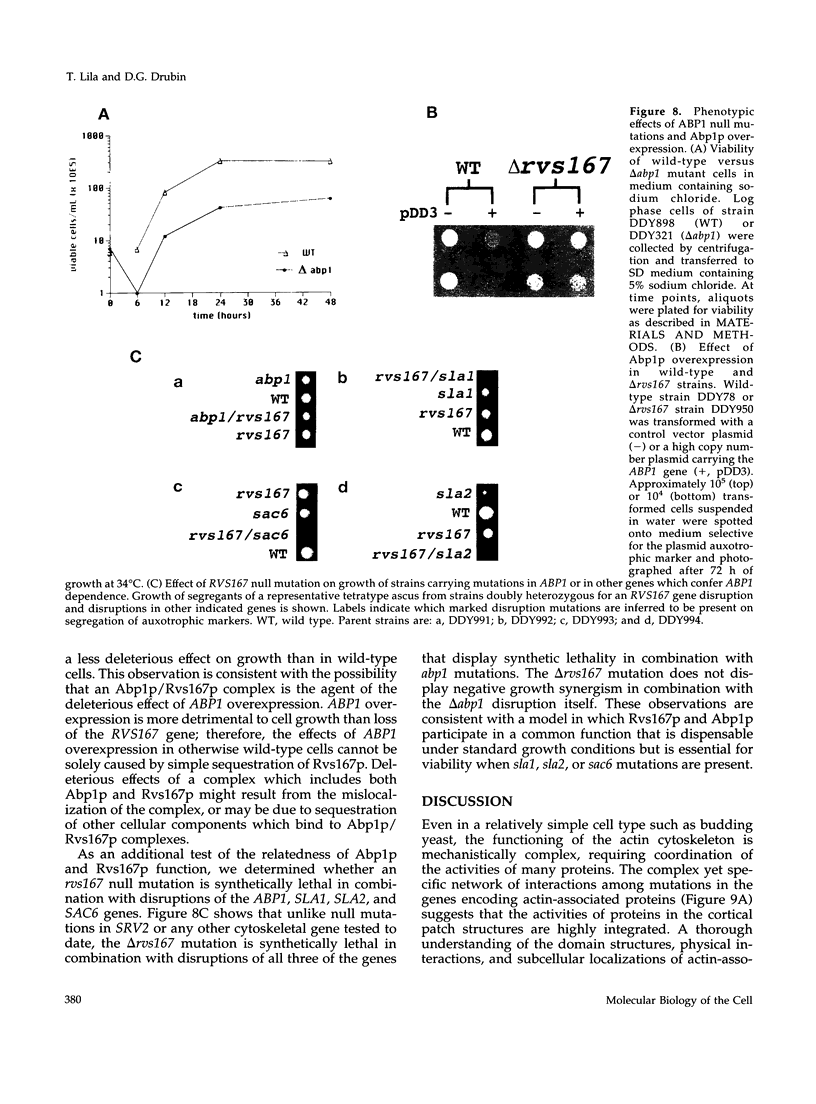
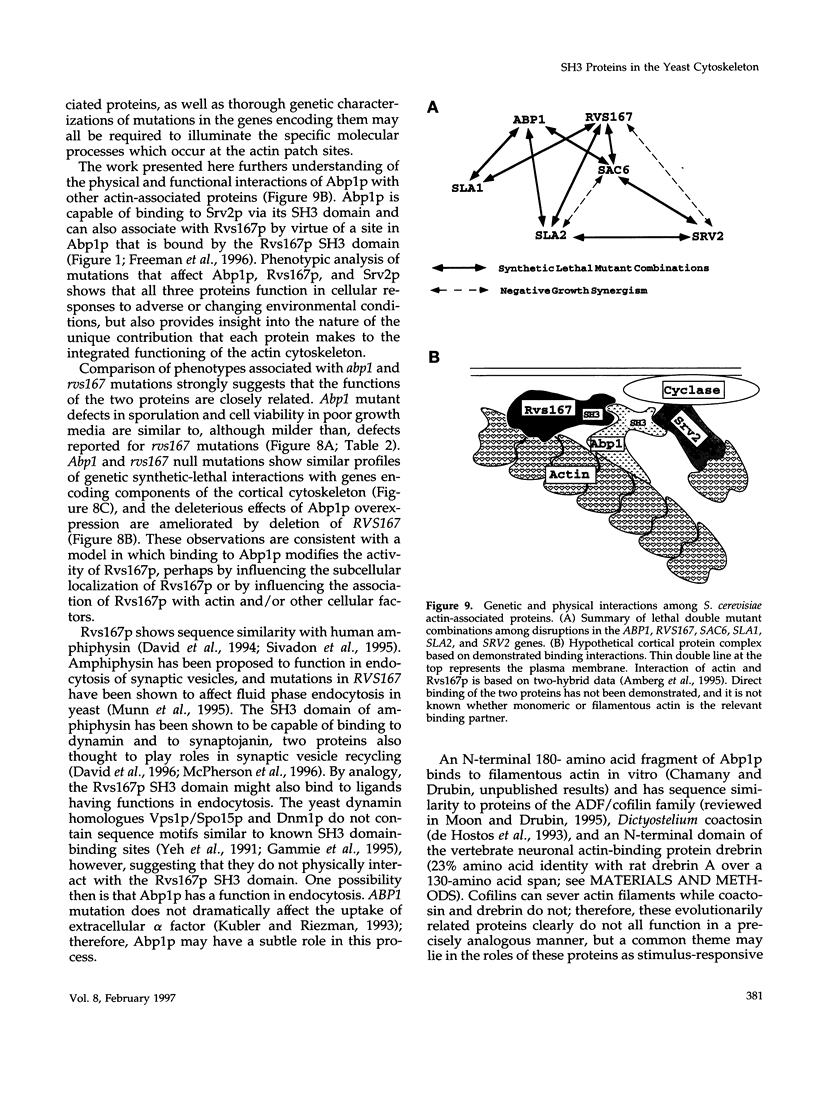
Abstract
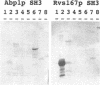
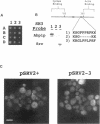
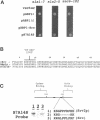
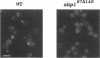
In a variety of organisms, a number of proteins associated with the cortical actin cytoskeleton contain SH3 domains, suggesting that these domains may provide the physical basis for functional interactions among structural and regulatory proteins in the actin cytoskeleton. We present evidence that SH3 domains mediate at least two independent functions of the Saccharomyces cerevisiae actin-binding protein Abp1p in vivo. Abp1p contains a single SH3 domain that has recently been shown to bind in vitro to the adenylyl cyclase-associated protein Srv2p. Immunofluorescence analysis of Srv2p subcellular localization in strains carrying mutations in either ABP1 or SRV2 reveals that the Abp1p SH3 domain mediates the normal association of Srv2p with the cortical actin cytoskeleton. We also show that a site in Abp1p itself is specifically bound by the SH3 domain of the actin-associated protein Rvs167p. Genetic analysis provides evidence that Abp1p and Rvs167p have functions that are closely interrelated. Abp1 null mutations, like rvs167 mutations, result in defects in sporulation and reduced viability under certain suboptimal growth conditions. In addition, mutations in ABP1 and RVS167 yield similar profiles of genetic "synthetic lethal" interactions when combined with mutations in genes encoding other cytoskeletal components. Mutations which specifically disrupt the SH3 domain-mediated interaction between Abp1p and Srv2p, however, show none of the shared phenotypes of abp1 and rvs167 mutations. We conclude that the Abp1p SH3 domain mediates the association of Srv2p with the cortical actin cytoskeleton, and that Abp1p performs a distinct function that is likely to involve binding by the Rvs167p SH3 domain. Overall, work presented here illustrates how SH3 domains can integrate the activities of multiple actin cytoskeleton proteins in response to varying environmental conditions.
Full text
PDF






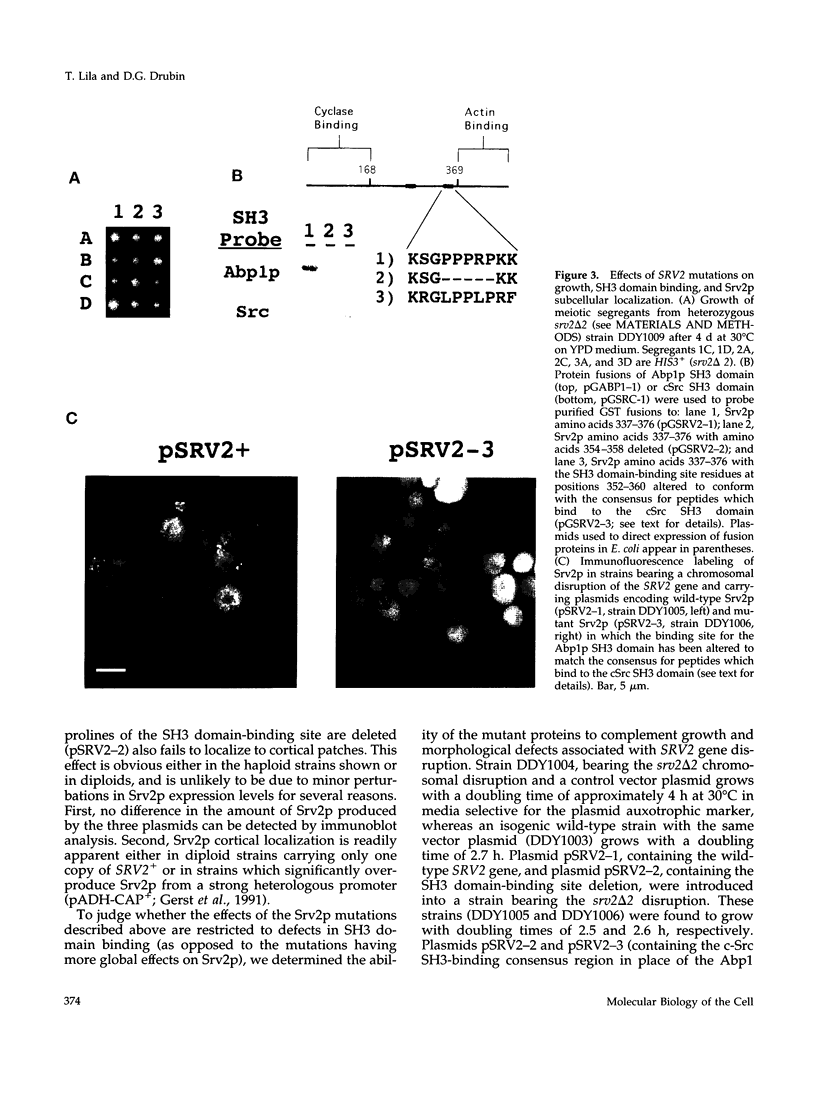

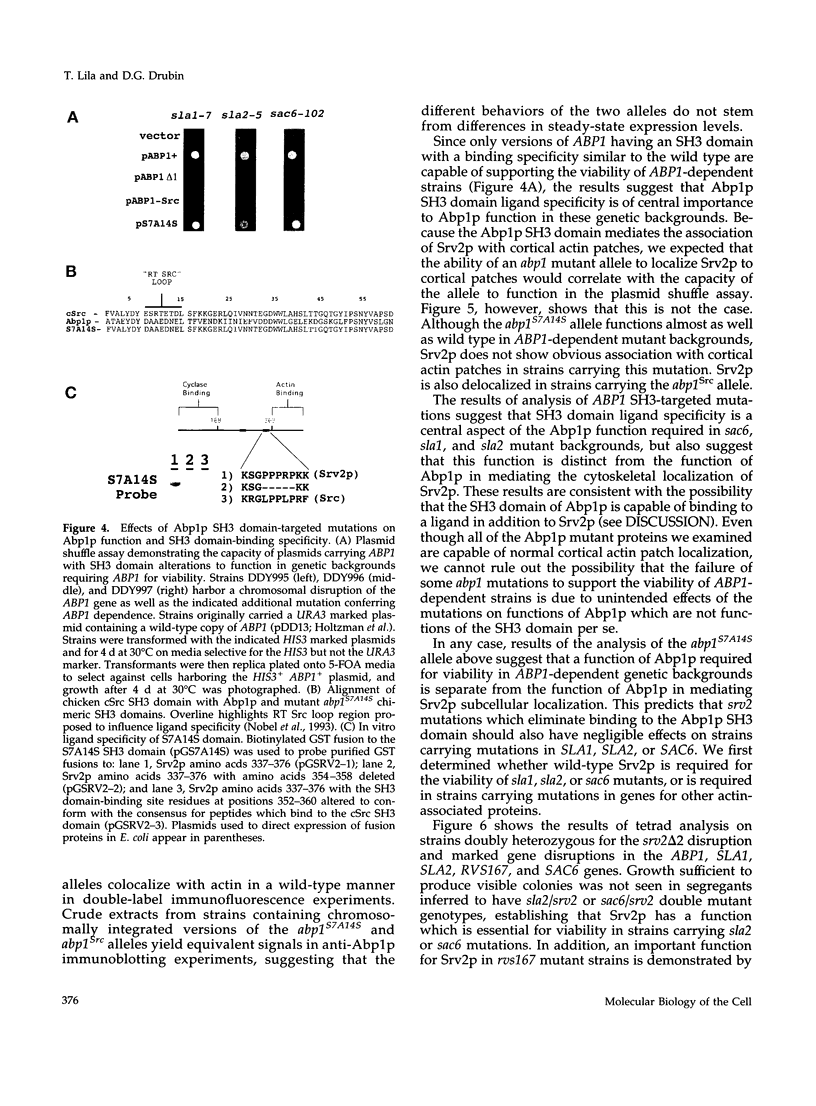
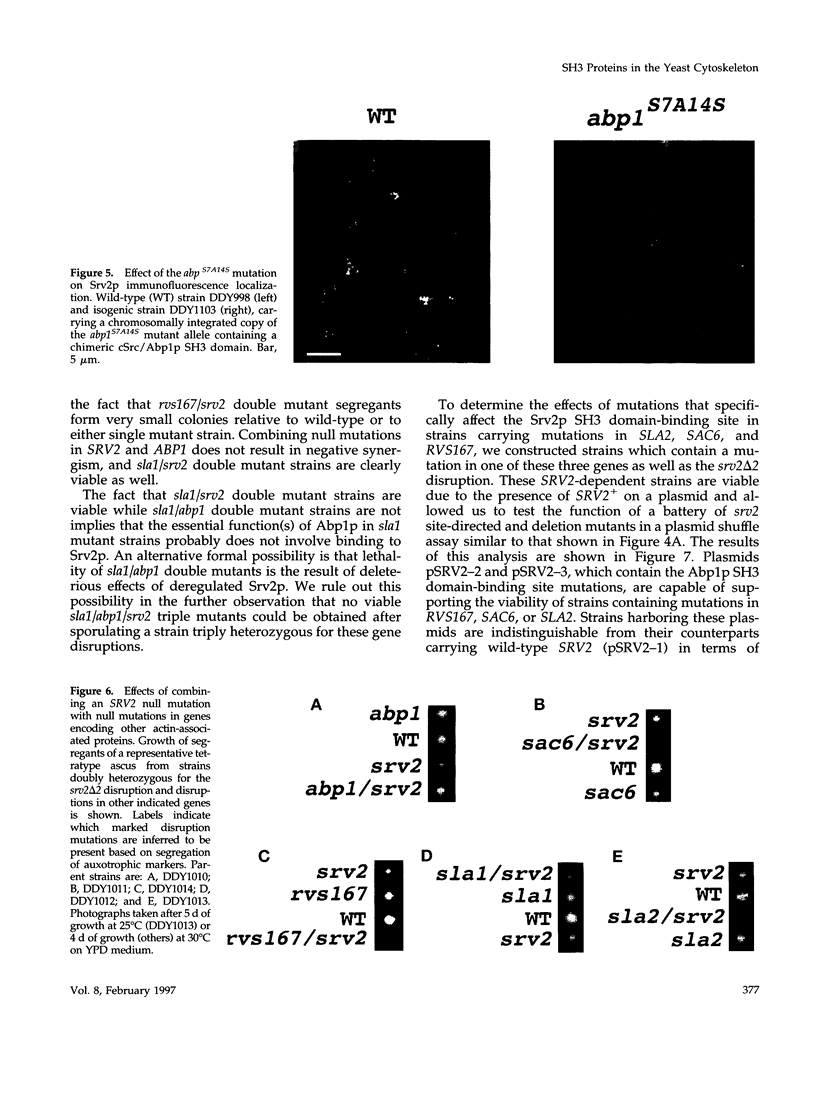
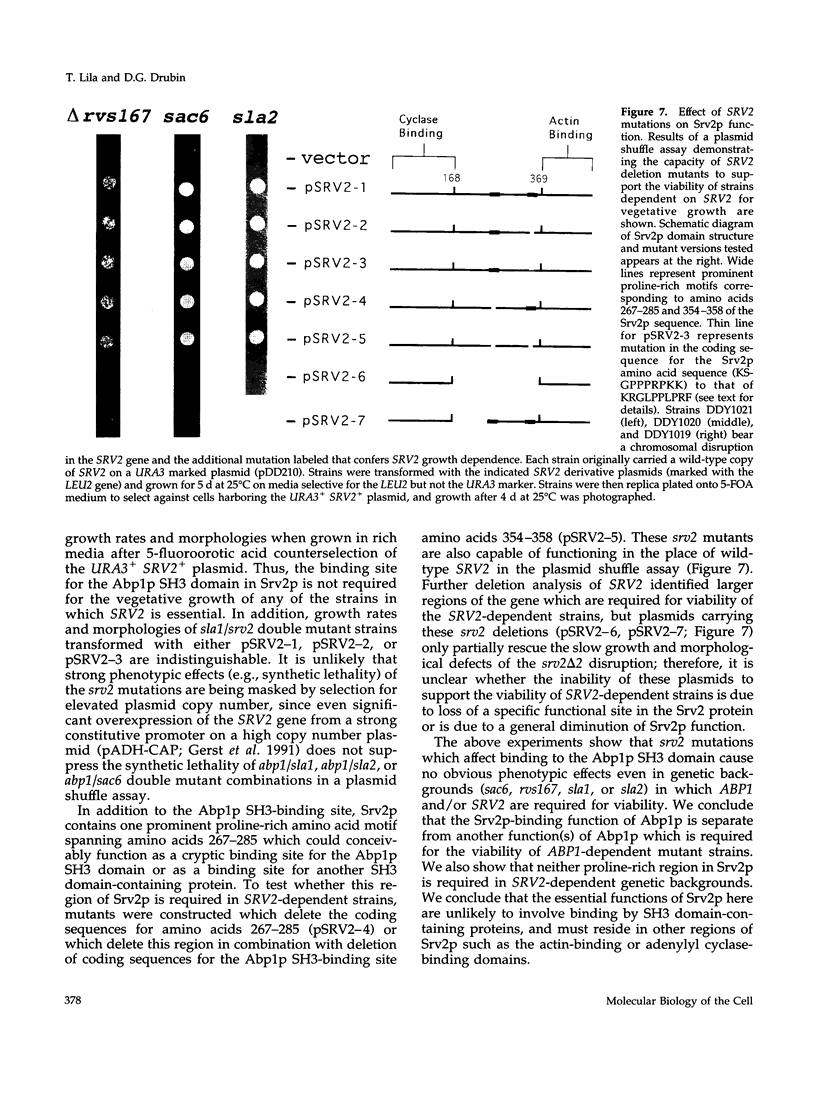





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams A. E., Botstein D., Drubin D. G. A yeast actin-binding protein is encoded by SAC6, a gene found by suppression of an actin mutation. Science. 1989 Jan 13;243(4888):231–233. doi: 10.1126/science.2643162. [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amberg D. C., Basart E., Botstein D. Defining protein interactions with yeast actin in vivo. Nat Struct Biol. 1995 Jan;2(1):28–35. doi: 10.1038/nsb0195-28. [DOI] [PubMed] [Google Scholar]
- Asada H., Uyemura K., Shirao T. Actin-binding protein, drebrin, accumulates in submembranous regions in parallel with neuronal differentiation. J Neurosci Res. 1994 Jun 1;38(2):149–159. doi: 10.1002/jnr.490380205. [DOI] [PubMed] [Google Scholar]
- Ayscough K. R., Drubin D. G. ACTIN: general principles from studies in yeast. Annu Rev Cell Dev Biol. 1996;12:129–160. doi: 10.1146/annurev.cellbio.12.1.129. [DOI] [PubMed] [Google Scholar]
- Baroni M. D., Monti P., Alberghina L. Repression of growth-regulated G1 cyclin expression by cyclic AMP in budding yeast. Nature. 1994 Sep 22;371(6495):339–342. doi: 10.1038/371339a0. [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993 Jul 11;21(14):3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer F., Urdaci M., Aigle M., Crouzet M. Alteration of a yeast SH3 protein leads to conditional viability with defects in cytoskeletal and budding patterns. Mol Cell Biol. 1993 Aug;13(8):5070–5084. doi: 10.1128/mcb.13.8.5070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement W. M., Wirth J. A., Mooseker M. S. Cloning and mRNA expression of human unconventional myosin-IC. A homologue of amoeboid myosins-I with a single IQ motif and an SH3 domain. J Mol Biol. 1994 Oct 21;243(2):356–363. doi: 10.1006/jmbi.1994.1662. [DOI] [PubMed] [Google Scholar]
- Bender L., Lo H. S., Lee H., Kokojan V., Peterson V., Bender A. Associations among PH and SH3 domain-containing proteins and Rho-type GTPases in Yeast. J Cell Biol. 1996 May;133(4):879–894. doi: 10.1083/jcb.133.4.879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., Trueheart J., Natsoulis G., Fink G. R. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. doi: 10.1016/0076-6879(87)54076-9. [DOI] [PubMed] [Google Scholar]
- Broach J. R., Deschenes R. J. The function of ras genes in Saccharomyces cerevisiae. Adv Cancer Res. 1990;54:79–139. doi: 10.1016/s0065-230x(08)60809-x. [DOI] [PubMed] [Google Scholar]
- Broach J. R. Ras-regulated signaling processes in Saccharomyces cerevisiae. Curr Opin Genet Dev. 1991 Oct;1(3):370–377. doi: 10.1016/s0959-437x(05)80302-8. [DOI] [PubMed] [Google Scholar]
- Chenevert J., Corrado K., Bender A., Pringle J., Herskowitz I. A yeast gene (BEM1) necessary for cell polarization whose product contains two SH3 domains. Nature. 1992 Mar 5;356(6364):77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- Cicchetti P., Mayer B. J., Thiel G., Baltimore D. Identification of a protein that binds to the SH3 region of Abl and is similar to Bcr and GAP-rho. Science. 1992 Aug 7;257(5071):803–806. doi: 10.1126/science.1379745. [DOI] [PubMed] [Google Scholar]
- David C., McPherson P. S., Mundigl O., de Camilli P. A role of amphiphysin in synaptic vesicle endocytosis suggested by its binding to dynamin in nerve terminals. Proc Natl Acad Sci U S A. 1996 Jan 9;93(1):331–335. doi: 10.1073/pnas.93.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David C., Solimena M., De Camilli P. Autoimmunity in stiff-Man syndrome with breast cancer is targeted to the C-terminal region of human amphiphysin, a protein similar to the yeast proteins, Rvs167 and Rvs161. FEBS Lett. 1994 Aug 29;351(1):73–79. doi: 10.1016/0014-5793(94)00826-4. [DOI] [PubMed] [Google Scholar]
- Desfarges L., Durrens P., Juguelin H., Cassagne C., Bonneu M., Aigle M. Yeast mutants affected in viability upon starvation have a modified phospholipid composition. Yeast. 1993 Mar;9(3):267–277. doi: 10.1002/yea.320090306. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Miller K. G., Botstein D. Yeast actin-binding proteins: evidence for a role in morphogenesis. J Cell Biol. 1988 Dec;107(6 Pt 2):2551–2561. doi: 10.1083/jcb.107.6.2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drubin D. G., Mulholland J., Zhu Z. M., Botstein D. Homology of a yeast actin-binding protein to signal transduction proteins and myosin-I. Nature. 1990 Jan 18;343(6255):288–290. doi: 10.1038/343288a0. [DOI] [PubMed] [Google Scholar]
- Drubin D. G., Nelson W. J. Origins of cell polarity. Cell. 1996 Feb 9;84(3):335–344. doi: 10.1016/s0092-8674(00)81278-7. [DOI] [PubMed] [Google Scholar]
- Fedor-Chaiken M., Deschenes R. J., Broach J. R. SRV2, a gene required for RAS activation of adenylate cyclase in yeast. Cell. 1990 Apr 20;61(2):329–340. doi: 10.1016/0092-8674(90)90813-t. [DOI] [PubMed] [Google Scholar]
- Field J., Vojtek A., Ballester R., Bolger G., Colicelli J., Ferguson K., Gerst J., Kataoka T., Michaeli T., Powers S. Cloning and characterization of CAP, the S. cerevisiae gene encoding the 70 kd adenylyl cyclase-associated protein. Cell. 1990 Apr 20;61(2):319–327. doi: 10.1016/0092-8674(90)90812-s. [DOI] [PubMed] [Google Scholar]
- Flynn D. C., Leu T. H., Reynolds A. B., Parsons J. T. Identification and sequence analysis of cDNAs encoding a 110-kilodalton actin filament-associated pp60src substrate. Mol Cell Biol. 1993 Dec;13(12):7892–7900. doi: 10.1128/mcb.13.12.7892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman N. L., Chen Z., Horenstein J., Weber A., Field J. An actin monomer binding activity localizes to the carboxyl-terminal half of the Saccharomyces cerevisiae cyclase-associated protein. J Biol Chem. 1995 Mar 10;270(10):5680–5685. doi: 10.1074/jbc.270.10.5680. [DOI] [PubMed] [Google Scholar]
- Freeman N. L., Lila T., Mintzer K. A., Chen Z., Pahk A. J., Ren R., Drubin D. G., Field J. A conserved proline-rich region of the Saccharomyces cerevisiae cyclase-associated protein binds SH3 domains and modulates cytoskeletal localization. Mol Cell Biol. 1996 Feb;16(2):548–556. doi: 10.1128/mcb.16.2.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie A. E., Kurihara L. J., Vallee R. B., Rose M. D. DNM1, a dynamin-related gene, participates in endosomal trafficking in yeast. J Cell Biol. 1995 Aug;130(3):553–566. doi: 10.1083/jcb.130.3.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst J. E., Ferguson K., Vojtek A., Wigler M., Field J. CAP is a bifunctional component of the Saccharomyces cerevisiae adenylyl cyclase complex. Mol Cell Biol. 1991 Mar;11(3):1248–1257. doi: 10.1128/mcb.11.3.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst J. E., Rodgers L., Riggs M., Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gieselmann R., Mann K. ASP-56, a new actin sequestering protein from pig platelets with homology to CAP, an adenylate cyclase-associated protein from yeast. FEBS Lett. 1992 Feb 24;298(2-3):149–153. doi: 10.1016/0014-5793(92)80043-g. [DOI] [PubMed] [Google Scholar]
- Goodson H. V., Anderson B. L., Warrick H. M., Pon L. A., Spudich J. A. Synthetic lethality screen identifies a novel yeast myosin I gene (MYO5): myosin I proteins are required for polarization of the actin cytoskeleton. J Cell Biol. 1996 Jun;133(6):1277–1291. doi: 10.1083/jcb.133.6.1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottwald U., Brokamp R., Karakesisoglou I., Schleicher M., Noegel A. A. Identification of a cyclase-associated protein (CAP) homologue in Dictyostelium discoideum and characterization of its interaction with actin. Mol Biol Cell. 1996 Feb;7(2):261–272. doi: 10.1091/mbc.7.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greengard P., Valtorta F., Czernik A. J., Benfenati F. Synaptic vesicle phosphoproteins and regulation of synaptic function. Science. 1993 Feb 5;259(5096):780–785. doi: 10.1126/science.8430330. [DOI] [PubMed] [Google Scholar]
- Gumbiner B. M. Cell adhesion: the molecular basis of tissue architecture and morphogenesis. Cell. 1996 Feb 9;84(3):345–357. doi: 10.1016/s0092-8674(00)81279-9. [DOI] [PubMed] [Google Scholar]
- Hildebrand J. D., Taylor J. M., Parsons J. T. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996 Jun;16(6):3169–3178. doi: 10.1128/mcb.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtzman D. A., Yang S., Drubin D. G. Synthetic-lethal interactions identify two novel genes, SLA1 and SLA2, that control membrane cytoskeleton assembly in Saccharomyces cerevisiae. J Cell Biol. 1993 Aug;122(3):635–644. doi: 10.1083/jcb.122.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kübler E., Riezman H. Actin and fimbrin are required for the internalization step of endocytosis in yeast. EMBO J. 1993 Jul;12(7):2855–2862. doi: 10.1002/j.1460-2075.1993.tb05947.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauffenburger D. A., Horwitz A. F. Cell migration: a physically integrated molecular process. Cell. 1996 Feb 9;84(3):359–369. doi: 10.1016/s0092-8674(00)81280-5. [DOI] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. A cell cycle checkpoint monitors cell morphogenesis in budding yeast. J Cell Biol. 1995 May;129(3):739–749. doi: 10.1083/jcb.129.3.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lew D. J., Reed S. I. Morphogenesis in the yeast cell cycle: regulation by Cdc28 and cyclins. J Cell Biol. 1993 Mar;120(6):1305–1320. doi: 10.1083/jcb.120.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui Y., Matsui R., Akada R., Toh-e A. Yeast src homology region 3 domain-binding proteins involved in bud formation. J Cell Biol. 1996 May;133(4):865–878. doi: 10.1083/jcb.133.4.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer B. J., Eck M. J. SH3 domains. Minding your p's and q's. Curr Biol. 1995 Apr 1;5(4):364–367. doi: 10.1016/s0960-9822(95)00073-x. [DOI] [PubMed] [Google Scholar]
- McPherson P. S., Garcia E. P., Slepnev V. I., David C., Zhang X., Grabs D., Sossin W. S., Bauerfeind R., Nemoto Y., De Camilli P. A presynaptic inositol-5-phosphatase. Nature. 1996 Jan 25;379(6563):353–357. doi: 10.1038/379353a0. [DOI] [PubMed] [Google Scholar]
- Meriläinen J., Palovuori R., Sormunen R., Wasenius V. M., Lehto V. P. Binding of the alpha-fodrin SH3 domain to the leading lamellae of locomoting chicken fibroblasts. J Cell Sci. 1993 Jul;105(Pt 3):647–654. doi: 10.1242/jcs.105.3.647. [DOI] [PubMed] [Google Scholar]
- Mintzer K. A., Field J. Interactions between adenylyl cyclase, CAP and RAS from Saccharomyces cerevisiae. Cell Signal. 1994 Aug;6(6):681–694. doi: 10.1016/0898-6568(94)90050-7. [DOI] [PubMed] [Google Scholar]
- Mitchison T. J., Cramer L. P. Actin-based cell motility and cell locomotion. Cell. 1996 Feb 9;84(3):371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Moon A., Drubin D. G. The ADF/cofilin proteins: stimulus-responsive modulators of actin dynamics. Mol Biol Cell. 1995 Nov;6(11):1423–1431. doi: 10.1091/mbc.6.11.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munn A. L., Stevenson B. J., Geli M. I., Riezman H. end5, end6, and end7: mutations that cause actin delocalization and block the internalization step of endocytosis in Saccharomyces cerevisiae. Mol Biol Cell. 1995 Dec;6(12):1721–1742. doi: 10.1091/mbc.6.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble M. E., Musacchio A., Saraste M., Courtneidge S. A., Wierenga R. K. Crystal structure of the SH3 domain in human Fyn; comparison of the three-dimensional structures of SH3 domains in tyrosine kinases and spectrin. EMBO J. 1993 Jul;12(7):2617–2624. doi: 10.2210/pdb1shf/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noegel A. A., Luna J. E. The Dictyostelium cytoskeleton. Experientia. 1995 Dec 18;51(12):1135–1143. doi: 10.1007/BF01944731. [DOI] [PubMed] [Google Scholar]
- Pearson W. R., Lipman D. J. Improved tools for biological sequence comparison. Proc Natl Acad Sci U S A. 1988 Apr;85(8):2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollard T. D., Cooper J. A. Actin and actin-binding proteins. A critical evaluation of mechanisms and functions. Annu Rev Biochem. 1986;55:987–1035. doi: 10.1146/annurev.bi.55.070186.005011. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Adams A. E., Drubin D. G., Haarer B. K. Immunofluorescence methods for yeast. Methods Enzymol. 1991;194:565–602. doi: 10.1016/0076-6879(91)94043-c. [DOI] [PubMed] [Google Scholar]
- Pringle J. R., Preston R. A., Adams A. E., Stearns T., Drubin D. G., Haarer B. K., Jones E. W. Fluorescence microscopy methods for yeast. Methods Cell Biol. 1989;31:357–435. doi: 10.1016/s0091-679x(08)61620-9. [DOI] [PubMed] [Google Scholar]
- Read E. B., Okamura H. H., Drubin D. G. Actin- and tubulin-dependent functions during Saccharomyces cerevisiae mating projection formation. Mol Biol Cell. 1992 Apr;3(4):429–444. doi: 10.1091/mbc.3.4.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60(2-3):237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Shirao T., Kojima N., Obata K. Cloning of drebrin A and induction of neurite-like processes in drebrin-transfected cells. Neuroreport. 1992 Jan;3(1):109–112. doi: 10.1097/00001756-199201000-00029. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivadon P., Bauer F., Aigle M., Crouzet M. Actin cytoskeleton and budding pattern are altered in the yeast rvs161 mutant: the Rvs161 protein shares common domains with the brain protein amphiphysin. Mol Gen Genet. 1995 Feb 20;246(4):485–495. doi: 10.1007/BF00290452. [DOI] [PubMed] [Google Scholar]
- Stossel T. P., Chaponnier C., Ezzell R. M., Hartwig J. H., Janmey P. A., Kwiatkowski D. J., Lind S. E., Smith D. B., Southwick F. S., Yin H. L. Nonmuscle actin-binding proteins. Annu Rev Cell Biol. 1985;1:353–402. doi: 10.1146/annurev.cb.01.110185.002033. [DOI] [PubMed] [Google Scholar]
- Stöffler H. E., Ruppert C., Reinhard J., Bähler M. A novel mammalian myosin I from rat with an SH3 domain localizes to Con A-inducible, F-actin-rich structures at cell-cell contacts. J Cell Biol. 1995 May;129(3):819–830. doi: 10.1083/jcb.129.3.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan W., Theurkauf W. E. The cytoskeleton and morphogenesis of the early Drosophila embryo. Curr Opin Cell Biol. 1995 Feb;7(1):18–22. doi: 10.1016/0955-0674(95)80040-9. [DOI] [PubMed] [Google Scholar]
- Sun H. Q., Kwiatkowska K., Yin H. L. Actin monomer binding proteins. Curr Opin Cell Biol. 1995 Feb;7(1):102–110. doi: 10.1016/0955-0674(95)80051-4. [DOI] [PubMed] [Google Scholar]
- Tokiwa G., Tyers M., Volpe T., Futcher B. Inhibition of G1 cyclin activity by the Ras/cAMP pathway in yeast. Nature. 1994 Sep 22;371(6495):342–345. doi: 10.1038/371342a0. [DOI] [PubMed] [Google Scholar]
- Turner C. E., Miller J. T. Primary sequence of paxillin contains putative SH2 and SH3 domain binding motifs and multiple LIM domains: identification of a vinculin and pp125Fak-binding region. J Cell Sci. 1994 Jun;107(Pt 6):1583–1591. doi: 10.1242/jcs.107.6.1583. [DOI] [PubMed] [Google Scholar]
- Vojtek A. B., Cooper J. A. Identification and characterization of a cDNA encoding mouse CAP: a homolog of the yeast adenylyl cyclase associated protein. J Cell Sci. 1993 Jul;105(Pt 3):777–785. doi: 10.1242/jcs.105.3.777. [DOI] [PubMed] [Google Scholar]
- Wang J., Suzuki N., Kataoka T. The 70-kilodalton adenylyl cyclase-associated protein is not essential for interaction of Saccharomyces cerevisiae adenylyl cyclase with RAS proteins. Mol Cell Biol. 1992 Nov;12(11):4937–4945. doi: 10.1128/mcb.12.11.4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., Suzuki N., Nishida Y., Kataoka T. Analysis of the function of the 70-kilodalton cyclase-associated protein (CAP) by using mutants of yeast adenylyl cyclase defective in CAP binding. Mol Cell Biol. 1993 Jul;13(7):4087–4097. doi: 10.1128/mcb.13.7.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K., Knipfer M., Huang Q. Q., van Heerden A., Hsu L. C., Gutierrez G., Quian X. L., Stedman H. Human skeletal muscle nebulin sequence encodes a blueprint for thin filament architecture. Sequence motifs and affinity profiles of tandem repeats and terminal SH3. J Biol Chem. 1996 Feb 23;271(8):4304–4314. doi: 10.1074/jbc.271.8.4304. [DOI] [PubMed] [Google Scholar]
- Welch M. D., Holtzman D. A., Drubin D. G. The yeast actin cytoskeleton. Curr Opin Cell Biol. 1994 Feb;6(1):110–119. doi: 10.1016/0955-0674(94)90124-4. [DOI] [PubMed] [Google Scholar]
- Wu H., Parsons J. T. Cortactin, an 80/85-kilodalton pp60src substrate, is a filamentous actin-binding protein enriched in the cell cortex. J Cell Biol. 1993 Mar;120(6):1417–1426. doi: 10.1083/jcb.120.6.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh E., Driscoll R., Coltrera M., Olins A., Bloom K. A dynamin-like protein encoded by the yeast sporulation gene SPO15. Nature. 1991 Feb 21;349(6311):713–715. doi: 10.1038/349713a0. [DOI] [PubMed] [Google Scholar]
- Yu H., Chen J. K., Feng S., Dalgarno D. C., Brauer A. W., Schreiber S. L. Structural basis for the binding of proline-rich peptides to SH3 domains. Cell. 1994 Mar 11;76(5):933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- Zahner J. E., Harkins H. A., Pringle J. R. Genetic analysis of the bipolar pattern of bud site selection in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1996 Apr;16(4):1857–1870. doi: 10.1128/mcb.16.4.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Hostos E. L., Bradtke B., Lottspeich F., Gerisch G. Coactosin, a 17 kDa F-actin binding protein from Dictyostelium discoideum. Cell Motil Cytoskeleton. 1993;26(3):181–191. doi: 10.1002/cm.970260302. [DOI] [PubMed] [Google Scholar]