.1780390827. [DOI] [PubMed] [Google Scholar]
- 46.Brandt L, Hedberg H. Impaired phagocytosis by peripheral blood granulocytes in systemic lupus erythematosus. Scand J Haematol. 1969;6:348–353. doi: 10.1111/j.1600-0609.1969.tb02420.x. [DOI] [PubMed] [Google Scholar]
- 47.Vazquez-Doval J, Sanchez-Ibarrola A. Defective mononuclear phagocyte function in systemic lupus erythematosus: relationship of FcRII (CD32) with intermediate cytoskeletal filaments. J Investig Allergol Clin Immunol. 1993;3:86–91. [PubMed] [Google Scholar]
- 48.Ren Y, Tang J, Mok MY, Chan AW, Wu A, Lau CS. Increased apoptotic neutrophils and macrophages and impaired macrophage phagocytic clearance of apoptotic neutrophils in systemic lupus erythematosus. Arthritis Rheum. 2003;48:2888–2897. doi: 10.1002/art.11237. [DOI] [PubMed] [Google Scholar]
- 49.Hepburn AL, Lampert IA, Boyle JJ, Horncastle D, Ng WF, Layton M, Vyse TJ, et al. In vivo evidence for apoptosis in the bone marrow in systemic lupus erythematosus. Ann Rheum Dis. 2007;66:1106–1109. doi: 10.1136/ard.2006.065003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van Rossum AP, Limburg PC, Kallenberg CG. Activation, apoptosis, and clearance of neutrophils in Wegener's granulomatosis. Ann N Y Acad Sci. 2005;1051:1–11. doi: 10.1196/annals.1361.041. [DOI] [PubMed] [Google Scholar]