Abstract
Human DJ-1, a disease-associated protein that protects cells from oxidative stress, contains an oxidation-sensitive cysteine (C106) that is essential for its cytoprotective activity. The origin of C106 reactivity is obscure, due in part to the absence of an experimentally determined pKa value for this residue. We have used atomic resolution X-ray crystallography and UV spectroscopy to show that C106 has a depressed pKa of 5.4±0.1 and that the C106 thiolate accepts a hydrogen bond from a protonated glutamic acid sidechain (E18). X-ray crystal structures and cysteine pKa analysis of several site-directed substitutions at residue 18 demonstrate that the protonated carboxylic acid sidechain of E18 is required to maximally stabilize the C106 thiolate. A nearby arginine residue (R48) participates in a guanidinium stacking interaction with R28 from the other monomer in the DJ-1 dimer and elevates the pKa of C106 by binding an anion that electrostatically suppresses thiol ionization. Our results show that the ionizable residues (E18, R48 and R28) surrounding C106 affect its pKa in a way that is contrary to expectations based on the typical ionization behavior of glutamic acid and arginine. Lastly, a search of the Protein Data Bank (PDB) produces several candidate hydrogen bonded aspartic/glutamic acid-cysteine interactions, which we propose are particularly common in the DJ-1 superfamily.
Keywords: cysteine, pKa, atomic resolution X-ray crystallography, bond length analysis, Parkinsonism
Cysteine thiolates are potent nucleophiles that are used by many proteins for catalysis, metal binding, or to facilitate post-translational modification. However, the solution pKa value of cysteine (8.3) is not within the optimal physiological pH range of most proteins. To render cysteine residues reactive, the thiol pKa must be depressed by stabilization of the conjugate thiolate anion by the protein environment. Multiple types of stabilizing interactions can decrease cysteine pKa values, including electrostatic complementarity with nearby cations (1, 2), the α-helix macropole (3), and hydrogen bonding to the thiol (4).
The Parkinsonism-associated protein DJ-1 contains an oxidation-sensitive cysteine residue (C106 in human DJ-1) of unknown pKa that is required for the cytoprotective activity of the protein (5, 6). DJ-1 is a homodimeric protein of 189 a.a. monomers that is found both in the cytoplasm (7) and the mitochondria (5, 8) and protects cells from various types of oxidative stress (5, 9–18). Although C106 has been shown to be essential for the protective function of DJ-1 in cell culture (5, 16) and Drosophila melanogaster (6), the specific biochemical activity of the protein that requires this cysteine residue is uncertain. Identifying the structural determinants of C106 ionization (and hence reactivity) is an essential first step in understanding how this conserved cysteine residue contributes to the neuroprotective activity of DJ-1.
The reactive cysteine in human DJ-1 is one of the most highly conserved residues in the DJ-1 superfamily (19, 20). Crystal structures solved by several independent groups (21–25) show that C106 is located at a solvent-exposed sharp bend between a β-strand and an α-helix called the ‘nucleophile elbow’ (26). The backbone phi and psi angles for the nucleophile elbow cysteine fall in the unfavorable region of Ramachandran space for every structurally characterized member of the DJ-1 superfamily (27), possibly contributing to its reactivity. Furthermore, a computational study of structurally characterized proteins of the DJ-1 superfamily shows that functionally important residues with perturbed in silico pH titration profiles cluster around the reactive cysteine residue and can be used to classify distinct members of the superfamily (28). Of the residues in the environment of the conserved cysteine, a proximal glutamic acid (E18 in human DJ-1) is found in many members of the DJ-1 superfamily. The importance of this residue, however, remains poorly understood (27, 28).
In order to better understand the origin of cysteine reactivity in DJ-1, we have used a combination of structural and biochemical methods to show that C106 has a depressed pKa value of 5.4±0.1 and that the environment of C106 contains multiple ionizable residues that exert countervailing influence on the pKa of this residue. The conserved E18 residue has a protonated carboxylic acid sidechain that enhances cysteine ionization by donating a hydrogen bond to the Sγ atom of C106, while R28 and R48 interact through guanidinium stacking and bind an anion that suppresses C106 thiolate formation. The significance of the low pKa value for C106 for the proposed biochemical activities and oxidative regulation of DJ-1 is discussed.
Experimental Procedures
Protein expression and purification
Three forms of recombinant DJ-1 were used in this study; untagged DJ-1, DJ-1 bearing a non-cleavable C-terminal hexa-histidine tag, and DJ-1 bearing an N-terminal thrombin-cleavable hexa-histidine tag. DJ-1 with a non-cleavable C-terminal hexa-histidine tag was expressed in the bacterial expression vector pET21a bearing the DJ-1 gene cloned between the NdeI and XhoI sites. This construct results in eight vector-derived amino acids (LEHHHHH) appended to the C-terminus of the expressed protein and was used for DJ-1 crystallization. For the cysteine pKa experiments, DJ-1 with a N-terminal thrombin-cleavable hexa-histidine tag was expressed in the bacterial expression vector pET15b bearing the DJ-1 gene cloned between the NdeI and XhoI sites. After tag removal, this construct results in recombinant protein with three vector-derived amino acids (GSH) at the N-terminus of the protein and is called “tag-cleaved” DJ-1 throughout the remainder of the paper. For all recombinant proteins, BL21(DE3) E. coli (Novagen) containing the appropriate DJ-1 expression construct were grown in LB media supplemented with 100 µg/ml of ampicilin at 37°C with shaking and protein expression was induced by the addition of 0.1 mM IPTG. The induced culture was incubated at 20°C with shaking overnight and harvested by centrifugation the next day. Cell pellets were stored at −80°C until needed.
Hexa-histidine tagged recombinant DJ-1 was purified by Ni2+-NTA chromatography using His-Select resin (Sigma) following the manufacturer’s instructions. For C-terminal His tagged protein, the Ni2+-NTA purification was followed by dialysis against storage buffer (25 mM HEPES pH=7.5, 100 mM KCl, 2 mM DTT) and passage over a Q anion exchange resin to remove contaminating nucleic acid. DJ-1 appears in the flow-through fraction, while contaminating nucleic acids remain bound to the Q column. For the N-terminal His-tagged DJ-1, the hexa-histidine tag was removed by thrombin cleavage at 4° C for 6–12 hours, followed by sequential passage over His-Select resin to remove any protein that retained the tag, benzamidine-sepharose resin to remove thrombin, and a final passage over a Q anion exchange resin to remove nucleic acid. Untagged DJ-1 was purified as previously described (29). For all preparations, purified DJ-1 runs as a single band with apparent molecular weight of 25 kDa on overloaded Biosafe (Bio-Rad) Coomassie-stained SDS-PAGE. DJ-1 was concentrated to 20 mg ml−1 (ε280=4000 M –1 cm−1) in storage buffer, frozen in liquid nitrogen, and stored at −80°C.
Crystallization, data collection, and processing
Crystals of untagged wtDJ-1 in space group P212121 with two molecules in the asymmetric unit (referred to as orthorhombic wt throughout the remainder of the paper) were grown using the hanging drop vapor equilibration method by mixing 2 µL of DJ-1 (20 mg/ml) supplemented with 2.5 mM epigallocatechin 3-gallate (EGCG) with 2 µL of reservoir solution (28% PEG 4000, 200 mM ammonium sulfate, 50 mM sodium acetate pH=6.0) and incubating at room temperature. Plate-like crystals appeared in 2–5 days and were cryoprotected by serial transfer through increasing concentrations of ethylene glycol in the reservoir solution to a final concentration of 25% v/v. The crystals were mounted in nylon loops and cryocooled by immersion into liquid nitrogen. EGCG facilitates crystal growth, although it does not appear in the final electron density maps.
Crystals of C-terminal His-tagged E18Q, E18N, E18L, and E18D DJ-1 were grown using the hanging drop vapor equilibration method by mixing 2 µL of DJ-1 (20 mg/ml)) with 2 µL of reservoir solution (1.4–1.6 M sodium citrate, 50 mM HEPES pH=7.5–8.0, 10 mM DTT) and incubating at room temperature for 3–5 days. Bipyramidal crystals in space group P3121 appeared in 1–5 days and were cryoprotected by serial transfer through solutions of sodium malonate pH=7.0 and 5 mM DTT to a final concentration of 3.4 M sodium malonate (30). All DJ-1 crystals were mounted in nylon loop and cryocooled by immersion into liquid nitrogen.
Diffraction data for orthorhombic wt, E18L, E18D, E18Q and E18N DJ-1 were collected at the Advanced Photon Source (APS), BioCARS beamline 14 BM-C using 0.9 Å incident X-rays and an ADSC Q315 detector. Single crystals maintained at 100 K were used for the collection of each dataset, and the data for E18Q, E18L and E18D DJ-1 were collected in two passes (a separate high and a low resolution pass) with differing exposure times and detector distances in order to avoid overloaded pixels for intense low resolution reflections. Because C106 is sensitive to radiation-induced damage, the beam intensity was attenuated and the crystal was exposed to X-rays for 10 seconds or less per 1° oscillation. Nevertheless, negative mFo-DFc difference electron density around the Sγ atom of C106 indicates that some X-ray induced damage has occurred in the E18D, E18L and E18Q datasets. Diffraction data were integrated and scaled using HKL2000 (31) and final data statistics are provided in Table 1.
Table 1.
Crystallographic Data and Model Statistics
| Data Statistics | Wild-type | E18L | E18Q | E18D | E18N |
|---|---|---|---|---|---|
| Wavelength (Å) | 0.900 | 0.900 | 0.900 | 0.900 | 0.900 |
| Space Group | P212121 | P3121 | P3121 | P3121 | P3121 |
| Unit Cell | |||||
| a (Å) | 43.91 | 74.70 | 74.78 | 74.83 | 75.03 |
| b (Å) | 85.77 | 74.70 | 74.78 | 74.83 | 75.03 |
| c (Å) | 98.83 | 74.88 | 75.44 | 74.78 | 75.05 |
| Resolution limits (Å) | 23-1.20 | 25-1.15 | 30-1.35 | 24-1.20 | 30-1.60 |
| Unique reflections | 114,729 | 85,989 | 53,400 | 75,880 | 32,697 |
| Completeness (%)a | 97.8 (96.0) | 100 (100) | 98.6 (98.7) | 99.9 (100) | 99.8 (100) |
| Mean redundancya | 5.3 (4.2) | 10.9 (8.0) | 7.1 (4.8) | 11.1 (7.6) | 11.2 (11.2) |
| Rmerge (%)a, b | 6.5 (70.2) | 6.8 (36.5) | 7.3 (60.7) | 6.9 (69.5) | 7.0 (57.4) |
| 〈 I 〉/〈σ(I)〉a | 24.3 (2.0) | 38.3 (6.0) | 25.2 (2.3) | 35.5 (2.9) | 33.1 (5.1) |
| Model Statistics | Wild-type | E18L | E18Q | E18D | E18N |
| Molecules in ASU | 2 | 1 | 1 | 1 | 1 |
| Number of modeled residues | 374 | 187 | 186 | 186 | 186 |
| Number of solvent molecules | 583 | 239 | 193 | 221 | 182 |
| Number of residues in dual conformations | 24 | 13 | 6 | 10 | 7 |
| Heteroatoms | 2 sulfate | 1 PEG400 | none | 1 malonate | none |
| Rwork (%)c | 13.1 | 11.5 | 15.3 | 13.1 | 15.8 |
| Rwork (%) for FO>4σ(FO)c,d | 10.9 | 10.7 | n.d. | 11.6 | n.d. |
| Rfree (%)e | 18.2 | 14.0 | 16.8 | 16.4 | 18.4 |
| Rfree (%) for FO>4σ(FO)e,d | 15.4 | 13.0 | n.d. | 14.8 | n.d. |
| Rall (%)f | 13.4 | 11.5 | 15.3 | 13.1 | 16.0 |
| Rall (%) for FO>4σ(FO)d | 10.7 | 10.7 | n.d. | 12.9 | n.d. |
| Mean protein Beq (Å2) | 13.5 | 15.0 | 16.4 | 15.9 | 17.4 |
| Mean protein anisotropyg | 0.40 | 0.52 | 1.00 | 0.46 | 1.00 |
| Mean solvent Beq (Å2) | 33.3 | 33.6 | 32.6 | 35.5 | 30.0 |
| Mean solvent anisotropyg | 0.36 | 0.39 | 1.00 | 0.39 | 1.00 |
| RMS bond length deviation (Å) | 0.013 | 0.015 | 0.013 | 0.014 | 0.007 |
| RMS angle length deviation (Å)d | 0.031 | 0.031 | n.d. | 0.030 | n.d. |
| RMS bond angle deviation (degrees)h | n.d. | n.d. | 1.37 | n.d. | 1.06 |
| RMS chiral volume deviation (Å3)d | 0.077 | 0.096 | 0.088 | 0.091 | 0.095 |
Values in parenthesis indicate the statistics in the highest resolution shells. For the wild-type and E18D DJ-1 datasets, this range is 1.24-1.20 Å. For the E18L DJ-1 dataset, the highest resolution shell is 1.19-1.15 Å. For E18Q DJ-1, it is 1.40-1.35 Å and for E18N DJ-1, it is 1.66-1.60 Å.
, where i is the ith observation of a reflection with indices h,k,l and angle brackets indicate the average over all i observations.
, where Fchkl is the calculated structure factor amplitude with index h,k,l and Fohkl is the observed structure factor amplitude with index h,k,l.
A statistic that is reported for structures refined in SHELXL; n.d., not determined.
Rfree is calculated as Rwork, where the Fohkl are taken from a test set comprising 5% of the data that were excluded from the refinement.
Rall is calculated as Rwork, where the Fohkl include all measured data (including the Rfree test set).
Anisotropy is calculated as the ratio of the smallest to the largest eigenvalues of the refined ADP tensor and is calculated using PARVATI.
A statistic that is reported for structures refined in REFMAC5; n.d., not determined.
Crystal structure refinement
The structure of orthorhombic wtDJ-1 was solved by molecular replacement with human DJ-1 (PBD code 1P5F) (25) as a search model in PHASER (32) as implemented in the CCP4 suite of programs (33). The structures of E18L, E18Q, E18D and E18N DJ-1 in space group P3121 were refined using human DJ-1 (1P5F) as the starting model (25). Diffraction data that extend to atomic resolution (dmin≤1.2 Å) were collected for orthorhombic wt, E18L, and E18D DJ-1, while the data for E18Q and E18N DJ-1 were of lower resolution at 1.35 Å and 1.6 Å, respectively. SHELX-97 was used for the restrained least-squares refinement of the atomic resolution structures against a weighted intensity-based residual target function (34). The refinements for E18Q, and E18N DJ-1 were performed using REFMAC5 against an amplitude-based maximum likelihood target function (35). All refinements included both stereochemical and displacement parameter restraints and excluded a test set of 5% of randomly chosen reflections that were sequestered and used for the calculation of the Rfree value (36). Bulk solvent models were used to allow inclusion of all measured data (excluding the test set) in all refinements and anisotropic scaling was used in the REFMAC5 refinements.
Manual adjustments to the models were guided by inspection of 2mFO-DFC and mFO-DFC electron density maps in the program COOT (37), followed by additional cycles of refinement. For orthorhombic wt, E18L and E18D DJ-1, anisotropic atomic displacement parameters (ADPs) were refined, resulting in approximately a 5% decrease in both R and Rfree compared to the isotropic displacement parameter model. Due to the lower resolution of the E18Q, and E18N DJ-1 datasets, only the isotropic displacement parameter models were used for these structures. For all refinements, hydrogen atoms were introduced in the riding positions on all atoms for which there is an unambiguous assignment of hydrogen geometry.
Final model quality was assessed with PROCHECK (38), MolProbity (39) and the validation tools in COOT (37). In every DJ-1 structure, the backbone torsion angles for C106 were in the unfavorable region of Ramachandran space. This cysteine residue is always found in marginal or unfavorable regions in the Ramachandran plots of DJ-1 superfamily proteins (40). The refined ADP models for orthorhombic wt, E18L, and E18D DJ-1 were analyzed using the PARVATI server (41). Final model statistics are provided in Table 1.
Atomic resolution bond length analysis
The protonation state of E18 was determined using an analysis of the changes in the bond length between heavier atoms that occurs upon protonation of one of the atoms (42, 43). To allow bond length determination for E18 that was minimally biased by crystallographic restraints, the refined 1.2 Å resolution structure of orthorhombic wtDJ-1 was subjected to an additional ten additional cycles of conjugate gradient least squares refinement in SHELX-97 (34) after removing all geometric (DFIX, DANG, and CHIV) restraints for E18. The resulting model was then subjected to a single cycle of blocked (BLOC 1), unrestrained full matrix least squares refinement and inversion of the least-squares matrix to determine the estimated standard uncertainties (esu) of the bond lengths in E18. We note that the final model deposited in the PDB (PDB code 2OR3) was refined with bond length and angle restraints; the unrestrained refinement was used only to determine the protonation state of E18 during analysis of the structure.
Spectrophotometric cysteine pKa determination
Samples of tag-cleaved DJ-1 in storage buffer (25 mM HEPES, 100 mM KCl, and 2 mM DTT) were centrifugally desalted using PD-6 resin (Bio-Rad) that had been equilibrated with freshly degassed water. Solutions with pH values in the range of 3.3–7.4 were prepared using an equimolar sodium citrate and sodium phosphate double buffer system adjusted to the desired pH by addition of small volumes (0.5–1 µL) of 500 mM NaOH. The double buffer was used at a concentration of 10 mM for each buffer component and the pH of each freshly prepared buffer used in the experiment was measured at its working concentration using a micro pH electrode after the addition of 20–30 µM DJ-1. Titrations of wtDJ-1 in 10 mM double buffer supplemented with 100 mM NaCl showed no effect of increased ionic strength on the pKa of C106 in wtDJ-1 (Supporting Information S1).
Ionization of the cysteine thiol was monitored by absorption of the thiolate anion at 240 nm (44, 45) using a Cary 50 UV-visible spectrometer (Varian) and a 1 cm pathlength quartz cuvette. Freshly desalted DJ-1 in water was diluted 10–15 fold (to a final concentration of 20–30 µM DJ-1) into 10 mM double buffer and the absorption of the sample at 240 and 280 nm was measured after correction for the absorption of the buffer alone. The extinction coefficient at 240 nm (ε240) was calculated using the ratio of absorbance at 280 and 240 nm with the following equation:
| (1) |
Where A240/A280 is the ratio of the absorbance of the protein at 240 nm and 280 nm, ε280 is the known extinction coefficient of DJ-1 at 280 nm (4000 M−1 cm−1) and ε240 is the extinction coefficient at 240 nm. Using the A240/A280 ratio normalizes all measurements to the amount of protein present in the cuvette. We note that this method assumes that the ε280 value for DJ-1 is not pH-dependent, which is justified by the behavior of C106S DJ-1 (Figure 1B).
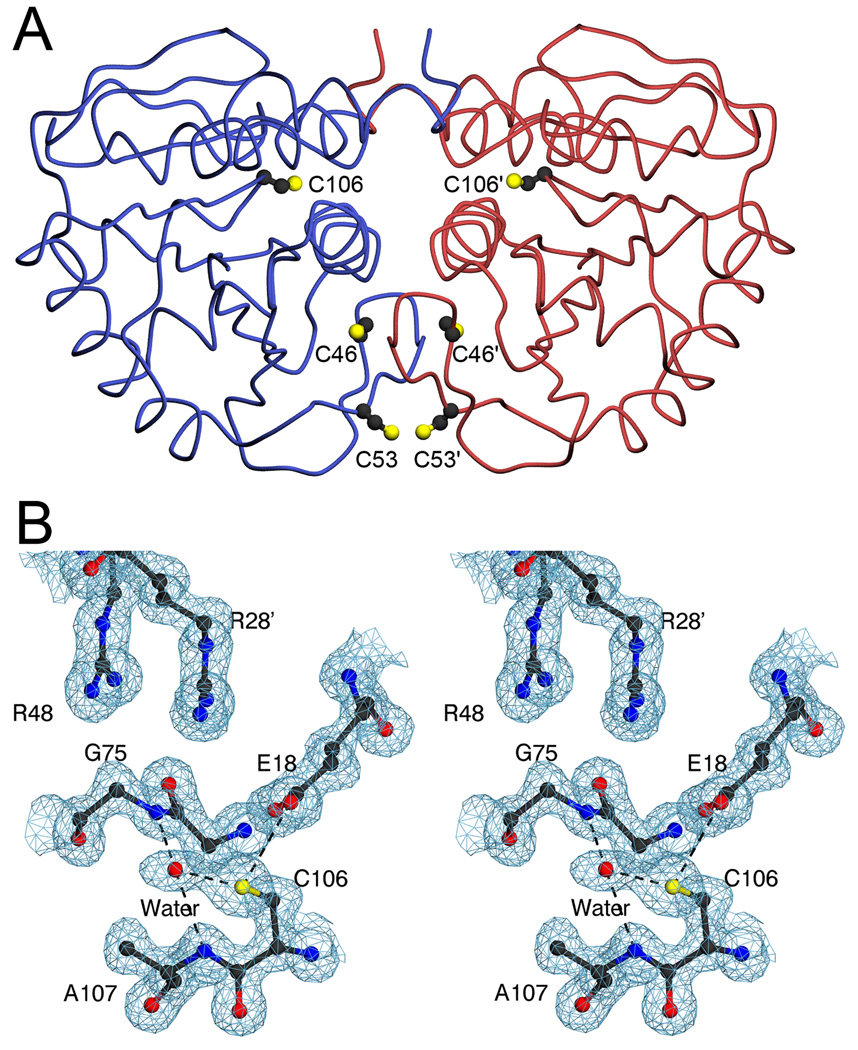
Figure 1.
Cysteine residues in human DJ-1. In Panel A, the DJ-1 dimer is represented with one monomer in blue and the other in red. The three cysteine residues in DJ-1 are shown as ball-and-stick and symmetry-related residues are labeled with a prime. Both C106 and C53 are solvent exposed, while C46 is buried. In Panel B, the residues composing the environment of C106 are shown in stereo view with 2mFo-DFc electron density contoured at 1σ (blue). The thiol of C106 makes two direct hydrogen bonds with surrounding atoms (dashed); one to E18 and the other to an ordered water molecule. The ordered water is bound in the G75/A107 “amide pocket” and is stabilized by two hydrogen bonds with the peptide backbone. This bound water molecule is not always seen in crystal structures of DJ-1 and may initiate the oxidation of C106. The figure was created with POVscript+(61).
The titrations for tag-cleaved wild-type and C106S DJ-1 were performed in triplicate and average ε240 values with associated standard deviations from three trials for each protein sample were calculated. The data were plotted as a function of pH using SigmaPlot (Systat Software) and the pKa was determined by fitting a version of the Henderson-Hasselbalch equation to the data:
| (2) |
Where is the observed extinction coefficient of the protein at 240 nm as a function of pH, is the extinction coefficient of the protein at 240 nm when the cysteine residue is in the thiol form, is the extinction coefficient of the protein at 240 nm when the cysteine residue is in the thiolate form, and pH is the measured pH of the buffer. Nonlinear regression was performed using SigmaPlot and the reported errors on the pKa values are those obtained from the fit procedure.
Search of the PDB for aspartic/glutamic acid-cysteine interactions
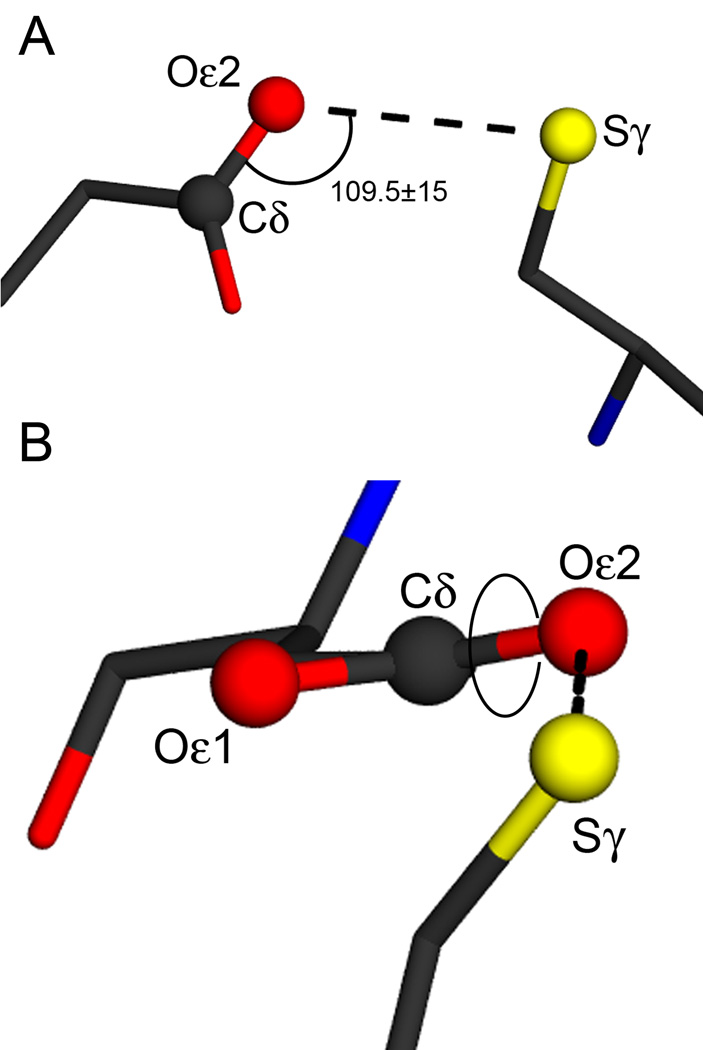
A list of all non-redundant X-ray crystal structures determined at a resolution of 3.0 Å or better was obtained by searching the 4/15/08 release of the PDB using the RCSB search tool (46). Non-redundant structures were defined as those sharing less than 90% sequence identity with the sequences of other deposited structures and the resulting list (11,765 entries) included only the highest resolution example of a set of similar proteins. This set of structures was searched for all instances in which one of the carboxylate oxygen atoms from aspartic (Oδ1 and Oδ2) or glutamic (Oε1 and Oε2) acid occurred within 4.0 Å of the Sγ atom of a cysteine residue using a custom-written search routine. Additional geometric selection criteria were applied to select for candidate hydrogen bonds between D/E and C by including the hydrogen bond angle and a “virtual” dihedral angle that includes the hydrogen bond. The hydrogen bond angle was defined as the angle spanned by the C(δ/γ)-O(δ/ε;1/2)–Sγ atoms, where the C(δ/γ) and O(δ/ε;1/2) atoms belong to carboxylic acid group of glutamic or aspartic acid. The virtual dihedral angle was defined as the dihedral spanned by the O(δ/ε;1/2)–C(δ/γ)-O(δ/ε;2/1)–Sγ atoms, where the first three atoms belong to the carboxylic acid group of aspartic or glutamic acid. The target hydrogen bond angle was 109.5±15° and the target dihedral angle values were 0±20° and 180±20°, corresponding to the cis and trans orientations of this dihedral. For each set of values, the later number indicates the accepted deviation from the target. By convention, clockwise rotation is a positive displacement for the dihedral angle.
Results
C106 has a depressed pKa
Human DJ-1 contains three cysteine residues (C46, C53 and C106), two of which (C106 and C53) are solvent-exposed and thus would be more likely to form reactive thiolates (Fig 1A). The most oxidation-sensitive cysteine in human DJ-1 is C106 (5, 47), and its environment is rich in ionizable residues that could perturb the pKa of this functionally essential residue (Figure 1B). The pKa of C106 was monitored by measuring the increased absorbance of UV light at 240 nm resulting from formation of the thiolate anion (44, 45). The pH-dependence of the molar extinction coefficient of DJ-1 at 240 nm (ε240) displays a transition with pKa=5.4±0.1 and a total increase of 3500 M−1 cm−1 (Fig 2A), consistent with the reported extinction coefficient of 4000 M−1 cm−1 for a single thiolate at 240 nm (44). This increase in ε240 is due solely to ionization of C106, as demonstrated by the absence of a pH-dependent transition for C106S DJ-1 (Fig 2A). The pKa of C106 in unchanged when measured in 10 mM double buffer supplemented with 100 mM NaCl (Supporting Information Figure S1), demonstrating that the pKa of C106 is 5.4±0.1 in solutions of physiologically relevant ionic strength.
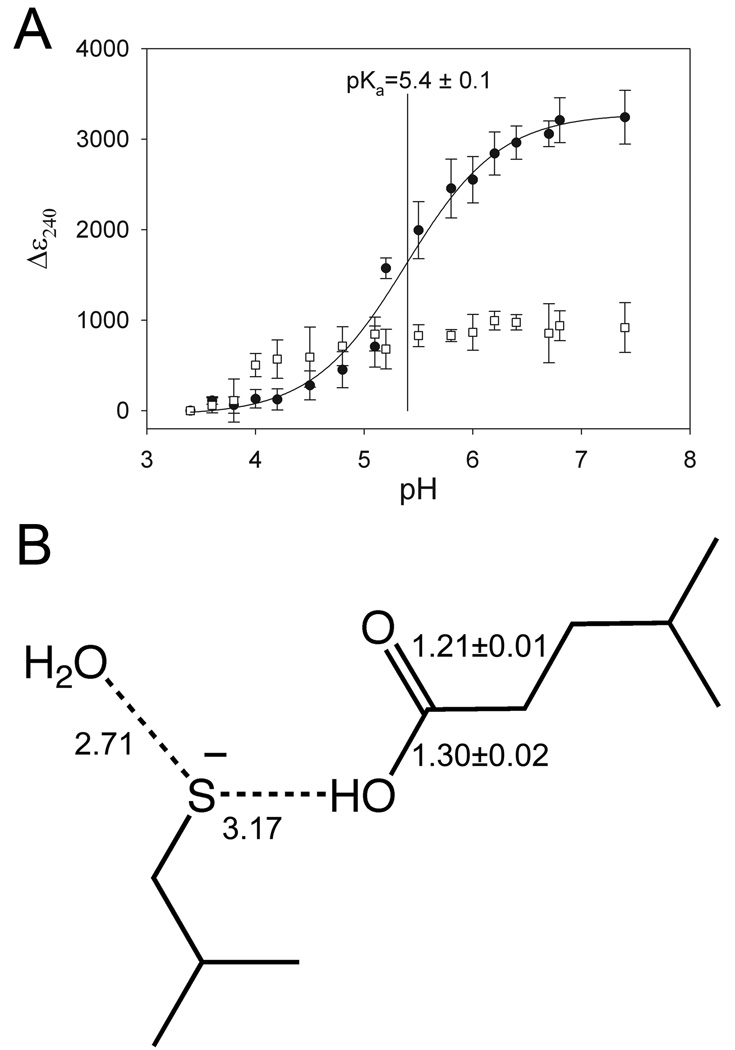
Figure 2.
C106 has a depressed pKa and accepts a hydrogen bond from a protonated E18. In Panel A, the change in the extinction coefficient at 240 nm (Δε240) is measured as a function of pH to monitor formation of the cysteine thiolate anion. The Δε240 value is calculated by subtraction of the ε240 measured at the lowest pH from all other measured ε240 values. Wild-type DJ-1 (filled circles) exhibits a Δε240 transition corresponding to a pKa of 5.4±0.1, where mean data and associated standard deviations for triplicate measurements are shown. The curve shows the nonlinear regression fit of equation 2 to the measured data. C106S DJ-1 (open squares) shows no transition, indicating that ionization of C106 is solely responsible for the observed transition in wtDJ-1. In Panel B, unrestrained bond length refinement of a 1.2 Å resolution crystal structure of wtDJ-1 indicates that E18 is constitutively protonated and donates a hydrogen bond to C106. The refined bond lengths and estimated standard uncertainties for the carbon-oxygen bonds of E18 are shown, and hydrogen bonds are indicated with dashed lines. Distances are given in Ångstroms and the values for molecule B in the DJ-1 dimer are shown. Panel B was created with ChemDraw (CambridgeSoft).
The depression of the pKa value of C106 from the solution value of 8.3 to the measured value of 5.4 corresponds to a −4.0 kcal mol−1stabilization of the thiolate ion by the local environment of C106 in DJ-1, calculated as follows:
| (3) |
Where is the difference in the Gibbs free energy of ionization of the cysteine thiol in the protein environment compared with solution and is a measure of the degree of thiolate stabilization by the protein environment. In equation 3, ΔG0(protein) is the free energy of ionization of the cysteine residue in the protein environment, ΔG0(solution) is the free energy of ionization of the cysteine residue in solution, pKa(protein) is the pKa of the thiol in the protein environment, pKa(solution) is the pKa of the thiol in solution, R is the gas constant, and T is the temperature (K). In the environment of C106, the carboxylic acid sidechain of E18 is the only residue that is oriented properly to hydrogen bond to the thiol of C106 (Figure 1B). Both of these residues are very highly conserved in the DJ-1 superfamily (19, 20), suggesting that the E18-C106 interaction is likely to be functionally important.
We note that an ordered water molecule bound in the G75/A107 “amide pocket” is apparent in this new crystal structure of DJ-1 and is within sub-van der Waals distance (2.5 Å in molecule A, 2.7 Å in molecule B) of the Sγ atom of C106 (Figure 1B and Figure 5). This sulfur-oxygen distance is too great to correspond to a cysteine-sulfenic acid, but too short to be a normal hydrogen bond between these atoms. We propose that this density is most consistent with a mixture of species, including the bound water (or other oxygen-containing species) and a minor contribution from C106-sulfenic acid. Therefore, we speculate that this may represent an early oxidation state for C106 that contains a mixture of stable intermediate species. This is consistent with a previously proposed oxidation sequence for the reactive cysteine residue in the E. coli DJ-1 homologue YajL (40).
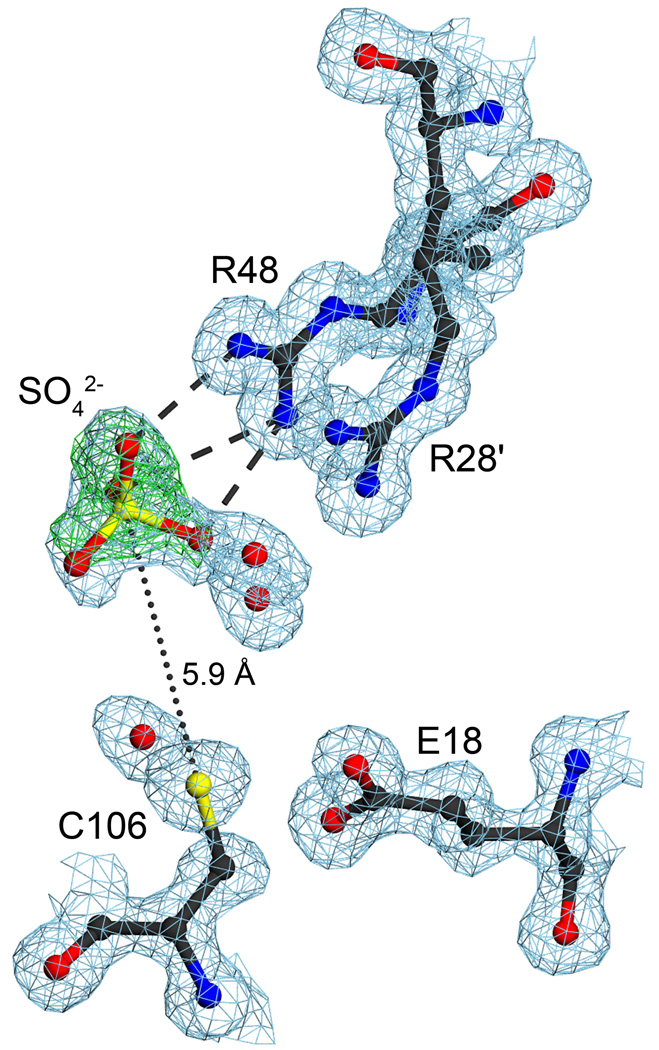
Figure 5.
Two proximal arginine residues bind an anion near C106. The environment of the R28’/R48 dyad is shown with 2mFo-DFc electron density is contoured at 1σ (blue) and mFo-DFc electron density contoured at 4σ (green). R28 and R48 participate in an unusual guanidinium stacking interaction that spans the DJ-1 dimer interface, with R28’ contributed by the other monomer in this view (indicated by a prime). A bound sulfate ion interacts with R48 through three direct hydrogen bonds to the guanidinium sidechain, illustrated as dashed lines. Positive difference electron density contoured at 4σ (green) was calculated prior to the inclusion of the sulfate ion in the model. The R28’/R48 anion binding site places a negative charge within 5.9 Å of C106. The ordered water bound near C106 in the G75/A107 “amide pocket” likely initiates oxidation of this residue. The figure was made with POVscript+(61).
E18 is protonated and donates a hydrogen bond to C106
Formation of a cysteine thiolate anion is electrostatically disfavored by nearby negative charges. It is therefore surprising that C106 has both a depressed pKa and makes a 3.2 Å hydrogen bond with E18 (Figure 1B). Bond length analysis of a new crystal structure of orthorhombic wtDJ-1 refined at 1.2 Å resolution shows that E18 is protonated (and thus uncharged), and donates a hydrogen bond to C106. The orthorhombic crystal form of DJ-1 has both molecules of the physiological DJ-1 dimer in the asymmetric unit, providing two independent views of E18. Unrestrained refinement of E18 bond lengths in SHELX-97 (see Materials and Methods) yields Cδ-Oε1 bond lengths of 1.241 Å (esu=0.021 Å) for molecule A and 1.213 Å (esu=0.012 Å) for molecule B. The Cδ-Oε2 bond lengths are 1.296 Å (esu=0.021 Å) for molecule A and 1.304 Å (esu=0.020 Å) for molecule B, demonstrating that the Oε2 atom of E18 is protonated (Figure 2B). These bond lengths compare well with the bond lengths for a fully protonated (uncharged) carboxylic acid, which are 1.21 Å for a carbon-oxygen double bond and 1.30 Å for a carbon-oxygen single bond from the Engh and Huber parameter set that is widely used in macromolecular refinement (48). The crystals were grown at pH=7.5, suggesting that E18 is constitutively protonated at physiological pH.
Hydrogen bond donation from E18 strongly influences C106 pKa
The influence of hydrogen bond donation by E18 on the pKa of C106 was determined using site-directed mutagenesis and spectrophotometric pKa determination. The E18-C106 hydrogen bond was eliminated by creating an E18L mutation, which elevates the pKa of C106 to 6.4±0.1 (Table 2). The elevation of the pKa of C106 by 1.0 unit in E18L DJ-1 corresponds to a loss of 1.4 kcal mol−1 of stabilization energy for the C106 thiolate compared to wtDJ-1 (1.4 kcal mol−1 =; see equation 3 and Table 2); approximately 1/3 of the total stabilization of C106 ionization by DJ-1 relative to free cysteine in solution. The 1.15 Å resolution crystal structure of E18L DJ-1 shows no major differences in the overall structure of DJ-1 or the local conformation of C106 occur in response to the E18L substitution (Figure 3A). Therefore, we conclude that the elevation of C106 pKa by 1.0 unit in E18L DJ-1 is due principally to the loss of a single hydrogen bond donated by E18.
Table 2.
C106 pKa Values and Thiolate Stabilization Free Energies in DJ-1
| Protein | C106 pKa | (kcal mol−1)a |
|---|---|---|
| wt | 5.4±0.1 | −4.0 |
| E18L | 6.4±0.1 | −2.6 |
| E18Q | 5.7±0.1 | −3.5 |
| E18D | 6.1±0.1 | −3.0 |
| E18N | 6.1±0.2 | −3.0 |
| R28Q | 5.4±0.1 | −4.0 |
| R48Q | 5.2±0.1 | −4.2 |
| R28Q/R48Q | 5.0±0.1 | −4.5 |
is the difference Gibbs free energy for ionization of the free cysteine thiol in solution and in the protein environment and is calculated using equation 3.
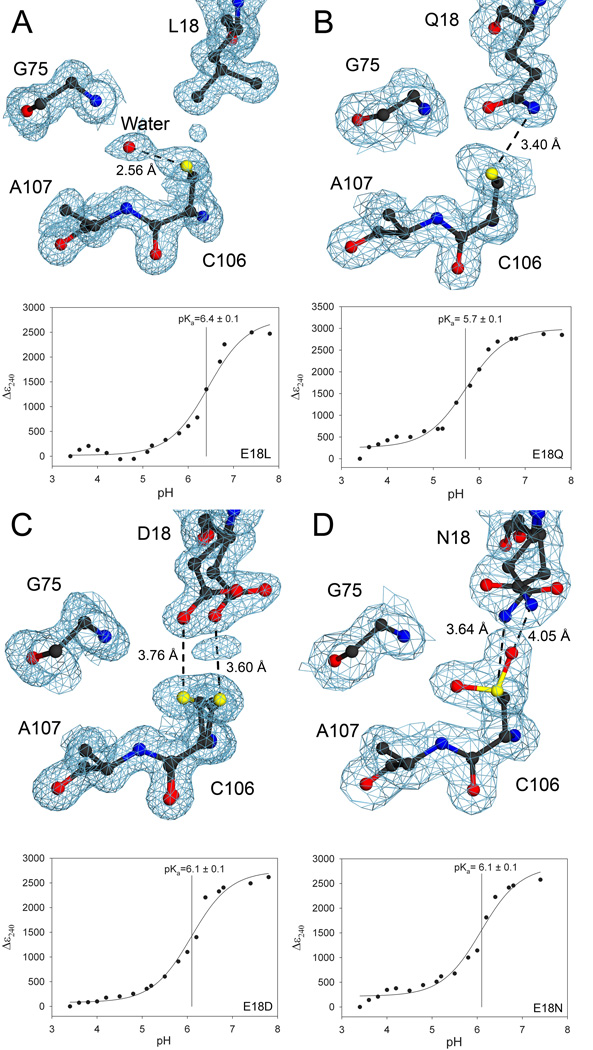
Figure 3.
Hydrogen bond donation by residue 18 contributes to pKa depression of C106. In all panels, 2mFo-DFc electron density is contoured at 1σ (blue) and the pH dependence of the Δε240 value is shown in the lower portion of each panel. The Δε240 value is calculated by subtraction of the ε240 measured at the lowest pH from all other measured ε240 values. For the Δε240 vs. pH plots, measured data are shown with the best-fit curve from equation 2. Panel A shows that eliminating the hydrogen bond between residue 18 and C106 with an E18L substitution elevates the C106 pKa value by 1.0 unit to pKa=6.4. Panel B shows that a structurally conservative E18Q substitution lengthens the hydrogen bond to Sγ of C106 by 0.2 Å and increases the C106 pKa value by 0.3 unit. Panel C shows that the E18D substitution results in spatially correlated discrete disorder at D18 and C106, lengthens the hydrogen bond by approximately 0.5 Å compared to wtDJ-1, and increases the C106 pKa value by 0.7 unit. Panel D shows that the E18N substitution leads to discrete disorder at N18, enhanced oxidation at C106, and also increases the C106 pKa value by 0.7 unit. All of the substitutions at residue 18 illustrate the requirement for a protonated glutamic acid at position 18 for maximal depression of C106 pKa. The figure was made with POVscript+(61).
To obtain a more complete understanding of the influence of E18 on the pKa of C106, we altered the strength of the hydrogen bond donated by residue 18 by generating E18Q, E18D, and E18N substitutions in DJ-1. Each of these engineered mutations at residue 18 elevates the pKa of C106 (Table 2). The most structurally conservative substitution, E18Q, increases the pKa by 0.3 units and lengthens the hydrogen bond to Sγ by 0.2 Å (Figure 3B). The electron density for C106 in E18Q DJ-1 is elongated at the Sγ atom, consistent either with conformational disorder or minor oxidation of the cysteine residue. In contrast to the well-ordered Q18 sidechain in E18Q DJ-1, the shorter sidechains of the E18D and E18N substitutions result in discrete conformational disorder and increase the hydrogen bond to C106 by approximately 0.4 Å compared to wtDJ-1 (Figure 3C,D). In addition, C106 is invariably oxidized to C106-sulfinic acid in crystals of E18N DJ-1 (Figure 3D), further lengthening the Nδ2-Sγ distance. We note that, in contrast to the oxidized crystalline protein, the E18N DJ-1 samples used for solution pKa determination are approximately 85% reduced as determined by electrospray mass spectrometry (UNL Mass Spectrometry Core). Oxidation of C106 to C106-sulfinic acid is well-established in DJ-1 and can occur during crystallization of initially reduced protein (5). This combined structural and C106 pKa comparison of the E18Q, E18N and E18D substitutions shows that even minor lengthening of the hydrogen bond donated by residue 18 significantly destabilizes the C106 thiolate, emphasizing the critical role of the E18-C106 interaction in DJ-1.
Interactions between acidic residues and cysteine in the PDB
The set of 11,765 non-redundant X-ray crystal structures determined at 3.0 Å resolution or better was extracted from the 4/15/08 release of the PDB and searched for every instance in which one of the carboxylate oxygen atoms of aspartic or glutamic acid was within 4.0 Å of the cysteine Sγ atom (see Experimental Procedures). This is a permissive screen for candidate D/E-C interactions and is likely to result in many false positives. We identified 6477 instances of proximal D/E-C pairs in 2349 unique structures, or roughly 20% of the models in the search set. With this set, we performed a more stringent search for all examples in which the hydrogen bond angle defined by the C(δ/γ)-O(δ/ε;1/2)-Sγ atoms was within 109.5±15° and the O(δ/ε;1/2)–C(δ/γ)-O(δ/ε;1/2)–Sγ dihedral angle was either 0±20° or 180±20° (Figure 4A,B). These geometric criteria have been previously established for hydrogen bonds involving protonated aspartic or glutamic acids, with the 0° and 180° targets for the dihedral angle corresponding to the favored cis and trans orientations (49). For members of the DJ-1 superfamily, this dihedral is near the cis orientation. This results in 235 D/E-C pairs occurring in 149 unique structures (Supporting Information Table S2), or approximately 1.3% of the original search set. All DJ-1 superfamily members of known structure that contain a putative E-C dyad are represented in this list except E. coli Hsp31 (PDB code 1N57 (50)), whose 25.7° H-bond dihedral angle only slightly exceeds our accepted deviation tolerance of 20°. Moreover, the E18-C106 interaction is correctly identified for human DJ-1 (PDB 2RK3 (51)). In total, the relatively small fraction of structures that returned a positive hit in our search suggests that hydrogen bonded D/E-C interactions are uncommon amongst all structures in the PDB, but common within the DJ-1 superfamily.
Figure 4.
Geometric parameters for carboxylic acid-cysteine hydrogen bonding. The E18-C106 interaction is shown in both panels, with the hydrogen bond indicated with the dotted line. Atoms involved in the definition of the hydrogen bond angle and dihedral are shown as balls and labeled. Panel A shows the definition of the hydrogen bond angle, which is set to a target value of 109.5±15° in the PDB search. Panel B shows the convention for defining the hydrogen bond dihedral angle, with the rotatable bond indicated by the circle. This dihedral angle has a value of 15° in human DJ-1 and target values of 0±20° (cis) and 180±20° (trans) were used in the PDB search. The figure was made with POVscript+(61)
R28 and R48 bind anions that elevate the pKa of C106
DJ-1 contains two highly conserved arginine residues (R28’ and R48) that participate in an unusual guanidinium stacking interaction that spans the dimer interface and is located near C106 (Figure 4). The prime notation (e.g. R28’) indicates a residue that is donated by the other monomer in the DJ-1 dimer. The charge state of these two residues is ambiguous: the high solution pKa of the arginine sidechain (pKa=12) would suggest that they are positively charged, however the atypical 3.6 Å stacking of the guanidinium groups of R28’ and R48 is electrostatically unfavorable for two positively charged groups (Figure 5). Despite the uncertain charge state of these residues, R28’ and R48 compose an anion binding site that is occupied by sulfate in both molecules in the ASU of the orthorhombic crystal structure of DJ-1 (Figure 5). The sulfur atom of the sulfate anion is 5.9 Å from the Sγ of C106, and this proximal negatively charged group is expected to suppress ionization of the C106 thiol.
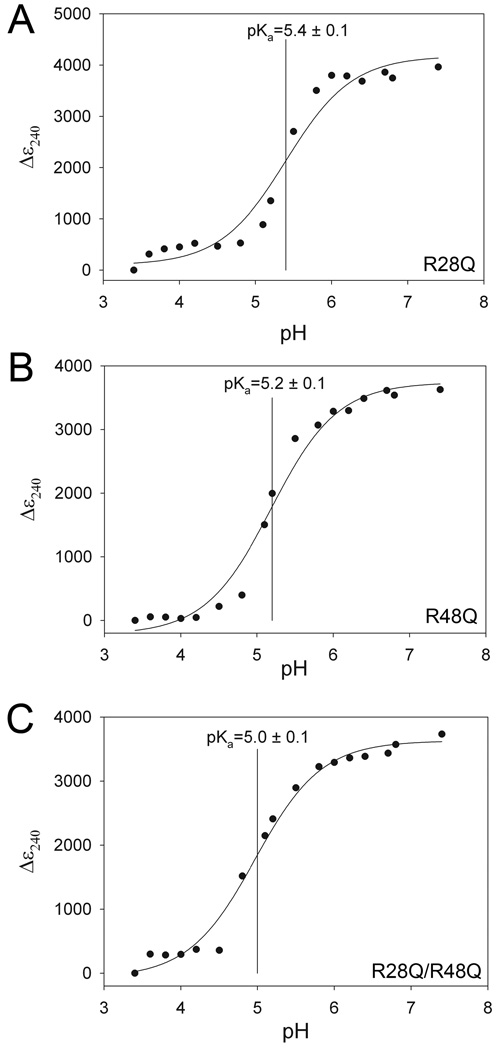
The bound sulfate anion makes direct hydrogen bonds only to R48, and mutation of R28’ to glutamine does not effect the pKa of C106 under these experimental conditions (Figure 6A). The R48Q substitution results in a 0.2 unit decrease in C106 pKa, while the double R28Q/R48Q substitution results in a larger 0.4 unit C106 pKa decrease. The depression of the C106 pKa value upon removal of two nearby basic residues is surprising and strongly suggests that the R48Q and R28Q/R48Q engineered substitutions disrupt the anion binding site near C106. The loss of this bound anion alleviates electrostatic suppression of thiolate formation, thereby decreasing the pKa of C106 (Figure 6, Table 2). The sulfate observed in the crystal structure is not present in the double buffer used in the C106 pKa titration, thus the R28/R48 anion binding site must be occupied by other anions in buffer, most likely phosphate. Intriguingly, the pH-dependent ε240 transition for both R28Q and R48Q is more cooperative than expected for a single ionization obeying the Henderson-Hasselbalch equation (Figure 6A,B), suggesting that the C106 pKa may be influenced by the ionization of other nearby groups. A plausible candidate is the E15/E16/D24’ cluster of acidic residues which make multiple hydrogen bonds to the R48/R28’ pair and may be perturbed by the single R to Q mutations being used in this study.
Figure 6.
The R28/R48 anion binding site elevates the pKa of C106. Substitution of glutamine for R28 (Panel A) results in no change in the pKa of C106, while a R48Q substitution reduces the C106 pKa by 0.2 unit and the R28Q/R48Q double substitution decreases the C106 pKa by 0.4 unit. Measured data are shown with the best-fit curve from equation 2. Both the R48Q and double R28Q/R48Q substitutions decrease the C106 pKa value, indicating that the bound anion observed in the crystal structure suppresses the ionization of C106 in wtDJ-1.
Discussion
The cytoprotective activity of DJ-1 requires C106 (5, 6), and our results establish that this residue has a low pKa of 5.4±0.1. Site directed mutation of residue 18 demonstrates that approximately one third of the total stabilization energy for the ionization of C106 results from the Sγ thiolate of C106 accepting a hydrogen bond from the constitutively protonated Oε2 atom of E18. It is noteworthy that the disordered E18D and E18N substitutions increase the C106 pKa value by comparable amounts (Table 2), despite the different electrostatic and hydrogen bonding character of these two substitutions (Table 2). Furthermore, the E18D and E18N substitutions have pKa values near that of the E18L mutant, which eliminates this hydrogen bond entirely. We conclude that the C106-E18 interaction in DJ-1 represents an unusual type of dyad that uses a protonated carboxylic acid moiety to depress the pKa of a reactive cysteine residue. Residues that are structurally equivalent to C106 and E18 in human DJ-1 are very highly conserved in other members of the DJ-1 superfamily, indicating that the glutamic acid-cysteine dyad is widely distributed in the superfamily (19, 20). We note that our current data do not preclude the possibility that the E18-C106 dyad represents a general acid-general base pair, although this is highly speculative in the absence of a well-established catalytic activity for DJ-1.
The influence of E18, R28 and R48 on the pKa of C106 contradicts expectations based on the typical ionization behavior of these residues in other proteins. In this regard, DJ-1 serves as a cautionary example that illustrates the potential limitations of computational approaches to determining pKa values, as a previous report suggested a pKa value of 11.38 for C106 based on computational prediction (52). Furthermore, our results show that when the hydrogen bond between E18 and C106 is eliminated by mutagenesis, the pKa of C106 is still 1.9 pH units below that of free cysteine. This suggests that additional factors contribute to the depressed pKa of C106 in DJ-1, possibly including the strained backbone conformation around this residue or the α-helix macropole of helix E (3). Future experimental studies of the structural determinants of depressed cysteine pKa values in DJ-1 and other systems are expected to aid in the development of improved algorithms for the computational prediction of cysteine pKa values.
Cysteine residues with depressed pKa values often serve catalytic roles (53–55). The biochemical function of DJ-1 is uncertain, and attempts to detect various enzymatic activities have been inconclusive (15, 25, 29, 52, 56). Most published reports agree, however, that C106 is exquisitely sensitive to oxidation and readily forms the C106-sulfinic acid (C106-SO2−) under physiological conditions (5, 29, 47, 52, 57). The 1.2 Å resolution crystal structure of oxidized DJ-1 containing C106-SO2− shows that one of the two sulfinic acid oxygen atoms of C106-SO2− makes an unusually short 2.47 Å hydrogen bond to the Oε2 atom of E18 (5). Therefore, the protonated carboxylic acid of E18 contributes both to rendering C106 reactive by depressing its pKa and also to the stabilization of a specific oxidized form of this cysteine residue. The C106-SO2− form of DJ-1 has been implicated in the neuroprotective function of the protein by several distinct mechanisms, including enhancing mitochondrial DJ-1 localization (5), regulating a redox-regulated chaperone activity of the protein (15, 29), and directly scavenging reactive oxygen species (17, 58). Although further research will be required to determine which, if any, of these activities is critical for DJ-1’s cytoprotective function, it is clear that the E18-C106 dyad plays a major role in directing the chemistry that can occur at C106 in DJ-1 and its many homologues.
A survey of structures in the PDB reveals that approximately 20% of the structures in our search list have at least one carboxylic oxygen atom of either glutamic and aspartic acid within 4 Å of the sulfur atom of cysteine. Of this set of possible D/E-C dyads, only 149 structures (1.3% of the original 11,765 structures) contain a candidate D/E-C interaction that satisfies more stringent angular and dihedral criteria for hydrogen bonding (see Experimental Procedures). Therefore, candidate D/E-C dyads that are structurally similar to the hydrogen bonded E18-C106 interaction observed in DJ-1 are rare. We note that we have used moderately stringent criteria for identifying candidate D/E-C interactions, and protonated acidic residues can influence cysteine reactivity in diverse structural environments that do not satisfy these criteria. In the well-studied example of E. coli thioredoxin, a buried aspartic acid residue (D26) with an elevated pKa acts as a general acid/base that facilitates thiol-disulfide exchange involving C32 in the enzyme active site (59, 60). While the E18-C106 interaction in DJ-1 does not appear to be involved in thiol-disulfide exchange, our results suggest that functionally important interactions between carboxylates and thiols are present in several types of proteins. It is difficult to speculate as to the frequency that a protonated acidic residue depresses the pKa of cysteine because it is impractical to determine the cysteine pKa values and carboxylic acid protonation states of all of the candidate D/E-C interactions listed in Supporting Information Table S2. We propose, however, that this type of interaction is common for residues that are structurally homologous to E18 and C106 in the large DJ-1 superfamily.
Supplementary Material
Figure S1 shows a titration of wtDJ-1 in a double buffer supplemented with 100 mM NaCl, demonstrating that increased ionic strength has no effect on the pKa of C106 under these conditions. Table S2 lists every example of a candidate D/E-C hydrogen bonded dyad in the 4/15/08 release of the PDB. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
This work was supported in part by a grant to M.A.W. from the American Parkinson’s Disease Association as well as a grant from the National Institutes of Health (P 20RR-17675). Use of the Advanced Photon Source was supported by the U.S. Department of Energy Basic Energy Sciences, Office of Science, under Contract No. W-31-109-Eng-38. Use of the BioCARS Sector 14 was supported by the National Institutes of Health, National Center for Research Resources, under grant number RR07707
We thank the staff of BioCARS 14-BMC at the Advanced Photon Source, Argonne, IL for beamline support, Dr. Hideaki Moriyama for support of the rotating anode X-ray source at the University of Nebraska, and Dr. Todd Holyoak (University of Kansas Medical Center) for helpful discussions.
ABBREVIATIONS
- ADP
anisotropic atomic displacement parameter
- DTT
dithiothreitol
- EGCG
epigallocatechin 3-gallate
- HEPES
4-(2-hydroxyethyl)-1-piperazine-ethane sulfonic acid
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- PEG
polyethylene glycol
- PDB
Protein Data Bank
- UV
ultraviolet
- wt
wild-type
Footnotes
Note: Refined model coordinates and experimental structure factors have been deposited in the PDB with accession codes 2OR3 (orthorhombic wtDJ-1), 3CY6 (E18Q), 3CYF (E18N), 3CZ9 (E18L) and 3CZA (E18D).
References
- 1.Kortemme T, Darby NJ, Creighton TE. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of protein disulfide isomerase. Biochemistry. 1996;35:14503–14511. doi: 10.1021/bi9617724. [DOI] [PubMed] [Google Scholar]
- 2.Jao SC, English Ospina SM, Berdis AJ, Starke DW, Post CB, Mieyal JJ. Computational and mutational analysis of human glutaredoxin (thioltransferase): probing the molecular basis of the low pKa of cysteine 22 and its role in catalysis. Biochemistry. 2006;45:4785–4796. doi: 10.1021/bi0516327. [DOI] [PubMed] [Google Scholar]
- 3.Kortemme T, Creighton TE. Ionisation of cysteine residues at the termini of model alpha-helical peptides. Relevance to unusual thiol pKa values in proteins of the thioredoxin family. J. Mol. Biol. 1995;253:799–812. doi: 10.1006/jmbi.1995.0592. [DOI] [PubMed] [Google Scholar]
- 4.Naor MM, Jensen JH. Determinants of cysteine pKa values in creatine kinase and alpha1-antitrypsin. Proteins. 2004;57:799–803. doi: 10.1002/prot.20261. [DOI] [PubMed] [Google Scholar]
- 5.Canet-Aviles RM, Wilson MA, Miller DW, Ahmad R, McLendon C, Bandyopadhyay S, Baptista MJ, Ringe D, Petsko GA, Cookson MR. The Parkinson's disease protein DJ-1 is neuroprotective due to cysteine-sulfinic acid-driven mitochondrial localization. Proc. Natl. Acad. Sci. USA. 2004;101:9103–9108. doi: 10.1073/pnas.0402959101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Meulener MC, Xu K, Thomson L, Ischiropoulos H, Bonini NM. Mutational analysis of DJ-1 in Drosophila implicates functional inactivation by oxidative damage and aging. Proc. Natl. Acad. Sci. USA. 2006;103:12517–12522. doi: 10.1073/pnas.0601891103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299:256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 8.Zhang L, Shimoji M, Thomas B, Moore DJ, Yu SW, Marupudi NI, Torp R, Torgner IA, Ottersen OP, Dawson TM, Dawson VL. Mitochondrial localization of the Parkinson's disease related protein DJ-1: implications for pathogenesis. Hum. Mol. Genet. 2005;14:2063–2073. doi: 10.1093/hmg/ddi211. [DOI] [PubMed] [Google Scholar]
- 9.Choi J, Sullards MC, Olzmann JA, Rees HD, Weintraub ST, Bostwick DE, Gearing M, Levey AI, Chin LS, Li L. Oxidative damage of DJ-1 is linked to sporadic Parkinson and Alzheimer diseases. J. Biol. Chem. 2006;281:10816–10824. doi: 10.1074/jbc.M509079200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li HM, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Association of DJ-1 with chaperones and enhanced association and colocalization with mitochondrial Hsp70 by oxidative stress. Free Radic. Res. 2005;39:1091–1099. doi: 10.1080/10715760500260348. [DOI] [PubMed] [Google Scholar]
- 11.Meulener M, Whitworth AJ, Armstrong-Gold CE, Rizzu P, Heutink P, Wes PD, Pallanck LJ, Bonini NM. Drosophila DJ-1 mutants are selectively sensitive to environmental toxins associated with Parkinson's disease. Curr. Biol. 2005;15:1572–1577. doi: 10.1016/j.cub.2005.07.064. [DOI] [PubMed] [Google Scholar]
- 12.Mitsumoto A, Nakagawa Y. DJ-1 is an indicator for endogenous reactive oxygen species elicited by endotoxin. Free Radic. Res. 2001;35:885–893. doi: 10.1080/10715760100301381. [DOI] [PubMed] [Google Scholar]
- 13.Park J, Kim SY, Cha GH, Lee SB, Kim S, Chung J. Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene. 2005;361:133–139. doi: 10.1016/j.gene.2005.06.040. [DOI] [PubMed] [Google Scholar]
- 14.Sekito A, Koide-Yoshida S, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. DJ-1 interacts with HIPK1 and affects H2O2-induced cell death. Free Radic. Res. 2006;40:155–165. doi: 10.1080/10715760500456847. [DOI] [PubMed] [Google Scholar]
- 15.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2:1764–1773. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5:213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Takahashi-Niki K, Niki T, Taira T, Iguchi-Ariga SM, Ariga H. Reduced anti-oxidative stress activities of DJ-1 mutants found in Parkinson's disease patients. Biochem. Biophys. Res. Commun. 2004;320:389–397. doi: 10.1016/j.bbrc.2004.05.187. [DOI] [PubMed] [Google Scholar]
- 18.Menzies FM, Yenisetti SC, Min KT. Roles of Drosophila DJ-1 in survival of dopaminergic neurons and oxidative stress. Curr. Biol. 2005;15:1578–1582. doi: 10.1016/j.cub.2005.07.036. [DOI] [PubMed] [Google Scholar]
- 19.Bandyopadhyay S, Cookson MR. Evolutionary and functional relationships within the DJ1 superfamily. BMC Evol. Biol. 2004;4 doi: 10.1186/1471-2148-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lucas JI, Marin I. A New evolutionary paradigm for the Parkinson disease gene DJ-1. Mol. Biol. Evol. 2006;24:551–561. doi: 10.1093/molbev/msl186. [DOI] [PubMed] [Google Scholar]
- 21.Honbou K, Suzuki NN, Horiuchi M, Niki T, Taira T, Ariga H, Inagaki F. The crystal structure of DJ-1, a protein related to male fertility and Parkinson's disease. J. Biol. Chem. 2003;278:31380–31384. doi: 10.1074/jbc.M305878200. [DOI] [PubMed] [Google Scholar]
- 22.Huai Q, Sun YJ, Wang HC, Chin LS, Li L, Robinson H, Ke HM. Crystal structure of DJ-1/RS and implication on familial Parkinson's disease. FEBS Lett. 2003;549:171–175. doi: 10.1016/s0014-5793(03)00764-6. [DOI] [PubMed] [Google Scholar]
- 23.Lee SJ, Kim SJ, Kim IK, Ko J, Jeong CS, Kim GH, Park C, Kang SO, Suh PG, Lee HS, Cha SS. Crystal structures of human DJ-1 and Escherichia coli Hsp31, which share an evolutionarily conserved domain. J. Biol. Chem. 2003;278:44552–44559. doi: 10.1074/jbc.M304517200. [DOI] [PubMed] [Google Scholar]
- 24.Tao X, Tong L. Crystal structure of human DJ-1, a protein associated with early onset Parkinson's disease. J. Biol. Chem. 2003;278:31372–31379. doi: 10.1074/jbc.M304221200. [DOI] [PubMed] [Google Scholar]
- 25.Wilson MA, Collins JL, Hod Y, Ringe D, Petsko GA. The 1.1-angstrom resolution crystal structure of DJ-1, the protein mutated in autosomal recessive early onset Parkinson's disease. Proc. Natl. Acad. Sci. USA. 2003;100:9256–9261. doi: 10.1073/pnas.1133288100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ollis DL, Cheah E, Cygler M, Dijkstra B, Frolow F, Franken SM, Harel M, Remington SJ, Silman I, Schrag J, Sussman JL, Verschueren KHG, Goldman A. The alpha/beta-hydrolase fold. Protein Engineering. 1992;5:197–211. doi: 10.1093/protein/5.3.197. [DOI] [PubMed] [Google Scholar]
- 27.Wilson MA, St Amour CV, Collins JL, Ringe D, Petsko GA. The 1.8-angstrom resolution crystal structure of YDR533Cp from Saccharomyces cerevisiae: A member of the DJ-1/ThiJ/Pfpl superfamily. Proc. Natl. Acad. Sci. USA. 2004;101:1531–1536. doi: 10.1073/pnas.0308089100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wei Y, Ringe D, Wilson MA, Ondrechen MJ. Identification of functional subclasses in the DJ-1 superfamily proteins. PLoS Comput. Biol. 2007;3:e10. doi: 10.1371/journal.pcbi.0030010. [DOI] [PubMed] [Google Scholar]
- 29.Zhou W, Zhu M, Wilson MA, Petsko GA, Fink AL. The oxidation state of DJ-1 regulates its chaperone activity toward alpha-synuclein. J. Mol. Biol. 2006;356:1036–1048. doi: 10.1016/j.jmb.2005.12.030. [DOI] [PubMed] [Google Scholar]
- 30.Holyoak T, Fenn TD, Wilson MA, Moulin AG, Ringe D, Petsko GA. Malonate: a versatile cryoprotectant and stabilizing solution for salt-grown macromolecular crystals. Acta Crystallogr. 2003;D59:2356–2358. doi: 10.1107/s0907444903021784. [DOI] [PubMed] [Google Scholar]
- 31.Otwinowski Z, Minor W. Methods in Enzymology. New York: Academic Press; 1997. Processing of X-ray diffraction data collected in oscillation mode; pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 32.McCoy AJ, Grosse-Kunstleve RW, Storoni LC, Read RJ. Likelihood-enhanced fast translation functions. Acta Crystallogr. 2005;D61:458–464. doi: 10.1107/S0907444905001617. [DOI] [PubMed] [Google Scholar]
- 33.Collaborative Computational Project, Number 4. The CCP4 Suite - Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 34.Sheldrick GM. A short history of SHELX. Acta Crystallogr A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 35.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Brunger AT. Free R-Value - A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 37.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 38.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- 39.Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson MA, Ringe D, Petsko GA. The atomic resolution crystal structure of the YajL (ThiJ) protein from Escherichia coli: A close prokaryotic homologue of the Parkinsonism-associated protein DJ-1. J. Mol. Biol. 2005;353:678–691. doi: 10.1016/j.jmb.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 41.Merritt EA. Expanding the model: anisotropic displacement parameters in protein structure refinement. Acta Crystallogr. 1999;D55:1109–1117. doi: 10.1107/s0907444999003789. [DOI] [PubMed] [Google Scholar]
- 42.Coates L, Erskine PT, Crump MP, Wood SP, Cooper JB. Five atomic resolution structures of endothiapepsin inhibitor complexes: Implications for the aspartic proteinase mechanism. J. Mol. Biol. 2002;318:1405–1415. doi: 10.1016/s0022-2836(02)00197-3. [DOI] [PubMed] [Google Scholar]
- 43.Erskine PT, Coates L, Mall S, Gill RS, Wood SP, Myles DAA, Cooper JB. Atomic resolution analysis of the catalytic site of an aspartic proteinase and an unexpected mode of binding by short peptides. Prot. Sci. 2003;12:1741–1749. doi: 10.1110/ps.0305203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Noda LH, Kuby SA, Lardy HA. Properties of thiolesters: Kinetics of hydrolysis in dilute aqueous media. J. Am. Chem. Soc. 1953;75:913–917. [Google Scholar]
- 45.Polgar L. Spectrophotometric determination of mercaptide ion, an activated form of SH-group in thiol enzymes. FEBS Lett. 1974;38:187–190. [Google Scholar]
- 46.Deshpande N, Addess KJ, Bluhm WF, Merino-Ott JC, Townsend-Merino W, Zhang Q, Knezevich C, Xie L, Chen L, Feng Z, Green RK, Flippen-Anderson JL, Westbrook J, Berman HM, Bourne PE. The RCSB Protein Data Bank: a redesigned query system and relational database based on the mmCIF schema. Nucleic Acids Res. 2005;33:D233–D237. doi: 10.1093/nar/gki057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kinumi T, Kimata J, Taira T, Ariga H, Niki E. Cysteine-106 of DJ-1 is the most sensitive cysteine residue to hydrogen peroxide-mediated oxidation in vivo in human umbilical vein endothelial cells. Biochem. Biophys. Res. Commun. 2004;317:722–728. doi: 10.1016/j.bbrc.2004.03.110. [DOI] [PubMed] [Google Scholar]
- 48.Engh RA, Huber R. Accurate bond and angle parameters for X-Ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- 49.Nie B, Stutzman J, Xie A. A vibrational spectral maker for probing the hydrogen-bonding status of protonated Asp and Glu residues. Biophys. J. 2005;88:2833–2847. doi: 10.1529/biophysj.104.047639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quigley PM, Korotkov K, Baneyx F, Hol WGJ. The 1.6-angstrom crystal structure of the class of chaperones represented by Escherichia coli Hsp31 reveals a putative catalytic triad. Proc. Natl. Acad. Sci. USA. 2003;100:3137–3142. doi: 10.1073/pnas.0530312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lakshminarasimhan M, Maldonado MT, Zhou W, Fink AL, Wilson MA. Structural impact of three Parkinsonism-associated missense mutations on human DJ-1. Biochemistry. 2008;47:1381–1392. doi: 10.1021/bi701189c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andres-Mateos E, Perier C, Zhang L, Blanchard-Fillion B, Greco TM, Thomas B, Ko HS, Sasaki M, Ischiropoulos H, Przedborski S, Dawson TM, Dawson VL. DJ-1 gene deletion reveals that DJ-1 is an atypical peroxiredoxin-like peroxidase. Proc. Natl. Acad. Sci. U S A. 2007;104:14807–14812. doi: 10.1073/pnas.0703219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shipton M, Kierstan MP, Malthouse JP, Stuchbury T, Brocklehurst K. The case for assigning a value of approximately 4 to pKa-i of the essential histidine-cysteine interactive systems of papain, bromelain and ficin. FEBS Lett. 1975;50:365–368. doi: 10.1016/0014-5793(75)80529-1. [DOI] [PubMed] [Google Scholar]
- 54.Lo Bello M, Parker MW, Desideri A, Polticelli F, Falconi M, Del Boccio G, Pennelli A, Federici G, Ricci G. Peculiar spectroscopic and kinetic properties of Cys-47 in human placental glutathione transferase. Evidence for an atypical thiolate ion pair near the active site. J. Biol. Chem. 1993;268:19033–19038. [PubMed] [Google Scholar]
- 55.Darby NJ, Creighton TE. Characterization of the active site cysteine residues of the thioredoxin-like domains of protein disulfide isomerase. Biochemistry. 1995;34:16770–16780. doi: 10.1021/bi00051a027. [DOI] [PubMed] [Google Scholar]
- 56.Olzmann JA, Brown K, Wilkinson KD, Rees HD, Huai Q, Ke H, Levey AI, Li L, Chin LS. Familial Parkinson's disease-associated L166P mutation disrupts DJ-1 protein folding and function. J. Biol. Chem. 2004;279:8506–8515. doi: 10.1074/jbc.M311017200. [DOI] [PubMed] [Google Scholar]
- 57.Mitsumoto A, Nakagawa Y, Takeuchi A, Okawa K, Iwamatsu A, Takanezawa Y. Oxidized forms of peroxiredoxins and DJ-1 on two-dimensional gels increased in response to sublethal levels of paraquat. Free Radic. Res. 2001;35:301–310. doi: 10.1080/10715760100300831. [DOI] [PubMed] [Google Scholar]
- 58.Aleyasin H, Rousseaux MW, Phillips M, Kim RH, Bland RJ, Callaghan S, Slack RS, During MJ, Mak TW, Park DS. The Parkinson's disease gene DJ-1 is also a key regulator of stroke-induced damage. Proc. Natl. Acad. Sci. U S A. 2007;104:18748–18753. doi: 10.1073/pnas.0709379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chivers PT, Prehoda KE, Volkman BF, Kim BM, Markley JL, Raines RT. Microscopic pKa values of Escherichia coli thioredoxin. Biochemistry. 1997;36:14985–14991. doi: 10.1021/bi970071j. [DOI] [PubMed] [Google Scholar]
- 60.Chivers PT, Raines RT. General acid/base catalysis in the active site of Escherichia coli thioredoxin. Biochemistry. 1997;36:15810–15816. doi: 10.1021/bi971504l. [DOI] [PubMed] [Google Scholar]
- 61.Fenn TD, Ringe D, Petsko GA. POVScript+: a program for model and data visualization using persistence of vision ray-tracing. J. Appl. Crystallogr. 2003;36:944–947. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 shows a titration of wtDJ-1 in a double buffer supplemented with 100 mM NaCl, demonstrating that increased ionic strength has no effect on the pKa of C106 under these conditions. Table S2 lists every example of a candidate D/E-C hydrogen bonded dyad in the 4/15/08 release of the PDB. This material is available free of charge via the Internet at http://pubs.acs.org.